Abstract
Human immunodeficiency virus type 1 (HIV-1) gene expression is regulated by both cellular transcription factors and Tat. The ability of Tat to stimulate transcriptional elongation is dependent on its binding to TAR RNA in conjunction with cyclin T1 and CDK9. A variety of other cellular factors that bind to the HIV-1 long terminal repeat, including NF-κB, SP1, LBP, and LEF, are also important in the control of HIV-1 gene expression. Although these factors have been demonstrated to regulate HIV-1 gene expression by both genetic and biochemical analysis, in most cases a direct in vivo demonstration of their role on HIV-1 replication has not been established. Recently, the efficacy of RNA interference in mammalian cells has been shown utilizing small interfering RNAs (siRNAs) to result in the specific degradation of host mRNAs and decreases the levels of their corresponding proteins. In this study, we addressed whether siRNAs directed against either HIV-1 tat or reverse transcriptase or the NF-κB p65 subunit could specifically decrease the levels of these proteins and thus alter HIV-1 replication. Our results demonstrate the specificity of siRNAs for decreasing the expression of these viral and cellular proteins and inhibiting HIV-1 replication. These studies suggest that RNA interference is useful in exploring the biological role of cellular and viral regulatory factors involved in the control of HIV-1 gene expression.
The regulation of human immunodeficiency virus type 1 (HIV-1) gene expression involves the interplay of both viral and cellular factors (32, 46, 76). In a variety of cells that are permissive for HIV-1 infection, transcriptional initiation from the HIV-1 long terminal repeat (LTR) is efficient but requires the HIV-1 Tat protein in order to generate full-length transcripts (48, 57). Tat binding to the TAR RNA element, which extends between +1 and +60 in all HIV-1 transcripts, is required for efficient HIV-1 transcriptional elongation and the generation of high levels of HIV-1 virions (9, 21, 38, 73, 75). Both direct and indirect interactions of Tat with a variety of cellular factors are critical for its function (45).
Tat binding to TAR is facilitated by its association with Tat-associated kinase (TAK) (40), which is comprised of the CDK9 kinase subunit and its binding partner cyclin T1 (46, 61, 80, 86, 91). The assembly of this complex facilitates CDK9 phosphorylation of the RNA polymerase II carboxy-terminal domain and also the transcriptional elongation factors SPT5 and SPT4 (10, 14, 28, 29, 43, 46, 53, 56, 61, 71, 80, 84, 86, 88, 91). Both genetic and biochemical evidence demonstrates the critical roles of these cellular proteins in mediating Tat activation.
A variety of cellular transcriptional factors that bind to the HIV-1 LTR are also critical for regulating HIV-1 gene expression. One of the best studied of these factors is the NF-κB family, which binds to two regulatory elements in the HIV-1 LTR (35, 64). Members of the NF-κB family, which include p105/50, p100/52, p65/RelA, c-Rel, and RelB, contain a Rel homology domain that mediates their heterodimerization and homodimerization and their DNA binding properties (3). Recent studies suggest that the NF-κB p65 subunit, which is a key component required for NF-κB transcriptional activation, can also bind to TAK to stimulate HIV-1 transcriptional elongation (4, 81). Thus, it is likely that, during the early phases of HIV-1 infection of activated T lymphocytes, NF-κB binding to the HIV-1 LTR serves to generate at least some full-length transcripts to result in the synthesis of Tat, which then potently stimulates HIV-1 transcriptional elongation.
NF-κB proteins are sequestered in the cytoplasm of most of the cells, where they are bound to a family of inhibitory proteins known as IκB (2). Treatment of cells with a variety of stimuli, including the cytokines tumor necrosis factor alpha and interleukin 1, stimulates the activity of kinases that phosphorylate IκB on amino-terminal serine residues, resulting in its ubiquitination and degradation by proteasome (15, 20, 49, 50, 63). This process leads to the nuclear translocation of the NF-κB proteins, which then bind to sites in the HIV-1 LTR (35, 64), in addition to binding to promoter elements of cellular genes that are involved in the control of the immune and inflammatory response (33, 34, 51). In addition to NF-κB a variety of other factors bind to the HIV-1 LTR, including SP1 (8, 37, 69), TBP (62, 66), LEF (79), and LBP (47, 83), and other factors that have been identified to interact with these proteins, including CBP/p300 (7, 41), Tat-SF1 (54, 60, 89, 90), and TFIIH (30, 67). It would be useful to have cell lines that lack each of these factors in order to better understand their role in regulating HIV-1 replication.
RNA interference (RNAi) that utilizes small interfering RNAs (siRNAs) provides a methodology for further addressing the role of cellular and viral regulatory factors in the HIV-1 life cycle. RNAi is an evolutionarily conserved mechanism that is operative in insects, nematodes, plants, and mammalian cells (12, 16, 17, 23-26, 52, 82). In this process, sequence-specific posttranscriptional silencing is initiated by the introduction into cells of double-stranded, annealed sense and antisense RNAs that are homologous to the sequence of the silenced gene (5). The ultimate mediators of RNAi-mediated degradation are 21-mer siRNAs that are generated by RNase III cleavage of double-stranded RNAs that have been introduced into cells and may extend up to several hundred nucleotides (36, 87). For adaptation of RNAi to mammalian cells, 21-mer sense and antisense RNA oligonucleotides homologous to a portion of the gene of interest are synthesized and annealed and introduced into cells by transfection (12, 23). These RNAs bind specifically to the cellular mRNA of interest and activate an RNA degradation process that leads to an 80 to 90% decreases in the levels of the corresponding protein. Thus, RNAi can be used to silence the gene of interest but not other genes (74). The use of 21-mer RNAs in mammalian cells, rather than of longer RNAs that are utilized in other species, avoids the activation of double-stranded RNA dependent protein kinase PKR and nonspecific RNases that nonspecifically silence gene expression (6, 12, 23).
In this study, we utilized RNAi to address several specific points. First, we asked whether a viral gene such as the HIV-1 tat could be inhibited by RNAi during both early and late phases of HIV-1 infection. Second, we addressed whether RNAi-mediated decreases in the NF-κB p65 subunit could alter HIV-1 replication. Finally, we addressed whether inhibition of another viral gene, in this case HIV-1 reverse transcriptase (RT), could also alter viral replication. The results of these studies suggest that RNAi is a useful technique to address the complex interactions of viral and cellular regulatory proteins involved in the control of HIV-1 replication.
MATERIALS AND METHODS
Cell lines.
293T and HeLa-CD4-LTR-β-gal (MAGI; obtained from the NIH AIDS Research and Reference Reagent Program, catalog no. 1470) cells were cultured and maintained in Dulbecco's minimal essential medium containing 10% fetal bovine serum (FBS) (Gibco-BRL) and 1% glutamine (Gibco-BRL) with 1% antibiotic solution (penicillin and streptomycin; Gibco-BRL). The MAGI cells were detached from the tissue culture plates using sterile phosphate-buffered saline (PBS) containing 1 mM EDTA. Jurkat cells were cultured in RPMI culture medium containing 10% FBS.
Plasmids.
A plasmid containing the HIV-1 NL4.3 proviral DNA (pNL4.3) was a gift from J. Victor Garcia, University of Texas Southwestern Medical Center, Dallas, Tex. The plasmid pTat-GFP was generated by ligating a HindIII and BamHI fragment from HIV-1 tat encoding 1 to 71 amino acids (amplified from a plasmid bearing the HIV-1 HXB2 proviral DNA using the primer pair 5′-CTCGAGCTCAAGCTTATGGAGCCAGT-AGATCCTAGA-3′ and 5′-CGGTGGATCCTGCTTTGATAGAGAAACT-TGATG-3′) into the corresponding sites in the plasmid pEGFPN-1 (Clontech, Palo Alto, Calif.). The tat gene was then subjected to DNA sequencing.
siRNA sequences.
The sequence of the sense strand of the siRNAs oligonucleotides is shown for HIV-1 tat (tat1, 5′-UAUGGCAGGAAGAAGCGGA-3′; tat2, 5′-CUAGAGCCCUGGAAGC-AUC-3′; and tat3, 5′-GAAGCGGAGACAGCGACGA-3′), HIV-1 RT (RT1, 5′-GAGACACCAGGGAUUAGAU-3′; and RT2, 5′-UGAGACACCAGGGAUU-AGA-3′), p65 (5′-GCCCUAUCCCUUUACGUCA-3′), and human T-cell leukemia type 1 (HTLV-1) tax (5′-GAUGGACGCGUUAUCGGCU-3′). All these RNAs and the corresponding antisense RNA oligonucleotides were synthesized. An overhang of two thymidine residues (dTdT) was included at the 3′ end of all RNA oligonucleotides. The RNA oligonucleotides were synthesized at Dharmacon Research Inc. (Lafayette, Colo.). The RNA oligonucleotides were dissolved in Tris-EDTA (10 mM Tris-HCl, pH 8.0, and 1 mM EDTA) as 200 μM solutions and were stored at −20°C. Double-stranded siRNA molecules were generated by mixing the corresponding pair of sense and antisense RNA oligonucleotides in annealing buffer (30 mM HEPES-KOH, pH 7.9; 100 mM potassium acetate, and 2 mM magnesium acetate) at 20 μM and by then incubating the reaction mixture at 95°C for 2 min, followed by gradual cooling to room temperature. The siRNAs were then aliqoted and stored at −20°C.
Generation of HIV-1 virions.
