Abstract
We investigated the effect of substrate binding on the mechanical stability of mouse dihydrofolate reductase using single-molecule force spectroscopy by atomic force microscopy. We find that under mechanical forces dihydrofolate reductase unfolds via a metastable intermediate with lifetimes on the millisecond timescale. Based on the measured length increase of ∼22 nm we suggest a structure for this intermediate with intact substrate binding sites. In the presence of the substrate analog methotrexate and the cofactor NADPH lifetimes of this intermediate are increased by up to a factor of two. Comparing mechanical and thermodynamic stabilization effects of substrate binding suggests mechanical stability is dominated by local interactions within the protein structure. These experiments demonstrate that protein mechanics can be used to probe the substrate binding status of an enzyme.
In many physiological systems like muscle or the cytoskeleton, mechanical properties and stability of proteins are important for protein function. Single-molecule techniques have made such material properties accessible on the level of individual protein domains (1,2). But even with proteins whose function is not primarily mechanical, single-molecule protein unfolding experiments may yield valuable information about the molecular conformation. In this study we investigate whether ligand binding to the enyzme dihydrofolate reductase (DHFR) alters the mechanical stability of the enzyme and how single-molecule mechanical experiments can be used to report on the binding status of individual enzymes.
DHFR reduces dihydrofolic acid to tetrahydrofolic acid in the presence of the cofactor NADPH. This enzymatic reaction is an important step in nucleotide synthesis. There are a variety of drugs targeting DHFR that have gained great importance in chemotherapy and in the treatment of autoimmune diseases; among these is methotrexate (MTX), which binds competitively to the folate binding site of DHFR. DHFR is well studied in classical folding experiments (3) and it is known that MTX binding greatly enhances the thermodynamic stability of the enzyme (4). Mechanical stabilization of the enzyme is hence likely although a direct link between unfolding forces and thermodynamic stability cannot be established (5). For our study we chose mouse DHFR because it has been used to study the mitochondrial protein import motor that unfolds proteins before import (6). DHFR without bound substrate is readily imported into mitochondria whereas binding of MTX completely stops the import (7). Mechanical stabilization of the enzyme by substrate binding seems especially interesting in this context.
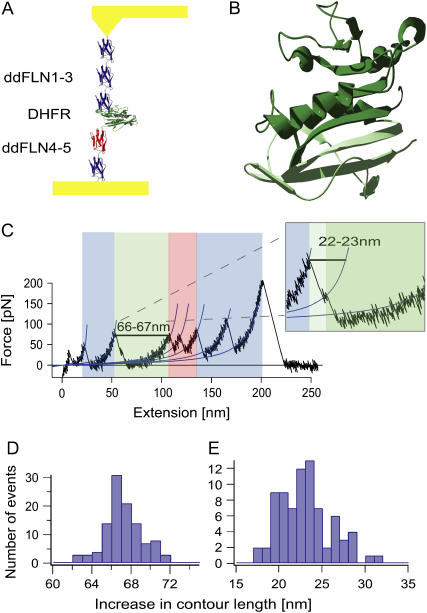
To investigate the mechanical stability of single mouse DHFR enzyme molecules we inserted DHFR into the rod of the actin cross-linking protein Ddfilamin (see Fig. 1 A). The Ddfilamin rod consists of five immunoglobulin domains and has been mechanically characterized in a series of previous studies (8). In our experiment the Ddfilamin domains serve as handles to contact DHFR at its termini. A typical force curve is shown in Fig. 1 C. All features known from Ddfilamin unfolding can be observed in the force curve. The unfolding events marked in blue reflect unfolding of Ig domains in the Ddfilamin rod whereas the red events reflect unfolding of Ig domain 4 that unfolds via a stable intermediate (8). In contrast to the pure Ddfilamin rod we now consistently observe an additional unfolding event (marked in green) where the polypeptide chain lengthens by 66–67 nm (see Fig. 1 D).
FIGURE 1.
(A) Schematic representation of the Ddfilamin-DHFR protein construct, stretched between a gold surface and a gold-coated cantilever tip. (B) Proposed structure of the intermediate state (dark green). The C-terminal part shown in light green unfolds during the first unfolding event. (C) Typical force versus extension curve. The unfolding pattern of the Ddfilamin domains is shown in blue (and in red for domain 4), whereas the unfolding event of DHFR is shown in green. The inset shows a magnification in which the intermediate state is clearly visible. (D) Increase in contour length for the complete unfolding of DHFR. (E) Increase in contour length for the unfolding of DHFR from the native state to the intermediate state.
This gain in length is exactly the expected value for an unfolding event where the 186 amino acids folded in a DHFR domain go from a folded into a completely stretched conformation. Closer inspection of the DHFR unfolding event shows substructure within the unfolding event (cf. inset in Fig. 1 C). Before completely relaxing to the unfolded state the cantilever dwells for ∼3 ms at an intermediate level. This is indication for a metastable mechanical unfolding intermediate (9). Analysis of the length gain from the folded to the intermediate conformation (Fig. 1 E) allows us to identify the amino acid residues that form the intermediate structure. The 22 ± 3 nm gain corresponds to 55–65 amino acid residues detaching from either the N- or C-terminus. Given the large structural change within the tightly folded core of DHFR induced by detachment of 55–65 residues from the N-terminus, the most likely candidate is detachment of the five C-terminal β-sheets (marked in light green in Fig. 1 B). The remainder of 121–131 amino acids (marked in dark green) will still be able to form intact binding sites for both substrates.
