Abstract
Actinobacillus pleuropneumoniae is the etiological agent of porcine pleuropneumonia. Tetracycline is used for therapy of this disease, and A. pleuropneumoniae carrying the tet(B) gene, coding for an efflux protein that reduces the intercellular tetracycline level, has been described previously. Of the 46 tetracycline-resistant (Tcr) Spanish A. pleuropneumoniae isolates used in this study, 32 (70%) carried the tet(B) gene, and 30 of these genes were associated with plasmids. Eight (17%) isolates carried the tet(O) gene, two (4%) isolates carried either the tet(H) or the tet(L) gene, and all these genes were associated with plasmids. This is the first description of these tet genes in A. pleuropneumoniae. The last two Tcr isolates carried none of the tet genes examined. Except for tet(O)-containing plasmids, the other 34 Tcr plasmids were transformable into an Escherichia coli recipient. Two plasmids were completely sequenced. Plasmid p11745, carrying the tet(B) gene, was 5,486 bp and included a rep gene, encoding a replication-related protein, and two open reading frames (ORFs) with homology to mobilization genes of Neisseria gonorrhoeae plasmid pSJ7.4. Plasmid p9555, carrying the tet(L) gene, was 5,672 bp and, based on its G+C content, consisted of two regions, one of putative gram-positive origin containing the tet(L) gene and the other comprising four ORFs organized in an operon-like structure with homology to mobilization genes in other plasmids of gram-negative bacteria.
Porcine pleuropneumonia caused by Actinobacillus pleuropneumoniae is a worldwide disease of swine that manifests as severe outbreaks, resulting in important economic losses, among fattening and finishing pigs (1). Clinical signs include dyspnea, fever, reduced appetite, and rapidly occurring death, with an elevated mortality rate. A cough and, in peracute cases, a frothy and bloodstained nasal discharge can also be observed. In addition, the lung lesions are characteristic in acute cases, with consolidation and raised cherry-red or black areas of necrotizing and hemorrhagic pneumonia, with interlobular edema on the diaphragmatic lobe and fibrinous pleuritis (27). The change from traditional continuous swine production to the modern “all in/all out” industry has contributed to new epidemiological situations (20).
Tetracyclines are often used for therapy of this disease (2). Tetracyclines belong to a family of broad-spectrum antibiotics that include tetracycline, doxycycline, and other semisynthetic derivatives (4). Tetracyclines have been widely used in veterinary medicine for prophylaxis and growth promotion in food animals. Numerous studies have described the effects of long-term usage of subtherapeutic doses of tetracyclines, which include increased levels of resistant gut bacteria or pathogens (24). The use of antibiotics as growth promoters has been discouraged by the European Union (Council Directive 70/524 EEC, 1977); however, consumption figures suggest that the use of tetracyclines in veterinary practice is still high compared with use of other classes of antibiotics (30).
Several studies have reported the high incidence of tetracycline resistance (Tcr) in A. pleuropneumoniae. Forty-two of the 71 (59.2%) isolates collected in Spain in the period 1992 to 1993 were resistant to oxytetracycline (11), whereas 55 of 60 (90%) A. pleuropneumoniae strains isolated from pigs in Taiwan during 1985 to 1993 showed tetracycline resistance (3). Previous work identified the tet(B) gene in 8 of the 17 (47%) Tcr A. pleuropneumoniae isolates collected in Norway between 1986 and 1992 (34). However, the data of that study suggested that other tet genes may be present in these isolates.
The aim of the present study was to determine the type and genomic location of tet genes carried in a collection of Spanish Tcr A. pleuropneumoniae isolates recovered from different farms of the “Castilla-León” region between 1997 and 2001.
MATERIALS AND METHODS
Isolation and serotyping of A. pleuropneumoniae.
Forty-six Tcr A. pleuropneumoniae strains isolated from the “Castilla y León” region of Spain between 1997 and 2001 were studied. The isolates were grown on chocolate agar plates with gonococcus (GC) agar (Oxoid, Basingstoke, United Kingdom) supplemented with 5% sheep blood (bioMerieux, Marcy l'Etoile, France), 12 μg/ml of NAD (Sigma-Aldrich Química., Madrid, Spain), and Kellogg's supplement solution (0.22 M d-glucose, 0.03 M l-glutamine, 0.001 M ferric nitrate, and 0.02 M cocarboxylase) (Sigma-Aldrich Química, Madrid, Spain) as previously described (5). The plates were incubated at 37°C under a 5% CO2 atmosphere. The isolates were serotyped as previously described (11).
Growth of Escherichia coli strains.
Escherichia coli S17-1 (31), DH5α (28), and IT1022 (9) were grown on Luria-Bertani (LB) agar (Pronadisa S.A., Madrid, Spain) supplemented with the appropriate antibiotics (300 μg of streptomycin/ml, 20 μg of nalidixic acid/ml, or 10 μg of tetracycline/ml).
Antimicrobial susceptibility testing.
