Abstract
This study on Tomato bushy stunt virus identified and defined three previously unknown regulatory sequences involved in RNA accumulation that are located within the 3′-proximal nested movement protein genes p22 and p19. The first is a 16-nucleotide (nt) element termed III-A that is positioned at the very 3′ end of p22 and is essential for RNA accumulation. Approximately 300 nt upstream of III-A resides an ∼80-nt inhibitory element (IE) that is obstructive to replication only in the absence of a third regulatory element of ∼30 nt (SUR-III) that is positioned immediately upstream of III-A. Inspection of the nucleotide sequences predicted that III-A and SUR-III can form looped hairpins. A comparison of different tombusviruses showed, in each case, conservation for potential base pairing between the two predicted hairpin-loops. Insertion of a spacer adjacent to the predicted hairpins had no or a minimal effect on RNA accumulation, whereas an insertion in the putative III-A loop abolished genomic RNA multiplication. We conclude that the sequences composing the predicted III-A and SUR-III hairpin-loops are crucial for optimal RNA accumulation and that the inhibitory effect of IE surfaces when the alleged interaction between SUR-III and III-A is disturbed.
Tomato bushy stunt virus (TBSV), the type member of the genus Tombusvirus in the family Tombusviridae, is a small icosahedral virus with a positive-sense single-stranded RNA genome of approximately 4.8 kb that encodes five major open reading frames (ORFs) (Fig. 1) (19). The genomic RNA (gRNA) functions as an mRNA for the translation of two 5′-proximal ORFs, p33 and p92, encoding two replication-associated proteins, P33 and its readthrough product P92 (27, 43). The tombusvirus coat protein is translated from p41 on subgenomic RNA1 (sgRNA1), and P22 and P19 are expressed from p22 and p19 on sgRNA2 (21-23, 36). TBSV P22 is required for cell-to-cell movement (7, 10, 42), and P19 is involved in host-specific systemic invasion and symptom development (6, 38, 41, 42) and is active as a suppressor of gene silencing (31, 32, 44).
FIG. 1.
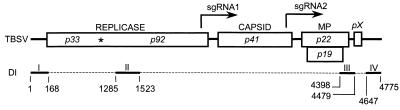
Schematic diagrams of the TBSV genome (top) and DI (bottom). Open boxes denote ORFs of genes identified by the molecular mass, in kilodaltons, of the encoded proteins, whose functions are indicated above the boxes. MP indicates movement proteins expressed from the overlapping p22 and p19 ORFs. The function of pX is unknown, but its RNA sequences have a host-dependent effect on viral RNA accumulation (39). The solid lines between ORFs indicate presumed untranslated regions. The transcription start sites for sgRNA1 and -2 are indicated by right-angled arrows above the TBSV genome. The asterisk inside the TBSV genome indicates the readthrough amber stop codon of p33 for expression of p92. Regions I through IV represent the conserved motifs present in DIs. The dashed lines between these regions indicate sequences that are deleted from the TBSV genome during DI formation. The nucleotide positions are based on an infectious cDNA clone of the wild-type genome, pTBSV-100 (19), and a typical infectious DI cDNA, B10 (24).
Tombusviruses can serve as helper viruses for satellite RNAs (1, 2), and they are notorious for the accumulation of defective interfering RNAs (DIs) (13, 17, 20, 24). DIs are spontaneous deletion mutants derived from the helper genome, and they are successfully amplified by trans-acting helper replicase proteins (20, 24). DIs have been valuable tools with which to identify RNA elements involved in viral accumulation (3, 16, 17). TBSV DIs are composed of four noncontiguous RNA regions (I through IV) from the helper genome (Fig. 1) (45). Regions I and IV correspond to the 5′- and 3′-untranslated regions of the TBSV genome, and regions II and III represent internal regions of p92 and a segment at the 3′ end of the overlapping p19 and p22 ORFs, respectively (Fig. 1) (33, 45-47). These features are also common for DIs derived from other tombusviruses, such as Cymbidium ringspot virus (CymRSV) and Cucumber necrosis virus (CNV) (9, 12, 13, 16, 17).
Regions I, II, and IV of DIs represent sequences essential for RNA amplification (3, 33, 45, 49), and while region III is dispensable for DI replication of TBSV, it is required for CymRSV DI accumulation (3, 17, 33). The length of TBSV DI region III varies from 46 to 81 nucleotides (nt), depending on the DI isolate (3, 20, 47). Since DIs are thought to have evolved in a secondary-structure-dependent manner (18, 47, 48), the differences in sizes or requirements for region III among tombusviruses could be a reflection of the accessibility of alternative DI structures for trans-acting factors. Likewise, the biological activity of region III may also be exerted in a context-dependent manner because, in contrast to its dispensability for replication of TBSV DIs, region III is essential in cis for amplification of the helper genome (33). This is reminiscent of our observation that an element within the replicase gene differentially affects DI versus genome amplification (29). Another example involves the different effects of a region downstream of region III, termed pX, that showed host-specific activities for RNA accumulation despite its dispensability for DI replication (39). On the basis of these cumulative observations on context-dependent effects, it appears necessary to consider RNA elements adjacent to regions I through IV in order to fully understand their biological activity in the context of the TBSV genome.
This study was performed to investigate the activity of region III through mutational analysis of RNA within and upstream of region III. An essential region was defined at the 5′ end of region III (III-A), and another ancillary element was identified positioned immediately upstream of region III (SUR-III). Sequence analyses indicated that both SUR-III and III-A can form looped hairpin structures with complementary sequences in the loops. The potential for loop-mediated base pairing is conserved among tombusviruses, and insertion of a spacer in the putative loop of III-A abolished TBSV RNA accumulation. A third element (inhibitory element [IE]) was identified ∼300 nt upstream of region III that inhibited RNA accumulation but only in the absence of SUR-III. These results suggest that structurally mediated interactions occur among the three interdependent elements to govern RNA accumulation.
MATERIALS AND METHODS
Viral constructs.