293T cells at 50 to 70% confluence were transiently transfected with 10 μg of pNL4.3 vector per 100-mm-diameter plate using 25 μl of Genejuice (Novagen). Three days posttransfection, the culture supernatant was collected and subjected to centrifugation followed by filtration. HIV-1 virions in the supernatants were quantitated using a p24 antigen enzyme-linked immunosorbent assay (ELISA) (Perkin-Elmer Life Sciences, Boston, Mass.).
Transfections of siRNAs.
Twenty-four hours before siRNA transfection, the MAGI cells were seeded onto six-well plates (Corning) in Dulbecco's minimal essential medium containing 10% FBS with no antibiotics. Approximately 106 cells were plated per six-well plate to give 30 to 50% confluence at the time of transfections. The siRNAs were transfected to a final concentration of 50 nM using Oligofectamine (Invitrogen) according to the manufacturer's recommendations. The siRNAs were incubated with the cells for 3 or 6 days, and the cells were processed for Western blot analysis. For tat-specific RNA interference, the MAGI cells were transfected with 2.0 μg of pTat-GFP/well by using 3.0 μl of GeneJuice (Novagen) approximately 24 h after siRNA transfection without changing the medium. The cells were allowed to grow for another 48 h, and whole-cell extracts were prepared.
To transfect Jurkat cells with siRNAs, 2 × 105 cells were washed with serum- and antibiotic-free RPMI medium, resuspended in 50 μl of serum- and antibiotic-free RPMI medium, and added to a well in a 96-well tissue culture dish. A preincubated mixture of 100 pmol of siRNA in optiMEM medium and 0.8 μl of Oligofectamine (50 μl in total volume) was added to Jurkat cells and incubated overnight at 37°C. Cells were washed once and were resuspended in 200 μl of 10% FBS containing RPMI medium.
HIV-1 infection.
DNase I-treated HIV-1 virions equivalent to 10 to 50 ng of p24 antigen were used for infection of MAGI and Jurkat cells. The virions were mixed with 20 μg of DEAE-Dextran/ml (stock solution, 10 mg/ml) and were added to the MAGI or Jurkat cell medium 12 to 24 h after transfection of the siRNAs. Culture supernatants were collected 1, 3, and 6 days following infection. On day 3, the cells were washed twice with PBS, fresh medium was added, and the culture supernatants were collected 3 days later. In several experiments, the siRNA-transfected and HIV-1-infected cells were also split on days 3 and 6 following infection and were grown for an additional 7 to 10 days. The concentration of virus in the culture supernatants was determined using the p24 antigen ELISA.
Analysis of HIV-1-specific reverse-transcribed DNA.
Twenty-four hours after HIV-1 infection of MAGI cells, the cells were washed two times with sterile PBS and total cytoplasmic nucleic acids were obtained by modified Hirt lysis as described earlier (39). Chromosomal DNA concentrations of Hirt lysates were normalized by “hot” PCR amplification targeted to the mitochondrial DNA-encoded cytochrome c oxidase II (cyt-oxi II) gene as described previously (39). Following normalization of Hirt lysate DNA for their cyt-oxi II content, 2 to 5 μl of each lysate was directly assayed for HIV-1 DNA by 30 cycles of hot PCR as previously described (39). A 143-bp, “strong-stop” DNA fragment that corresponds to the R-U5 sequences was amplified using primers for sense (5′-GCTAACTAGGG-AACCCACTGCTT-3′) (+43/+65) and antisense (5′-CTGCTAGAGATTTTT-CCACACTGAC-3′) (+182/+158). The primer pair sense (5′-TGGCGTGC-CCTCAGATGCTG-3′) (−49/−30) and antisense (5′-AAGCAGTGGGTTCCCT-AGTTAG-3′) (+62/+42), which amplify a “jump” DNA fragment (111 bp) and correspond to R and U3 sequences, and sense (5′-CAAGTAGTGTGTGCCCGTCTGTT-3′) (+96/+118) and antisense (5′-CGAGAGAGCTCCTCTGGTTCTAC-3′) (+234/+212), which amplify a full-length DNA fragment (138 bp) that contains the 5′-untranslated and U5 sequence, were also used to amplify Hirt lysate DNA. One of the primers was 32P labeled and used in hot PCR. The PCR amplification was initiated with a 95°C denaturation for 5 min, and the DNA was amplified for 30 cycles with each cycle consisting of two steps at 93°C for 1 min and 65°C for 2 min. To amplify the full-length DNA fragment, an initial 5 cycles at 93°C for 1 min, 40°C for 30 s, and 72°C for 1 min was included, followed by 30 cycles of amplification as discussed earlier. All PCRs were subjected to electrophoresis in polyacrylamide gels and autoradiography.
RNA isolation.
RNA from HIV-1 virion-infected MAGI cells was isolated using Trizol reagent (Gibco-BRL) as per the manufacturer's recommendations. Nucleic acids were precipitated overnight and recovered by 15,000 × g centrifugation at 4°C for 15 min. The pellet was washed with 70% ethanol, centrifuged as before, and resuspended in 50 μl of Tris-EDTA containing 10 mM Tris, pH 7.8, and 0.1 mM EDTA. Approximately 4 μg of total RNA in a 10-μl reaction mixture was treated with RNase-free DNase I (Gibco-BRL) for 15 min at room temperature followed by inactivation with the addition of EDTA and the heat inactivation at 70°C for 10 min. Reverse transcription of tat, HIV-1 leader sequences, and the α-actin gene was performed separately in either the presence or absence of Moloney murine leukemia virus RT (Gibco-BRL). A 10-μl reaction mixture containing 2 to 4 μl of RNA (0.25 to 1 μg), RT buffer (Gibco-BRL), 10 mM dithiothreitol, 200 U of Moloney murine leukemia virus RT, 0.1 mM deoxynucleoside triphosphates, 0.01% bovine serum albumin, 10 to 15 U of RNasin, and 3 μg of random primers (Gibco-BRL) was incubated for 2 h at 37°C. Subsequently, 1 to 3 ml of RT reaction mixture was amplified separately by primers specific for HIV-1 tat (forward, 5′-CTCGAGCTCAAGCTTA-TGGAGCCAGTAGATCCTAGA-3′; and reverse, 5′-CGGTGGATCCTGC-TTTGATAGAGAAACTTGATG-3′), HIV-1 leader RNA extending from +31 to +259 (forward, 5′-CTGGGAGCTCTCTGGCTAAC-3′; and reverse, 5′-CTTCAGCAAGCCGAGTCCTG-3′), or α-actin (forward, 5′-GGCATCA-TCACCAACTGGG-3′; and reverse, 5′-CAGGGCCACGTAGCACAG-3′) genes using Taq polymerase (Gibco-BRL) and buffer D (Invitrogen). Following a 95°C denaturation for 5 min, 30 cycles (25 cycles for HIV leader RNA) of amplification was performed at 93°C for 1 min, 55°C for 30 s, and 72°C for 1 min. The PCR products were visualized after agarose gel electrophoresis and photographed.
Analysis of HIV-1 proviral DNA.
MAGI cells transfected with siRNA were infected with HIV-1 NL4.3 virus and split into six-well plates (1:7 ratio) on day 3 postinfection. The cells were grown for another 7 days, and chromosomal DNA was then prepared. Cell pellets were further processed for genomic DNA isolation using the Wizard Genomic DNA purification kit (Promega, Madison, Wis.). Subsequently, 40 ng of genomic DNA was amplified using primer pairs specific for HIV-1 gag (forward, 5′-GCGGGGGAGAATTAGATCGAT-3′; and reverse, 5′-CTCTTCCTCTATC TTGTCTAA-3′) and the cellular α-actin gene (forward, 5′-GGCATCATCA CCAACTGGG-3′; and reverse, 5′-CAGGGCCACGTAGCACAG-3′) using Platinum Taq polymerase (Gibco-BRL) and buffer D (Invitrogen). Following a 95°C denaturation for 5 min, 35 cycles of amplification was performed at 93°C for 1 min, 55°C for 30 s, and 72°C for 1 min. To amplify the HIV-1 gag gene, an initial 5 cycles at 93°C for 1 min, 37°C for 30 s, and 72°C for 1 min was included followed by 30 cycles of amplification as discussed earlier. The PCR products were visualized after agarose gel electrophoresis and photographed following ethidium bromide staining. The pNL4.3 DNA (1 or 4 ng) was amplified as a control.
Western blot analysis.
Whole-cell extracts were prepared from various siRNA-transfected and HIV-1-infected MAGI cells 6 days postinfection. Cell extracts were also prepared from MAGI cells transfected with siRNA alone or transfected with siRNA followed by transfection of pTat-GFP. Extracts (equivalent of 20 to 30 μg) of protein were subjected to electrophoresis on sodium dodecyl sulfate-10% polyacrylamide gels, transferred onto nitrocellulose membranes, and analyzed for the relative levels of both HIV-1-specific and cell-specific proteins. For the detection of HIV-1 proteins, the blots were probed with a 1:10,000 dilution of human HIV-1 immunoglobulin (NIH ARRP no. 3957) and a 1:5,000 dilution of horseradish peroxidase-linked sheep anti-human immunoglobulin (Amersham). Visualization of proteins was carried out using enhanced chemiluminescence (Amersham). These blots were reprobed with either p65, green fluorescent protein (GFP), or α-actin antibodies (Santa Cruz Biotechnology, Santa Cruz, Calif.). The blots were scanned using ChemiImager (Alpha Innotech Corp., San Leandro, Calif.) to determine the relative intensity of each band.
β-Galactosidase staining of cells.