Bacteria were grown overnight on chocolate agar plates and resuspended in sterilized saline solution, and the turbidity was adjusted to a 0.5 McFarland standard. The MICs of tetracycline and doxycycline were determined by broth microdilution assays in accordance with CLSI (formerly NCCLS) guidelines (23) in veterinary fastidious medium supplemented with 0.025% NAD (Sigma-Aldrich Química, Madrid, Spain). The microplates (Sensititre VAV5; Trek Diagnostic Systems, England) were incubated at 37°C for 24 h, and the MIC of each antimicrobial agent was the lowest concentration that inhibited visible growth. A. pleuropneumoniae ATCC 27090 (serovar 3) was included in the study as a quality control.
Extraction of DNA.
Genomic DNA was obtained as previously described (6, 29). Plasmid DNA was prepared from 100 ml broth culture using a Midi plasmid preparation and purification kit (QIAGEN S.A., Courtaboeuf, France).
DNA-DNA hybridization.
Whole-cell bacterial dot blots were prepared, and DNA-DNA hybridization was performed as previously described with 32P-labeled probes (19) or with chemiluminescently labeled probes by using the CDP-Star system according to the manufacturer's instructions (Roche Diagnostics, S.L., Barcelona, Spain). tet genes—tet(A), tet(B), tet(C), tet(D), tet(E), tet(G), tet(H), tet(K), tet(L), tet(M), tet(O), tet(S), tet(Y), and tet(29)—were detected by DNA-DNA dot blot hybridization and/or PCR assays as previously described (21). Positive and negative controls were included in each assay. To determinate the location of tet genes, Southern blotting of uncut genomic and plasmid DNA was performed according to reference 28.
PCR detection of tet genes.
The presence of the various tet genes was confirmed using PCR assays with primers specific for each gene and by sequencing of the PCR products. The sequences of the primers and their annealing temperatures have been described previously and are available upon request (see the supplemental material). The PCR mixture consisted of 10 μl of DNA template (10 pg/μl), 4 U of Taq DNA polymerase (Bioline, London, United Kingdom), 1.5 mM MgCl2, 10 μl of 10× PCR amplification buffer [160 mM (NH4)2SO4, 670 mM Tris-HCl (pH 8.8), 0.1% Tween 20], 40 pmol of each primer, 0.2 mM each deoxynucleoside triphosphate (Bioline, London, United Kingdom), and double-distilled water to a final volume of 100 μl. DNA was first denatured at 95°C for 5 min and then subjected to 30 cycles of amplification under the following conditions: denaturation at 95°C for 1 min, annealing for 1 min, and extension at 72°C for 1 min. After the final cycle, the reactions were terminated by an extra run at 72°C for 10 min, and the reaction products were then kept at 4°C until analysis.
Electrotransformation.
Competent E. coli S17-1 cells were prepared as previously described (28) and electroporated with 100 ng of A. pleuropneumoniae plasmid DNA. Electrotransformation was performed in a Gene Pulser II electroporation system (Bio-Rad, Foster City, CA) by applying an electric pulse as described by the manufacturer (2.5 kV, 25 μFa, 800 Ω) (8). Transformants were selected on LB agar supplemented with tetracycline (10 μg/ml) and streptomycin (300 μg/ml).
Mobilization experiments.
Conjugal transfer of the tetracycline resistance phenotype was assayed by mating A. pleuropneumoniae isolates carrying the various plasmids to an E. coli S17-1 recipient. A. pleuropneumoniae isolates APP11745 [tet(B)], APP13142 [tet(O)], APP9956 [tet(H)], and APP9555 [tet(L)], or E. coli S17-1 transformants carrying the Tcr plasmid p9555, were used as donors, and E. coli DH5α was used as the recipient. The bacteria were grown at 37°C overnight and mixed in a donor-to-recipient ratio of 1:8. The resultant suspension was then placed on a 0.45-μm HA membrane (Millipore) and incubated at 37°C overnight. Transconjugants were selected on LB agar plates containing 10 μg of tetracycline and 20 μg of nalidixic acid (Nx) per ml. Tcr Nxr colonies were tested for streptomycin resistance in order to confirm that they were true transconjugants. Transfer frequency was expressed as the number of transconjugants per recipient cell obtained after plating on selective medium.
DNA sequencing and analyses.
Plasmids p11745 and p9555 were amplified using primers tetBoutF (AAA GAG TCA TCA GCA AGG TGC T)-tetBoutR (TAT GCG GTG AAA TCT CTC CTG C) and tetLoutF (TTA CTT GAT CAA AGG TTG TT)-tetLoutR (AAT CAT TTG CAA TAT CAG GT), respectively, which are complementary to the tet(B) and the tet(L) gene, respectively. The PCR amplicons were sequenced by following a primer-walking strategy at the “Servicio de Secuenciación de DNA HUMV—Unican” of the “Universidad de Cantabria” using a Beckman CEQ 2000XL apparatus. Contiguous sequences were assembled using the ContigExpress Vector NTI Suite 5.5 (Informax, Bethesda, Md.). Homology searches were performed with BLAST and ORF Finder tools at the website of the National Center for Biotechnology Information (Bethesda, MD).
