Abstract
An unusual In0-like class 1 integron containing a common region that includes the putative recombinase gene named orf513 (CR1) and blaCTX-M-2 was characterized from Escherichia coli. The integron contained an unusual gene cassette array, estX-aadA1, embedded between the 5′-conserved segment (5′-CS) and 3′-CS1 regions and was flanked by mer-Tn21 sequences downstream of the tni truncated module. This element constitutes one of the few examples of CR1-bearing class 1 integrons that has been fully characterized.
The CTX-M enzymes are among the most widespread extended-spectrum β-lactamases of Ambler class A (5). Five clusters of CTX-M β-lactamases (CTX-M-1, CTX-M-2, CTX-M-8, CTX-M-9, and CTX-M-25) have been described on the basis of their amino acid sequences (http://www.lahey.org/studies/webt.htm). Chromosomal genes from Kluyvera species have been identified as progenitors of each CTX-M group, and mobilization seems to have occurred by the association of these genes to CR1, ISECp1, or phage-related elements (3, 5, 16, 25). The blaCTX-M-2 and blaCTX-M-9 genes have been mainly associated with class 1 integrons containing CR1. Their backbone structure consists on the 5′-conserved segments (5′-CS) and 3′-CS flanking variable gene cassette arrays, CR1, several antibiotic resistance genes that do not resemble gene cassettes, and a second partial copy of the 3′-CS designated 3′-CS2 (2, 5, 24). The sequences upstream of the 5′-CS and beyond the second copy of qacEΔ1 at the 3′-CS2 have been described only for In6 and In34 (19). The aim of this work was to characterize the genetic environment of blaCTX-M-2 in Escherichia coli strain VS27, one of the few CTX-M-2-producing isolates described in Spain.
E. coli VS27 (resistant to β-lactams and streptomycin, sulfonamide, tetracycline, nalidixic acid, and ciprofloxacin) was recovered from the feces of a healthy volunteer without recent hospitalization or antibiotic exposure in 2003 (29). Transfer of blaCTX-M-2 by broth and filter-mating methods using E. coli BM21R (nalidixic acid and rifampin resistant, lactose fermentation positive, and plasmid free) or E. coli HB101 (kanamycin and azide resistant, lactose fermentation negative, and plasmid free) as the recipient strain was unsuccessful. The whole-plasmid profile determined by the Kado and Liu method using E. coli V517 and E. coli NCTC 50192 as control strains for the estimation of plasmid sizes consisted of three plasmids of 70, 40, and 5 kb and three plasmids of less than 3 kb (27). Hybridization of plasmid DNA (26) with an intragenic blaCTX-M-2 probe labeled and detected by ECL kits according to the manufacturer's instructions (Amersham Life Sciences, Uppsala, Sweden) was negative.