Cells were washed twice with PBS and were then fixed with 3.7% formaldehyde in PBS for 10 min at room temperature. Cells were then washed with PBS to remove traces of formaldehyde and were treated with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) solution (0.2% X-Gal, 2 mM MgCl2, 5 mM K4Fe[CN]6 · 3H2O, and 5 mM K3Fe[CN]6 prepared in PBS) for 1 to 16 h at 37°C. When the blue-stained cells were visible, X-Gal solution was removed and cells were covered with PBS and photographed.
RESULTS
siRNA inhibition of the expression of the NF-κB p65 subunit and Tat.
It is important to better understand the role of specific viral and cellular genes that regulate the HIV-1 life cycle. A variety of CD4-positive human T-lymphocyte cell lines, including Jurkat, CEM, HUT78, SupT, and U297 in addition to non-T-cell lines such as HeLa-CD4 and HeLa-CD4-β-gal (MAGI), have been used to study different aspects of the HIV-1 life cycle. These later cells, which stably express CD4 receptors on the cell surface, are capable of being infected by HIV-1. Since they contain an HIV-1 LTR fused to the β-galactosidase gene, infectious virus producing Tat can transactivate the LTR-β-galactosidase reporter and increase β-galactosidase activity. Thus, staining of MAGI cells to determine β-galactosidase activity makes these cells an excellent indicator to determine the number of HIV-1 infectious particles (55).
RNAi with siRNAs has been successfully utilized in a number of recent studies in cultured mammalian cells to reduce the expression of specific cellular genes (12, 23, 31). In this study, RNAi was utilized in MAGI cells to assay how inhibition of the expression of the NF-κB p65 subunit and of the HIV-1 Tat and RT proteins alters HIV-1 replication. MAGI cells were utilized because their high transfection efficiency with RNA oligonucleotides permitted us to establish the efficacy of RNAi in the study of HIV-1 replication.
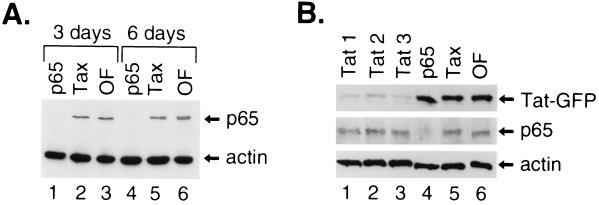
First, the degree and the duration of siRNA inhibition of p65 andTat expression were addressed. As a control in these studies, siRNA specific to the HTLV-1 tax gene was used. MAGI cells were transfected with these siRNA oligonucleotides, and the cells were harvested at either 3 or 6 days posttransfection. Cell lysates were prepared and then subjected to Western blot analysis. This analysis indicated that the p65 levels in these extracts were decreased in the presence of p65-specific siRNA (Fig. 1A, lanes 1 and 4), whereas the levels of p65 were not affected by transfection of either tax siRNA (Fig. 1A, lanes 2 and 5) or Oligofectamine alone (Fig. 1A, lanes 3 and 6). Interestingly, the RNAi-mediated decreases in p65 levels could be observed for almost 6 days following a single transfection. These siRNAs had no significant effect on the expression levels of another endogenous gene, α-actin (Fig. 1A, lanes 1 to 6). These results indicate the specificity of siRNA in decreasing p65 expression.
FIG. 1.
Effect of siRNAs on inhibiting gene expression. (A) MAGI cells were transfected with 50 nM siRNAs directed against either the NF-κB p65 subunit (lanes 1 and 4), HTLV-1 tax (lanes 2 and 5), or Oligofectamine alone (OF, lanes 3 and 6). Cells were harvested at either 3 days (lanes 1 to 3) or 6 days (lanes 4 to 6) posttransfection, and the lysates (20 μg) were subjected to Western blot analysis using antibodies directed against p65 (top gel) or α-actin (lower gel). (B) MAGI cells were transfected with a 50 nM concentration of each of three siRNAs directed against HIV-1 tat (tat1, tat2, or tat3, lanes 1 to 3), p65 (lane 4), tax (lane 5), or Oligofectamine alone (lane 6) for 24 h. The cells were then transfected with 2 μg of a CMV expression vector expressing Tat-GFP. Cells were harvested 48 h later, and Western blot analysis was performed on these lysates (20 μg) with antibodies directed against GFP (top gel). The Western blot was reprobed with antibodies directed against p65 (middle gel) and α-actin (bottom gel).
Previous studies demonstrated that transfection of luciferase-specific siRNAs could result in the decreased expression of a transfected luciferase reporter gene (23). Next, we sought to determine whether siRNA could be utilized to decrease the levels of Tat expressed from a cytomegalovirus (CMV) expression vector. Due to the inability of Western blot analysis with Tat-specific antibodies to detect expression of a transfected HIV-1 tat gene in MAGI cells, we generated a CMV expression vector, pTat-GFP, which expressed a GFP fusion with Tat. The pTat-GFP vector was transfected into MAGI cells that had been previously transfected with each of three specific siRNAs directed against tat or siRNAs directed against either p65 or tax. Western blot analysis with antibody directed against GFP indicated reduced expression of Tat-GFP for each of the three different tat-specific siRNAs, with tat1 and tat3 being two or three times more effective than tat2 (Fig. 1B, lanes 1 to 3). In contrast, there was no detectable decrease in Tat-GFP levels in the presence of siRNA directed against p65 (Fig. 1B, lane 4), tax (Fig. 1B, lane 5), or Oligofectamine alone (Fig. 1B, lane 6). Tat-GFP expression was reduced up to 70 to 80% by tat-specific siRNAs, compared to that with p65 or Tax siRNAs or Oligofectamine alone. The expression of p65 was decreased only in the presence of the p65-specific siRNA (Fig. 1B, lane 4), whereas α-actin levels were not affected by transfection of any of these siRNAs (Fig. 1B, lanes 1 to 6). These results demonstrate the specificity of siRNAs in decreasing the expression of both endogenous and exogenously introduced genes.
Inhibition of HIV-1 replication by siRNAs directed against HIV-1 tat and the NF-κB p65 subunit.
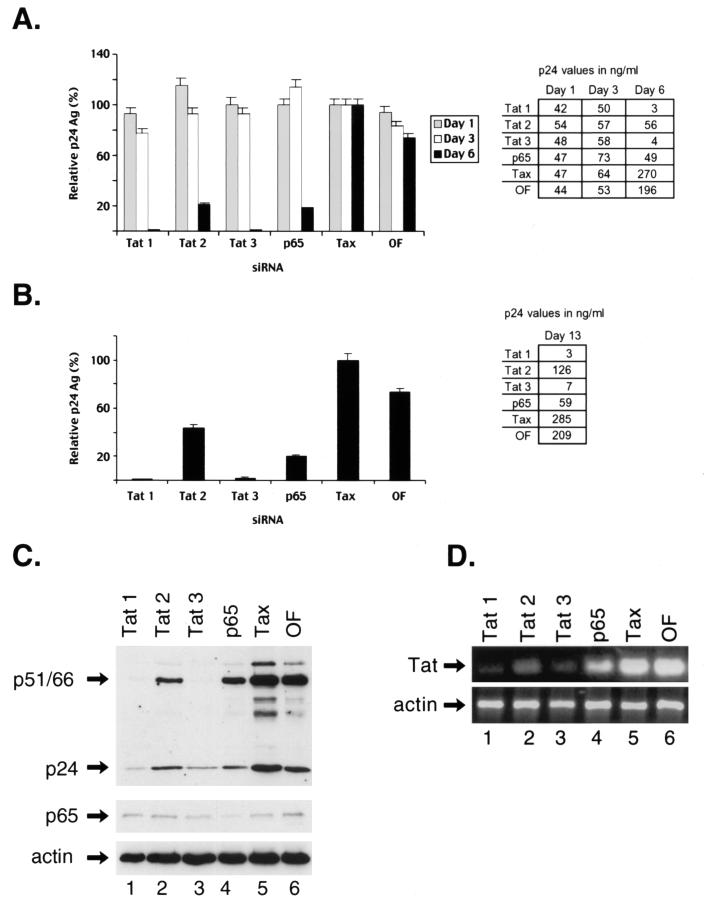
It is well established that both the NF-κB and HIV-1 Tat proteins play an important role in regulating HIV-1 gene expression (1, 19, 27, 64). Thus, it is likely that the siRNA-mediated decreases in the levels of these proteins could potentially inhibit HIV-1 replication. MAGI cells were transfected with siRNAs directed against either p65, tat, or tax for 24 h. These cells were then infected with the HIV-1 NL4.3 strain, and culture supernatants were collected at either day 1, 3, or 6 following infection and were assayed for p24 antigen levels using ELISA. The relative amounts of virus produced in the presence of siRNAs directed against either tat, p65, or Oligofectamine relative to tax are shown (Fig. 2A). The amounts of virus released into the culture supernatants of cells that were treated with various siRNAs were similar at 1 (Fig. 2A, gray bars) and 3 days postinfection (Fig. 2A, white bars), indicating that RNAi-mediated decreases in the levels of the targeted proteins at these times did not significantly alter viral production. However, a significant decrease in HIV-1 replication was observed when supernatants were collected at 6 days post-HIV-1 infection in MAGI cells that were transfected with siRNAs directed against either tat or p65 but not with siRNA directed against tax or Oligofectamine alone (Fig. 2A, black bars). The cells transfected with the tat1 and tat3 siRNAs demonstrated more than a 99% decrease in HIV-1 production compared to those transfected with tat2 siRNA, which exhibited an 80% decrease in HIV-1 production (Fig. 2A). The reason for the differences in the efficacy of these siRNAs was unclear. The siRNA directed against p65 did not result in as great an inhibition of HIV-1 production as that found for the cells transfected with the tat1 and tat3 siRNAs. These results indicate that HIV-1 infection can be markedly inhibited by transfection of MAGI cells with siRNAs directed against either tat or p65.