The molecular biology protocols used here followed those described by Sambrook et al. (37). Throughout this report, plasmids will be designated with a p subnotation, which will be omitted when discussing the corresponding transcripts or viral RNA. Plasmid pTBSV-100, a full-length infectious cDNA clone of a TBSV cherry isolate (19), was used to generate the mutants described in this report. For the construction of pTHB19, a reverse primer (5′-GGCTTCGAATTAGTGATGGTGATGGTGATGCTCGAAGGTTTGAGTACC-3′) was designed to add 18 nt at the BstBI site (underlined) located at nt 4385, followed by a PCR in combination with the forward primer (seq22A; 5′-GTGTCCTGCGAGGGGCC-3′) to amplify a TBSV fragment between nt 3819 and 4385. For cloning of the PCR product, pKAN157ΔEcoRV (6) was used, which contains the BamHI (nt 2439)-to-SphI (nt 4801) fragment of pTBSV-100 but in which an internal EcoRV fragment (nt 3410 to nt 4101) was deleted to remove a BstBI site at nt 4036. This plasmid and the PCR product were both digested with HpaI and BstBI and then ligated to generate an intermediate construct, pKAN157ΔEcoRV-HB19. Subsequently, the HpaI-to-SalI fragment of pKAN157ΔEcoRV-HB19 was used to replace the same restriction fragment of pTBSV-100 to generate pTHB19. To construct pTHS19, a SacI site (underlined in the sequences below) was generated at the 3′ end of the p19 ORF of pHS49 (39) by oligonucleotide-directed mutagenesis with a primer (5′-GAAGGCGAGCTCGACAGACTC-3′) to produce pHS49/19S. The NcoI-to-SalI fragment of pTBSV-100 was replaced with the same fragment of pHS49/19S to generate an intermediate construct, pTS19, that contains the SacI site at the 3′ end of p19. To add 18 nt at the SacI site, a reverse primer (5′-GGCGAGCTCTTAGTGATGGTGATGGTGATGCTCGCCTTCTTTTTCG-3′) was used for a PCR with forward primer seq22A (positioned 20 nt upstream of the 5′ end of p22) to amplify the fragment between nt 3819 and 4403. This PCR product was digested with NcoI and SacI to replace the NcoI-to-SacI fragment of pTS19 to produce pTHS19. Construct pD5, with an 18-nt insertion at the 3′ end of p22, was constructed by the same method as that used for pTHS19. The SacI site was generated at the 3′ end of the p22 ORF of pHS49 with a primer (5′-CAGACTCTTCAGTCGAGCTCTGAGTTTGTGGAGATGAG-3′) to produce pHS49/22S. The NcoI-to-SalI fragment of pTBSV-100 was replaced with the same fragment of pHS49/22S to generate pTS22, which contains a SacI site at the 3′ end of p22. The HpaI-to-SacI fragment of the intermediate construct pTS22 was replaced with a PCR product that was amplified from the template pTBSV-100. This was performed with a reverse primer (5′-CGGGAGCTCTCAGTGATGGTGATGGTGATGGACTGAAGAGTCTG-3′) containing an 18-nt insertion at the SacI site at the 3′ end of p22 in combination with forward primer seq22A. The 18-nt insertion of each mutant was confirmed by sequencing.
The deletion mutants used in this study were generated by digestion of pTBSV-100, pTS19, pTHS19, or pD5 with restriction enzymes, followed by treatment with the DNA polymerase I large fragment (Klenow treatment) to fill in 3′-recessed termini or to remove 3′ overhangs, followed by blunt-end ligation with T4 DNA ligase. Constructs pT/NBg, pT/NHp, pT/NSl, and pT/HSl were derived from pTBSV-100 by restriction enzyme digestion with NcoI and BsgI for pT/NBg, NcoI and HpaI for pT/NHp, NcoI and SalI for pT/NSl, and HpaI and SalI for pT/HSl. Constructs pT/NSc, pT/ScSl, and pT/HSc were obtained by restriction digestion of pTS19 with NcoI and SacI for pT/NSc, SacI and SalI for pT/ScSl, and HpaI and SacI for pT/HSc. Constructs pT/BgSc and pT/ESc were generated by restriction digestion of pTHS19 with BsgI and SacI for pT/BgSc and EcoRI and SacI for pT/ESc. pD5/NSc was obtained by NcoI-and-SacI digestion of pD5. To generate pT/NBt, plasmid pKAN157 (6), which contains the BamHI-to-SphI fragment of pHS157 (42), was digested with NcoI and BstBI to generate an intermediate construct, pKAN157/NBt, from which the PflMI-to-SalI fragment was used to replace the PflMI-to-SalI fragment of pTBSV-100 to produce pT/NBt. For the construction of pT/BtSc, first the SacI site was inserted into plasmid pKAN157ΔEcoRV by replacing the HpaI-to-SalI fragment with the HpaI-to-SalI fragment of pTS19 that produced an intermediate plasmid, pKAN157ΔEcoRV/SacI. Plasmid pKAN157ΔEcoRV/SacI was then digested with BstBI and SacI to generate pKAN157ΔEcoRV/BtSc, the HpaI-to-SphI fragment of which was used to replace the HpaI-to-SphI fragment of pTBSV-100 to produce pT/BtSc. Constructs pT/AlFo and pT/AlSc were serendipitously obtained upon screening for pT/BgSc, which was generated by BsgI-and-SacI digestion of pTHS19. Constructs pT/BtSc-NBg and pT/HSc-NBg were obtained by NcoI-and-BsgI digestion of pT/BtSc and pT/HSc, respectively. Construct pT/HSc-NEc was obtained by NcoI-and-EcoRI digestion of pT/HSc. Sequencing of all of the mutant constructs was done to confirm the deletions and fusions.
In vitro transcription and translation.
The templates used for in vitro transcription were prepared by SmaI digestion of all pTBSV-100-derived constructs. The in vitro transcription reaction mixture (5 μl of 5× T7 RNA polymerase reaction buffer, 2.5 μl of 5 mM nucleotides, 1.5 μl of 100 mM dithiothreitol, 0.5 μl of RNasin [Promega], 0.5 μl of T7 RNA polymerase [Gibco], 1 μl of SmaI-digested template [200 to 500 ng/μl], 15 μl of distilled H2O [dH2O]) was prepared and incubated at 37°C for 1 h. An aliquot of in vitro transcription products was analyzed by 1% agarose gel electrophoresis.
In vitro translation with wheat germ extract was performed with 1-μl transcripts, and the reaction mixture was incubated at 25°C for 1 h as described by the manufacturer (Promega, Madison, Wis.). The in vitro translation products were analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, followed by fixing in 200 ml of fixer (450 ml of methanol, 100 ml of acetic acid, 450 ml of dH2O) and drying of the gel at 75°C for 1 h, which was then used for autoradiography.
Isolation and transfection of protoplasts and inoculation of plants.
Protoplast preparation was slightly modified in comparison with that in our previous studies (40, 42, 43), as outlined in this section. Fully expanded (but tender) leaves of Nicotiana benthamiana and N. tabacum or 5- to 6-day-old cucumber var. Straight 8 cotyledons were sliced into 3- to 4-mm strips. These were digested at 25°C in 20 ml of an enzyme mixture containing 0.3 g of cellulase (Calbiochem), 0.05 g of pectinase (Calbiochem), and 0.15 g of bovine serum albumin (Sigma) in 10% mannitol for 5 to 6 h with gentle shaking (60 rpm) in the dark. The digested leaf material was strained through nylon mesh into 30-ml centrifuge tubes, and the protoplasts were harvested by centrifugation (600 rpm [IEC], 5 min). The harvested protoplasts were washed with 10% mannitol and collected by centrifugation, followed by resuspension in 10% mannitol. A 20% sucrose solution was gently placed beneath the protoplast suspension. After centrifugation (600 rpm [IEC]) for 5 min, the protoplast band was removed from the top of the 20% sucrose cushion. These concentrated protoplasts were diluted to 105/ml and used for transfection with in vitro-generated transcripts as described previously (40) and incubated in a growth chamber under fluorescent lighting at 25°C for the periods of time specified in this report. For bioassays with plants, the in vitro-generated TBSV RNAs were mixed in RNA inoculation buffer (40) and then used to rub inoculate plants.
Northern blot analysis.
