Abstract
The transcription and splicing of human immunodeficiency virus type 1 (HIV-1) mRNA in primary blood monocyte-derived macrophages (MDM) and CD4+ peripheral blood lymphocytes (PBL) were compared to determine whether any differences might account for the slower noncytopathic infection of cells of the macrophage lineage. The expression of regulatory mRNAs during acute infection of MDM was delayed by about 12 h compared to that of PBL. In each cell type, an increase in spliced viral mRNAs slightly preceded virus production from the culture. Following the peak of productive infection, there was a proportional decrease in the expression of all regulatory mRNAs detected in PBL. In MDM, a dramatic additional decrease specifically in the tat mRNA species heralded a reduction in virus production. This decline in tat mRNA was reflected by a concomitant decrease in Tat activity in the cells and occurred with the same kinetics irrespective of the age of the cells when infected. Addition of exogenous Tat protein elicited a burst of virus production from persistently infected MDM, suggesting that the decrease in virus production from the cultures is a consequence of the reduction in tat mRNA levels. Our results show that modulation of HIV-1 mRNAs in macrophages during long-term infection, which is dependent on the period of infection rather than cell differentiation or maturation, results in a selective reduction of Tat protein levels at the commencement of a persistent, less productive phase of infection. Determination of the mechanism of this mRNA modulation may lead to novel targets for control of replication in these important viral reservoirs.
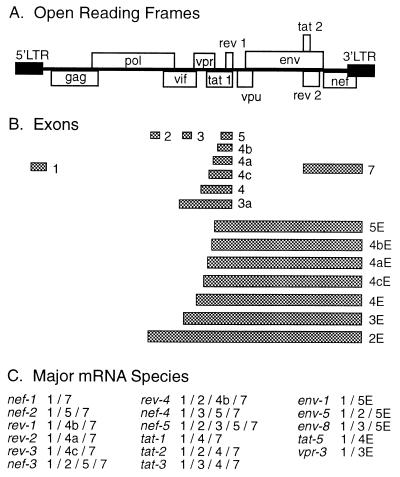
The human immunodeficiency virus type 1 (HIV-1) genomic RNA undergoes a distinctly complex series of multiple splicing events to produce an array of 22 different 4.0-kb mRNAs for the Env, Vpu, Tat, Vpr, and Vif proteins and 22 different 1.8-kb mRNAs for the Tat, Rev, Vpr, and Nef proteins (38, 44, 47, 48). These seven viral proteins, encoded by more than 40 alternatively spliced mRNAs, regulate virus replication and important host cell functions and are crucial for HIV-1 infectivity and in vivo pathogenesis. Alterations to the balanced splicing of these HIV mRNAs can have a dramatic impact on viral infectivity and pathogenesis (44).
The large array of alternatively spliced HIV-1 mRNA primarily results from the differential use of six competing alternative 3′ splice acceptor (SA) sites (A3, A4c, A4b, A4a, A5, and A7) (4, 28, 44, 47) and the use of two upstream noncoding exons (exons 2 and 3) that alter the 5′ untranslated region (Fig. 1). RNA elements called exonic splice silencers (ESS) have been identified in the splice control regions of the genomic RNA that bind cellular heterogeneous nuclear ribonucleoproteins (hnRNPs) of the A, B, and H groups (9, 14, 30) and act to suppress the splicing of adjacent 3′ SA sites, such as in the second exon of tat (Fig. 1B, exon 4), the second tat/rev coding exon (exon 7), and noncoding exon 3 downstream of Vpr splice site A2 (2, 6, 57). Other elements called exonic splice enhancers (1, 57) bind serine/arginine-rich proteins such as ASF/SF2 and SC35 (59, 63) to enhance the splicing of neighboring exons, such as the third tat exon (Fig. 1B, exon 7). The high level of conservation of 5′ SD and 3′ SA sequences among different viral isolates of diverse origins suggests that complex splice regulation is important for efficient viral replication (45, 49).
FIG. 1.
Combinations of exons that are spliced from the HIV RNA genome join to form the alternatively spliced mRNAs of HIV-1. (A) Schematic representation of the HIV-1 genome. Open boxes show locations of the open reading frames that encode the viral proteins. (B) HIV-1 exons are generated by the use of different combinations of SAs and splice donors during RNA splicing. The exons, represented as bars, are numbered as described by Muesing et al. (38). (C) The exon contents of the major 1.8- and 4.0-kb classes of spliced transcripts detected in MDM and PBL are shown in abbreviated form, e.g., nef-1 combines exons 1 and 7 and is annotated as 1/7.
The role of the complex HIV-1 RNA-processing control elements in modulating the course of HIV-1 infection of CD4+ T lymphocytes and cells of the macrophage lineage is unclear. Many studies have demonstrated that blood monocytes and tissue macrophages display a characteristically slower, nonlytic, chronic course of infection, in contrast to primary CD4+ T lymphocytes, in which HIV-1 infection is typically rapid, lytic, and highly productive of new virions (11, 41, 46). Some studies have suggested that the diverse outcomes of HIV-1 infection are due, in part, to different processing of viral RNA by these cells (32, 37, 43, 45).
However, most of these studies examining RNA splicing during HIV-1 replication have used T-cell lines, primary CD4+ T lymphocytes, or monocytic cell lines in their analyses (32, 43, 45, 47). Relatively little is known about the control of RNA splicing in acutely or chronically infected primary cells of the macrophage lineage, although these cells are primary targets of HIV-1 infection and are important in the initial transmission event and in maintaining the infection (25, 54, 55). Freshly isolated monocytes are poorly susceptible to infection in vitro but become increasingly susceptible during differentiation in culture over 5 to 7 days to a macrophage phenotype (52, 53), at least in part because of the increased expression of the CCR5 coreceptor (40). Because of the inherent difficulties of obtaining tissue macrophages for study, these in vitro-derived macrophages are often used as a model of their in vivo counterparts. In this study, we examined whether modulation of the large array of alternatively spliced HIV-1 regulatory transcripts correlates with the differential viral replication and expression observed following in vitro infection of primary monocyte-derived macrophages (MDM) compared to peripheral blood lymphocytes (PBL). This work increases our understanding of HIV-1 infection in macrophages and has the potential to lead to novel therapeutic targets for the control of replication in these important reservoirs.
MATERIALS AND METHODS
Isolation of lymphocytes and monocytes from blood.
PBL and monocytes were isolated from HIV-1-seronegative blood cell packs (Red Cross Blood Bank, Melbourne, Australia) by density gradient centrifugation and plastic adherence in 175-cm2 petri dishes as previously described (52). The nonadherent PBL were resuspended in RPMI 1640 medium (Gibco BRL, Grand Island, N.Y.) with 10% fetal calf serum, 2 mM l-glutamine (ICN Biomedicals Inc., Costa Mesa, Calif.), and 50 μg of gentamicin per ml (RF10) and then stimulated with phytohemagglutinin (2.5 μg/ml; Wellcome Diagnostics, Dartford, United Kingdom) for 2 to 3 days prior to infection with HIV-1. Infected PBL were maintained in flasks in RF10 with the addition of interleukin-2 (10 U/ml; Roche, Castle Hill, Australia), and the medium was changed twice weekly. The monocytes were detached from the petri dishes and usually maintained adherent in 24-well plates (Nunc, Naperville, Ill.) in Iscove's medium (Gibco BRL) supplemented with 10% AB/Rh+ human serum, glutamine, and gentamicin (IH10) at a concentration of 106 monocytes/well. They were then washed thoroughly with phosphate-buffered saline (PBS) and infected 5 to 7 days after isolation, by which time they had differentiated to a macrophage phenotype and were maximally permissive to infection with R5 strains of HIV-1. In one experiment, monocyte-derived macrophages were infected at approximately weekly intervals until 4 weeks after isolation. MDM cultures were given weekly medium changes and maintained under endotoxin-free conditions throughout.