FIG. 2.
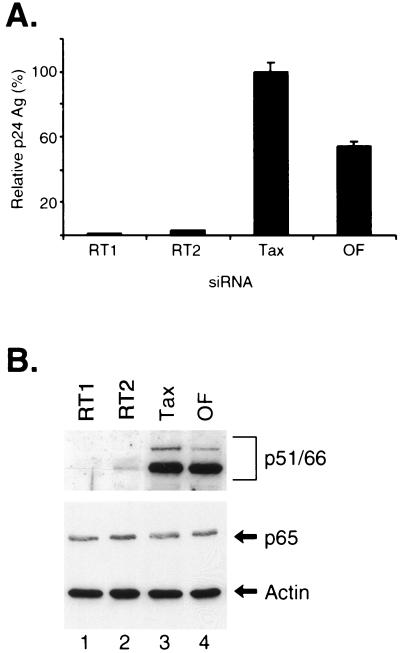
Inhibition of HIV-1 replication by siRNAs. (A) MAGI cells were transfected with 50 nM concentrations of siRNAs directed against either HIV-1 tat (tat1, tat2, and tat3), p65, or HTLV-1 tax or with Oligofectamine alone (OF). Twenty-four hours later, the cells were infected with HIV-1 NL4.3 and cell culture supernatants were collected on day 1 (gray bars), 3 (white bars), or 6 (black bars) postinfection and were assayed for p24 antigen (Ag). The p24 antigen values are expressed as the percent values relative to those seen with Tax siRNA. The actual p24 antigen values in the culture supernatants are shown in the adjacent table. (B) MAGI cells transfected with various siRNAs for 24 h prior to HIV-1 infection were split on the 3rd day postinfection, and culture supernatants were collected on day 10 (day 13 postinfection) and were assayed for p24 antigen. The p24 antigen values were expressed as the percentage relative to that seen with tax siRNA. (C) MAGI cells were transfected with siRNAs directed against HIV-1 tat (tat1, lane 1; tat2, lane 2; and tat3, lane 3), p65 (lane 4), HTLV-1 tax (lane 5), or Oligofectamine alone (OF, lane 6) for 24 h and were then infected with HIV-1 for 6 days. Whole-cell lysates from these cells were prepared and analyzed by Western blotting. The membrane was first probed with HIV-1-specific antisera and then reprobed with antibodies directed against p65 and α-actin. The positions of the HIV-1-specific proteins p24 antigen and p51/p66 (RT) are indicated. (D) RNA was prepared from the HIV-1-infected MAGI cells and was subjected to RT-PCR amplification to determine the relative levels of HIV-1 tat RNA expression. The α-actin RNA was also amplified in these samples as a control.
Next, we investigated the duration of siRNA inhibition of HIV-1 replication. For this analysis, MAGI cells were first transfected with various siRNAs for 24 h and then these cells were infected with HIV-1. The cells were then split on the 3rd day following infection and were analyzed for p24 antigen levels at 13 days postinfection (Fig. 2B). A single transfection of HIV-1 tat1 and tat3 siRNAs led to a >95% decrease in p24 antigen levels, while tat2 and p65 siRNAs resulted in a 60 to 80% decrease in HIV-1 gene expression that lasted for at least 13 days (Fig. 2B). The cells were not assayed for longer times, since the MAGI cells that were transfected with either tax siRNA or Oligofectamine alone showed extensive syncytial formation indicative of HIV-1 cytopathicity with >90% syncytium formation, compared to <5% seen in cells transfected with tat- or p65-specific siRNAs.
Finally, we determined the relative expression levels of HIV-1-specific proteins in siRNA-transfected MAGI cells at 6 days post-HIV-1 infection. Western blot analysis with HIV-1-specific antisera demonstrated marked decreases in the levels of HIV-1- specific proteins, including RT (p51/p66) and p24 (Fig. 2C, top gel), which correlated with the decrease in HIV-1 virions in supernatants from MAGI cells (Fig. 2A). There were decreased levels of HIV-1-specific proteins in the presence of all three of the tat siRNAs (Fig. 2C, lanes 1 to 3, top gel), as well as of p65 siRNA (Fig. 2C, lane 4, top gel), compared to the HIV-1 protein levels seen in MAGI cells transfected with tax siRNA (Fig. 2C, lane 5, top gel) or Oligofectamine alone (Fig. 2C, lane 6, top gel). The tat1 and tat3 siRNAs resulted in significantly more inhibition of HIV-1 protein expression than did either tat2 or p65 siRNA. The p65 siRNA specifically reduced the expression of p65 (Fig. 2C, middle gel), and none of the siRNAs affected the expression of α-actin (Fig. 2C, bottom gel).
Finally, we analyzed the effect of these siRNAs on the expression of the tat gene. RT-PCR analysis of total RNA isolated from these samples revealed the down-regulation of HIV-1 tat RNA in the presence of all three of these tat siRNAs (Fig. 2D, lanes 1 to 3), as well as of p65 siRNA (Fig. 2D, lane 4), compared to the levels of this RNA in MAGI cells transfected with tax siRNA (Fig. 2D, lane 5) or Oligofectamine alone (Fig. 2D, lane 6). None of the siRNAs affected the levels of α-actin RNA (Fig. 2D, lower gel). These results further corroborate the specificity of RNAi in down-regulating the expression of HIV-1-specific proteins.
Effects of siRNA on early stages of HIV-1 infection.
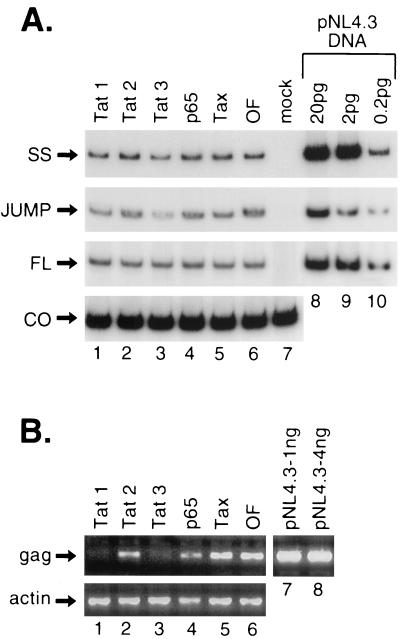
Next we explored whether transfection of siRNAs prior to HIV-1 infection altered an early step in the HIV-1 infectious cycle. Following HIV-1 binding and internalization, the initiation of reverse transcription occurs. The effect of various siRNAs on the HIV-1 reverse transcription was examined next. MAGI cells were transfected with siRNAs for 24 h and were then infected with HIV-1 for 24 h. Cytoplasmic DNA was prepared from these cells by Hirt lysis, and the level of HIV-1-specific reverse-transcribed strong-stop, jump, or full-length DNA was then analyzed. PCR amplification of HIV-1-specific strong-stop, jump, or full-length DNA demonstrated that transfection of siRNAs directed against either tat, p65, tax, or Oligofectamine alone prior to HIV-1 infection did not result in significant inhibition of the initiation or completion of HIV-1 reverse transcription (Fig. 3A, lanes 1 to 6, top three gels). For example, quantitation of the levels of product for strong-stop and jump DNA indicated that tat1 siRNA reduced the amount of this product by 30%, while tat3 siRNA reduced the amount of this product by 20%. None of these siRNAs resulted in decreases in the synthesis of the full-length product. No strong-stop DNA was detected in mock-infected cytoplasmic DNA (Fig. 3A, lane 7, top three gels). Different amounts of NL4.3 plasmid DNA were used as additional controls (Fig. 3A, lanes 8 to 10, top three gels). The cytoplasmic DNA used for PCR amplification was normalized using the mitochondrial cytochrome c oxidase gene (Fig. 3A, lanes 1 to 7, lower gel). These results indicate that an early step in the HIV-1 infection cycle was not significantly affected by transfection of the siRNAs and that the major effects of these siRNAs likely occur subsequent to HIV-1 reverse transcription.
FIG. 3.
Effect of siRNAs on different steps in the HIV-1 life cycle. (A) MAGI cells were transfected with siRNAs directed against either HIV-1 tat (tat1, lane 1; tat2, lane 2; and tat3, lane 3), p65 (lane 4), HTLV-1 tax (lane 5), or Oligofectamine alone (OF, lane 6) for 24 h and were then infected with HIV-1 for another 24 h. Hirt lysates were prepared from these cells, and the cytoplasmic DNA was amplified with oligonucleotide primers to assay for either HIV-1-specific strong-stop (SS) (+43/+65 and +182/+158) (top gel), first-strand jump (−49/−30 and +93/+70) (second gel), full-length (FL) viral DNA (+96/+118 and +234/+212) (third gel), or the mitochondrial cytochrome c oxidase (CO) gene (bottom gel). As controls, cytoplasmic DNA from mock-infected (lane 7) and different amounts of NL4.3 plasmid DNA (20, 2, and 0.2 pg, lanes 8 to 10) were also amplified. (B) Chromosomal DNA was prepared from MAGI cells that were transfected with siRNAs directed against either HIV-1 tat (tat1, lane 1; tat2, lane 2; and tat3, lane 3), p65 (lane 4), HTLV-1 tax (lane 5), or Oligofectamine alone (OF, lane 6) for 24 h and were then infected with HIV-1 for 10 days. Portions of HIV-1 gag (top gel) and cellular α-actin (lower gel) genes were amplified as detailed in Materials and Methods. As controls, 1 ng (lane 7) and 4 ng (lane 8) of NL4.3 plasmid DNA were also amplified.