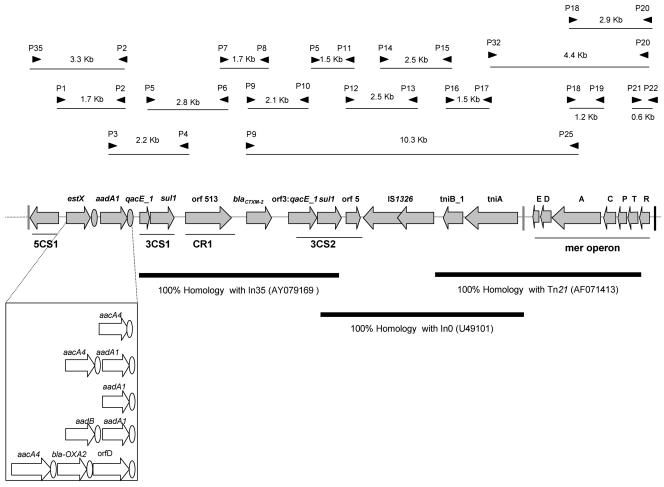
Although blaCTX-M-2 has been previously associated with class 1 integrons bearing CR1, these elements have been only partially characterized (2, 3, 23). An overlapping PCR assay based on the sequence of In35 containing blaCTX-M-2 and Tn21, often associated with class 1 integrons, was designed (GenBank accession numbers AY079169 and AF071413) (3, 12, 13) (Fig. 1). PCR assays were performed in volumes of 50 μl with a mixture containing 1.5 mM MgCl2, 0.2 mM of each deoxynucleoside triphosphate, 0.1 μM of each primer, and 1.5 units of Taq DNA polymerase (AmpliTaq Gold; PE Applied Biosystems, Norwalk, Conn.) for 12 min at 94°C and for 35 cycles at 94°C (1 min), 56 to 65°C (1 to 2 min), and 72°C (1 to 3 min) followed by a final step for 10 min at 72°C for standard PCR assays and with a mixture containing 2.5 mM MgCl2, 0.1 μM of each primer, and 2.5 units of Takara LA Taq polymerase (Takara Bio Inc, Shiga, Japan) for 1 min at 94°C and for 35 cycles of 96°C (20 s), 60°C (1 min), and 72°C (3 to 5 min) followed a final step for 10 min at 72°C for long PCRs (>3 kb). Amplified products were purified using the QIAquick PCR purification kit (QIAGEN) and sequenced on an ABI Prism 377 automated sequencer (PE Applied Biosystems). The oligonucleotide sequences used in PCR assays are listed in Table 1.
FIG. 1.
Schematic representation of the blaCTX-M-2 genetic loci. A comparison with other gene array cassettes located within the 5′-CS-3′-CS1 region described to date is represented at the bottom of the left side (2). The location of the primers used for the identification of the element by PCR-overlapping assay are represented with black arrows. Vertical bars symbolize inverted repeats of the integron (gray) or Tn21 (black). Gray shaded open reading frames represent the 20,032-bp region fully sequenced, with 15,882 bp corresponding to In117. Circles represent 59-bp elements of the corresponding gene cassettes. GenBank accession numbers are in parentheses.
TABLE 1.
Oligonucleotides used in this study
| Primer no. | Primer | Sequence | Positions | GenBank accession no. | Reference or source |
|---|---|---|---|---|---|
| 1 | 5′-CS | 5′-GGCATCCAAGCAGCAAG-3′ | 5298-5314 | AF071413 | 11 |
| 2 | 3′-CS | 5′-AAGCAGACTTGACCTGAT-3′ | 6306-6289 | AF071413 | 11 |
| 3 | aadA1F | 5′-GCTGGCCGTGCATTTGTACG-3′ | 5487-5506 | AF071413 | This study |
| 4 | ORF513rF1R | 5′-GAGCTCTGCACCATCCCAC-3′ | 527-507 | AY079169.1 | This study |