Viral strains and infection assays.
Replicate wells of 106 MDM or cultures of PBL at 2 × 106 cells/ml were infected for kinetic studies with the prototype R5 strain, HIV-1Ba-L (obtained from the AIDS Research and Reference Reagent Program, National Institute of Allergy and Infectious Diseases, National Institutes of Health; high-titer virus stocks produced in peripheral blood mononuclear cells [PBMC] [24]), or with a recombinant R5 strain, HIV-1NL(AD8) [supernatant from SW480 cells transfected with pNL(AD8), a stable, full-length molecular clone of pNL4-3 in which the envelope gene has been replaced with that from the macrophage-tropic ADA isolate (58)]. Cells were incubated with virus for 2 h at a multiplicity of infection (MOI) of 0.1 to 1 infectious particle per cell (assessed by determining the 50% tissue culture-infective dose in MDM), followed by thorough washing with PBS. Virus production was monitored by assaying for virion-associated reverse transcriptase (RT) activity by using [α-33P]TTP incorporation into an oligo(dT)-poly(A) template (micro-RT assay) (60). To one long-term-infected culture, Tat protein (obtained through the AIDS Research and Reference Reagent Program, National Institute of Allergy and Infectious Diseases, National Institutes of Health, and contributed by J. Brady [7]), 13-phorbol-12-myristate acetate (PMA; Sigma Chemical Co., St. Louis, Mo.), or tumor necrosis factor alpha (TNF-α) was added in an attempt to reactivate virus production. Tat (200 ng/well in the presence of 100 μg of protamine sulfate per ml to enhance uptake [19]), PMA (100 ng/ml), and TNF-α (100 ng/ml) were added to the culture medium 4 to 5 weeks after infection. Cells were then monitored for a further 2 weeks with a medium change but no replenishment of additives after the first week. Also, to cultures from four different donors, recombinant soluble CD4 (rsCD4; 50 μg/ml; a generous gift from Glaxo-SmithKline, King of Prussia, Pa. [13, 53]) was added immediately after infection with DNase-treated Ba-L (10 U of RNase-free DNase [Roche] per ml of virus stock in 10 mM MgCl2 for 30 min at room temperature) and HIV-1 replication was assessed by monitoring the accumulation of viral DNA by PCR as described previously (33, 54).
Bioassay for HIV-1 Tat activity.
An assay system consisting of a set of two adenovirus-luciferase reporter vectors, of which one is responsive to HIV-1 Tat protein activity (Ad-HIVluc) and the other is a control vector that is not Tat responsive because of a deletion of the TAR site (Ad-HIVΔTARluc), was used to assess Tat activity in HIV-infected MDM and PBMC cultures (5). The vectors were grown and titrated in 293 cells as previously described (5), and then each was used at an equivalent MOI of 100 to superinfect triplicate wells of 96-well plates containing 105 MDM or PBMC from three separate donors at twice weekly intervals for 6 and 3 weeks, respectively, following HIV-1 infection. The cells were washed with PBS 48 h after adenovirus superinfection and then lysed in 20 μl of cell culture lysis reagent (CCLB; Promega) per well and stored at −20°C until analyzed for luciferase activity at the completion of the experiment. Luciferase activity was measured by mixing 10 μl of cell lysate (undiluted for PBMC lysates and diluted 1:10 in CCLB for MDM lysates) with 50 μl of luciferase assay reagent (Promega) and immediate detection of the emitted light in a Triathler luminometer (Perkin-Elmer, Branchburg, N.J.). The presence of Tat activity in this bioassay is indicated by threefold or greater luciferase activity in Ad-HIVluc-infected cells than in those infected at the same time point with Ad-HIVΔTARluc, which measures the background level of Tat-independent transactivation of the HIV-1 long terminal repeat (LTR) in the cells.
PCR for luciferase DNA.
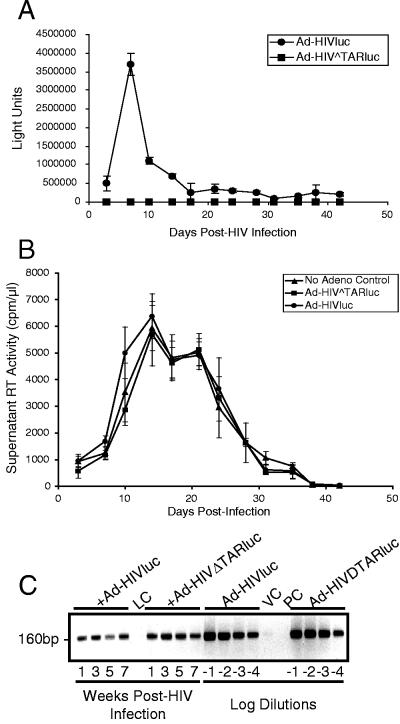
The above-described CCLB lysates (5 μl of each replicate pooled) were digested with proteinase K (10 to 15 μg; Roche) at 60°C for 1 h and then at 95°C for 10 min. The lysates were then clarified by centrifugation at 13,000 × g for 2 min, and 5 μl was used in a PCR with primers specific for the firefly (Photinus pyralis) luciferase (luc) gene (luc-5′, GAAGGTTGTGGATCTGGATACC, positions 1621 to 1642; luc-3′, AGCTATGTCTCCAGAATGTAGCC, positions 1780 to 1758 [15]). The primers were used at a final concentration of 0.4 μM in reaction mixtures containing 1.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, and 1 U of Taq polymerase for 25 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min. The 160-bp PCR products were analyzed by electrophoresis in 2% agarose gels and stained with ethidium bromide, and band intensities upon UV illumination were compared by phosphorimaging (Fuji LAS1000).
Preparation of mRNA and cDNA.
Poly(A)+ RNA was purified at various time points postinfection with magnetic oligo(dT)25 Dynabeads (Dynal, Oslo, Norway) at a concentration of 25 μl of beads/106 cell equivalents. The cells were lysed, incubated for 10 min at room temperature with the oligo(dT)25 beads, and washed in accordance with the manufacturer's instructions. The RNA was then reverse transcribed to cDNA with a 25-μl reaction mixture containing the bead-mRNA complex, 1× avian myeloblastosis virus RT buffer, 1 mM deoxynucleoside triphosphates, 1 U of RNasin per μl, and 1 U of avian myeloblastosis virus RT per μl (all from Promega Corp., Madison, Wis.). Samples were incubated for 1 h at 42°C, the RT mixture was removed, and the beads were resuspended in 100 μl of elution solution (2 mM EDTA) before being heated to 95°C for 5 min to remove the mRNA. The cDNA libraries attached to the beads were stored in 10 mM Tris-HCl-1 mM EDTA, pH 7.6, at 4°C for use in subsequent reverse transcription-PCR analyses.
Standardization and amplification of HIV-1 spliced mRNA.
To ensure a standard input of cDNA in HIV-specific PCRs, 105 cell equivalents of cDNA attached to beads was serially diluted to less than 1 cell equivalent and PCR amplified with primers specific for β-actin, BA1/4 (26). PCR products (360 bp) were detected by electrophoresis in 2% agarose and ethidium bromide staining. Band intensities for all samples were compared by phosphorimaging as described above, and equivalent amounts of the cDNA were used in a subsequent PCR for viral RNA.