Nucleotide sequence accession numbers.
The complete sequences of plasmids p11745 and p9555, which contain the tet(B) and tet(L) genes, respectively, have been deposited in GenBank under accession no. DQ176855 and AY359464. The sequences of the tet regions of the A. pleuropneumoniae isolates have been deposited in GenBank under accession no. AY987963 for tet(O) and AY987962 for tet(H).
RESULTS
Serotyping of A. pleuropneumoniae.
The 46 Tcr A. pleuropneumoniae isolates were serotyped. Among the five serotypes detected, serovar 2 and serovar 4 were predominant (six isolates each), followed by serotypes 6 and 7 (two isolates each); one isolate belonged to serotype 1. Twenty-nine of the isolates were nontypeable.
Distribution of the tet genes.
Of the 46 Tcr A. pleuropneumoniae isolates tested, 44 (96%) carried one of the tet genes tested. These included 32 (70%) isolates carrying the tet(B) gene, 8 (17%) isolates carrying the tet(O) gene, and 2 isolates each (4% each) carrying the tet(H) or the tet(L) gene (Table 1). None of the 44 isolates carried more than one tet gene (Table 1).Two Tcr isolates, as well as A. pleuropneumoniae ATCC 27090, did not carry any of the 14 tet genes examined.
TABLE 1.
A. pleuropneumoniae isolates and tet genes carried
| No. of isolatesa | Serovar | MIC (μg/ml) of:
|
tet gene | Location | Plasmid size (kb) | Transformation of E. coli | |
|---|---|---|---|---|---|---|---|
| Tetracycline (0.25-128 μg/ml) | Doxycycline (0.25-32 μg/ml) | ||||||
| 5 | 2 | 64 | 16 | B | Plasmid | 5.5 | + |
| 1 | 2 | 64 | 32 | B | Plasmid | 5.5 | + |
| 4 | 4 | 64 | 16 | B | Plasmid | 5.5 | + |
| 1 | 7 | 64 | 16 | B | Plasmid | 5.5 | + |
| 19 | Nontypeable | 16-64 | 4-32 | B | Plasmid | 5.5 | + |
| 2 | Nontypeable | 16-32 | 2-8 | B | Not done | − | |
| 2 | 6 | 32-64 | 32 | O | Plasmid | 6 | − |
| 1 | 7 | 64 | 32 | O | Plasmid | 6 | − |
| 5 | Nontypeable | 64 | 32 | O | Plasmid | 6 | − |
| 1 | 4 | 32 | 16 | H | Plasmid | 5.7 | + |
| 1 | Nontypeable | 64 | 16 | H | Plasmid | >12 | + |
| 1 | 4 | 32 | 8 | L | Plasmid | 5.6 | + |
| 1 | 1 | 64 | 8 | L | Plasmid | >12 | + |
| 2 | Nontypeable | 16-64 | 2-16 | Unknown | |||
| 1b | 3 | 0.5 | 0.5 | No gene | |||
The total number of isolates was 47.
A. pleuropneumoniae ATCC 27090.
Characterization of the tet(B) genes.
To determine the location of the tet(B) genes, Southern blots of uncut genomic and plasmid DNA were hybridized with a tet(B) probe consisting of a 1-kb internal region of the gene isolated from E. coli IT1022 which contains Tn10. In 30 of the 32 tet(B)-positive isolates, their plasmids hybridized with the tet(B) probe, indicating a plasmid location. The location was verified by transforming the plasmids into E. coli and selecting for tetracycline resistance. The location of the tet(B) gene in the remaining two isolates is still under investigation.
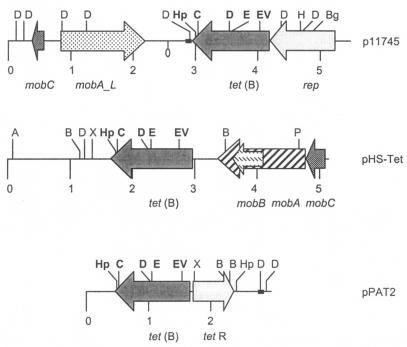
The plasmid size (5.5 kb) and indistinguishable restriction patterns with the HindIII and DraI endonucleases indicated that the 30 tet(B)-positive plasmids were closely related, if not identical. Plasmid p11745, from isolate APP11745, was selected and completely sequenced. This plasmid consisted of 5,486 bp and had a G+C content of 38.1%. The plasmid had four open reading frames (ORFs), with orf3 containing the 1,209-bp structural gene tet(B), which had 99% sequence identity at the DNA and amino acid levels with the tet(B) gene from Tn10 (GenBank accession no. AF162223). The tet(B) gene usually has a regulator gene, tetR(B), immediately upstream of the structural tet(B) gene; the two genes share a common promoter and are read in opposite directions (25). However, the tetR(B) gene was not present in plasmid p11745 (Fig. 1). A PCR assay for the tetR(B) gene gave a PCR product of the correct size for the control, but the tet(B)-positive A. pleuropneumoniae isolate did not produce a PCR product from the assay, nor was the sequence found on the plasmid. The 21 bp upstream of the tet(B) gene was identical to that found in Tn10 and included the putative ribosome binding site (RBS) sequence (AGAGAA) 7 bp upstream of the tet(B) gene. Beyond this sequence, −35 (TAAAAT) and −10 (TAAAAT) boxes resembling A. pleuropneumoniae consensus promoter sequences (7) were found. A putative Rho-independent transcription terminator located 35 bp downstream from the translational stop codon of the tet(B) gene was also identified. In the downstream region there was a 75-bp stretch showing identity with pPAT2 (Fig. 1).