| 5 | qacEΔ2 | 5′-ATCGCAATAGTTGGCGAAGT-3′ | 6383-6392 | AF071413 | This study |
| 6 | ORF513-4F | 5′-CTCGCTTGAGGCGTTGCAT-3′ | 2106-2088 | AY079169.1 | This study |
| 7 | ORF513-4R | 5′-ATGCAACGCCTCAAGCGAG-3′ | 2088-2106 | AY079169.1 | This study |
| 8 | CTX-M-2R/P2b | 5′-TCCCGACGGCTTTCCGCCTT-3′ | 3655-3637 | AY079169.1 | 31 |
| 9 | CTX-M-2F/P3 | 5′-ATGATGACTCAGAGCATTCG-3′ | 2823-2842 | AY079169.1 | 31 |
| 10 | qacEΔ1B | 5′-CAAGCTTTTGCCCATGAAGC-3′ | 5050-5031 | AY079169.1 | This study |
| 11 | orf5-R | 5′-AGTTCTAGGCGTTCTGCG-3′ | 8157-8140 | AF071413 | This study |
| 12 | orf5-F | 5′-CGATATCGACGAGGTTGTGC-3′ | 7712-7730 | AF071413 | This study |
| 13 | IS1326-F | 5′-TACCGGGTCTTATGACCGAGT-3′ | 10357-10337 | AF071413 | This study |
| 14 | IS1326-R | 5′-ACTGTCATAGCGGTTCACGTT-3′ | 9141-9161 | AF071413 | This study |
| 15 | tniBΔ1F | 5′-ATCATCGACCTGTCCCACCT-3′ | 13201-13182 | AF071413 | This study |
| 16 | tniBΔ1R | 5′-AGGTGGGACAGGTCGATGAT-3′ | 13182-13201 | AF071413 | This study |
| 17 | tniAF | 5′-TCGTGCGGAGATCATCAGTCC-3′ | 14821-14801 | AF071413 | This study |
| 18 | merA1 | 5′-ACCATCGGCGGCACCTGCGT-3′ | 17597-17578 | AF071413 | 13 |
| 19 | merA5 | 5′-ACCATCGTCAGGTAGGGGAACAA-3′ | 16360-16382 | AF071413 | 13 |
| 20 | merR1 | 5′-GCGGATTTGCCTCCACGTTGA-3′ | 19278-19260 | AF071413 | 13 |
| 21 | merT1 | 5′-CCAGGCAGCAGGTCGATGCAAG-3′ | 19055-19076 | AF071413 | 13 |
| 22 | Tn21IR/38 | 5′-GGGCACCTCAGAAAACGGAAA-3′ | 19669-19649 | AF071413 | 14 |
| 23 | TnpR-Fa | 5′-ATGCTATGCACCACCACGG-3′ | 3376-3394 | AF071413 | 14 |
| 24 | intF1 | 5′-GGGTCAAGGATCTGGATTTCG-3′ | 4774-4754 | AF071413 | This study |
| 25 | merA6 | 5′-GCCGACCAGTTGTTCCCCTACCTGACG-3′ | 16391-16365 | AF071413 | 13 |
| 26 | merD1 | 5′-CGCACGATATGCACGCTCACCC-3′ | 16211-16233 | AF071413 | 13 |
| 27 | merA0 | 5′-GTCGCAGGTCATGCCGGTGATTTT-3′ | 178950-17974 | AF071413 | 13 |
| 28 | merP1 | 5′-GGCTATCCGTCCAGCGTCAA-3′ | 18520-18501 | AF071413 | 13 |
| 29 | merC1 | 5′-CATCGGGCTGGGCTTCTTGAG-3′ | 18361-18351 | AF071413 | 13 |
| 30 | merC2 | 5′-CATCGTTCCTTATTCGTGTGG-3′ | 17987-18007 | AF071413 | 13 |
| 31 | IRIn2R | 5′-TGGTGCAGTCGTCTTCTGAAAA-3′ | 15012-15033 | AF071413 | 14 |
| 32 | tniAR | 5′-GGACTGATGATCTCCGCACGA-3′ | 14801-14821 | AF071413 | This study |
| 33 | IRTn21Fa | 5′-GGGTCGTCTCAGAAAACGG-3′ | 1-38 | AF071413 | This study |
| 34 | TnpR-Ra | 5′-CCGTGGTGGTGCATAGCAT-3′ | 3394-3376 | AF071413 | This study |
| 35 | IRIn2F | 5′-TTTCAGAAGACGGCTGCACTG-3′ | 4046-4066 | AF071413 | 14 |
Primers 23, 33, and 34 were used in combination with primers 24 and 35 in order to characterize the 5′ end of In117 using appropriate controls.