Protoplasts were collected by centrifugation, and total RNAs were extracted by resuspension of protoplasts in 300 μl of RNA extraction buffer as described previously (40). After two phenol-chloroform extractions, 100 μl of 10 M ammonium acetate and 800 μl of ice-cold ethanol were added to the aqueous phase, which was placed at −70°C for 1 h to precipitate RNA. The precipitated total RNA was collected by centrifugation, resuspended in dH2O, and electrophoresed through 1% agarose for Northern blot analysis. The gels were photographed after staining with ethidium bromide to assess the approximate amount of total RNAs loaded in each well. After overnight capillary transfer of RNAs from agarose gels onto nylon membrane, the membrane was hybridized with a TBSV-specific probe for 10 h at 65°C in hybridization buffer containing 2× SSPE (300 mM NaCl, 20 mM NaH2PO4 [pH 7.4], 2 mM EDTA) and 1% SDS. The membrane was washed twice with buffer containing 2× SSPE and 0.1% SDS for 5 min at room temperature and twice with buffer containing 0.2× SSPE and 1% SDS for 5 min at 65°C. An X-ray film was placed on the membrane and exposed at −70°C.
RESULTS
A 5′ portion of region III (III-A) is required for viral RNA accumulation.
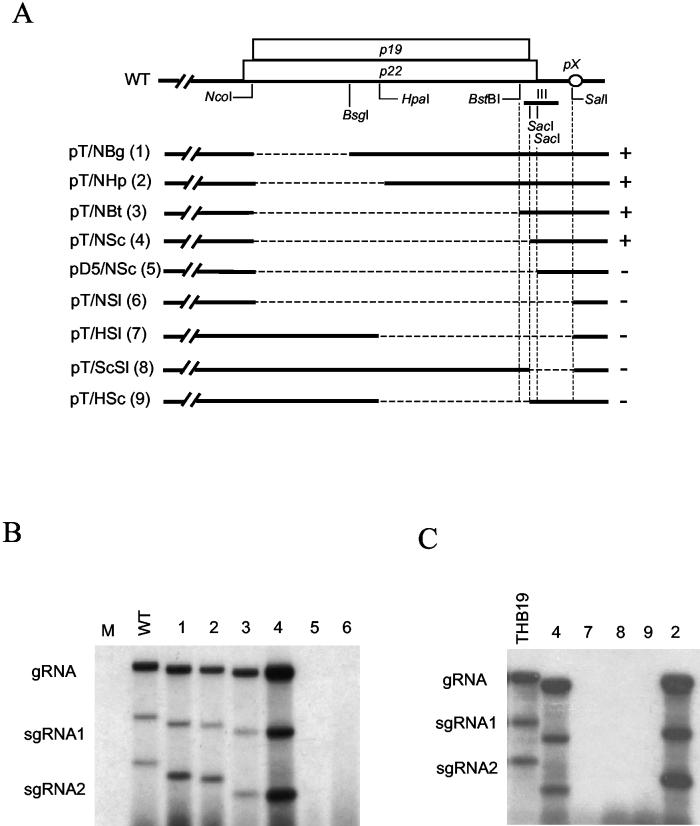
To evaluate the importance of region III (Fig. 1, nt 4398 to 4479) and adjacent sequences on TBSV replication, an extensive mutational analysis was performed and RNA accumulation of the mutants was determined in protoplasts. Results in Fig. 2 showed that essentially the entire p22 and p19 coding region is dispensable for RNA accumulation (e.g., Fig. 2B, lanes 3 and 4). The second observation was that deletions affecting pX (nt 4492 to 4699), such as those in pT/NSl (Δnt 3890 to 4499), pT/HSl (Δnt 4172 to 4497), and pT/ScSl (Δnt 4400 to 4498), prohibited efficient viral RNA accumulation (Fig. 2B and C, lanes 6, 7, and 8), as expected on the basis of previous results (39).
FIG. 2.
Effects of deletions on viral RNA accumulation. (A) Schematic diagram of deletions. The overlapping p19 and p22 ORFs are indicated with two open boxes, and for clarity, the remainder of the genome is omitted. The small open circle represents the pX region. Restriction enzyme sites are indicated below p19 and p22. The exact deletion sites of each mutant are listed in the text. The approximate position of region III is indicated by the small bar beneath the 3′ ends of p19 and p22. The dashed lines indicate deleted regions. The numbers in parentheses correspond to lane designations in panels B and C. A plus or minus sign indicates the ability or inability of each mutant to replicate in protoplasts (as shown in panels B and C and based on reproducible repeated tests). (B and C) Northern blot analysis of deletion mutants with total RNAs prepared from N. benthamiana protoplasts at 20 hpi. The relative positions of gRNA and sgRNAs are indicated on the left. M and WT represent samples from mock-transfected and wild-type-transfected protoplasts, respectively, and THB19 is a positive control (see later sections and Fig. 6).
The deletions in pD5/NSc (Δnt 3890 to 4420) and pT/AlFo (Δnt 3895 to 4479) did not affect the pX region, yet their accumulation was also below detectable levels (Fig. 2B, lane 5, and 3B, lanes 7 and 8), revealing the importance of region III for RNA accumulation. Comparison of results obtained with transcripts from pT/NSc (Δnt 3890 to 4404) and pD5/NSc (Δnt 3890 to 4420) (Fig. 2B, lanes 4 and 5) revealed that removal of only 16 nt from the 5′ half of region III (between the two SacI sites in Fig. 2A) severely compromised viral RNA accumulation. Therefore, this first set of experiments provided new evidence that the 5′ portion of region III, from here on referred to as III-A, is essential for gRNA accumulation.
Construct-dependent effects of SUR-III.
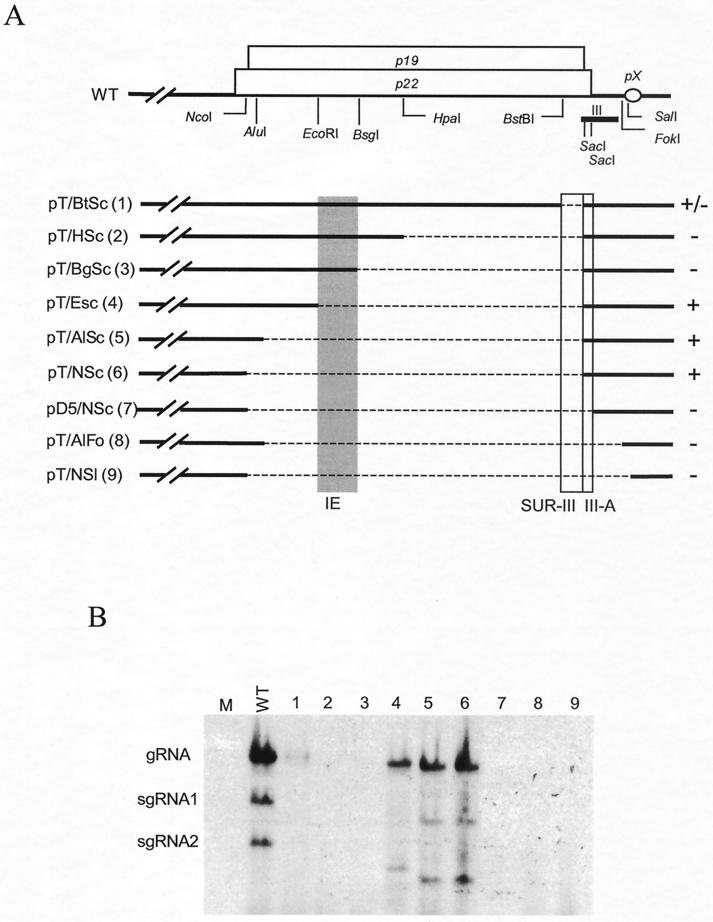
On the basis of results shown in Fig. 2, it was anticipated that the small deletion between nt 4368 and 4404 in pT/BtSc (Fig. 3A), comprising the extreme 5′ 6 nt of region III and sequences upstream of region III (defined as SUR-III), would not prohibit RNA accumulation because the essential sequences of region III (including III-A) and pX were present. However, the unexpected result was that RNA accumulation was compromised for pT/BtSc (Fig. 3B, lane 1) and transcripts from pT/HSc and pT/BgSc (Fig. 3B, lanes 2 and 3) were replication defective. This suggested an important role for SUR-III. However, such an effect was less dramatic for other constructs with larger deletions (Fig. 3A), which produced RNA that replicated effectively, albeit less efficiently than wild-type RNA (Fig. 3B, lanes 4, 5, and 6). These results led to the conclusion that the 36-nt SUR-III (located between nt 4368 and 4404) is important for RNA accumulation but that its contribution could be quantitatively influenced by construct-dependent features.