FIG. 1.
Plasmid p11745 from A. pleuropneumoniae compared to plasmids pHS-Tet from H. parasuis and pPAT2 from P. aerogenes. Numbers indicate size in kilobases. Striped and stippled arrows correspond to plasmid mobilization genes (mobA, mobA_L, mobB, and mobC). Shaded arrows correspond to tetracycline resistance genes or replication genes [tet(B), tet(R), or rep]. The putative transcriptional terminator for p11745 tet(B) is depicted. Black boxes represent the 99-bp region of homology between p11745 and pPAT2. Restriction sites are abbreviated as follows: B (BclI), Bg (BglII), C (ClaI), D (DraI), E (EcoRI), EV (EcoRV), H (HindIII), Hp (HpaI), K (KpnI), P (PstI), and X (XbaI). Boldface indicates coincident restriction sites.
Three other ORFs (orf1, orf2, and orf4) were identified in plasmid p11745 (Table 2). The orf4 gene encoded a replication-related protein, whereas orf1 and orf2 coded for mobilization-related proteins, which had 70% and 56% homology, respectively, with the MobC and MobA_L proteins of the Neisseria gonorrhoeae plasmid pSJ7.4 (GenBank accession no. AY221996) (Fig. 1) (26). The functional map of p11745 was compared with those of two other tet(B)-positive plasmids of Pasteurellaceae, pHS-Tet from Haemophilus parasuis (GenBank accession no. AY862435) (18) and pPAT2 from Pasteurella aerogenes (GenBank accession no. AY278685) (15). DNA homology between p11745 and the other two plasmids was limited to the tet(B) genes (Fig. 1).
TABLE 2.
Plasmid p11745 ORFsa
| ORF | Gene | % G+C | Size (aa) | Identity of predicted protein
|
|
|---|---|---|---|---|---|
| % Identity | Closest predicted protein (GenBank accession no.) | ||||
| 1 | mobC | 33.7 | 97 | 70 | MobC of Neisseria gonorrhoeae pSJ7.4 (AAO45531) |
| 2 | mobA_L | 39.2 | 450 | 56 | MobA of Neisseria gonorrhoeae pSJ7.4 (AAO45530) |
| 3 | tet(B) | 42.9 | 411 | 99 | TetB of Haemophilus parasuis pHS-Tet (AAW51465) |
| 4 | rep | 35.1 | 337 | 47 | Replication protein of Mannheimia varigena pMVSCS1 (NP_573540) |
See Fig. 1.
Characterization of the tet(O) genes.
Eight isolates carried the tet(O) gene (Table 1), which was associated with a 6-kb plasmid in each isolate. From plasmid p13142, 2,489 bp was sequenced, which included the complete tet(O) gene (GenBank accession no. AY987963) and shared 99% identity at the DNA and amino acid levels with the tet(O) gene from Campylobacter jejuni (GenBank accession no. AY394561) (33). The upstream (270-bp) and downstream (299-bp) sequences were identical to that of Campylobacter coli and included a putative RBS sequence 12 bp upstream of the start codon. The −35 and −10 regions corresponding to the putative tet(O) promoter were identified.
Characterization of the tet(H) genes.
The tet(H) genes were located on a 5.6-kb plasmid (p9956) (isolate APP9956) and a >12-kb plasmid (p12494) (isolate APP12494). One-kilobase PCR amplicons from these plasmids were sequenced, and their DNA and amino acid sequences were 100% identical to those of the tet(H) gene from the Pasteurella plasmid pPAT1 (GenBank accession no. AJ245947) (14). The tet(H) structural gene and the tetR(H) repressor gene were identified in a 1,179-bp region by partial sequencing (3 kb) of plasmid p9956. These gene sequences were identical to the tet(H) gene from plasmid pPAT1 but were 24 bp shorter than the complete tet(H) gene (12). This deletion produced a truncated structural tet protein that was missing the last 8 residues in the C terminus. The deleted protein is functional, since the tetracycline MIC was 32 μg/ml for isolate APP9956, and it is also functional in plasmid pPAT1 (14). The tetR(H) gene was 624 bp long, and its DNA and amino acid sequences were 100% identical with those of the tetR(H) gene from Pasteurella plasmids (14).
Characterization of the tet(L) genes.