Analysis of the 15,882-bp sequence between inverted repeats of Tn402 located upstream of intI1 and downstream of tniA revealed the presence of an integron belonging to the In0 group that we called In117. The 5′-CS region includes a copy of the integrase intI1 with a Pc promoter identical to that of In1 in R46 (GenBank accession number AY046276), consisting of TGGACA(−35) and TAAACT(−10) hexamers separated by 17 bp (22). This Pc promoter is of intermediate strength in In1, and although it has been described less frequently than Pc promoters of weak or strong strength, it is increasingly being found in specific class 1 integrons carrying blaIMP, blaVIM, or blaGES (10, 28, 30). A gene cassette array, estX-aadA1, within the 5′-CS-3′-CS1 region was identified. The deduced amino acid sequence of the estX gene displayed 90% amino acid identity with sat-1 of Tn1825 conferring resistance to streptothricin, 40% amino acid identity with proteins annotated as putative esterases or hydrolases of the α/β fold superfamily, and 42% amino acid identity with a protein encoded in E. coli multiresistance plasmids. The estX gene was initially considered to be a sat cassette because of its similarity with sat-1; however, Partridge and Hall have demonstrated that sat-1 resulted from the fusion of estX and sat-2 genes, suggesting a change in the nomenclature of these genes (18). The estX-aadA1 gene cassette combination has not been previously linked to CTX-M-2-producing isolates, although it has been associated with class 1 integrons from Shigella sonnei clinical strains, and estX has been found in different class 1 and class 2 integrons from community isolates at different locations (1, 4, 8).
The 5,585-bp region from 3′-CS1 to the second copy of qacEΔ1 in 3′-CS2 showed 100% homology with that of In35 (GenBank accession number AY079169). The extent of 3′-CS2 was identified as a 6,900-bp sequence with 100% homology with class 1 integron In0 (GenBank accession number U49101). This sequence includes the typical 3′-CS (qacEΔ1, sul1, and orf5) followed by the insertion sequence IS1326, a member of the IS21 family, and a truncated tni module of Tn402 (7). Although several members of the IS21 family are widely distributed, IS1326 remains associated with the class 1 integron lineage In0-In2-In5 (7, 19, 22), which differ one from another in the promoter of intI1 and in the truncated tni module sequences originating from the insertion of IS1326 and further deletion events. Tn21 sequences (left inverted repeats and tnpR) upstream of intI1 were not detected. Interestingly, amplification with primers specific for the mer locus and inverted repeats of Tn21 and the integron showed the presence of mer-Tn21 sequences downstream of tniA (Fig. 1) (12, 32).
Our results revealed the presence of blaCTX-M-2 in a defective transposon derivative of the Tn402 family and constitute, besides In34 and In6, one of the few examples of class 1 integrons containing CR1 in which the structure beyond the 3′-CS2 has been established (19). In0, In2, and In5 are Tn402 derivatives located in plasmids and/or transposons, often in mercury resistance transposons such as Tn21. These transposons are considered to be a worldwide disseminated population composed of a few variants shared by gram-negative environmental and clinical bacteria (32). The absence of a Tn21-like transposition module upstream of intI1 was not surprising, since Tn21 subgroup transposons are frequently inactivated or yield mosaic structures by exchanging transposition modules by recombination at the res site (15, 20, 32). The great polymorphism within the 5′-CS-3′-CS1 region of integrons carrying blaCTX-M-2 (2) suggests recombinatorial exchange either among cassettes of different class 1 integrons or among CR1 and class 1 integrons containing the 3′-CS1 (6, 17, 19, 21).
The presence of blaCTX-M-2 in Spain increases the diversity of blaCTX-M genes described in our area, already epidemic for those of CTX-M-9 (blaCTX-M-9 and blaCTX-M-14) and CTX-M-1 (blaCTX-M-1, blaCTX-M-3, blaCTX-M-10, blaCTX-M-15, and blaCTX-M-32) clusters (9; unpublished results). The genetic elements containing blaCTX-M-2 may fuel the dissemination of this gene, as has recently occurred for carbapenemase genes in Europe (28) or blaCTX-M-9 in Spain (unpublished results), both of which are associated with composite transposon platforms.
Nucleotide sequence accession number.
The sequence for In117 was deposited in the GenBank database under accession number DQ125241.
Acknowledgments
A.V. was funded by a fellowship from the Fondo de Investigaciones Sanitarias of Spain (PI020943-2002). This work was partially supported by research grants from Ministerio de Ciencia y Tecnología of Spain (SAF 2003-09285), the European Commission (LSHM-CT-2003-503335), and the Red Española de Investigación en Patología Infecciosa (REIPI-ISCIII-C03/14).