Following standardization of the cDNA, we performed semiquantitative analyses for the 1.8-kb class of HIV-1 RNA with 1 μM primers Odp45 and Odp32 as described before (44) and for the 4.0-kb mRNA with primers Odp45 and Odp84 (5′-TCATTGCCACTGTCTTCTGCTCT-3′) in a hot-start PCR (25 cycles of 95°C for 1.5 min, 55°C for 1 min, and 72°C for 2.5 min with a final 7 min at 72°C). One microliter of each amplification product was radiolabeled by performing a single round of PCR as before but with the addition of 10 μCi of [α-32P]dCTP. The labeled samples were subsequently analyzed by electrophoresis through a 6% polyacrylamide-urea gel at sufficient wattage to maintain a temperature of 65°C. Bands were visualized by autoradiography and/or phosphorimaging (Fuji FLA2000). We have shown previously that this protocol preserves the relative proportions of the 1.8- and 4.0-kb classes of HIV-1 RNA (44).
RESULTS
Contribution of multiple rounds of HIV-1 replication to infection in primary MDM and PBL cultures.
Since the kinetic accumulation of spliced HIV-1 RNA might be confounded by asynchronous secondary infection, we used both a relatively high multiplicity of virus for infection (MOI of 1) and blocked subsequent rounds of infection in cultures from four donors with rsCD4. The degree of asynchronous infection of MDM was measured by comparing the relative abundance of gag viral DNA to that of cellular HLA-DQ-α DNA in the presence and absence of blocking concentrations of rsCD4. The amounts and ratios of gag and HLA-DQ-α DNA were similar in both treated and untreated MDM cultures and remained relatively constant from 48 h after infection (data not shown), demonstrating maximal synchronous infection of susceptible cells with minimal virus superinfection or spread after initial infection.
MDM and PBL differ in the kinetic expression and splicing of HIV-1 mRNA during acute infection.
The splicing of HIV-1 mRNA during acute infection of primary PBL and MDM with Ba-L was investigated (Fig. 2). A 12-h delay in the first appearance of HIV-1 mRNA from MDM (24 h), compared to that of PBL (12 h; Fig. 2A), results from the delayed establishment of HIV-1 infection in the former cell type. Both primary cell types expressed the same mRNA species. However, significant differences in the relative abundance of particular isoforms were observed between the cell types during the first 48 h. In MDM, nef-1 (Fig. 1C, 1/7), nef-2 (1/5/7), rev-1 (1/4b/7), and tat-1 (1/4/7) were the most abundant spliced isoforms of HIV-1 1.8-kb mRNA at 48 h (Fig. 2A, right side). nef-2 and rev-1 appeared simultaneously at 24 h postinfection, and the other tat, rev, and nef multiply spliced mRNAs (Fig. 1C) were detectable at 48 h postinfection. MDM expressed tat-1 mRNA at a significantly higher level than the other tat mRNAs. In contrast, in the PBL cultures, only a subset of spliced HIV-1 mRNA species, nef-2, rev-1, and rev-2 (1/4a/7), that predominate at 48 h were detected 12 h postinfection (Fig. 2A, left side). Other mRNAs prominent in PBL at 48 h postinfection, most notably, nef-1, tat-1, and tat-2 (1/2/4/7), were proportionally underrepresented at earlier time points. PBL synthesized high levels of several other isoforms of nef and rev (i.e., rev-2, rev-3, rev-4, nef-3, nef-4, and nef-5) that were detected at relatively low levels in infected MDM. Similar mRNA profiles were found following infection of cells from six different donors.
FIG. 2.
The kinetic expression of alternatively spliced 1.8-kb viral mRNA during early HIV infection of PBL and MDM. Poly(A)+ RNA was extracted from cells at 2-h intervals for the first 12 h and then at 24 and 48 h after infection with Ba-L. Semiquantitative PCR with primers Odp45 and Odp32 for multiply spliced transcripts (A) was performed on cDNA standardized by β-actin PCR (B). In our reverse transcription-PCR assay, the detection limits of 1.8- and 4.0-kb mRNA transcripts were 102 and 103 HIV-infected cells, respectively. The RNA patterns shown are representative of results of six experiments.
Alternative splicing of HIV mRNA during chronic infection in MDM and PBL.
We next examined the splicing of HIV-1 mRNA in primary PBL and MDM during chronic infection over a 3-week period (Fig. 3). Duplicate cultures of MDM or a single culture of PBL from each of two donors were infected for sequential analysis of spliced HIV-1 RNA at intervals of 3 or 4 days. Reverse transcription-PCR analysis of the different alternatively spliced 1.8- and 4.0-kb HIV-1 RNAs showed that the relative proportions of the various species were similar over this time (Fig. 3A and B), indicating that there was little difference between PBL and MDM in the cell-controlled splicing of HIV-1 RNA.
FIG. 3.
Spliced HIV-1 mRNA accumulates in similar patterns in lymphocytes and macrophages. Duplicate cultures of MDM and a single culture of PBL were infected with Ba-L for sequential analysis of spliced HIV RNA. PBL were maintained at 2 × 106 cells/ml, and RNA was extracted from 5 × 105 cells that were removed from the PBL infection or from 106 replicate infected MDM at intervals of 3 or 4 days over a 3-week period. (A) Expression of 1.8-kb viral mRNA in PBL and MDM from the same donor infected with HIV-1Ba-L. Semiquantitative PCR with primers Odp45 and Odp32 was performed with standardized amounts of cDNA (C) attached to magnetic beads. (B) Viral 4.0-kb mRNA profile of the same samples as in panel A following amplification with primers Odp45 and Odp84. Note that the gel in panel B was exposed four times longer than that in panel A for detection of minor bands. Patterns shown for 1.8- and 4.0-kb RNAs were similar in two donor lymphocyte and monocyte populations. (D) Virus production from the cultures shown in panels A and B as assessed by supernatant RT activity. M, molecular size markers (sizes are in base pairs); U, uninfected-cell control; −ve, no-cDNA control.
The kinetic expression of HIV-1 RNA differed between the MDM and PBL cultures. The production of spliced 1.8- and 4.0-kb HIV-1 RNAs in PBL (Fig. 3A and B, respectively) peaked at 7 to 10 days after infection (cell donor dependent) and then gradually declined in a uniform manner until day 21, by which time the viability of the lymphocytes had significantly diminished. The time of maximum RNA production in PBL typically preceded the peak of supernatant RT activity by 3 or 4 days (Fig. 3D). The decline in production of HIV-1 RNA after 7 to 10 days in PBL resulted from a proportionate decrease in HIV-1-infected cells because of both their death and their failure to divide in the culture (42, 62). This is reflected in virus production from these cells, which declined by days 17 to 21 (Fig. 3D), although there was still significant expression of the standard pattern of alternatively spliced HIV-1 mRNAs at this time (Fig. 3A and 4B).
FIG. 4.
Selective reduction in the abundance of HIV-1 tat mRNA marks the beginning of virus dormancy in MDM during long-term HIV-1 infection. MDM were infected with Ba-L after 5 days in culture, and samples for RNA and virus production were taken approximately weekly for 5 to 6 weeks. (A) Virus production was assessed by determining the virion-associated RT activity in culture supernatants. (B) Northern blot analysis of total RNA from a representative culture. Purified mRNAs from the same culture as in panel A were first standardized with β-actin PCR (D), and then equivalent amounts were amplified with primers Odp45 and Odp32 for multiply spliced viral transcripts (C, left side). A similar analysis was performed with MDM infected with NL(AD8) (C, right side). M, molecular size markers (kilobases or base pairs). Results similar to those shown were found for 10 donor MDM populations infected with Ba-L and several others with NL(AD8) and 676 (not shown).
As observed in acute infection, HIV-1 mRNA accumulated with much slower kinetics in infected MDM than in PBL during chronic infection (Fig. 3A and B). In contrast to PBL, both the overall level of viral 1.8-kb mRNA and virus production in MDM continued to increase until day 21 (Fig. 3A and D). HIV-1 infection of MDM, which are nondividing, does not result in significant cell death (11). The slight lag in the detection and the relatively lower abundance of the 4.0-kb mRNA compared to the 1.8-kb RNA in MDM (four times longer exposure for Fig. 3B than for Fig. 3A) reflects the requirement for Rev prior to the cellular accumulation of the 4.0-kb mRNAs that contain the Rev-responsive element.