Two isolates (APP9555 and APP13213) carried the tet(L) genes, which were located on plasmids, p9555 and p13123, of 5.6 and >12 kb, respectively. The 0.9-kb amplicons were sequenced and were 100% identical at the DNA and amino acid levels to the tet(L) gene from Bacillus subtilis (GenBank accession no. AY129652). Plasmid p9555 was completely sequenced and consisted of 5,672 bp with a G+C content of 39.4%. The plasmid had five ORFs with significant homology to proteins of known function (Fig. 2; Table 3). orf2 was the tet(L) structural gene and consisted of 1,356 bp which had 99% identity at the DNA and amino acid levels to other plasmid-borne tet(L) genes, including the Mannheimia plasmid pCCK3259 (EMBL database accession no. AJ966516) (17) and the Bacillus cereus plasmid pJHI (GenBank accession no. AY129652). The tet(L) gene in plasmid p9555 is preceded by a RBS sequence (AGAAGG) at positions 417 to 422 but lacks the translational attenuator which is found upstream in other tet(L) gene sequences along with the promoter sequence (Fig. 2). The intergenic region between orf1 and the tet(L) gene does not present significant homologies with DNA sequences registered in databases. However, −35 (TTAGCT) and −10 (TTTAAT) regions resembling A. pleuropneumoniae consensus promoter sequences at positions 3109 to 3114 and 3140 to 3145 are potential promoters for tet(L) gene expression (7). A putative Rho-independent transcription terminator was found 9 bp downstream of the translational stop codon. This terminator is very similar to that previously found in all tet(L) genes of gram-positive bacteria (GenBank accession no. AY129652).
FIG. 2.
(A) Plasmid p9555 from A. pleuropneumoniae compared to plasmids pCCK3259 from Mannheimia haemolytica and pHS-Tet from H. parasuis. Numbers indicate size in kilobases. Striped and stippled arrows correspond to plasmid mobilization genes (mobA, mobB, and mobC). Shaded arrows correspond to tetracycline resistance genes [tet(B) and tet(L)]. The putative transcriptional terminator (oval) for the p9555 tet(L) gene and the potential recombination site (diamond) detected in this plasmid are depicted. Restriction sites are abbreviated as follows: B (BclI), Bg (BglII), C (ClaI), D (DraI), E (EcoRI), EV (EcoRV), H (HindIII), Hp (HpaI), K (KpnI), P (PstI), and X (XbaI). The pCCK3259 357-bp deletion is represented by a black box flanked by the 10-bp direct repeats. (B) Potential recombination site (boxed) downstream of tet(L) in p9555 and comparison with the corresponding sequences of plasmid pTB19 from Geobacillus stearothermophilus and plasmid pAB2 from Mannheimia haemolytica. The comparison includes the following bases in their respective plasmid sequences: bp 3222 to 3268 in the G. stearothermophilus pTB19 sequence (GenBank accession no. M63891), bp 1971 to 2017 in the A. pleuropneumoniae p9555 sequence (GenBank accession no. AY359464), and bp 3229 to 3274 in the M. haemolytica pAB2 sequence (GenBank accession no. Z21724).
TABLE 3.
Plasmid p9555 ORFsa
| ORF | Gene | % G+C | Size (aa) | Identity of predicted protein
|
|
|---|---|---|---|---|---|
| % Identity | Closest predicted protein (GenBank accession no.) | ||||
| 1 | repA | 34.8 | 67 | 50 | RepB of Lactococcus lactis plasmid pFX2 (Q04138) |
| 2 | tet(L) | 35.5 | 451 | 99 | TetL of Bacillus cereus plasmid pJH1 (AAN05731) |
| 3 | mobC | 49.4 | 76 | 91 | MobC of Mannheimia haemolytica plasmid pMHSCS1 (CAC20662) |
| 4 | mobA | 43.7 | 468 | 89 | MobA of Haemophilus parasuis plasmid pHS (AAW51466) |
| 5 | mobB | 45.1 | 160 | 89 | MobB of Haemophilus parasuis plasmid pHS (AAW51467) |
See Fig. 2A.
Four other orfs (orf1 and orf3 to orf5) were identified in plasmid p9555. The orf1 gene codes for a truncated 67-amino-acid replication-related protein, whereas the other three showed homology at the DNA and amino acid levels with previously described mobilization-related genes organized in an operon structure (Fig. 2) (22). A potential recombination site was found downstream of the tet(L) gene (Fig. 2).
Two regions were identified in plasmid p9555. The first region consists of the tet(L) gene and has a G+C content of 35%. The second contains the mob genes (mobC, mobA, and mobB), with a G+C content of 40 to 45%, and is indistinguishable from the recently described Mannheimia plasmid pCCK3259 (17) with the exception of an additional 363-bp region that is absent in pCCK3259 (Fig. 2). This 363-bp region is flanked by 10-bp direct repeats which could possibly be easily deleted. The mob region of plasmid p9555 is highly homologous (>90%) to the mob region of the H. parasuis plasmid pHS-Tet (Fig. 2).
Transfer of A. pleuropneumoniae plasmids.