We thank Hatch Stokes (Macquarie University, Sydney, Australia) for kindly providing us control strains for different class 1 integrons.
REFERENCES
- 1.Ahmed, A. H., H. Nakano, and T. Shimamoto. 2005. Molecular characterization of integrons in non-typhoid Salmonella serovars isolated in Japan: description of an unusual class 2 integron. J. Antimicrob. Chemother. 55:371-374. [DOI] [PubMed] [Google Scholar]
- 2.Arduino, S. M., M. Catalano, B. E. Orman, P. H. Roy, and D. Centrón. 2003. Molecular epidemiology of orf513-bearing class 1 integrons in multiresistant clinical isolates from Argentinean hospitals. Antimicrob. Agents Chemother. 47:3945-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arduino, S. M., P. H. Roy, G. A. Jacoby, B. E. Orman, S. A. Pineiro, and D. Centron. 2002. blaCTX-M-2 is located in an unusual class 1 integron (In35) which includes Orf513. Antimicrob. Agents Chemother. 46:2303-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bischoff, K. M., D. G. White, M. E. Hume, T. L. Poole, and D. J. Nisbet. 2005. The chloramphenicol resistance gene cmlA is disseminated on transferable plasmids that confer multiple-drug resistance in swine Escherichia coli. FEMS Microbiol. Lett. 243:285-291. [DOI] [PubMed] [Google Scholar]
- 5.Bonnet, R. 2004. Growing group of extended-spectrum beta-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd, D., A. Cloeckaert, E. Chaslus-Dancla, and M. R. Mulvey. 2002. Characterization of variant Salmonella genomic island 1 multidrug resistance regions from serovars Typhimurium DT104 and Agona. Antimicrob. Agents Chemother. 46:1714-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, H., H. Stokes, and R. Hall. 1996. The integrons In0, In2, and In5 are defective transposon derivatives. J. Bacteriol. 178:4429-4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeLappe, N., F. O'Halloran, S. Fanning, G. Corbett-Feeney, T. Cheasty, and M. Cormican. 2003. Antimicrobial resistance and genetic diversity of Shigella sonnei isolates from Western Ireland, an area of low incidence of infection. J. Clin. Microbiol. 41:1919-1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hernández, J. R., L. Martínez-Martínez, R. Cantón, T. M. Coque, A. Pascual, and the Spanish Group for Nosocomial Infections (GEIH). 2005. Escherichia coli and Klebsiella pneumoniae producing extended-spectrum β-lactamases in Spain: a nationwide study. J. Clin. Microbiol. 49:2122-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laraki, N., M. Galleni, I. Thamm, M. L. Riccio, G. Amicosante, J. M. Frère, and G. M. Rossolini. 1999. Structure of In31, a blaIMP-containing Pseudomonas aeruginosa integron phyletically related to In5, which carries an unusual array of gene cassettes. Antimicrob. Agents Chemother. 43:890-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levesque, C., L. Piche, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liebert, C., R. M. Hall, and A. O. Summers. 1999. Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63:507-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liebert, C. A., J. Wireman, T. Smith, and A. O. Summers. 1997. Phylogeny of mercury resistance (mer) operons of gram-negative bacteria isolated from the fecal flora of primates. Appl. Environ. Microbiol. 63:1066-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mazel, D., B. Dychinco, V. A. Webb, and J. Davies. 2000. Antibiotic resistance in the ECOR collection: integrons and identification of a novel aad gene. Antimicrob. Agents Chemother. 44:1568-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noguchi, N., J. Katayama, and M. Sasatsu. 2000. A transposon carrying the gene mphB for macrolides 2′-phosphotransferase II. FEMS Microbiol. Lett. 192:175-178. [DOI] [PubMed] [Google Scholar]