Marked alteration of HIV-1 RNA profile as MDM enter a nonproductive phase during long-term infection.
We next examined viral RNA processing in MDM that were maintained in culture for 6 weeks following infection (Fig. 4). We sought to determine whether the diminished production of HIV-1 in MDM after a peak of infection at around 3 weeks was due to an alteration in the profile of spliced viral mRNA. MDM cultures infected with Ba-L produced little or no detectable virus in the supernatant after 28 days (Fig. 4A) and very little viral RNA as determined by Northern blotting of total cellular RNA (Fig. 4B). Reverse transcription-PCR analysis of the spliced HIV-1 RNA showed that the peak of RT activity coincided with, or slightly followed, the peak in abundance of viral RNA occurring between days 14 and 21 (Fig. 4A and C). It was not until after the peak of productive infection that any variation in the RNA profile was observed. We found a significant decrease in the relative proportion of tat-specific mRNA at times later than 21 days, compared to that of all other alternatively spliced mRNA.
The most striking change in RNA processing during long-term infection of MDM was the dramatic decrease observed in the relative accumulation of tat-1. The tat-1 mRNA was one of the most abundant mRNA species at the peak of infection (Fig. 4C, days 14 to 21) but became a very minor species by the end of the culture period. The decline in tat-1 was coincident with the decrease in productive infection in MDM cultures and was in marked contrast to the continued abundance of the other major species, nef-1, nef-2, rev-1, and rev-2. The decline in each of the tat mRNA species indicates a selective decrease in the viral RNA spliced at SA3, located at the 5′ end of exon 4. This exon contains the translation initiation site of Tat (Fig. 1). The selective decrease in tat mRNA species was observed during infection of each of 10 MDM donors with Ba-L and several more with NL(AD8) (Fig. 4C) and the primary isolate HIV-1676 (52) (data not shown); however, the relative decrease in tat differed between donors, ranging from 60-fold (Fig. 4) to 10-fold (see Fig. 8). This selective decrease in tat species was not observed in lymphocytes in which, after the peak in RNA production at 7 to 10 days, all spliced transcripts declined proportionally until day 21 (Fig. 4A). As was found in the PBL cultures, the three-exon form of nef mRNA (nef-2) was the predominant viral mRNA expressed throughout the course of infection in MDM, along with the two three-exon forms of rev mRNA (rev-1 and rev-2). The two-exon Nef mRNA (nef-1) was not as prevalent as previously reported (45).
FIG. 8.
A burst of HIV-1 production results from addition of exogenous Tat to persistently infected MDM. MDM from one donor were infected with Ba-L 7 days after isolation and maintained for 4 to 5 weeks before the addition of Tat protein (200 ng/well), TNF-α (100 ng/ml), or PMA (100 ng/ml) to the culture medium. The culture was then monitored for virion production and viral RNA expression for a further 2 weeks. (A) RT activity in supernatant from a representative culture infected for 5 weeks before any additions to the culture medium were made. The solid line represents the RT activity in supernatant from cells maintained in culture medium alone for 7 weeks following infection. Dotted lines represents the RT activity in the same cells after the addition of Tat, TNF-α, or PMA, respectively, on day 35 (arrow). (B) Viral 1.8-kb RNA (top) and β-actin mRNA (bottom) in cells from the culture shown in panel A. The left side of each panel represents mRNA from cells to which no Tat was added and that were lysed 7 to 49 days after infection. The right side represents RNAs from cells lysed 1, 7, and 14 days after addition of Tat (day 35 postinfection). U, RNA from uninfected cells; C, control infected cells to which protamine sulfate only was added.
Altered proportions of spliced viral mRNA in long-term-infected MDM is not dependent on the age of the cells.
The marked specific reduction in Tat-encoding mRNAs during long-term infection in MDM could conceivably be due to changes induced in the cells by long-term culture. To determine whether this is the case or whether it arises as a consequence of infection, MDM from the same donor were infected with Ba-L at approximately weekly intervals over a 4-week period following isolation of the cells from blood. Samples were then taken weekly for 5 weeks and analyzed for virion-associated RT production and viral mRNA expression as above.
While MDM maintained in culture for a month were still permissive to infection with HIV-1, the amount of virus produced at peak infection was smaller and the kinetics of replication were more delayed with increasing time in culture before infection (Fig. 5A). When mRNA expression of the major 1.8-kb viral mRNA species over the course of infection was compared between the cells infected at different times after isolation, only the tat mRNAs again showed a pattern that correlated with virus production from the cells, i.e., an initial increase followed by a dramatic decline approximately 1 week before virus production waned. The tat species (tat-1, -2, and -3) again decreased markedly over the final 2 weeks of culture relative to their peak expression level around 3 weeks after infection in this donor, irrespective of when the cells were infected (Fig. 5B). In contrast, the rev and nef mRNAs remained at similar relative proportions throughout the course of infection. The patterns of viral mRNA expression were remarkably similar regardless of how long after isolation the cells were infected, with only the overall abundance somewhat reduced with increasing time in culture before infection (not shown), correlating with virus production from the cells (Fig. 5A).
FIG. 5.
HIV-1 mRNA profile during long-term infection of MDM is the same irrespective of the time in culture before infection. MDM from a single donor were infected with Ba-L at weekly intervals over 4 weeks, and then samples were taken weekly for a further 5 weeks after infection for analysis of virus production and mRNA abundance. (A) Kinetics of virus production as assessed by RT activity of culture supernatants from MDM infected 1 and 4 weeks after isolation, respectively, are shown. (B) Relative proportions of tat-1, rev-1, and nef-1 over time in cells infected 1 and 4 weeks after isolation. Expression of particular viral mRNA species in the same cells from which virus production was measured as shown in panel A were determined as a proportion of the total 1.8-kb mRNA by phosphorimaging analysis, and then this proportion was related to that present at peak expression at 21 days postinfection. wk, week.
Declining tat mRNA abundance results in dramatically lower Tat protein activity in long-term-infected MDM.
To determine if the observed decrease in tat mRNA expression after peak virus production in MDM resulted in a corresponding decrease in the level of Tat activity in these cells, monocytes from three different donors were cultured in 96-well plates and infected 5 to 7 days after isolation. Triplicate wells were then superinfected with recombinant adenovirus vectors containing the firefly luciferase gene under the control of either the wild-type HIV-1 LTR (Ad-HIVluc) or an LTR with TAR deleted (Ad-HIVΔTARluc) (5) at twice-weekly intervals for 6 weeks. This system is highly responsive to Tat and allows one to determine Tat-specific HIV-1 LTR-mediated gene expression, which increases linearly with increasing levels of HIV-1 infection in a wide variety of cell types (5). In addition, the Ad-HIVΔTARluc recombinant virus controls for any effects of adenovirus superinfection that are not due to Tat. Forty-eight hours after addition of the recombinant adenoviruses, the cells were lysed and luciferase activity in the lysates was measured. Luciferase activity in HIV-infected MDM superinfected with Ad-HIVluc, a specific measure of Tat-dependent transactivation of the HIV-1 LTR (19), was very high, peaked 1 to 2 weeks after HIV infection (donor variable), and then declined dramatically over subsequent weeks to near background levels by 2 to 3 weeks (Fig. 6A). Virus production from the MDM cultures showed a pattern similar to that of Tat activity, except for the expected delay between peak Tat activity and maximal virus production (Fig. 6B). This delay was between 2 and 7 days, depending on the donor culture. In contrast, the luciferase activity within the lysates from MDM infected with Ad-HIVΔTARluc, which indicates the level of Tat-independent LTR-driven transcription in the cells, remained relatively constant throughout the course of the infection (Fig. 6A). This was not due to a lower superinfection efficiency of the Ad-HIVΔTARluc compared to Ad-HIVluc virus, however, as the luciferase gene levels determined by PCR in the MDM lysates remained similar throughout (Fig. 6C). Replicate wells of HIV-infected MDM superinfected with either of the recombinant adenoviruses, or to which adenovirus was not added, produced essentially identical HIV growth curves (Fig. 6B).