FIG. 3.
Delineation of RNA elements upstream of region III affecting viral RNA accumulation. (A) Schematic diagram of deletions introduced into p19 and p22. Symbols are the same as in Fig. 2. The shaded bar depicts the 79-nt segment (IE). The 36-nt RNA segment (SUR-III) and the 5′ portion of region III (III-A) are marked with open boxes near the 3′ ends of p19 and p22. The plus and minus signs on the right of the diagram of each deletion mutant summarize the viral RNA accumulation based on several reproducible repeats. The numbers in parentheses correspond to lane designations in panel B. The +/− indicates a very low level of viral RNA accumulation. (B) Example of Northern blot analysis of deletion mutants with total RNAs prepared from N. benthamiana protoplasts at 20 hpi. M and WT represent samples from mock-transfected and wild-type-transfected protoplasts, respectively.
Identification of a defined IE.
Comparison of accumulation of RNAs for pT/NSc and pT/HSc (Fig. 2B, lanes 4 and 9) suggested that the construct dependence of SUR-III is controlled by sequences located between the NcoI and HpaI sites. Removal of these sequences, as in pT/NBg, pT/NHp, pT/NBt, and pT/NSc (Fig. 2B, lanes 1 to 4), restored replication. To more precisely define the position of the region between the NcoI and HpaI sites responsible for the inhibitory effect in the absence of SUR-III, a series of deletions were introduced that gradually extended upstream from the HpaI site toward the NcoI site (Fig. 3A). As mentioned above, accumulation of viral RNAs for pT/BtSc (Δnt 4368 to 4404), pT/HSc (Δnt 4172 to 4404), and pT/BgSc (Δnt 4117 to 4404) was very low or below detectable levels (Fig. 3B, lanes 1 to 3). However, introduction of an additional deletion that extended farther toward the 5′ end of p19 and p22, as in pT/ESc (Δnt 4038 to 4404), pT/AlSc (Δnt 3895 to 4404), and pT/NSc (Δnt 3890 to 4404), restored viral RNA accumulation (Fig. 3B, lanes 4 to 6). The constructs tested in lanes 7 through 9 of Fig. 3B represent negative controls confirming the importance of region III-A and pX. The results shown in Fig. 3 revealed that the 79-nt segment between nt 4038 and 4117 inhibits viral RNA accumulation. However, the effect of this IE only surfaced in the absence of SUR-III.
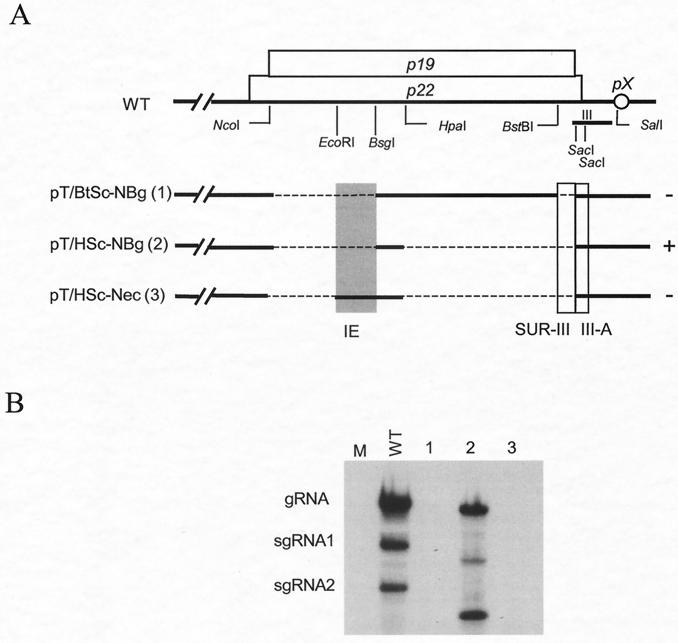
Since the replication-positive mutants in Fig. 3 varied in the composition of the 5′ p22/p19 sequences, the activity of the IE was verified with mutants containing identical sequences except for the presence or absence of the IE (Fig. 4A). The results (Fig. 4B, lanes 2 and 3) confirmed that the ∼80-nt IE was compromising RNA accumulation in the absence of SUR-III. It was anticipated that the mutant pT/BtSc-NBg (Δnt 3891 to 4119 and Δnt 4368 to 4404) would be replication competent since it retained region III (including III-A) and was devoid of the IE. However, this mutant failed to accumulate (Fig. 4B, lane 1), suggesting that the retention of the sequences upstream of nt 4368 and other than IE may also exert additional inhibitory activity. Our focus at this juncture was to define the role and activity of IE, and therefore, inhibitory effects of other regions were not pursued. However, as elaborated in the Discussion, some effects may not be caused by the presence of inhibitory sequences per se but rather could be attributable to the absence of sequences able to establish beneficial alternative structures. Finally, repositioning of the IE near the 5′ end of the coat protein ORF (by insertion into pT/NSc; Fig. 2) had no noticeable negative effect (data not shown), indicating that the effect of the IE is position dependent.
FIG. 4.
Effect of the 79-nt IE on TBSV accumulation. (A) Schematic diagram of deletions introduced, with restriction enzyme sites indicated below p19 and p22; symbols are the same as in the previous figures. The numbers in parentheses correspond to lane designations in panel B. (B) Example of Northern blot analysis of deletion mutants with total RNAs prepared from transfected N. benthamiana protoplasts at 20 hpi. M and WT represent samples from mock-transfected and wild-type-transfected protoplasts, respectively.
Collectively, the results support the conclusion that, in the absence of SUR-III, the ∼300-nt upstream IE sequence has a strong negative effect on RNA accumulation and that this inhibitory activity is exerted in a position- and context-dependent manner.
SUR-III and III-A can potentially interact in many tombusviruses.
Results in previous sections illustrated that there is an absolute requirement for region III-A for optimal RNA accumulation and that SUR-III plays a critical auxiliary role. We hypothesized that III-A forms a functional structural unit with SUR-III and that deletion of SUR-III resulted in aberrant structures that exposed the inhibitory effect of IE (and perhaps other elements). This “communication” hypothesis was supported by the observation that inversion of sequences upstream of region III (including SUR-III but not III-A) abolished RNA accumulation (data not shown).