The quantity of HIV-1 proviral DNA integrated into the host genome was then analyzed (Fig. 3B). Chromosomal DNA was purified from siRNA-transfected MAGI cells that had been infected with HIV-1 for 10 days. This time postinfection was chosen to allow for several cycles of reinfection of the target cells. DNA prepared from these cells was used to amplify a portion of the HIV-1 gag and cellular α-actin genes (Fig. 3B). The relative content of HIV-1 proviral DNA in cells transfected with siRNAs directed against either tat1, tat2, tat3, or p65 was compared with that of cells transfected with tax siRNA or Oligofectamine. Significant differences in the content of integrated HIV-1 DNA reflected in the amount of the HIV-1 gag gene was observed in cells containing tat (Fig. 3B, lanes 1 to 3, top gel) and p65 (Fig. 3B, lane 4, top gel) siRNAs, compared to MAGI cells that were transfected with tax siRNA (Fig. 3B, lane 5, top gel) or were treated with Oligofectamine alone (Fig. 3B, lane 6, top gel). There was no change in the levels of the α-actin gene in MAGI cells transfected with these siRNAs (Fig. 3B, lower gel). The decreased amounts of integrated HIV-1 DNA indicate that the production of small amounts of HIV-1 virions leads to the reinfection of fewer target cells.
Effect of tat and p65 siRNAs transfected following HIV-1 infection.
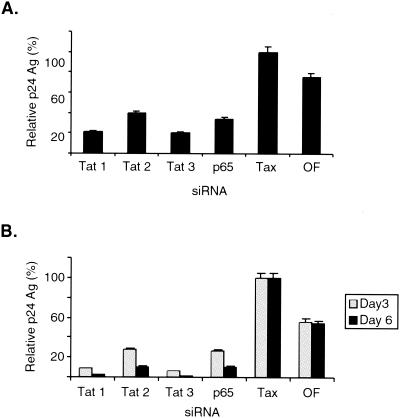
In the previous experiments, MAGI cells were transfected with siRNAs prior to HIV-1 infection. To address whether these siRNAs could reduce gene expression following HIV-1 infection, MAGI cells were infected with HIV-1 for 12 h prior to transfection of the various siRNAs. The culture supernatants were collected at 6 days following siRNA transfection and were assayed for relative p24 antigen levels. These results indicated that p65 and tat siRNAs, but not tax siRNA or Oligofectamine alone, inhibited viral replication by 60 to 80%, respectively, when introduced into MAGI cells following HIV-1 infection (Fig. 4A).
FIG. 4.
Transfection of tat and p65 siRNAs following HIV-1 infection. (A) MAGI cells were infected with HIV-1 for 12 h and were then transfected with siRNAs directed against either HIV-1 tat (tat1, tat2, or tat3), p65, HTLV-1 tax, or Oligofectamine alone (OF). Culture supernatants were collected at 6 days following siRNA transfection and were assayed for p24 antigen (Ag). (B) MAGI cells infected with HIV-1 for 6 days were split and were then transfected with siRNAs directed against HIV-1 tat (tat1, tat2, or tat3), p65, HTLV-1 tax, or Oligofectamine (OF) alone. Culture supernatants were collected at 3 and 6 days following siRNA transfection and were assayed for p24 antigen (Ag). In both parts of the figure, the p24 values are expressed as the percentage relative to that seen with tax siRNA.
In addition, we assayed whether transfection of siRNAs directed against tat and p65 could inhibit HIV-1 replication in MAGI cells that were previously infected with HIV-1. For this purpose, MAGI cells were infected with HIV-1 for 6 days and these cells were split prior to transfection of the various siRNAs. Culture supernatants from these HIV-1-infected cells were then collected on days 3 and 6 post-siRNA transfection, and the relative p24 antigen levels were then determined (Fig. 4B). These results indicated that siRNAs directed against tat and p65 but not against tax or Oligofectamine alone could inhibit HIV-1 replication from 80 to 95% when introduced into cells after 6 days of HIV-1 infection (Fig. 4B).
RT siRNAs can inhibit HIV-1 replication.
RT is a primary target for antiviral drugs that inhibit HIV-1 replication (18). We designed two siRNAs against the HIV-1 RT gene and tested whether they could inhibit HIV-1 replication in MAGI cells. MAGI cells were transfected for 24 h with either of these two siRNAs (RT1 and RT2) or tax siRNA and were then infected with HIV-1 (Fig. 5A). The culture supernatants were assayed for p24 antigen levels at 6 days postinfection. Transfection of both the RT1 and RT2 siRNAs markedly inhibited HIV-1 replication in the MAGI cells by >90%, compared to transfection of tax siRNA or Oligofectamine alone (Fig. 5A). Western blot analysis of whole-cell extracts from these cells demonstrated a specific decrease in the p51/p66 RT proteins following transfection of either of these RT siRNAs (Fig. 5B, top gel). There was no change in the levels of p65 and actin in the presence of these siRNAs (Fig. 5B, lower gel).
FIG. 5.
Inhibition of HIV-1 replication by siRNAs specific for HIV-1 RT. (A) MAGI cells were transfected with siRNAs directed against HIV-1 RT (RT1 or RT2), HTLV-1 tax, or Oligofectamine (OF) alone for 24 h, and the cells were then infected with HIV-1. Culture supernatants were collected at 6 days following siRNA transfection and were assayed for p24 antigen levels and expressed as the percentage relative to that seen with tax siRNA. (B) Whole-cell extracts were prepared from the MAGI cells that were transfected with siRNAs directed against HIV-1 RT (RT1, lane 1; or RT2, lane 2), HTLV-1 tax (lane 3), or Oligofectamine alone (OF, lane 4) for 24 h and then were infected with HIV-1 for 6 days. Western blot analysis was performed with HIV-1-specific antisera, and then the blot was reprobed with antibodies against p65 and α-actin.
Tat, RT, and p65 siRNAs inhibit β-galactosidase activity in HIV-1-infected MAGI cells.
MAGI cells contain a stably integrated HIV-1 LTR fused to the β-galactosidase gene (55). HIV-1 infection of these cells leads to increased expression of β-galactosidase due to the expression of Tat. The activity of β-galactosidase can be detected by staining of these cells with an X-Gal-containing reaction mixture. Thus, MAGI cells can be used as indicator cells to identify the level of HIV-1 infectious particles.
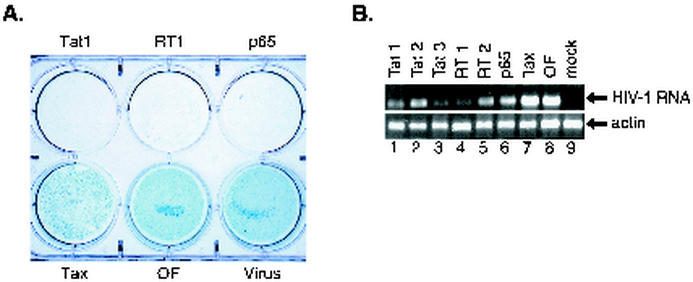
To further extend the studies of the effects of siRNAs on HIV-1 replication, these siRNAs were transfected into MAGI cells and, 24 h later, these cells were infected with HIV-1. These MAGI cells were then stained with the X-Gal-containing solution to determine β-galactosidase activity. More than 90% of the infected cells that were transfected with either tax siRNA or Oligofectamine alone or were infected with HIV-1 without other treatment demonstrated marked β-galactosidase positivity (Fig. 6A). In contrast, only a few cells had β-galactosidase activity when the MAGI cells were transfected with siRNAs directed against either tat, RT, or p65 (Fig. 6A). These results indicate that inhibition of HIV-1 replication in the presence of siRNAs directed against either tat, RT, or p65 resulted in very low levels of β-galactosidase expression.
FIG. 6.
β-Galactosidase staining of MAGI cells. MAGI cells were transfected with 50 nM siRNAs directed against either HIV-1 tat, RT, p65, HTLV-1 tax, or Oligofectamine alone (OF). Twenty-four hours later, the cells were infected with HIV-1 NL4.3. (A) On day 6 postinfection, the cells were washed and stained for β-galactosidase as described in Materials and Methods and photographed. Cells infected with HIV-1 in the absence of other treatment (control) are also shown. (B) At the same time postinfection, RNA was prepared from these HIV-1-infected MAGI cells and subjected to RT-PCR amplification of HIV-1 leader sequences with oligonucleotide primers to generate a fragment extending from +31 to +259 in order to determine relative levels of HIV-1 mRNA expression. The α-actin mRNA was also amplified from these samples as a control.
Next, we determined whether these siRNAs altered the expression of an RNA sequence extending between residues +31 and +259 that is present in all spliced and nonspliced HIV-1-encoded RNAs. RNAs prepared from these siRNA-transfected HIV-1-infected MAGI cells were analyzed by RT-PCR using oligonucleotide primers that would detect this RNA sequence. Decreased levels of HIV-1 mRNA were found in cells transfected with the tat1, tat2, and tat3 siRNAs in addition to the RT1, RT2, and p65 siRNAs but not Tax siRNA. There was no change in the levels of actin mRNA in these samples (Fig. 6B). These results suggest that siRNAs directed against different regions of the HIV-1 genome can result in decreased expression of both nonspliced and spliced HIV-1 mRNAs.
siRNAs directed against tat, RT, and p65 inhibit HIV-1 replication in Jurkat cells.