FIG. 6.
Tat activity declines rapidly to near background levels during persistent HIV-1 infection in MDM. Tat activity during long-term HIV-1 infection of MDM was determined by superinfection with recombinant adenoviruses containing the wild-type HIV-1 LTR or the HIV-1 LTR with TAR deleted controlling the expression of a luciferase reporter gene (Ad-HIVluc and Ad-HIVΔTARluc, respectively). (A) Luciferase activity was measured in cells lysed 48 h after superinfection and expressed as arbitrary light units. Lysates were diluted 10-fold to fall within the range measurable by the luminometer used. Background levels of cells not superinfected with adenovirus vectors were <8,000 light units. (B) Supernatant RT activity in the replicate MDM cultures from panel A superinfected with Ad-HIVluc or Ad-HIVΔTARluc or maintained as nonsuperinfected controls (No Adeno Control). (C) PCR for luciferase DNA in 1-, 3-, 5-, and 7-week cell lysates from which luciferase activity was determined as described in panel A (left side) and in log dilutions of the standardized vector stocks (right side). LC, nonsuperinfected cell lysate control; VC, virus control; PC, PCR control. The examples shown are representative of the results obtained with samples from three donors.
Tat activity in infected PBMC cultures from three donors also increased over the first week of infection and then declined gradually over the next week as the cells became less viable and cell numbers decreased because of cytopathicity (Fig. 7A). This was mirrored by the RT activity in the culture supernatant (Fig. 7B), with Tat activity again preceding virus production by several days. Despite higher levels of HIV-1 replication in PBMC cultures (Fig. 6B and 7B), the peak luciferase activity in these cells was more than 1,000-fold lower than that found in MDM cultures (Fig. 6A and 7A), most likely because of the low expression level of the adenovirus receptor on lymphocytes (36).
FIG. 7.
Tat activity in HIV-1-infected PBMC. An analysis similar to that performed with MDM and shown in Fig. 6 was also done with PBMC cultures infected for 2 weeks. (A) Tat activity as determined by luciferase activity in superinfected cells (note that the lysates did not need to be diluted 1 in 10 as did the macrophages lysates in Fig. 6). (B) Virus production from the cultures. (C) Luciferase gene PCR in cell lysates and adenovirus stocks. The results shown are representative of the results obtained with samples from three donors. No Adeno Control, no-adenovirus control; LC, nonsuperinfected cell lysate control; PC, PCR control.
Exogenous Tat rescues virus production from long-term-infected MDM.
Given the low levels of tat mRNA and Tat protein activity in MDM from about 3 to 4 weeks after HIV infection, the decline in productive infection in these cells might result from suboptimal levels of Tat protein. When Tat protein was added to an MDM culture 2 to 3 weeks after the peak in virus expression at a concentration shown previously to induce HIV-1 production from the latently infected promonocytic cell line U1 (17), a spike in supernatant RT activity was found (Fig. 8A). RT activity in the culture supernatant began to rise 3 days after Tat addition, peaked at 10 days, and then began to decline again by 14 days (Fig. 8A). In the absence of exogenous Tat, RT activity declined from low to undetectable levels over the same 2-week period. This reactivation of virus production is likely to be in response to the provision of Tat to cells shown above to have very low Tat activity (Fig. 6A) and not result solely from any lipopolysaccharide (LPS) contaminating the protein preparation since we were unable to induce virus replication from long-term-infected macrophage cultures with LPS treatment (1 ng/ml), despite several attempts (unpublished data). Analysis of 1.8-kb mRNA within MDM again showed a decreased abundance of tat mRNA after the peak of infection. Following the addition of Tat protein, there was no change in the RNA profile from that found in control cells without exogenous Tat (Fig. 8B). Significantly, the levels of the tat mRNA species remained consistently low.
As found previously with the latently infected U1 and ACH2 cell lines (8, 22, 23), a similar but lower burst of virus production was induced by the addition of PMA or TNF-α to long-term-infected MDM (Fig. 8A). Activation by these factors occurs, at least in part, through translocation of the transcription factor NF-κB to the nucleus (27) and is independent of Tat.
DISCUSSION
The genome length mRNA of HIV-1 contains many features, such as multiple alternative splice sites and exonic splice enhancers and ESS, that could modulate RNA splicing to change the outcome of viral infection. In this study, we examined the complex splicing profile of HIV-1 mRNA during infection of primary cells of the lymphocyte and macrophage lineages from the first hours of infection through to late times, when virus production wanes. Unlike earlier studies of the kinetic accumulation of HIV-1 mRNAs (31, 32, 45), the reverse transcription-PCR method that we used could accurately measure the relative proportion of each of the alternative mRNA isoforms.
We found that HIV-1 1.8-kb mRNA accumulated 12 h earlier in lymphocytes than in MDM. This comparative delay in MDM agrees with the 12-h delay observed in one study of U937 cells (39) but not the 4-h delay reported in another study (32). It also agrees with our previous work in which we found that in fully susceptible MDM, compared to freshly isolated monocytes that are resistant to infection, reverse transcription and integration are detectable by 12 and 24 h postinfection, respectively (52). We determined that a fixed relative proportion of each of the 1.8-kb mRNAs appears simultaneously in MDM. Other studies that employed specific probes (32, 45) concluded that the nef mRNAs were the first expressed during infection; however, our results show that this interpretation stems from the relatively larger proportion of nef mRNA than the rev and tat RNAs. Primary lymphocytes and MDM produced the same array of spliced mRNAs; however, MDM had a lower relative abundance of the minor tat, rev, and nef isoforms that include noncoding exon 2 and/or 3. The most highly spliced nef mRNA, nef-1, was detected with lower abundance during infection of MDM than during infection of PBL and was not readily detected until late time points in a spreading infection of MDM. The requirement for Rev protein explains the delay in accumulation of the 4.0-kb series of HIV-1 mRNA compared to the 1.8-kb mRNAs (16, 20, 29, 34); however, a greater delay was evident in MDM than in PBL.
While there were the above-described relatively minor differences between lymphocytes and MDM during the first few weeks of infection, a much more significant difference was found in long-term-infected MDM just prior to and following the peak of viral infection in the macrophages, namely, a dramatic specific decrease in tat mRNAs greater than that found for any of the other spliced mRNAs. This specific decrease in tat was never observed in lymphocytes and was found to occur in macrophages as a consequence of infection rather than cell differentiation or maturation in culture. MDM are predominantly nondividing, and their expression of cellular mRNA, as indicated by the expression of the housekeeping gene β-actin, varies little, even over 8 weeks or more in culture, regardless of the HIV-1 status of the cells. Therefore, this specific decrease in tat mRNA was not due to a decline in cell numbers or viability but appears to coincide with the entry of MDM into a state of chronic or persistent infection with minimal virus production (at least at the level of sensitivity of a micro-RT assay). These persistently infected, long-term-cultured macrophages were still capable of producing virus, however, since addition of Tat protein was able to increase virus production from cells, reinforcing the potentially important role of low Tat levels in the long-term, nonproductive HIV-1 infection of MDM. Reactivation of virus production by Tat was achieved without altering the profile of viral RNA splicing from that seen at the time of Tat addition. This suggests that diminished splicing or stability of tat-specific mRNAs during long-term infection of MDM results in insufficient levels of Tat protein for efficient virion production, which is supported by the rapid decrease in Tat activity in these cells, determined with adenovirus vectors, after the first week or two of infection to near background levels in MDM infected for a month or more. As expected, this rapid decrease was not found in infected lymphocyte cultures that continue to produce virus while the cells remain viable.