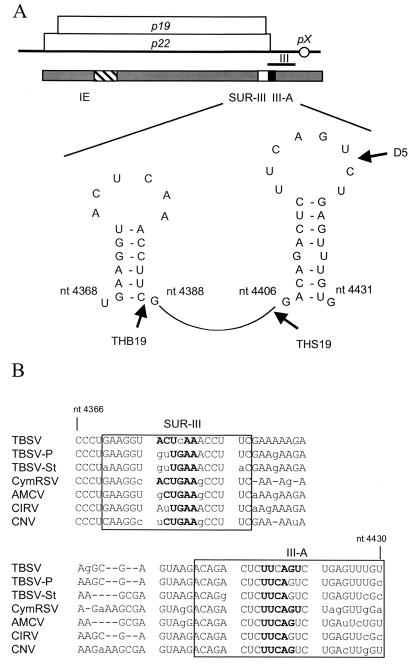
To further examine the possibility that region III-A and SUR-III might “communicate,” a sequence and putative structure analysis was performed (26, 51). This predicted two hairpin structures, one in SUR-III and the other in III-A, that could potentially interact with each other because of complementary nucleotide sequences in the loop portion of each predicted hairpin (Fig. 5A). To determine if this is a general feature of tombusviruses, the corresponding regions were analyzed for CymRSV, CNV, Artichoke mottle crinkle virus (AMCV), Carnation Italian ringspot virus (CIRV), and two other TBSV isolates (pepper [TBSV-P] and statice [TBSV-St]). Even though there was some nucleotide sequence variation among these tombusviruses, in all cases, SUR-III could potentially interact with region III-A because of the presence of a complementary nucleotide sequence (Fig. 5B) and all had the predicted ability to form SUR-III and III-A hairpin structures (data not shown).
FIG. 5.
Schematic representation of the predicted structure of RNA segments affecting TBSV RNA accumulation. (A) Symbols are the same as in the previous figures. The predicted hairpin structures residing in the 36-nt SUR-III segment and III-A are shown with nucleotide positions. The 18-nt insertions at or adjacent to restriction enzyme sites at positions 4385, 4402, and 4421 of the pTHB19, pTHS19, and pD5 constructs, respectively, are indicated by thick arrows. (B) Nucleotide sequence comparison of region III and the upstream region for different tombusviruses (TBSV cherry isolate, TBSV-P, TBSV-St, CymRSV, AMCV, CIRV, and CNV). The GenBank accession numbers of the viruses are as follows: U80935 for TBSV-P, AJ249740 for TBSV-St, X15511 for CymRSV, X62493 for AMCV, X85215 for CIRV, and M25270 for CNV. The sequence similarities between the TBSV cherry isolate and the other tombusviruses are 93% for TBSV-P, 85% for TBSV-St, 77% for CymRSV, 87% for AMCV, 80% for CIRV, and 77% for CNV. The boxed regions indicate the predicted hairpins of SUR-III (top) and III-A (bottom); nucleotides in bold represent the complementary sequences in the predicted hairpin-loops. The lowercase letters represent the nucleotide sequence differences between the tombusviruses compared. As previously reported (7, 39), the sequence of our laboratory version of the pTBSV-100 cDNA clone contains a few differences from those in the database.
The interpretation of our experimental results within the context of the predicted structures was that the putative hairpin-loop in region III-A was absolutely essential and that its activity was mediated through its interaction with SUR-III, although firm conclusions must await structural analyses and generation of compensatory mutants. Furthermore, in the absence of SUR-III, the hairpin-loop of III-A failed to function properly and the inhibitory effect of IE became noticeable. This illustrated that the predicted hairpin-loop of region III-A plays a critical and dominant role in virus accumulation.
Insertional mutants reveal the critical and specific importance of sequences predicted to form the hairpin-loop of III-A.
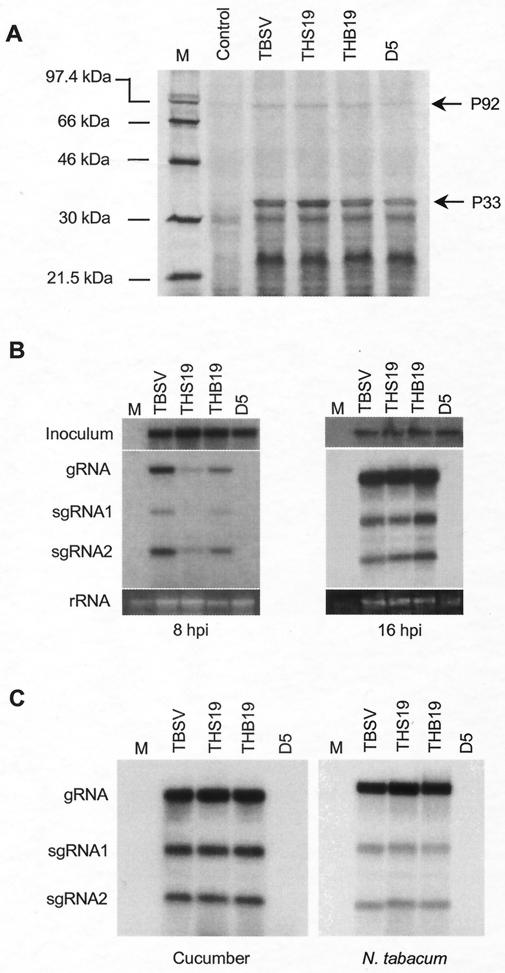
To empirically illustrate the importance of the III-A sequences predicted to form the hairpin-loop, we were aided by results of experiments with mutants that had been generated for a separate parallel study focusing on P19 and P22 protein purification (data not shown). In this project, 18 nt were inserted at or near the 3′ terminus of p19 or p22 (Fig. 5A) to add six-histidine codons at or near the carboxyl terminus of P19 (pTHB19 and pTHS19) or P22 (pD5). The His tag insertions are located just downstream of the SUR-III hairpin in pTHB19, just upstream of the III-A hairpin in pTHS19, and in the hairpin-loop of III-A in pD5 (Fig. 5A). In vitro translation assays showed that translation of replicase proteins P33 and P92 occurred for all of the mutants (Fig. 6A), suggesting that any effect on RNA accumulation observed was not due to compromised production of replicase proteins.
FIG. 6.
Effects of spacer insertions on RNA accumulation. (A) In vitro translation assay in wheat germ extracts with in vitro-generated transcripts of pTHB19, pTHS19, and pD5 (see Fig. 5 for site of insertion). The molecular sizes of markers (lane M) are indicated on the left. The positions of P33 and P92 are indicated on the right. (B) Northern blot analyses of total RNAs prepared from transfected N. benthamiana protoplasts at 8 and 16 hpi. To permit comparison of the amounts of RNA used for inoculum and loading, RNA was harvested at 0 h posttransfection and hybridized with a TBSV-specific probe (shown at the top); at the bottom are rRNAs from protoplasts harvested at 8 and 16 hpi. The positions of gRNA and sgRNAs are indicated on the left. M represents a sample from mock-transfected protoplasts. (C) Northern blot analyses of total RNAs prepared from transfected cucumber and N. tabacum protoplasts at 16 hpi.