The tet(B)-, tet(H)-, and tet(L)-positive plasmids were electroporated into E. coli S17-1, and Tcr transformants were shown to carry the different tetracycline resistance plasmids, indicating that these plasmids replicate and that their associated tet genes are expressed in E. coli. In contrast, no Tcr transformants occurred when plasmids from isolates carrying the tet(O) gene were electroporated into E. coli S17-1.
Mobilization of plasmids p11745, p13142, p9956, and p9555 from their original A. pleuropneumoniae isolates into E. coli S17-1 was examined, but no transconjugants were obtained. However, analysis of the plasmid p9555 sequence revealed the existence of a mob operon encoding MobA, a relaxase belonging to the ColE1 superfamily of relaxases (10), suggesting that p9555 is mobilizable. Plasmids encoding relaxases of this family can be mobilized with the assistance of IncP plasmid conjugation machinery. Mobilization of p9555 was assayed from E. coli S17-1, which carried a derivative of the conjugative RP4 plasmid integrated into the chromosome, to the E. coli DH5α recipient, and Tcr colonies were obtained at a frequency of 10−3 per recipient. The PCR assay with tet(L)-specific primers determined that all 10 Tcr transconjugants carried p9555, confirming the transfer of p9555 from E. coli S17-1 to E. coli DH5α.
DISCUSSION
This is the first description of the tet(L), tet(H), and tet(O) genes in A. pleuropneumoniae or the genus Actinobacillus. All three of these tet genes were on small plasmids, as were the majority (94%) of the tet(B) genes. This is also the first time the tet(O) gene has been identified on small nonconjugative plasmids (4). The tet(O) gene is of gram-positive origin but has been identified in gram-negative Campylobacter, Neisseria, and Megasphaera spp. and has been associated with conjugative plasmids in some of these isolates (http://faculty.washington.edu/marilynr/) (25). In the eight tet(O)-positive A. pleuropneumoniae isolates, the gene is on a 6-kb nonmobilizable plasmid.
Among the 46 Tcr A. pleuropneumoniae isolates analyzed, 32 (70%) carried tet(B). The tet(B) gene is widespread among gram-negative bacteria; it has been found in 24 other genera (http://faculty.washington.edu/marilynr/) and is the only tet gene previously identified in the A. pleuropneumoniae literature (34). However, unlike most tet(B)-containing bacteria, none of the tet(B)-positive A. pleuropneumoniae isolates appeared to carry a tetR(B) gene on the small 5-kb nonmobilizable plasmid. Similar findings have been reported for a Neisseria meningitidis isolate recovered from Japan (32), three strains of Haemophilus parainfluenzae (13), and an H. parasuis 5.1-kb plasmid carrying tet(B) (GenBank accession no. NC006828) (18). All the 5-kb nonmobilizable tet(B) plasmids had identical restriction patterns, suggesting that they were highly related or identical to each other. The RBS and transcription terminator sequences of the tet(B) gene are conserved in A. pleuropneumoniae tet(B) plasmids, whereas the original promoter was not found. Instead, upstream of the tet(B) gene there were two sequences that resemble A. pleuropneumoniae promoters (Fig. 1). Therefore, the absence of tetR(B) and sequence features of the tet(B) upstream region suggest that the promoter and transcriptional regulatory signals have been adapted for expression in A. pleuropneumoniae.
Sequence analysis of tet(B)-positive p11745 indicated that this plasmid codes for RepB, a replication-related protein homologous to a family of proteins involved in initiation of plasmid replication and widespread in plasmids of both gram-negative and gram-positive bacteria. Plasmid p11745 was transformed into both E. coli and H. parasuis (unpublished results). The development of plasmid vectors based on the p11745 replication region for use in Pasteurellaceae is currently in progress in our laboratory.
Plasmid p9956 carried the gram-negative tet(H) gene, which has previously been found as part of the small composite transposon Tn5706 on plasmids or in the chromosome of Pasteurella, Mannheimia spp., Acinetobacter radioresistens, and Moraxella spp. (15, 16, 21). The tet regions of p9956 and the Pasteurella plasmid pPAT1 (14) are identical and, in contrast to tet(B) plasmids, include the regulator gene tetR(H) and a truncated copy of tet(H) which is functional in both A. pleuropneumoniae and E. coli.
Sequence analysis of p9555 suggests that this plasmid was built by insertion of a region of gram-positive origin containing tet(L) into a Pasteurellaceae plasmid. The p9555 tet(L) was homologous to previously sequenced tet(L) genes found in plasmids of several low-GC-content gram-positive bacteria. The upstream region of tet(L) lacks the regulatory sequences characteristic of gram-positive tet(L) determinants but contains two 6-bp boxes resembling the A. pleuropneumoniae consensus promoter. However, the translational terminator downstream of tet(L) is conserved. Thus, as in the case of tet(B) of p11745, sequences upstream of tet(L) seem to be suited for its expression in A. pleuropneumoniae. The tet(L) gene has been found in other gram-negative bacteria such as Salmonella, Morganella, Fusobacterium, and Veillonella spp. (http://faculty.washington.edu/marilynr/). The presence of a mob operon including a ColE1 superfamily relaxase indicated that p9555 can be mobilized in E. coli if transfer functions from a conjugative IncP plasmid are provided. Mobilization of the plasmids in the E. coli strains using a donor-containing plasmid, RP4, demonstrated that p9555 Mob proteins are functional. This plasmid could have been transmitted to A. pleuropneumoniae from other bacteria inhabiting the respiratory tracts of pigs and encoding the appropriate transfer machinery for assisted mobilization.