Several groups examining the chronically infected cell lines U1 and ACH2, which have been used extensively as models of HIV-1 latency (22, 23, 37, 43), have shown that mutations within tat and TAR, respectively, are responsible for the dormant infections seen within these cells (8, 17, 18). Analysis of tat cDNAs from U1 cells showed two distinct forms of mutated tat, each represented in one of two integrated DNA copies. One tat cDNA was shown to lack an AUG initiation codon, while the other had a point mutation (H13L) that caused a severe reduction in transcriptional elongation in Tat reporter assays (18). In a fashion analogous to this report of long-term-infected macrophages, the addition of Tat protein to the culture medium led to activation of HIV-1 production from U1 cells (8). In contrast, in the ACH-2 cell line, normal activity was not rescued by addition of Tat protein to the culture medium (8), showing that the point mutation in TAR in these cells was sufficient to explain their transcriptional dormancy (18).
CEM T cells chronically infected with HIV-1 gradually become latently infected over a 4- to 6-week period. The viral genes influencing the rate of shutdown in these cells were mapped, and the 3′ region of the LTR was found to be the major determinant and the tat/rev/vpu region was found to contribute to a lesser extent (50). Since one of the five possible mutations in the LTR affecting HIV shutdown was in TAR, these findings also suggest that disruption of the Tat-TAR transcriptional axis can induce and maintain viral latency. In MDM, mutations in TAR would not explain our findings since the cultures remain responsive to exogenous Tat.
The diminished accumulation of Tat in long-term-infected MDM may result from altered RNA splicing, possibly induced by changes in cellular proteins that bind to the known ESS elements in the tat gene. Such events could reduce the expression of Tat sufficiently to severely disrupt Tat-TAR transcription and shut off virion production. The tat gene contains three ESS elements (ESS2, ESS2p, and ESS3) (2, 30, 57) that require the binding of hnRNPs of the A, B, or H group for their function of inhibiting splicing at adjacent upstream splice sites (9, 14, 30). These cellular factors and their role in HIV-1 splicing regulation have mostly been identified with HeLa cells. In addition, members of the serine/arginine-rich family of splicing factors have also been implicated in splicing regulation, usually by increasing splicing via exonic splice enhancer elements (59). Differential antagonism between these positive and negative regulators helps determine alternative exon usage (63). Although it is likely that such an interplay of these factors is also involved in alternative splicing regulation in macrophages, there is no direct evidence of this, nor whether this balance moves toward suppression of splicing during persistent infection in these long-lived cells.
The observed decline in tat mRNA and, subsequently, Tat activity to suboptimal levels for efficient virus production in macrophages could also be explained by the differential stability of tat mRNA relative to that of other HIV-1 transcripts during long-term infection of these cells. Various cellular genes are known to be regulated by controlling the rate at which their mRNAs decay, providing the cell with flexibility for rapid changes in their expression. Several cytokine-, growth factor-, and proto-oncogene-encoding mRNAs, such as granulocyte-macrophage colony-stimulating factor, TNF-α, gamma interferon, interleukin-2, c-fos, c-myc, and c-jun, are regulated by differential RNA stability (reviewed in reference 61), while several viruses, such as herpesviruses and hepatitis C virus, also use this strategy to control the levels and kinetics of viral and cellular gene expression (21, 56). Whether this is a mechanism used by HIV-1 to regulate its replication in macrophages is not known.
Whatever mechanism is responsible for viral shutoff in macrophages, it appears to be induced in response to infection rather than as a consequence of differentiation or maturation of the cells per se. Irrespective of whether MDM are infected with HIV-1 after a week or so in culture, as is the common practice with this culture system, or maintained for a month or more before they are infected, the same pattern of tat mRNA decline several weeks after infection is seen. This might suggest that accumulation in these cells of a particular viral protein or proteins, perhaps Tat itself, leads to cellular changes that affect either the splicing regulation of tat mRNA or its relative stability. The resultant chronically, but nonproductively, infected macrophage may well be significant as a viral reservoir and an important means by which the virus can persist in the body.
Although we have been able to show that HIV-1 persists in peripheral blood monocytes in vivo (54), evidence of this pattern of chronic infection in monocytes or macrophages from tissue sites of HIV-infected individuals is lacking, most probably because of the difficulty in obtaining sufficient infected cells for such an analysis. We and others have, however, shown that low-level replication does continue in monocytes despite years of effective antiretroviral therapy (54, 64) and have speculated that this reflects continued local replication of HIV-1 in the tissue macrophage reservoir (12, 51), which is generally considered to be an important source from which virus can rapidly rebound (3, 10, 35). Our demonstration of this pattern of infection in the most commonly used in vitro model of HIV-1 infection in macrophages suggests that regulation of viral mRNA processing may prove to be an important mechanism by which macrophages become long-lived reservoirs of HIV-1, where dormant or low-level expression of virus assists in the evasion of immune clearance.
Determination of the mechanism of HIV-1 mRNA regulation in macrophages, whether it involves altered splicing or differential stability or another mechanism, may uncover novel therapeutic targets that can be used against these critical cells. Currently available drugs are, at best, poorly efficacious against chronically infected cells, and alternative treatments and approaches are required that will be active against all reservoirs of HIV-1, will restrict viral replication in and transmission from these cells, and will lead eventually to their elimination from the body. In the shorter term, the ability to control replication in macrophages should greatly assist with the problem of rapid rebound after cessation or interruption of even the best currently available regimens.
Acknowledgments
This work was supported by National Health and Medical Research Council grants 990772 (S.S.) and 970558 and 111700 (D.F.J.P.), Commonwealth AIDS Research grant 92/09416 (S.S. and S.M.C.), and the Burnet Institute Research Fund. H.P.M. was the recipient of a Commonwealth AIDS Research grant Ph.D. scholarship.
We thank Martin Stoltzfus for many useful discussions and valuable suggestions for the manuscript.