Deletion mutants described in previous sections were not suitable for verification that the effects on RNA accumulation also affect infectivity on whole plants because the movement protein ORFs were affected, but the insertion mutants could be tested on plants. The results of these in planta experiments (not shown) will briefly be summarized. In vitro-generated transcripts of pD5 were not infectious to whole plants. However, at 4 to 6 days postinoculation with the wild-type and pTHB19 RNAs, symptoms were observed on the upper leaves of N. benthamiana and spinach plants. These plants developed a lethal systemic necrosis at about 10 to 15 days postinoculation. THS19 induced both less severe and delayed symptoms in N. benthamiana than in plants infected with the wild type and THB19. On spinach plants, THS19 failed to establish a systemic infection although symptoms were observed on inoculated leaves. In summary, the results showed that (i) insertion of sequences adjacent to the predicted hairpin of SUR-III had no measurable effects, (ii) the insertion of a spacer upstream of the hairpin of region III-A interfered with the infectivity, and (iii) the insertion in the loop of region III-A was detrimental for infection.
Some of the effects observed in plants could be caused by the influence of mutations on the activity of the P22 or P19 protein or its dosage or by host-specific effects of RNA mutations on infectivity (6, 38, 39, 42). Therefore, to specifically examine the effects on RNA accumulation, each mutant construct was tested in protoplasts. A previous study suggested that it is important to also measure RNA accumulation early after infection (30); therefore, total RNAs were extracted from transfected N. benthamiana protoplasts at 8 and 16 h postinoculation (hpi) (Fig. 6B). Although the level of THB19 viral RNA accumulation was somewhat reduced at 8 hpi, it was comparable to that of wild-type TBSV at 16 hpi (Fig. 6B), which indicated that the insertion adjacent to the SUR-III hairpin did not exert any substantial negative effect on replication. In the case of THS19, with the insertion adjacent to the III-A hairpin, viral RNA accumulation at 8 hpi was reproducibly much lower than that of the wild type or THB19. However, at 16 hpi, this distinction was no longer apparent (Fig. 6B). The delayed accumulation of THS19 RNA was also observed in cucumber protoplasts (data not shown). These observations indicate that the insertion just upstream of the III-A hairpin affects the efficiency of TBSV RNA accumulation, which may serve to explain its influence on the progression of the disease in whole plants.
To rule out host-associated effects, each mutant construct was also tested in protoplasts prepared from cucumber and N. tabacum (Fig. 6C). The results obtained with these protoplasts were essentially the same as those obtained with N. benthamiana protoplasts (Fig. 6B). In all cases, viral RNA accumulation of D5, in which the 18 nt were inserted in the hairpin-loop of III-A, was undetectable (Fig. 6B and C). This demonstrated that the sequences composing the predicted hairpin-loop of III-A were essential for RNA accumulation in different hosts. This conclusion agrees with the impact of the insertion on infectivity in whole plants.
DISCUSSION
Three newly identified cis elements influence TBSV RNA accumulation.
The results of this study identified three new elements on the TBSV genome that affect RNA accumulation: III-A, SUR-III, and IE (Fig. 5). All of the experiments supported the notion that there is an absolute requirement for region III-A, whereas SUR-III appears to be necessary for III-A to function properly. In the absence of SUR-III, the inhibitory effect of the upstream IE became prominent. We have not yet addressed whether the elements identified are active on the plus- or minus-sense RNA, or both, but have limited the scope of our study to defining their positions in relation to other known elements.
The importance of III-A reported here for gRNA accumulation is supported by experiments showing that the 5′ half of region III is more critical than its 3′ half for efficient DI accumulation (33). Earlier studies with various gRNA mutants of CNV (34, 35) and CymRSV (8) also produced results that, in the context of our new findings, can now be interpreted as supporting the notion that SUR-III and III-A are critical for viral RNA accumulation of tombusviruses. Our results also confirm previous findings that sequences upstream of position 4388 (BstBI site, Fig. 1 to 4) of TBSV are dispensable for replication (32, 40, 42). However, in those studies, mutants were used that had retained SUR-III and/or were devoid of the IE and therefore our newly reported activity of the IE and its dependence for activity on the absence of SUR-III had gone unnoticed.
Why is the IE inhibitory in the absence of SUR-III? Structural analysis revealed the possible presence of a highly ordered secondary structure in the ∼80-nt IE segment (data not shown). Havelda et al. (18) showed that the replicase proteins of CymRSV failed to dissolve a particular double-stranded RNA structure introduced into a CymRSV DI. It is possible that a stable secondary structure within the IE becomes established in the absence of SUR-III. This structure might not be amenable for the replication-related proteins to proceed because of the absence of the proper neighboring sequences necessary to relieve this higher-order secondary structure. This is reminiscent of our recent observation that a region within the replicase gene can also exert an inhibitory effect on DI accumulation but only when present in a specific orientation (29).
Because the IE exerts its inhibitory activity only in the absence of SUR-III, this suggests that, in the wild-type genome, the presence of SUR-III somehow neutralizes the inhibitory activity exerted by the IE region. Although sequence and computer-assisted structure predictions did not show any support for a direct interaction between these two segments, the possibility remains that the interaction is mediated by other viral or cellular components.
Our results suggested that the adjacent III-A and SUR-III segments form a single functional RNA unit and, in the absence of SUR-III, the IE region exerts a dominant-negative effect on the function of III-A. This scenario explains how the deletion of IE has a positive effect, yet the question of how III-A can function in the absence of its proposed partner, SUR-III, remains. For example, mutants pT/NSc, pT/AlSc, pT/ESc, and pT/HSc-NBg (lacking the 79-nt IE) are also devoid of SUR-III (Fig. 3 and 4); nevertheless, they accumulate RNA to relatively high levels. This provoked the hypothesis that, in the absence of the IE, region III-A (which is normally used for the interaction with SUR-III) could form alternative cooperative interactions. Sequence analysis of replication-positive mutants such as pT/NSc, pT/AlSc, pT/ESc, and pT/HSc-NBg predicted the presence of complementary nucleotide sequences (data not shown) that have the potential to interact with III-A, and this mimics the interaction that normally occurs with SUR-III. Conversely, even when all of the elements necessary for replication are predicted to be present, RNA accumulation can still be compromised (e.g., pT/BtSc-NBg; Fig. 4), suggesting the formation of alternative disruptive structures. Similar events were previously noticed when deletions affecting the 5′-untranslated region of tombusviruses stimulated the formation of alternative replication-competent structures (49).
Our results favor a model in which optimal TBSV RNA accumulation involves an interaction of III-A with SUR-III. However, in the absence of SUR-III, alternative interactions are established by III-A. Dependent on the context, these interactions can be conducive to RNA accumulation or, in the presence of the IE, these alternative associations are detrimental.
Interaction between III-A and SUR-III.
Computer-assisted structural analysis predicted two hairpin structures, one in SUR-III and the other in III-A, that harbor complementary nucleotide sequences on their loop portions, potentially allowing the formation of a tertiary structure (Fig. 5A). The importance of complementary sequences in SUR-III and III-A is supported by sequence comparisons revealing that region III and its upstream RNA elements are conserved among tombusviruses (Fig. 5B). In addition, the results obtained with the insertion mutants in protoplasts of several hosts (Fig. 6) also strongly support the model in which the hairpin-loop of III-A performs a critical function. Although chemical structure analyses need to be performed to conclusively prove the existence of the hairpin and the loops, the cumulative results conclusively demonstrate that a critical component of region III-A is represented by sequences in the predicted hairpin-loop. The suspected interaction between SUR-III and III-A would not be unusual in TBSV biology; for example, refined mapping and mutagenesis experiments provided strong supportive evidence that defined RNA elements on the genome interact for control of sgRNA transcription (4, 5, 32, 50). The effects of distal sequences and/or structural elements on ribosomal frameshifting, transcription, and translation have been investigated in detail for Barley yellow dwarf virus (15, 25, 28). These studies also illustrated the importance of proper interaction between RNA elements. Studies with Tobacco mosaic virus have illustrated the importance of stem-loop structures for transcription of sgRNA (14).