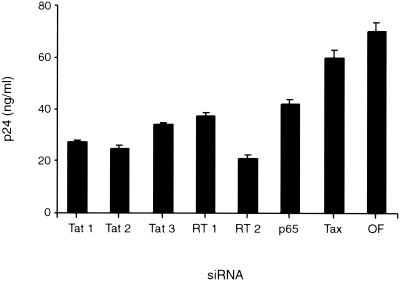
It was important to address whether siRNAs directed against tat, RT, and p65 could inhibit HIV-1 replication in T cells. For these studies, we assayed whether transfection of these siRNAs into Jurkat cells altered HIV-1 replication. Given the low efficiency of transfection of lymphocytes by RNAs, 20 times more siRNA was required to demonstrate these effects in Jurkat cells compared to what was required for MAGI cells. The siRNAs were transfected into Jurkat cells, and 16 h later these cells were infected with HIV-1; 3 days postinfection the levels of p24 antigen were measured. This analysis revealed a result similar to the results obtained with MAGI cells: siRNAs directed against different regions of tat, RT, and p65 reduced the amount of p24 antigen by two- or threefold compared to siRNA directed against Tax or mock transfection (Fig. 7). Transfection of these siRNAs did not alter the cell numbers for the 3 days of this analysis. Thus, siRNAs can inhibit HIV-1 replication in both T-cell and non-T-cell lines.
FIG. 7.
SiRNAs directed against tat, RT, and p65 inhibit HIV-1 replication in Jurkat cells. Jurkat cells (n = 2 × 105) were transfected with Oligofectamine containing 100 pmol of siRNAs corresponding to either tat1, tat2, and tat3; p65 and RT1 and -2; Tax or mock transfected. After 16 h, the cells were washed, resuspended in RPMI medium, and infected with HIV-1 NL4.3, and p24 antigen levels were determined by ELISA at 3 days postinfection.
DISCUSSION
In this study, we utilized RNAi to establish the efficacy of this technique for investigating HIV-1 gene expression and replication. We found that siRNAs directed against HIV-1 tat and RT and the p65 NF-κB subunit could result in specific decreases in the corresponding expression of these proteins. Thus, RNAi provides a useful tool for better dissecting the complex interplay between viral and cellular factors that regulate HIV-1 gene expression.
HIV-1 Tat is a key regulator of viral gene expression (45). In this study, we demonstrated that RNAi-mediated decreases in Tat expression at various times post-HIV-1 infection result in marked effects on its replication. Moreover, we were able to demonstrate that multiple regions of the tat gene could be targeted by RNAi and markedly decrease its expression. Such results will be important if RNAi is ever used for therapeutic purposes, given that tat has a high degree of conservation among different viral isolates and is not subject to frequent sequence variation. Similar inhibitory effects on HIV-1 replication were seen when siRNAs were targeted to the HIV-1 RT gene. RNAi-mediated degradation of multiple HIV-1 genes could potentially be utilized to inhibit HIV-1 replication. In addition, RNAi will be useful in better characterizing the functional interactions of various HIV-1 gene products during its replication.
NF-κB plays an important role in stimulating HIV-1 gene expression during T-cell activation (64). Various heterodimeric combinations of the NF-κB proteins, including p50/p65 and p52/p65, predominantly bind to two elements in the HIV-1 LTR to activate its gene expression. Although there has been conflicting evidence on whether NF-κB sites are essential for HIV-1 replication, it is clear that NF-κB plays an important role in the HIV-1 life cycle (13, 59, 68, 72). In the present study, we demonstrate that decreases in the expression of the NF-κB p65 subunit reduce HIV-1 gene expression and replication. Although the effects of reducing p65 on HIV-1 replication were significant, they did not result in as marked a degree of inhibition of HIV-1 gene expression as seen with siRNA directed against tat. However, these results indicate that, in non-T-cells, a reduction in p65 levels inhibits HIV-1 replication.
The use of siRNA directed against the NF-κB p65 subunit resulted in reduced HIV-1 replication. These results indicate that inhibition of the expression of cellular factors that bind to the HIV-1 LTR can decrease HIV-1 replication. However, it is important to note that the NF-κB proteins are critical for the regulation of immune function (51). For example, these proteins regulate the expression of a variety of genes encoding cytokines and cytokine receptors, chemokines, cell adhesion molecules, and cell surface receptors that are critical for T- and B-cell function. In mice, the targeted disruption of the gene encoding p65 results in reduced T- and B-cell proliferation without major defects in T- and B-cell maturation (22). These studies indicate that it will be important to monitor immune function if p65 siRNAs are utilized in clinical settings to treat HIV-1 infection.
The results presented here establish RNAi as a technique that will be useful in addressing the in vivo roles of cellular factors involved in HIV-1 replication. For example, a variety of transcriptional elongation factors, such as SPT5/SPT4 and NELF, has been implicated in HIV-1 gene regulation (42, 53, 70, 77, 78, 84, 85). Other HIV-1 LTR binding factors, such as SP1, LEF, and LBP, can also be targeted by RNAi in order to further address their role in the HIV-1 life cycle. Finally, additional proteins that have been reported to have roles in the HIV-1 life cycle, such as TFIIH and TFIID, can be investigated with RNAi. For example, the Tsg101 protein, which functions in vacuolar protein sorting, was demonstrated to be important in HIV-1 budding from host cells using siRNA analysis (31).
After this work was submitted, several manuscripts were published that demonstrated that transfection of siRNAs directed against other cellular and viral factors could inhibit HIV-1 replication. In one study, siRNAs directed against either CD4 or the gag gene were able to prevent HIV-1 replication in both MAGI cells and H9 cells (65). In another study, siRNAs directed against TAR, Vif, or a GFP gene inserted into nef were found to significantly reduce HIV-1 replication (44). Finally siRNA produced from an expression plasmid containing a polymerase III promoter was found to inhibit HIV-1 rev and to lead to decreased virus production (58). Thus, the results of these studies are in good agreement with the results presented in our analysis.
One question that arises from all of these studies is the step in the HIV-1 life cycle at which siRNA inhibits replication. The two leading possibilities are that siRNAs target HIV-1 genomic RNA at an early step following viral entry and/or that siRNA targets prespliced viral RNA produced following viral integration. The previously discussed studies and our study indicate that siRNAs can silence HIV-1 gene expression after viral integration. It is also probable that siRNA can inhibit HIV-1 at a step prior to integration. However, our results suggest that the major effect of this step is subsequent to HIV-1 reverse transcription, while a previous study suggested that this inhibition occurred prior to reverse transcription (44). Differences in the experimental conditions, including the time that the assays were performed and the amount of virus used, likely account for these different results. In our analysis, we were able to obtain prolonged suppression of HIV-1 replication in MAGI cells extending at least 13 days. The prolonged time of suppression of viral replication seen in our study compared to that found in a previous study is likely due to the more efficient transfection of siRNAs in MAGI cells than in H9 cells (65). The recent demonstration that siRNAs can be produced from RNA polymerase III promoters such as H1 and U6 suggests that both macrophages and primary T cells may also be efficiently targeted by using these promoters in an attempt to inhibit HIV-1 replication (11, 58). In summary, our data and those of other studies indicate that siRNAs can lead to the inhibition of HIV-1 replication likely by effects both prior to and following viral integration.
The studies presented here are useful in establishing a system in which to better investigate cellular factors that are involved in the control of HIV-1 gene expression. Moreover, transfection of siRNAs into cells avoids the potential cellular toxicity due to the chronic expression of these siRNAs. Thus, RNAi provides an interesting approach to better characterize the role of specific cellular and viral factors involved in regulating HIV-1 gene expression.
Acknowledgments
We thank Sharon Johnson for preparing the manuscript, Alex Herrera for assistance with the figures, and Victor Garcia-Martinez for helpful discussions.
This work was supported by grants from the NIH and the Veterans Administration.