REFERENCES
- 1.Amendt, B. A., D. Hesslein, J. Chang, and C. M. Stoltfus. 1994. Presence of negative and positive cis-acting RNA splicing elements within and flanking the first Tat-coding exon of human immunodeficiency virus type 1. Mol. Cell. Biol. 14:3960-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amendt, B. A., Z.-H. Si, and C. M. Stoltzfus. 1995. Presence of exon splicing silencers within human immunodeficiency virus type 1 tat exon 2 and tat-rev exon 3: evidence for inhibition mediated by cellular factors. Mol. Cell. Biol. 15:4606-4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aquaro, S., E. Balestra, A. Cenci, M. Francesconi, R. Calio, and C. F. Peron. 1997. HIV infection in macrophages: role of long-lived cells and related therapeutical strategies. J. Biol. Regul. Homeost. Agents 11:69-73. [PubMed] [Google Scholar]
- 4.Arrigo, S. J., S. Weitsman, J. A. Zack, and I. S. Chen. 1990. Characterization and expression of novel singly spliced RNA species of human immunodeficiency virus type 1. J. Virol. 64:4585-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Axelrod, J. H., and A. Honigman. 1999. A sensitive and versatile bioluminescence bioassay for HIV type 1 based on adenoviral vectors. AIDS Res. Hum. Retrovir. 15:759-767. [DOI] [PubMed] [Google Scholar]
- 6.Bilodeau, P. S., J. K. Domsic, A. Mayeda, A. R. Krainer, and C. M. Stoltzfus. 2001. RNA splicing at human immunodeficiency virus type 1 3′ splice site A2 is regulated by binding of hnRNP A/B proteins to an exonic splicing silencer element. J. Virol 75:8487-8497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bohan, C. A., F. Kashanchi, B. Ensoli, L. Buonaguro, K. A. Boris-Lawrie, and J. N. Brady. 1992. Analysis of Tat transactivation of human immunodeficiency virus transcription in vitro. Gene Expr. 2:391-407. [PMC free article] [PubMed] [Google Scholar]
- 8.Cannon, P., S. H. Kim, C. Ulich, and S. Kim. 1994. Analysis of Tat function in human immunodeficiency virus type 1-infected low-level-expression cell lines U1 and ACH-2. J. Virol. 68:1993-1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caputi, M., A. Mayeda, A. R. Krainer, and A. M. Zahler. 1999. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 18:4060-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chun, T. W., R. T. Davey, Jr., M. Ostrowski, J. Shawn Justement, D. Engel, J. I. Mullins, and A. S. Fauci. 2000. Relationship between pre-existing viral reservoirs and the re-emergence of plasma viremia after discontinuation of highly active anti-retroviral therapy. Nat. Med. 6:757-761. [DOI] [PubMed] [Google Scholar]
- 11.Crowe, S., J. Mills, and M. S. McGrath. 1987. Quantitative immunocytofluorographic analysis of CD4 surface antigen expression and HIV infection of human peripheral blood monocyte/macrophages. AIDS Res. Hum. Retrovir. 3:135-145. [DOI] [PubMed] [Google Scholar]
- 12.Crowe, S. M., and S. Sonza. 2000. HIV-1 can be recovered from a variety of cells including peripheral blood monocytes of patients receiving highly active antiretroviral therapy: a further obstacle to eradication. J. Leukoc. Biol. 68:345-350. [PubMed] [Google Scholar]
- 13.Deen, K. C., J. S. McDougal, R. Inacker, G. Folena-Wasserman, J. Arthos, J. Rosenberg, P. J. Maddon, R. Axel, and R. W. Sweet. 1988. A soluble form of CD4 (T4) protein inhibits AIDS virus infection. Nature 331:82-84. [DOI] [PubMed] [Google Scholar]
- 14.Del Gatto-Konczak, F., M. Olive, M.-C. Gesnel, and R. Breathnach. 1999. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol. Cell. Biol. 19:251-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Wet, J. R., K. V. Wood, M. DeLuca, D. R. Helenski, and S. Subramani. 1987. Firefly luciferase gene: structure and expression in mammalian cells. Mol. Cell. Biol. 7:725-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emerman, M., R. Vazeux, and K. Peden. 1989. The rev gene product of the human immunodeficiency virus affects envelope-specific RNA localization. Cell 57:1155-1165. [DOI] [PubMed] [Google Scholar]
- 17.Emiliani, S., W. Fischle, M. Ott, C. Van Lint, C. A. Amella, and E. Verdin. 1998. Mutations in the tat gene are responsible for human immunodeficiency virus type 1 postintegration latency in the U1 cell line. J. Virol. 72:1666-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emiliani, S., C. Van Lint, W. Fischle, P. Paras, Jr., M. Ott, J. Brady, and E. Verdin. 1996. A point mutation in the HIV-1 Tat responsive element is associated with postintegration latency. Proc. Natl. Acad. Sci. USA 93:6377-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feinberg, M. B., D. Baltimore, and A. D. Frankel. 1991. The role of Tat in the human immunodeficiency virus life cycle indicates a primary effect on transcriptional elongation. Proc. Natl. Acad. Sci. USA 88:4045-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Felber, B. K., M. Hadzopoulou-Cladaras, C. Cladaras, T. Copeland, and G. N. Pavlakis. 1989. The rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc. Natl. Acad. Sci. USA 86:1495-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng, P., D. N. Everly, Jr., and G. S. Read. 2001. mRNA decay during herpesvirus infections: interaction between a putative viral nuclease and a cellular translation factor. J. Virol. 75:10272-10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Folks, T. M., J. Justement, A. Kinter, C. A. Dinarello, and A. S. Fauci. 1987. Cytokine-induced expression of HIV-1 in a chronically infected promonocyte cell line. Science 238:800-802. [DOI] [PubMed] [Google Scholar]
- 23.Folks, T. M., J. Justement, A. Kinter, S. Schnittman, J. Orenstein, G. Poli, and A. S. Fauci. 1988. Characterization of a promonocyte clone chronically infected with HIV and inducible by 13-phorbol-12-myristate acetate. J. Immunol. 140:1117-1122. [PubMed] [Google Scholar]
- 24.Gartner, S., P. Markovits, D. M. Markovitz, M. H. Kaplan, R. C. Gallo, and M. Popovic. 1986. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science 233:215-219. [DOI] [PubMed] [Google Scholar]
- 25.Granelli-Piperno, A., E. Delgado, V. Finkel, W. Paxton, and R. M. Steinman. 1998. Immature dendritic cells selectively replicate macrophage-tropic (M-tropic) human immunodeficiency virus type 1, while mature cells efficiently transmit both M- and T-tropic virus to T cells. J. Virol. 72:2733-2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grassi, G., G. Pozzato, M. Moretti, and M. Giacca. 1995. Quantitative analysis of hepatitis C virus RNA in liver biopsies by competitive reverse transcription and polymerase chain reaction. J. Hepatol. 23:403-411. [DOI] [PubMed] [Google Scholar]
- 27.Griffin, G. E., K. Leung, T. M. Folks, S. Kunkel, and G. J. Nabel. 1989. Activation of HIV gene expression during monocyte differentiation by induction of NF-κB. Nature 339:70-73. [DOI] [PubMed] [Google Scholar]
- 28.Guatelli, J. C., T. R. Gingeras, and D. D. Richman. 1990. Alternative splice acceptor utilization during human immunodeficiency virus type 1 infection of cultured cells. J. Virol. 64:4093-4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hammarskjold, M. L., J. Heimer, B. Hammarskjold, I. Sangwan, L. Albert, and D. Rekosh. 1989. Regulation of human immunodeficiency virus env expression by the rev gene product. J. Virol. 63:1959-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacquenet, S., A. Mereau, P. S. Bilodeau, L. Damier, C. M. Stoltzfus, and C. Branlant. 2001. A second exon splicing silencer within human immunodeficiency virus type 1 tat exon 2 represses splicing of Tat mRNA and binds protein hnRNP H. J. Biol. Chem. 276:40464-40475. [DOI] [PubMed] [Google Scholar]
- 31.Kim, S. Y., R. Byrn, J. Groopman, and D. Baltimore. 1989. Temporal aspects of DNA and RNA synthesis during human immunodeficiency virus infection: evidence for differential gene expression. J. Virol. 63:3708-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klotman, M. E., S. Kim, A. Buchbinder, A. DeRossi, D. Baltimore, and F. Wong-Staal. 1991. Kinetics of expression of multiply spliced RNA in early human immunodeficiency virus type 1 infection of lymphocytes and monocytes. Proc. Natl. Acad. Sci. USA 88:5011-5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, T. H., F. J. Sunzeri, L. H. Tobler, B. G. Williams, and M. P. Busch. 1991. Quantitative assessment of HIV-1 DNA load by coamplification of HIV-1 gag and HLA-DQ-α genes. AIDS 5:683-691. [DOI] [PubMed] [Google Scholar]