The tet(L)-carrying plasmids identified in bovine Mannheimia and Pasteurella isolates were virtually identical to p9555 (more than 99% homologous) except in a 363-bp region absent in pCCK3259, suggesting that plasmid pCCK3259 might be a deleted derivative of p9555 (17). Supporting this hypothesis are the dates of isolation of the A. pleuropneumoniae strain in 1998 and the Mannheimia isolates in 2003 (17), with plasmid pCCK3259 created from the parental plasmid p9555 by a 363-bp deletion mediated by the 10-bp repeats flanking the 363-bp stretch. The distinct geographical origins of bacteria carrying p9555 and pCCK3259 could reflect the fact that movement of pigs between different European countries is a common practice for pig breeding in Europe.
The two A. pleuropneumoniae Tcr plasmids sequenced in this study, p11745 and p9555, encoded mobilization-related proteins. The recent finding of a plasmid carrying a tet(B) gene and a mob region in an isolate of H. parasuis (18), another respiratory swine pathogen, suggests that mobilizable plasmids are vehicles for active DNA exchange among bacteria inhabiting the upper respiratory tracts of pigs, contributing to the spread of resistance genes.
Our data suggest that the number of different tet genes in A. pleuropneumoniae has expanded. What impact this might have on therapy of this disease is not clear, but continued monitoring of this important animal pathogen is warranted.
Supplementary Material
Acknowledgments
We thank Stefan Schwarz for helpful discussions and aid in sequence analysis.
This work was supported by a grant from the Spanish Ministry for Science and Technology (AGL2002-04585). M. Blanco was a recipient of fellowships from the University of León and the University of Cantabria.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Bosse, J. T., H. Janson, B. J. Sheehan, A. J. Beddek, A. N. Rycroft, J. S. Kroll, and P. R. Langford. 2002. Actinobacillus pleuropneumoniae: pathobiology and pathogenesis of infection. Microbes Infect. 4:225-235. [DOI] [PubMed] [Google Scholar]
- 2.Bousquet, E., H. Morvan, I. Aitken, and J. H. Morgan. 1997. Comparative in vitro activity of doxycycline and oxytetracycline against porcine respiratory pathogens. Vet. Rec. 141:37-40. [DOI] [PubMed] [Google Scholar]
- 3.Chang, C. F., T. M. Yeh, C. C. Chou, Y. F. Chang, and T. S. Chiang. 2002. Antimicrobial susceptibility and plasmid analysis of Actinobacillus pleuropneumoniae isolated in Taiwan. Vet. Microbiol. 84:169-177. [DOI] [PubMed] [Google Scholar]
- 4.Chopra, I., and M. C. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung, W. O., C. Werckenthin, S. Schwarz, and M. C. Roberts. 1999. Host range of the ermF rRNA methylase gene in bacteria of human and animal origin. J. Antimicrob. Chemother. 43:5-14. [DOI] [PubMed] [Google Scholar]
- 6.De la Puente-Redondo, V. A., N. Garcia del Blanco, C. B. Gutierrez-Martin, J. Navas Mendez, and E. F. Rodriquez Ferri. 2001. Detection and subtyping of Actinobacillus pleuropneumoniae strains by PCR-RFLP analysis of the tbpA and tbpB genes. Res. Microbiol. 151:669-681. [DOI] [PubMed] [Google Scholar]
- 7.Doree, S. M., and M. H. Mulks. 2001. Identification of an Actinobacillus pleuropneumoniae consensus promoter structure. J. Bacteriol. 183:1983-1989. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fassler, J. S., I. Tessman, and E. S. Tessman. 1985. Lethality of the double mutations rho rep and rho ssb in Escherichia coli. J. Bacteriol. 161:609-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Francia, M. V., A. Varsaki, M. P. Garcillan-Barcia, A. Latorre, C. Drainas, and F. de la Cruz. 2004. A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol. Rev. 28:79-100. [DOI] [PubMed] [Google Scholar]
- 11.Gutierrez, C. B., J. I. Rodriguez Barbosa, R. I. Tascon, L. Costa, P. Riera, and E. F. Rodriguez Ferri. 1995. Serological characterization and antimicrobial susceptibility of Actinobacillus pleuropneumoniae strains isolated from pigs in Spain. Vet. Rec. 137:62-64. [DOI] [PubMed] [Google Scholar]