In a next set of experiments we studied the effect of substrate binding on the stability of DHFR and the mechanical unfolding intermediate. We could not detect any mechanical difference between the substrate free protein and the protein with NADPH bound (see Supplementary Material). To improve statistics of our measurements we therefore pooled those data together. We analyzed both the forces of the major unfolding peak and the lifetime of the unfolding intermediate in the absence of substrate as well as in the presence of MTX and in the simultaneous presence of the substrates MTX and NADPH (see Fig. 2, A and B). Binding of MTX was independently verified by the characteristic spectroscopic ultraviolet shift. We found the major unfolding peak was unchanged at all conditions with an average of ∼60 pN (insets in Fig. 2, C–E). In contrast to the unfolding force we did observe an effect of substrate binding on the stability of the intermediate. The lifetimes under the different substrate conditions are shown in the left panels of Fig. 2, C–E). While MTX alone increases the average lifetime of the intermediate under force by only 25%, in the presence of both MTX and NADPH the lifetimes double. The fits in Fig. 2, C–E, are Monte Carlo simulations with a potential width of 2 Å taking into account the distribution of forces acting on the intermediate (for details see Supplementary Material). The best fit values for the lifetimes under zero load condition are shown in Table 1.
FIGURE 2.
(A, B) Force versus time curves for DHFR without substrate (A) and with both substrates (B). The arrows mark the unfolding force and intermediate lifetime; panels C–E show the lifetimes of the intermediate state for different substrate conditions. The blue markers represent the experimental data with N1/2-error bars, whereas the green lines are Monte Carlo simulations of the experiment. Unfolding force distributions for the native state are shown as insets.
TABLE 1.
Intermediate state lifetimes at zero force (Monte Carlo simulations with a potential width of 2 Å)
| Lifetime of the intermediate state at zero force (ms) | |
|---|---|
| DHFR, DHFR + NADPH | 8 ± 2 |
| DHFR + MTX | 10 ± 2 |
| DHFR + MTX + NADPH | 16 ± 3 |
Surprisingly the effects of substrate binding on mechanical stability of the protein are overall moderate. The average force of the major unfolding peak was even unchanged at all conditions. In contrast, equilibrium experiments have shown drastic stabilization of DHFR upon binding of MTX. This seeming discrepancy can be resolved by considering that our mechanical experiments occur in nonequilibrium. Obviously the interactions determining the average unfolding force of 60 pN reside in the C-terminal part of the protein that detaches during the transition from folded to the intermediate. This part is distant from all substrate binding sites and hence an influence of substrate binding is not expected. The influence on the stability of the intermediate is stronger but only binding of both substrates leads to a significant effect. This is in accord with studies on the titin domain I27 showing that the interactions determining mechanical protein stability are local (2). In contrast, equilibrium free energy samples over all interactions within the protein structure. The location of force application to the protein is therefore likely of great importance. In the proposed intermediate structure (dark green part in Fig. 1 B) the N- and C-terminus is not in direct contact with the bound MTX and a weak effect is in perfect agreement. We anticipate that force application close to the substrate binding sites can be used to tune the sensitivity of structural stability on ligand binding.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Supplementary Material
References
- 1.Rief, M., M. Gautel, F. Oesterhelt, J. M. Fernandez, and H. E. Gaub. 1997. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 276:1109–1112. [DOI] [PubMed] [Google Scholar]
- 2.Marszalek, P. E., H. Lu, H. Li, M. Carrion-Vazquez, A. F. Oberhauser, K. Schulten, and J. M. Fernandez. 1999. Mechanical unfolding intermediates in titin modules. Nature. 402:100–103. [DOI] [PubMed] [Google Scholar]
- 3.Touchette, N. A., K. M. Perry, and C. R. Matthews. 1986. Folding of dihydrofolate reductase from Escherichia coli. Biochemistry. 25:5445–5452. [DOI] [PubMed] [Google Scholar]
- 4.Chunduru, S. K., V. Cody, J. R. Luft, W. Pangborn, J. R. Appleman, and R. L. Blakley. 1994. Methotrexate-resistant variants of human dihydrofolate reductase. Effects of Phe31 substitutions. J. Biol. Chem. 269:9547–9555. [PubMed] [Google Scholar]
- 5.Li, H., A. F. Oberhauser, S. B. Fowler, J. Clarke, and J. M. Fernandez. 2000. Atomic force microscopy reveals the mechanical design of a modular protein. Proc. Natl. Acad. Sci. USA. 97:6527–6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neupert, W., and M. Brunner. 2002. The protein import motor of mitochondria. Nat. Rev. Mol. Cell Biol. 3:555–565. [DOI] [PubMed] [Google Scholar]
- 7.Eilers, M., and G. Schatz. 1986. Binding of a specific ligand inhibits import of a purified precursor protein into mitochondria. Nature. 322:228–232. [DOI] [PubMed] [Google Scholar]
- 8.Schwaiger, I., A. Kardinal, M. Schleicher, A. A. Noegel, and M. Rief. 2004. A mechanical unfolding intermediate in an actin-crosslinking protein. Nat. Struct. Mol. Biol. 11:81–85. [DOI] [PubMed] [Google Scholar]
- 9.Dietz, H., and M. Rief. 2004. Exploring the energy landscape of GFP by single-molecule mechanical experiments. Proc. Natl. Acad. Sci. USA. 101:16192–16197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.