- 34.Malim, M. H., J. Hauber, S. Y. Le, J. V. Maizel, and B. R. Cullen. 1989. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 338:254-257. [DOI] [PubMed] [Google Scholar]
- 35.Martin, J. C., and J. C. Brandes. 1999. Cells of the monocyte-macrophage lineage and pathogenesis of HIV-1 infection. J. Acquir. Immune Defic. Syndr. 22:413-429. [DOI] [PubMed] [Google Scholar]
- 36.Mentel, R., G. Dopping, U. Wegner, W. Seidel, H. Liebermann, and L. Dohner. 1997. Adenovirus-receptor interaction with human lymphocytes. J. Med. Virol. 51:252-257. [PubMed] [Google Scholar]
- 37.Michael, N. L., P. Morrow, J. Mosca, M. Vahey, D. S. Burke, and R. R. Redfield. 1991. Induction of human immunodeficiency virus type 1 expression in chronically infected cells is associated primarily with a shift in RNA splicing patterns. J. Virol. 65:1291-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muesing, M. A., D. H. Smith, C. D. Cabradilla, C. V. Benton, L. A. Lasky, and D. J. Capon. 1985. Nucleic acid structure and expression of the human AIDS/lymphadenopathy retrovirus. Nature 313:450-458. [DOI] [PubMed] [Google Scholar]
- 39.Munis, J. R., R. S. Kornbluth, J. C. Guatelli, and D. D. Richman. 1992. Ordered appearance of human immunodeficiency virus type 1 nucleic acids following high multiplicity infection of macrophages. J. Gen. Virol. 73:1899-1906. [DOI] [PubMed] [Google Scholar]
- 40.Naif, H. M., S. Li, M. Alali, A. Sloane, L. Wu, M. Kelly, G. Lynch, A. Lloyd, and A. L. Cunningham. 1998. CCR5 expression correlates with susceptibility of maturing monocytes to human immunodeficiency virus type 1 infection. J. Virol. 72:830-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicholson, J. K., G. D. Cross, C. S. Callaway, and J. S. McDougal. 1986. In vitro infection of human monocytes with human T lymphotropic virus type III/lymphadenopathy-associated virus (HTLV-III/LAV). J. Immunol. 137:323-329. [PubMed] [Google Scholar]
- 42.Planelles, V., J. B. Jowett, Q. X. Li, Y. Xie, B. Hahn, and I. S. Chen. 1996. Vpr-induced cell cycle arrest is conserved among primate lentiviruses. J. Virol. 70:2516-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pomerantz, R. J., D. Trono, M. B. Feinberg, and D. Baltimore. 1990. Cells nonproductively infected with HIV-1 exhibit an aberrant pattern of viral RNA expression: a molecular model for latency. Cell 61:1271-1276. [DOI] [PubMed] [Google Scholar]
- 44.Purcell, D. F., and M. A. Martin. 1993. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J. Virol. 67:6365-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robert-Guroff, M., M. Popovic, S. Gartner, P. Markham, R. C. Gallo, and M. S. Reitz. 1990. Structure and expression of tat-, rev-, and nef-specific transcripts of human immunodeficiency virus type 1 in infected lymphocytes and macrophages. J. Virol. 64:3391-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salahuddin, S. Z., R. M. Rose, J. E. Groopman, P. D. Markham, and R. C. Gallo. 1986. Human T lymphotropic virus type III infection of human alveolar macrophages. Blood 68:281-284. [PubMed] [Google Scholar]
- 47.Schwartz, S., B. K. Felber, D. M. Benko, E. M. Fenyo, and G. N. Pavlakis. 1990. Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J. Virol. 64:2519-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwartz, S., B. K. Felber, E. M. Fenyo, and G. N. Pavlakis. 1990. Env and Vpu proteins of human immunodeficiency virus type 1 are produced from multiple bicistronic mRNAs. J. Virol. 64:5448-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith, J., A. Azad, and N. Deacon. 1992. Identification of two novel human immunodeficiency virus type 1 splice acceptor sites in infected T cell lines. J. Gen. Virol. 73:1825-1828. [DOI] [PubMed] [Google Scholar]
- 50.Song, S. K., H. Li, and M. W. Cloyd. 1996. Rates of shutdown of HIV-1 into latency: roles of the LTR and tat/rev/vpu gene region. Virology 225:377-386. [DOI] [PubMed] [Google Scholar]
- 51.Sonza, S., and S. M. Crowe. 2001. Reservoirs for HIV infection and their persistence in the face of undetectable viral load. AIDS Patient Care STDs 15:511-518. [DOI] [PubMed] [Google Scholar]
- 52.Sonza, S., A. Maerz, N. Deacon, J. Meanger, J. Mills, and S. Crowe. 1996. Human immunodeficiency virus type 1 replication is blocked prior to reverse transcription and integration in freshly isolated peripheral blood monocytes. J. Virol. 70:3863-3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sonza, S., A. Maerz, S. Uren, A. Violo, S. Hunter, W. Boyle, and S. Crowe. 1995. Susceptibility of human monocytes to HIV type 1 infection in vitro is not dependent on their level of CD4 expression. AIDS Res. Hum. Retrovir. 11:769-776. [DOI] [PubMed] [Google Scholar]
- 54.Sonza, S., H. P. Mutimer, R. Oelrichs, D. Jardine, K. Harvey, A. Dunne, D. F. Purcell, C. Birch, and S. M. Crowe. 2001. Monocytes harbor replication-competent, non-latent HIV-1 in patients on highly active antiretroviral therapy. AIDS 15:17-22. [DOI] [PubMed] [Google Scholar]
- 55.Soto-Ramirez, L. E., B. Renjifo, M. F. McLane, R. Marlink, C. O'Hara, R. Sutthent, C. Wasi, P. Vithayasai, V. Vithayasai, C. Apichartpiyakul, P. Auewarakul, V. Pena Cruz, D. S. Chui, R. Osathanondh, K. Mayer, T. H. Lee, and M. Essex. 1996. HIV-1 Langerhans' cell tropism associated with heterosexual transmission of HIV. Science 271:1291-1293. [DOI] [PubMed] [Google Scholar]
- 56.Spangberg, K., L. Wiklund, and S. Schwartz. 2000. HuR, a protein implicated in oncogene and growth factor mRNA decay, binds to the 3′ ends of hepatitis C virus RNA of both polarities. Virology 274:378-390. [DOI] [PubMed] [Google Scholar]
- 57.Staffa, A., and A. Cochrane. 1995. Identification of positive and negative splicing regulatory elements within the terminal tat-rev exon of human immunodeficiency virus type 1. Mol. Cell. Biol. 15:4597-4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Theodore, T. S., G. Englund, A. Buckler-White, C. E. Buckler, M. A. Martin, and K. W. Peden. 1996. Construction and characterization of a stable full-length macrophage-tropic HIV type 1 molecular clone that directs the production of high titers of progeny virions. AIDS Res. Hum. Retrovir. 12:191-194. [DOI] [PubMed] [Google Scholar]
- 59.Wang, J., S.-H. Xiao, and J. L. Manley. 1998. Genetic analysis of the SR protein ASF/SF2: interchangeability of RS domains and negative control of splicing. Genes Dev. 12:2222-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Willey, R. L., D. H. Smith, L. A. Lasky, T. S. Theodore, P. L. Earl, B. Moss, D. J. Capon, and M. A. Martin. 1988. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J. Virol. 62:139-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilusz, C. J., M. Wormington, and S. W. Peltz. 2001. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell. Biol. 2:237-246. [DOI] [PubMed] [Google Scholar]
- 62.Yu, X., M. F. McLane, L. Ratner, W. O'Brien, R. Collman, M. Essex, and T. H. Lee. 1994. Killing of primary CD4+ T cells by non-syncytium-inducing macrophage-tropic human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 91:10237-10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu, J., A. Mayeda, and A. R. Krainer. 2001. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol. Cell 8:1351-1361. [DOI] [PubMed] [Google Scholar]
- 64.Zhu, T., D. Muthui, S. Holte, D. Nickle, F. Feng, S. Brodie, Y. Hwangbo, J. I. Mullins, and L. Corey. 2002. Evidence for human immunodeficiency virus type 1 replication in vivo in CD14+ monocytes and its potential role as a source of virus in patients on highly active antiretroviral therapy. J. Virol. 76:707-716. [DOI] [PMC free article] [PubMed] [Google Scholar]