- 16.Oliver, A., T. M. Coque, D. Alonso, A. Valverde, F. Baquero, and R. Cantón. 2005. CTX-M-10 linked to a phage-related element is widely disseminated among Enterobacteriaceae in a Spanish hospital. Antimicrob. Agents Chemother. 49:1567-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Partridge, S., C. Collis, and R. Hall. 2002. Class 1 integron containing a new cassette, aadA10, associated to Tn1404 from R151. Antimicrob. Agents Chemother. 46:2400-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Partridge, S., and R. Hall. 2005. Correctly identifying the streptothricin resistance gene cassette. J. Clin. Microbiol. 43:4298-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Partridge, S., and R. Hall. 2003. In34, a complex In5 family class 1 integron containing orf513 and dfrA10. Antimicrob. Agents Chemother. 47:342-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Partridge, S., G. Recchia, H. W. Stokes, and R. Hall. 2001. Family of class 1 integrons related to In4 from Tn1696. Antimicrob. Agents Chemother. 45:3014-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Partridge, S., H. Brown, and R. Hall. 2002. Characterization and movement of the class 1 integron known as Tn2521 and Tn1405. Antimicrob. Agents Chemother. 46:1288-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Partridge, S., H. Brown, H. Stokes, and R. Hall. 2001. Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob. Agents Chemother. 45:1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Power, P., M. Galleni, J. Di Conza, J. A. Ayala, and G. Gutkind. 2005. Description of In116, the first blaCTX-M-2-containing complex class 1 integron found in Morganella morganii isolates from Buenos Aires, Argentina. J. Antimicrob. Chemother. 55:461-465. [DOI] [PubMed] [Google Scholar]
- 24.Sabaté, M., F. Navarro, E. Miró, S. Campoy, B. Mirelis, J. Barbé, and G. Prats. 2002. Novel complex sul1-type integron in Escherichia coli carrying blaCTX-M-9. Antimicrob. Agents Chemother. 46:2656-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saladin, M., V. T. Cao, T. Lambert, J. L. Donay, J. L. Herrmann, Z. Ould-Hocine, C. Verdet, F. Delisle, A. Philippon, and G. Arlet. 2002. Diversity of CTX-M beta-lactamases and their promoter regions from Enterobacteriaceae isolated in three Parisian hospitals. FEMS Microbiol. Lett. 209:161-168. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J, E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Threlfall, E. J., and N. Woodford. 1995. Plasmid profile typing and plasmid fingerprinting. Methods Mol. Biol. 46:225-236. [DOI] [PubMed] [Google Scholar]
- 28.Toleman, M. A., D. Biedenbach, D. Bennet, R. N. Jones, and T. R. Walsh. 2003. Genetic characterization of a novel metallo beta-lactamase gene, blaIMP-13, harboured by a novel Tn5051-type transposon disseminating carbapenemase genes in Europe: report from the SENTRY worldwide antimicrobial surveillance programme. J. Antimicrob. Chemother. 52:583-590. [DOI] [PubMed] [Google Scholar]
- 29.Valverde, A., T. M. Coque, M. P. Sánchez-Moreno, A. Rollán, F. Baquero, and R. Cantón. 2004. Dramatic increase in prevalence of fecal carriage of extended-spectrum β-lactamase-producing Enterobacteriaceae during nonoutbreak situations in Spain. J. Clin. Microbiol. 42:4769-4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wachino, J., Y. Doi, K. Yamane, N. Shibata, T. Yagi, T. Kubota, and Y. Arakawa. 2004. Molecular characterization of a cephamycin-hydrolyzing and inhibitor-resistant class A β-lactamase, GES-4, possessing a single G170S substitution in the Ω-loop. Antimicrob. Agents Chemother. 48:2905-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, H., S. Kelkar, W. Wu, M. Chen, and J. P. Quinn. 2003. Clinical isolates of Enterobacteriaceae producing extended-spectrum β-lactamases: prevalence of CTX-M-3 at a hospital in China. Antimicrob. Agents Chemother. 47:790-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yurieva, O., G. Kholodii, L. Minakhin, Z. Gorlenko, E. Kalyaeva, E. Mindlin, and V. Nikiforov. 1997. Intercontinental spread of promiscuous mercury-resistance transposons in environmental bacteria. Mol. Microbiol. 24:321-329. [DOI] [PubMed] [Google Scholar]