REFERENCES
- 1.Alcami, J., T. Lain de Lera, L. Folgueira, M. A. Pedraza, J. M. Jacque, F. Bachelerie, A. R. Noriega, R. T. Hay, D. Harrich, R. B. Gaynor, J. L. Virelizier, and F. Arenzana-Seisdedos. 1995. Absolute dependence on kappa B responsive elements for initiation and Tat-mediated amplification of HIV transcription in blood CD4 T lymphocytes. EMBO J. 14:1552-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baeuerle, P. A., and D. Baltimore. 1996. NF-κB: ten years after. Cell 87:13-20. [DOI] [PubMed] [Google Scholar]
- 3.Baldwin, A. S. 1996. The NF-κB and IκB proteins: new discoveries and insights. Annu. Rev. Immunol. 14:649-681. [DOI] [PubMed] [Google Scholar]
- 4.Barboric, M., R. M. Nissen, S. Kanazawa, N. Jabrane-Ferrat, and B. M. Peterlin. 2001. NF-κB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell 8:327-337. [DOI] [PubMed] [Google Scholar]
- 5.Bass, B. L. 2000. Double-stranded RNA as a template for gene silencing. Cell 101:235-238. [DOI] [PubMed] [Google Scholar]
- 6.Bass, B. L. 2001. RNA interference. The short answer. Nature 411:428-429. [DOI] [PubMed] [Google Scholar]
- 7.Benkirane, M., R. F. Chun, H. Xiao, V. V. Ogryzko, B. H. Howard, Y. Nakatani, and K. T. Jeang. 1998. Activation of integrated provirus requires histone acetyltransferase. p300 and P/CAF are coactivators for HIV-1 Tat. J. Biol. Chem. 273:24898-24905. [DOI] [PubMed] [Google Scholar]
- 8.Berkhout, B., and K.-T. Jeang. 1992. Functional roles for the TATA promoter and enhancers in basal and Tat-induced expression of the human immunodeficiency virus type 1 long terminal repeat. J. Virol. 66:139-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berkhout, B., R. H. Silverman, and K. T. Jeang. 1989. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell 59:273-282. [DOI] [PubMed] [Google Scholar]
- 10.Bieniasz, P. D., T. A. Grdina, H. P. Bogerd, and B. R. Cullen. 1998. Recruitment of a protein complex containing Tat and cyclin T1 to TAR governs the species specificity of HIV-1 Tat. EMBO J. 17:7056-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 12.Caplen, N. J., S. Parrish, F. Imani, A. Fire, and R. A. Morgan. 2001. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl. Acad. Sci. USA 98:9742-9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, B. K., M. B. Feinberg, and D. Baltimore. 1997. The κB sites in the human immunodeficiency virus type 1 long terminal repeat enhance virus replication yet are not absolutely required for viral growth. J. Virol. 71:5495-5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, D., Y. Fong, and Q. Zhou. 1999. Specific interaction of Tat with the human but not rodent P-TEFb complex mediates the species-specific Tat activation of HIV-1 transcription. Proc. Natl. Acad. Sci. USA 96:2728-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, Z. J., L. Parent, and T. Maniatis. 1996. Site-specific phosphorylation of IκBα by a novel ubiquitination-dependent protein kinase activity. Cell 84:853-862. [DOI] [PubMed] [Google Scholar]
- 16.Clemens, J. C., C. A. Worby, Simonson-Leff, M. Muda, T. Maehama, B. A. Hemmings, and J. E. Dixon. 2000. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl. Acad. Sci. USA 97:6499-6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cogoni, C. 2001. Homology-dependent gene silencing mechanisms in fungi. Annu. Rev. Microbiol. 55:381-406. [DOI] [PubMed] [Google Scholar]
- 18.Condra, J. H., M. D. Miller, D. J. Hazuda, and E. A. Emini. 2002. Potential new therapies for the treatment of HIV-1 infection. Annu. Rev. Med. 53:541-555. [DOI] [PubMed] [Google Scholar]
- 19.Dayton, A. I., J. G. Sodroski, C. A. Rosen, W. C. Goh, and W. A. Haseltine. 1986. The trans-activator gene of the human T cell lymphotropic virus type III is required for replication. Cell 44:941-947. [DOI] [PubMed] [Google Scholar]
- 20.DiDonato, J. A., M. Hayakawa, D. M. Rothwarf, E. Zandi, and M. Karin. 1997. A cytokine-responsive IκB kinase that activates the transcription factor NF-κB. Nature 388:548-554. [DOI] [PubMed] [Google Scholar]
- 21.Dingwall, C., I. Ernberg, M. J. Gait, S. M. Green, S. Heaphy, J. Karn, A. D. Lowe, M. Singh, M. A. Skinner, and R. Valerio. 1989. Human immunodeficiency virus 1 Tat protein binds trans-activation-responsive region (TAR) RNA in vitro. Proc. Natl. Acad. Sci. USA 86:6925-6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doi, T. S., T. Takahashi, O. Taguchi, T. Azuma, and Y. Obata. 1997. NF-κB RelA-deficient lymphocytes: normal development of T cells and B cells, impaired production of IgA and IgG1 and reduced proliferative responses. J. Exp. Med. 185:953-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 24.Elbashir, S. M., W. Lendeckel, and T. Tuschl. 2001. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15:188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elbashir, S. M., J. Martinez, A. Patkaniowska, W. Lendeckel, and T. Tuschl. 2001. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 20:6877-6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver, and C. C. Mello. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806-811. [DOI] [PubMed] [Google Scholar]
- 27.Fisher, A. G., M. B. Feinberg, S. F. Josephs, M. E. Harper, L. M. Marselle, G. Reyes, M. A. Gonda, A. Aldovini, C. Debouk, R. C. Gallo, and F. Wong-Staal. 1986. The trans-activator gene of HTLV-III is essential for virus replication. Nature 320:367-371. [DOI] [PubMed] [Google Scholar]
- 28.Fujinaga, K., R. Taube, J. Wimmer, T. P. Cujec, and B. M. Peterlin. 1999. Interactions between human cyclin T, Tat, and the transactivation response element (TAR) are disrupted by a cysteine to tyrosine substitution found in mouse cyclin T. Proc. Natl. Acad. Sci. USA 96:1285-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garber, M. E., P. Wei, V. N. KewalRamani, T. P. Mayall, C. H. Herrmann, A. P. Rice, D. R. Littman, and K. A. Jones. 1998. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 12:3512-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.García-Martínez, L. F., G. Mavankal, J. Neveu, W. Lane, D. Sigman, D. Ivanov, and R. B. Gaynor. 1997. Purification of a Tat associated kinase reveals a TFIIH complex that modulates HIV-1 transcription. EMBO J. 16:2836-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 32.Gaynor, R. B. 1995. Regulation of HIV-1 gene expression by the transactivator protein Tat. Curr. Top. Microbiol. Immunol. 193:51-77. [DOI] [PubMed] [Google Scholar]
- 33.Ghosh, S., and M. Karin. 2002. Missing pieces in the NF-kappaB puzzle. Cell 109:S81-S96. [DOI] [PubMed] [Google Scholar]
- 34.Ghosh, S., M. J. May, and E. B. Kopp. 1998. NF-κB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 16:225-260. [DOI] [PubMed] [Google Scholar]
- 35.Griffin, G. E., K. Leung, T. M. Folks, S. Kunkel, and G. J. Nabel. 1989. Activation of HIV gene expression during monocyte differentiation by induction of NF-κB. Nature 339:70-73. [DOI] [PubMed] [Google Scholar]
- 36.Hamilton, A. J., and D. C. Baulcombe. 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286:950-952. [DOI] [PubMed] [Google Scholar]
- 37.Harrich, D., J. Garcia, F. Wu, R. Mitsuyasu, J. Gonzalez, and R. Gaynor. 1989. Role of SP1-binding domains in in vivo transcriptional regulation of the human immunodeficiency virus type 1 long terminal repeat. J. Virol. 63:2585-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harrich, D., C. Hsu, E. Race, and R. B. Gaynor. 1994. Differential growth kinetics are exhibited by human immunodeficiency virus type 1 TAR mutants. J. Virol. 68:5899-5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harrich, D., C. Ulich, L. F. Garcia-Martinez, and R. B. Gaynor. 1997. Tat is required for efficient HIV-1 reverse transcription. EMBO J. 16:1224-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Herrmann, C. H., and A. P. Rice. 1995. Lentivirus Tat proteins specifically associate with a cellular protein kinase, TAK, that hyperphosphorylates the carboxyl-terminal domain of the large subunit of RNA polymerase II: candidate for a Tat cofactor. J. Virol. 69:1612-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hottiger, M. O., and G. J. Nabel. 1998. Interaction of human immunodeficiency virus type 1 Tat with the transcriptional coactivators p300 and CREB binding protein. J. Virol. 72:8252-8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ivanov, D., Y. T. Kwak, J. Guo, and R. B. Gaynor. 2000. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol. Cell. Biol. 20:2970-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ivanov, D., Y. T. Kwak, E. Nee, J. Guo, L. F. Garcia-Martinez, and R. B. Gaynor. 1999. Cyclin T1 domains involved in complex formation with Tat and TAR RNA are critical for tat-activation. J. Mol. Biol. 288:41-56. [DOI] [PubMed] [Google Scholar]
- 44.Jacque, J. M., K. Triques, and M. Stevenson. 2002. Modulation of HIV-1 replication by RNA interference. Nature 418:435-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jeang, K. T., H. Xiao, and E. A. Rich. 1999. Multifaceted activities of the HIV-1 transactivator of transcription, Tat. J. Biol. Chem. 274:28837-28840. [DOI] [PubMed] [Google Scholar]
- 46.Jones, K. A. 1997. Taking a new TAK on Tat transactivation. Genes Dev. 11:2593-2599. [DOI] [PubMed] [Google Scholar]
- 47.Jones, K. A., P. A. Luciw, and N. Duchange. 1988. Structural arrangements of transcription control domains within the 5′-untranslated leader regions of the HIV-1 and HIV-2 promoters. Genes Dev. 2:1101-1114. [DOI] [PubMed] [Google Scholar]