Since region III is conserved in DIs, it is conceivable that the proposed secondary structure of III-A specifically modulates DI formation through the interaction with a defined distal RNA element(s), resulting in template hopping of the replicase. Despite this possible involvement in directing DI formation, the functions of SUR-III and III-A in gRNA accumulation are unknown. However, an intriguing possibility is that III-A (perhaps in combination with its proposed interaction with SUR-III) may affect expression of an sgRNA for pX, a gene known to be important for RNA accumulation (39). However, region III is also conserved on DIs that lack pX; therefore, any such transcriptional effects must be specific for gRNA accumulation. An interesting addition to this scenario is the effect of deletions on sgRNA1 and sgRNA2 accumulation. Normally, the levels of sgRNA2 exceed those of sgRNA1 at time points at or beyond 16 hpi in N. benthamiana (e.g., Fig. 6B). However, the results in Fig. 2, 3, and 4 show various ratios of sgRNA2 to sgRNA1 for different mutants. It is therefore not unlikely that interactions involving the IE, SUR-III, and III-A might differentially influence sgRNA production. In this context, it is possible that SUR-III and III-A perform a function similar to that of 3′-regulatory sequences for other viruses (11) to control or time replication and/or transcription events.
Regardless of the precise mode of action, SUR-III and especially III-A are critical for optimal RNA accumulation of TBSV. The sequence analyses, the conservation of elements between tombusviruses, the results of mutagenesis experiments, and the host-independent nature of the elements all favor a strict sequence-mediated structure-function relationship.
Acknowledgments
We are very grateful to Karen-Beth G. Scholthof, T. Erik Mirkov, and Ellen Collisson for valuable suggestions during the experiments and the preparation of the manuscript and to Wenping Qiu, Sandrine Faure-Rabasse, Rustem Omarov, Jeff Batten, and Joanna Mirabile for help during the experiments.
This work was funded by the Texas Agricultural Experiment Station (TEX08387) and grants from USDA/CSREES-NRI-CGP (99-35303-8022), the Texas Higher Education Coordinating Board Advanced Technology Program (000517-0070-1999), and the S. R. Noble Foundation, Inc.
REFERENCES
- 1.Celix, A., J. Burgyan, and E. Rodriguez-Cerezo. 1999. Interactions between tombusviruses and satellite RNAs of tomato bushy stunt virus: a defect in satRNA B1 replication maps to ORF1 of a helper virus. Virology 262:129-138. [DOI] [PubMed] [Google Scholar]
- 2.Celix, A., E. Rodriguez-Cerezo, and F. Garcia-Arenal. 1997. New satellite RNAs, but no DI RNAs, are found in natural populations of tomato bushy stunt tombusvirus. Virology 239:277-284. [DOI] [PubMed] [Google Scholar]
- 3.Chang, Y. C., M. Borja, H. B. Scholthof, A. O. Jackson, and T. J. Morris. 1995. Host effects and sequences essential for accumulation of defective interfering RNAs of cucumber necrosis and tomato bushy stunt tombusviruses. Virology 210:41-53. [DOI] [PubMed] [Google Scholar]
- 4.Choi, I. R., M. Ostrovsky, G. Zhang, and K. A. White. 2001. Regulatory activity of distal and core RNA elements in tombusvirus subgenomic mRNA2 transcription. J. Biol. Chem. 276:41761-41768. [DOI] [PubMed] [Google Scholar]
- 5.Choi, I. R., and K. A. White. 2002. An RNA activator of subgenomic mRNA1 transcription in tomato bushy stunt virus. J. Biol. Chem. 277:3760-3766. [DOI] [PubMed] [Google Scholar]
- 6.Chu, M., B. Desvoyes, M. Turina, R. Noad, and H. B. Scholthof. 2000. Genetic dissection of tomato bushy stunt virus p19-protein-mediated host-dependent symptom induction and systemic invasion. Virology 266:79-87. [DOI] [PubMed] [Google Scholar]
- 7.Chu, M., J.-W. Park, and H. B. Scholthof. 1999. Separate regions on the tomato bushy stunt virus p22 protein mediate cell-to-cell movement versus elicitation of effective resistance responses. Mol. Plant-Microbe Interact. 12:285-292. [Google Scholar]
- 8.Dalmay, T., L. Rubino, J. Burgyan, A. Kollar, and M. Russo. 1993. Functional analysis of Cymbidium ringspot virus genome. Virology 194:697-704. [DOI] [PubMed] [Google Scholar]
- 9.Dalmay, T., G. Szittya, and J. Burgyan. 1995. Generation of defective interfering RNA dimers of cymbidium ringspot tombusvirus. Virology 207:510-517. [DOI] [PubMed] [Google Scholar]
- 10.Desvoyes, B., S. Faure-Rabasse, M.-H. Chen, J.-W. Park, and H. B. Scholthof. 2002. A novel plant homeodomain protein interacts in a functionally relevant manner with a virus movement protein. Plant Physiol. 129:1521-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dreher, T. 1999. Functions of the 3′-untranslated regions of positive strand RNA viral genomes. Annu. Rev. Phytopathol. 37:151-174. [DOI] [PubMed] [Google Scholar]
- 12.Finnen, R. L., and D. M. Rochon. 1995. Characterization and biological activity of DI RNA dimers formed during cucumber necrosis virus coinfections. Virology 207:282-286. [DOI] [PubMed] [Google Scholar]
- 13.Finnen, R. L., and D. M. Rochon. 1993. Sequence and structure of defective interfering RNAs associated with cucumber necrosis virus infections. J. Gen. Virol. 74:1715-1720. [DOI] [PubMed] [Google Scholar]
- 14.Grdzelishvili, V. Z., S. N. Chapman, W. O. Dawson, and D. J. Lewandowski. 2000. Mapping of the tobacco mosaic virus movement protein and coat protein subgenomic RNA promoters in vivo. Virology 275:177-192. [DOI] [PubMed] [Google Scholar]
- 15.Guo, L., E. Allen, and W. A. Miller. 2000. Structure and function of a cap-independent translation element that functions in either the 3′ or the 5′ untranslated region. RNA 6:1808-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Havelda, Z., and J. Burgyan. 1995. 3′ terminal putative stem-loop structure required for the accumulation of cymbidium ringspot viral RNA. Virology 214:269-272. [DOI] [PubMed] [Google Scholar]
- 17.Havelda, Z., T. Dalmay, and J. Burgyan. 1995. Localization of cis-acting sequences essential for cymbidium ringspot tombusvirus defective interfering RNA replication. J. Gen. Virol. 76:2311-2316. [DOI] [PubMed] [Google Scholar]
- 18.Havelda, Z., T. Dalmay, and J. Burgyan. 1997. Secondary structure-dependent evolution of cymbidium ringspot virus defective interfering RNA. J. Gen. Virol. 78:1227-1234. [DOI] [PubMed] [Google Scholar]
- 19.Hearne, P. Q., D. A. Knorr, B. I. Hillman, and T. J. Morris. 1990. The complete genome structure and synthesis of infectious RNA from clones of tomato bushy stunt virus. Virology 177:141-151. [DOI] [PubMed] [Google Scholar]
- 20.Hillman, B. I., J. C. Carrington, and T. J. Morris. 1987. A defective interfering RNA that contains a mosaic of a plant virus genome. Cell 51:427-433. [DOI] [PubMed] [Google Scholar]