- 12.Hansen, L. M., L. M. McMurry, S. B. Levy, and D. C. Hirsh. 1993. A new tetracycline resistance determinant, Tet H, from Pasteurella multocida specifying active efflux of tetracycline. Antimicrob. Agents Chemother. 37:2699-2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heuer, C., R. K. Hickman, M. S. Curiale, W. Hillen, and S. B. Levy. 1987. Constitutive expression of tetracycline resistance mediated by a Tn10-like element in Haemophilus parainfluenzae results from a mutation in the repressor gene. J. Bacteriol. 169:990-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kehrenberg, C., and S. Schwarz. 2000. Identification of a truncated, but functionally active tet(H) tetracycline resistance gene in Pasteurella aerogenes and Pasteurella multocida. FEMS Microbiol. Lett. 188:191-195. [DOI] [PubMed] [Google Scholar]
- 15.Kehrenberg, C., and S. Schwarz. 2001. Molecular analysis of tetracycline resistance in Pasteurella aerogenes. Antimicrob. Agents Chemother. 45:2885-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kehrenberg, C., S. A. Salmon, J. L. Watts, and S. Schwarz. 2001. Tetracycline resistance genes of Pasteurella multocida, Mannheimia haemolytica, Mannheimia glucosida and Mannheimia varigena from bovine and swine respiratory disease: intergeneric spread of the tet(H) plasmid pMHT1. J. Antimicrob. Chemother. 48:631-640. [DOI] [PubMed] [Google Scholar]
- 17.Kehrenberg, C., B. Catry, F. Haesebrouck, A. de Kruif, and S. Schwarz. 2005. tet(L)-mediated tetracycline resistance in bovine Mannheimia and Pasteurella isolates. J. Antimicrob. Chemother. 56:403-406. [DOI] [PubMed] [Google Scholar]
- 18.Lancashire, J. F., T. D. Terry, P. J. Blackall, and M. P. Jennings. 2005. Plasmid-encoded TetB tetracycline resistance in Haemophilus parasuis. Antimicrob. Agents Chemother. 49:1927-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luna, V. A., and M. C. Roberts. 1998. The presence of the tetO gene in a variety of tetracycline-resistant Streptococcus pneumoniae serotypes from Washington State. J. Antimicrob. Chemother. 42:613-619. [DOI] [PubMed] [Google Scholar]
- 20.Martineau, G. P. 1999. Una nueva manera de enfocar el manejo en banda. Anaporc 195:18-35. [Google Scholar]
- 21.Miranda, C. D., C. Kehrenberg, C. Ulep, S. Schwarz, and M. C. Roberts. 2003. Diversity of tetracycline resistance genes in bacteria from Chilean salmon farms. Antimicrob. Agents Chemother. 47:883-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naderer, M., J. R. Brust, D. Knowle, and R. M. Blumenthal. 2002. Mobility of a restriction-modification system revealed by its genetic contexts in three hosts. J. Bacteriol. 184:2411-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.NCCLS. 2002. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; approved standard, 2nd ed. NCCLS document M31-A2. NCCLS, Wayne, Pa.
- 24.Roberts, M. C. 2003. Tetracycline therapy: update. Clin. Infect. Dis. 36:462-467. [DOI] [PubMed] [Google Scholar]
- 25.Roberts, M. C. 1996. Tetracycline resistance determinants: mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol. Rev. 19:1-24. [DOI] [PubMed] [Google Scholar]
- 26.Rodriguez-Bonano, N. M., and L. J. Torres-Bauza. 2004. Molecular analysis of oriT and MobA protein in the 7.4 Kb mobilizable-lactamase plasmid pSJ7.4 from Neisseria gonorrhoeae. Plasmid 52:89-101. [DOI] [PubMed] [Google Scholar]
- 27.Rogers, R. J., L. E. Eaves, P. J. Blackall, and K. F. Truman. 1990. The comparative pathogenicity of four serovars of Actinobacillus (Haemophilus) pleuropneumoniae. Aust. Vet. J. 67:9-12. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Sangari, F. J. 1993. Ph.D. thesis. Universidad de Cantabria, Santander, Spain.
- 30.Schwarz, S., C. Kehrenberg, and T. J. Walsh. 2001. Use of antimicrobial agents in veterinary medicine and food animal production. Int. J. Antimicrob. Agents 17:431-437. [DOI] [PubMed] [Google Scholar]
- 31.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-790. [Google Scholar]
- 32.Takahashi, H., H. Watanabe, T. Kuroki, Y. Watanabe, and S. Yamai. 2002. Identification of tet(B), encoding high-level tetracycline resistance, in Neisseria meningitidis. Antimicrob. Agents Chemother. 46:4045-4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor, D. E. 1986. Plasmid-mediated tetracycline resistance in Campylobacter jejuni: expression in Escherichia coli and identification of homology with streptococcal class M determinant. J. Bacteriol. 165:1037-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wasteson, Y., D. E. Roe, K. Falk, and M. C. Roberts. 1996. Characterization of tetracycline and erythromycin resistance in Actinobacillus pleuropneumoniae. Vet. Microbiol. 48:41-50. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.