- 48.Kao, S. Y., A. F. Calnan, P. A. Luciw, and B. M. Peterlin. 1987. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature 330:489-493. [DOI] [PubMed] [Google Scholar]
- 49.Karin, M. 1999. How NF-κB is activated: the role of the IκB kinase (IKK) complex. Oncogene 18:6867-6874. [DOI] [PubMed] [Google Scholar]
- 50.Karin, M., and Y. Ben-Neriah. 2000. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 18:621-663. [DOI] [PubMed] [Google Scholar]
- 51.Karin, M., and A. Lin. 2002. NF-kappaB at the crossroads of life and death. Nat. Immunol. 3:221-227. [DOI] [PubMed] [Google Scholar]
- 52.Kennerdell, J. R., and R. W. Carthew. 1998. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell 95:1017-1026. [DOI] [PubMed] [Google Scholar]
- 53.Kim, J. B., and P. A. Sharp. 2001. Positive transcription elongation factor B phosphorylates hSPT5 and RNA polymerase II carboxyl-terminal domain independently of cyclin-dependent kinase-activating kinase. J. Biol. Chem. 276:12317-12323. [DOI] [PubMed] [Google Scholar]
- 54.Kim, J. B., Y. Yamaguchi, T. Wada, H. Handa, and P. A. Sharp. 1999. Tat-SF1 protein associates with RAP30 and human SPT5 proteins. Mol. Cell. Biol. 19:5960-5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kimpton, J., and M. Emerman. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J. Virol. 66:2232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kwak, Y. T., D. Ivanov, J. Guo, E. Nee, and R. B. Gaynor. 1999. Role of the human and murine cyclin T proteins in regulating HIV-1 Tat-activation. J. Mol. Biol. 288:57-69. [DOI] [PubMed] [Google Scholar]
- 57.Laspia, M. F., A. P. Rice, and M. B. Mathews. 1989. HIV-1 Tat protein increases transcriptional initiation and stabilizes elongation. Cell 59:283-292. [DOI] [PubMed] [Google Scholar]
- 58.Lee, N. S., T. Dohjima, G. Bauer, H. Li, M. J. Li, A. Ehsani, P. Salvaterra, and J. Rossi. 2002. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol. 20:500-505. [DOI] [PubMed] [Google Scholar]
- 59.Leonard, J., C. Parrott, A. J. Buckler-White, W. Turner, E. K. Ross, M. A. Martin, and A. B. Rabson. 1989. The NF-κB binding sites in the human immunodeficiency virus type 1 long terminal repeat are not required for virus infectivity. J. Virol. 63:4919-4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li, X.-Y., and M. R. Green. 1998. The HIV-1 Tat cellular coactivator Tat-SF1 is a general transcription elongation factor. Genes Dev. 12:2992-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mancebo, H. S. Y., G. Lee, J. Flygare, J. Tomassini, P. Luu, Y. Zhu, J. Peng, C. Blau, D. Hazuda, D. Price, and O. Flores. 1997. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 11:2633-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meisterernst, M., and R. Roeder. 1991. Family of proteins that interact with TFIID and regulate promoter activity. Cell 67:557-567. [DOI] [PubMed] [Google Scholar]
- 63.Mercurio, F., H. Zhu, B. W. Murray, A. Shevchenko, B. L. Bennett, J. Li, D. B. Young, M. Barbosa, and M. Mann. 1997. IKK-1 and IKK-2: cytokine-activated IκB kinases essential for NF-κB activation. Science 278:860-866. [DOI] [PubMed] [Google Scholar]
- 64.Nabel, G., and D. Baltimore. 1987. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature 326:711-713. [DOI] [PubMed] [Google Scholar]
- 65.Novina, C. D., M. F. Murray, D. M. Dykxhoorn, P. J. Beresford, J. Riess, S. K. Lee, R. G. Collman, J. Lieberman, P. Shankar, and P. A. Sharp. 2002. siRNA-directed inhibition of HIV-1 infection. Nat. Med. 8:681-686. [DOI] [PubMed] [Google Scholar]
- 66.Ou, S.-H. I., L. F. García-Martínez, E. J. Paulssen, and R. B. Gaynor. 1994. Role of flanking E box motifs on human immunodeficiency virus type 1 TATA element function. J. Virol. 68:7188-7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Parada, C. A., and R. G. Roeder. 1996. Enhanced processivity of RNA polymerase II triggered by Tat-induced phosphorylation of its carboxy-terminal domain. Nature 384:375-378. [DOI] [PubMed] [Google Scholar]
- 68.Perkins, N. D., A. B. Agranoff, E. Pascal, and G. J. Nabel. 1994. An interaction between the DNA-binding domains of RelA(p65) and Sp1 mediates human immunodeficiency virus gene activation. Mol. Cell. Biol. 14:6570-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Perkins, N. D., N. L. Edwards, C. S. Duckett, A. B. Agranoff, R. M. Schmid, and G. J. Nabel. 1993. A cooperative interaction between NF-κB and Sp1 is required for HIV-1 enhancer activation. EMBO J. 12:3551-3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ping, Y. H., and T. M. Rana. 2001. DSIF and NELF interact with RNA polymerase II elongation complex and HIV-1 Tat stimulates P-TEFb-mediated phosphorylation of RNA polymerase II and DSIF during transcription elongation. J. Biol. Chem. 276:12951-12958. [DOI] [PubMed] [Google Scholar]
- 71.Price, D. H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20:2629-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ross, E. K., A. J. Buckler-White, A. B. Rabson, G. Englund, and M. A. Martin. 1991. Contribution of NF-κB and Sp1 binding motifs to the replicative capacity of human immunodeficiency virus type 1: distinct patterns of viral growth are determined by T-cell types. J. Virol. 65:4350-4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Selby, M. J., E. S. Bain, P. A. Luciw, and B. M. Peterlin. 1989. Structure, sequence, and position of the stem-loop in TAR determine transcriptional elongation by Tat through the HIV-1 long terminal repeat. Genes Dev. 3:547-558. [DOI] [PubMed] [Google Scholar]
- 74.Sharp, P. A. 2001. RNA interference—2001. Genes Dev. 15:485-490. [DOI] [PubMed] [Google Scholar]
- 75.Sodroski, J., R. Patarca, C. Rosen, S. F. Wong, and W. Haseltine. 1985. Location of the trans-activating region on the genome of human T-cell lymphotropic virus type III. Science 229:74-77. [DOI] [PubMed] [Google Scholar]
- 76.Tang, H., K. L. Kuhen, and F. Wong-Staal. 1999. Lentivirus replication and regulation. Annu. Rev. Genet. 33:133-170. [DOI] [PubMed] [Google Scholar]
- 77.Wada, T., T. Takagi, Y. Yamaguchi, A. Ferdous, T. Imai, S. Hirose, S. Sugimoto, K. Yano, G. A. Hartzog, F. Winston, S. Buratowski, and H. Handa. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human SPT4 and SPT5 homologs. Genes Dev. 12:343-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wada, T., T. Takagi, Y. Yamaguchi, D. Watanabe, and H. Handa. 1998. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 17:7395-7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Waterman, M. L., and K. A. Jones. 1990. Purification of TCF-1α, a T-cell-specific transcription factor that activates the T-cell receptor Cα gene enhancer in a context-dependent manner. New Biol. 2:621-636. [PubMed] [Google Scholar]
- 80.Wei, P., M. E. Garber, S.-M. Fang, W. H. Fischer, and K. A. Jones. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92:451-462. [DOI] [PubMed] [Google Scholar]
- 81.West, M. J., A. D. Lowe, and J. Karn. 2001. Activation of human immunodeficiency virus transcription in T cells revisited: NF-κB p65 stimulates transcriptional elongation. J. Virol. 75:8524-8537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wianny, F., and M. Zernicka-Goetz. 2000. Specific interference with gene function by double-stranded RNA in early mouse development. Nat. Cell Biol. 2:70-75. [DOI] [PubMed] [Google Scholar]
- 83.Wu, F. K., J. A. Garcia, D. Harrich, and R. B. Gaynor. 1988. Purification of the human immunodeficiency virus type 1 enhancer and TAR binding proteins EBP-1 and UBP-1. EMBO J. 7:2117-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu-Baer, F., W. S. Lane, and R. B. Gaynor. 1998. Role of the human homologues of the yeast transcription factors SPT5 and SPT6 in HIV-1 Tat-activation. J. Mol. Biol. 277:179-197. [DOI] [PubMed] [Google Scholar]
- 85.Yamaguchi, Y., T. Takagi, T. Wada, K. Yano, A. Furuya, S. Sugimoto, J. Hasegawa, and H. Handa. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97:41-51. [DOI] [PubMed] [Google Scholar]
- 86.Yang, X. Z., M. O. Gold, D. N. Tang, D. E. Lewis, E. Aguilarcordova, A. P. Rice, and C. H. Herrmann. 1997. TAK, an HIV TAT-associated kinase, is a member of the cyclin-dependent family of protein kinases and is induced by activation of peripheral blood lymphocytes and differentiation of promonocytic cell lines. Proc. Natl. Acad. Sci. USA 94:12331-12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zamore, P. D., T. Tuschl, P. A. Sharp, and D. P. Bartel. 2000. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101:25-33. [DOI] [PubMed] [Google Scholar]
- 88.Zhou, M., M. A. Halanski, M. F. Radonovich, F. Kashanchi, J. Peng, D. H. Price, and J. N. Brady. 2000. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol. Cell. Biol. 20:5077-5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhou, Q., D. Chen, E. Pierstorff, and K. Luo. 1998. Transcription elongation factor P-TEFb mediates Tat activation of HIV-1 transcription at multiple stages. EMBO J. 17:3681-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhou, Q., and P. A. Sharp. 1996. Tat-SF1: cofactor for stimulation of transcriptional elongation by HIV-1 Tat. Science 274:605-610. [DOI] [PubMed] [Google Scholar]
- 91.Zhu, Y., T. Pe'ery, J. Peng, Y. Ramanathan, N. Marshall, T. Marshall, B. Amendt, M. B. Mathews, and D. H. Price. 1997. Transcription elongation factor P-TEFb is required for HIV-1 Tat transactivation in vitro. Genes Dev. 11:2622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]