- 21.Hillman, B. I., P. Hearne, D. M. Rochon, and T. J. Morris. 1989. Organization of tomato bushy stunt virus genome: characterization of the coat protein gene and the 3′ terminus. Virology 169:42-50. [DOI] [PubMed] [Google Scholar]
- 22.Johnston, J. C., and D. M. Rochon. 1996. Both codon context and leader length contribute to efficient expression of two overlapping open reading frames of a cucumber necrosis virus bifunctional subgenomic mRNA. Virology 221:232-239. [DOI] [PubMed] [Google Scholar]
- 23.Johnston, J. C., and D. M. Rochon. 1990. Translation of cucumber necrosis virus RNA in vitro. J. Gen. Virol. 71:2233-2241. [DOI] [PubMed] [Google Scholar]
- 24.Knorr, D. A., R. H. Mullin, P. Q. Hearne, and T. J. Morris. 1991. De novo generation of defective interfering RNAs of tomato bushy stunt virus by high multiplicity passage. Virology 181:193-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koev, G., B. R. Mohan, and W. A. Miller. 1999. Primary and secondary structural elements required for synthesis of barley yellow dwarf virus subgenomic RNA1. J. Virol. 73:2876-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 27.Oster, S. K., B. Wu, and K. A. White. 1998. Uncoupled expression of p33 and p92 permits amplification of tomato bushy stunt virus RNAs. J. Virol. 72:5845-5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paul, C. P., J. K. Barry, S. P. Dinesh-Kumar, V. Brault, and W. A. Miller. 2001. A sequence required for −1 ribosomal frameshifting located four kilobases downstream of the frameshift site. J. Mol. Biol. 310:987-999. [DOI] [PubMed] [Google Scholar]
- 29.Qiu, W., J.-W. Park, A. O. Jackson, and H. B. Scholthof. 2001. Retention of a small replicase gene segment in Tomato bushy stunt virus defective RNAs inhibits their helper-mediated trans-accumulation. Virology 281:51-60. [DOI] [PubMed] [Google Scholar]
- 30.Qiu, W., and H. B. Scholthof. 2001. Effects of inactivation of the coat protein and movement genes of tomato bushy stunt virus on early accumulation of genomic and subgenomic RNAs. J. Gen. Virol. 82:3107-3114. [DOI] [PubMed] [Google Scholar]
- 31.Qiu, W. P., J.-W. Park, and H. B. Scholthof. 2002. Tombusvirus P19-mediated suppression of virus induced gene silencing is controlled by genetic and dosage features that influence pathogenicity. Mol. Plant-Microbe. Interact. 15:269-280. [DOI] [PubMed] [Google Scholar]
- 32.Qu, F., and T. J. Morris. 2002. Efficient infection of Nicotiana benthamiana by Tomato bushy stunt virus is facilitated by the coat protein and maintained by p19 through suppression of gene silencing. Mol. Plant-Microbe Interact. 15:193-202. [DOI] [PubMed] [Google Scholar]
- 33.Ray, D., and K. A. White. 1999. Enhancer-like properties of an RNA element that modulates tombusvirus RNA accumulation. Virology 256:162-171. [DOI] [PubMed] [Google Scholar]
- 34.Reade, R., K. Delroux, K. Macdonald, T. L. Sit, S. A. Lommel, and D. Rochon. 2001. Spontaneous deletion enhances movement of a cucumber necrosis virus based chimera expressing the red clover necrotic mosaic virus movement protein gene. Mol. Plant Pathol. 2:13-25. [DOI] [PubMed] [Google Scholar]
- 35.Reade, R., Z. Wu, and D. Rochon. 1999. Both RNA rearrangement and point mutation contribute to repair of defective chimeric viral genomes to form functional hybrid viruses in plants. Virology 258:217-231. [DOI] [PubMed] [Google Scholar]
- 36.Rochon, D. M., and J. C. Johnston. 1991. Infectious transcripts from cloned cucumber necrosis virus cDNA: evidence for a bifunctional subgenomic mRNA. Virology 181:656-665. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Scholthof, H. B., B. Desvoyes, J. Kuecker, and E. Whitehead. 1999. Biological activity of two tombusvirus proteins translated from nested genes is influenced by dosage control via context-dependent leaky scanning. Mol. Plant-Microbe Interact. 12:670-679. [Google Scholar]
- 39.Scholthof, H. B., and A. O. Jackson. 1997. The enigma of pX: a host dependent cis-acting element with variable effects on tombusvirus RNA accumulation. Virology 237:56-65. [DOI] [PubMed] [Google Scholar]
- 40.Scholthof, H. B., T. J. Morris, and A. O. Jackson. 1993. The capsid protein gene of tomato bushy stunt virus is dispensable for systemic movement and can be replaced for localized expression of foreign genes. Mol. Plant-Microbe Interact. 6:309-322. [Google Scholar]
- 41.Scholthof, H. B., K.-B. G. Scholthof, and A. O. Jackson. 1995. Identification of tomato bushy stunt virus host-specific symptom determinants by expression of individual genes from a potato virus X vector. Plant Cell 7:1157-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scholthof, H. B., K.-B. G. Scholthof, M. Kikkert, and A. O. Jackson. 1995. Tomato bushy stunt virus spread is regulated by two nested genes that function in cell-to-cell movement and host-dependent systemic invasion. Virology 213:425-438. [DOI] [PubMed] [Google Scholar]
- 43.Scholthof, K.-B. G., H. B. Scholthof, and A. O. Jackson. 1995. The tomato bushy stunt virus replicase proteins are coordinately expressed and membrane associated. Virology 208:365-369. [DOI] [PubMed] [Google Scholar]
- 44.Voinnet, O., Y. M. Pinto, and D. C. Baulcombe. 1999. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. USA 96:14147-14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White, K. A. 1996. Formation and evolution of Tombusvirus defective interfering RNAs. Semin. Virol. 7:409-416. [Google Scholar]
- 46.White, K. A., and T. J. Morris. 1994. Enhanced competitiveness of tomato bushy stunt virus defective interfering RNAs by segment duplication or nucleotide insertion. J. Virol. 68:6092-6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.White, K. A., and T. J. Morris. 1994. Nonhomologous RNA recombination in tombusviruses: generation and evolution of defective interfering RNAs by stepwise deletions. J. Virol. 68:14-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.White, K. A., and T. J. Morris. 1995. RNA determinants of junction site selection in RNA virus recombinants and defective interfering RNAs. RNA 1:1029-1040. [PMC free article] [PubMed] [Google Scholar]
- 49.Wu, B., W. B. Vanti, and K. A. White. 2001. An RNA domain within the 5′ untranslated region of the tomato bushy stunt virus genome modulates viral RNA replication. J. Mol. Biol. 305:741-756. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, G., V. Slowinski, and K. A. White. 1999. Subgenomic mRNA regulation by a distal RNA element in a (+)-strand RNA virus. RNA 5:550-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zuker, M., D. H. Mathews, and D. H. Turner. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide, p. 11-43. In J. Baroiszewski and B. F. C. Clark (ed.), RNA biochemistry and bio/technology. Kluwer Academic Publishers, Dordrecht, The Netherlands.