Abstract
Parasite resistance to antimalarial drugs is a serious threat to human health, and novel agents that act on enzymes essential for parasite metabolism, such as proteases, are attractive targets for drug development. Recent studies have shown that clinically utilized human immunodeficiency virus (HIV) protease inhibitors can inhibit the in vitro growth of Plasmodium falciparum at or below concentrations found in human plasma after oral drug administration. The most potent in vitro antimalarial effects have been obtained for parasites treated with saquinavir, ritonavir, or lopinavir, findings confirmed in this study for a genetically distinct P. falciparum line (3D7). To investigate the potential in vivo activity of antiretroviral protease inhibitors (ARPIs) against malaria, we examined the effect of ARPI combinations in a murine model of malaria. In mice infected with Plasmodium chabaudi AS and treated orally with ritonavir-saquinavir or ritonavir-lopinavir, a delay in patency and a significant attenuation of parasitemia were observed. Using modeling and ligand docking studies we examined putative ligand binding sites of ARPIs in aspartyl proteases of P. falciparum (plasmepsins II and IV) and P. chabaudi (plasmepsin) and found that these in silico analyses support the antimalarial activity hypothesized to be mediated through inhibition of these enzymes. In addition, in vitro enzyme assays demonstrated that P. falciparum plasmepsins II and IV are both inhibited by the ARPIs saquinavir, ritonavir, and lopinavir. The combined results suggest that ARPIs have useful antimalarial activity that may be especially relevant in geographical regions where HIV and P. falciparum infections are both endemic.
Malaria is a major cause of morbidity and mortality, infecting 300 to 500 million and killing an estimated 2 million people annually (60). Three approaches to control the most important malaria parasite, Plasmodium falciparum, are vaccine development, vector control, and chemotherapy. An effective vaccine has yet to be developed and must await a better understanding of the targets for protective immunity, parasite antigenic variation, and the critical immune effector mechanisms. Vector control through insecticide spraying programs has eradicated the parasite from southern Europe, North America, and Australia and has reduced transmission elsewhere. However, vector ecology and rising insecticide resistance limit the effectiveness of this approach. Since its discovery in the 1940s, the 4-aminoquinoline drug chloroquine has been the most widely used antimalarial drug. However, the development of drug resistance has dramatically reduced its effectiveness in most regions where malaria is endemic (34), and resistance to other antimalarials is also developing.
The recent completion of the P. falciparum genome sequence is now enabling the identification of novel drug targets, and the increasing power of structure-based medicinal chemistry is facilitating rational drug design for as-yet-unexplored parasite targets (58). One example of successful rational structure-based drug design is the clinical development of antiretroviral drugs that block the action of the aspartyl protease of the human immunodeficiency virus (HIV) (1, 59). Two such antiretroviral protease inhibitors (ARPIs) have been reported to reduce in vitro cytoadherence of P. falciparum-infected erythrocytes to drug-exposed endothelial cells (37), a process linked in vivo with parasite virulence. More recently, we and others have reported that ARPIs have antiparasitic activity against P. falciparum in vitro (40, 51). We now report the in vivo activity of ARPI combinations at clinically relevant concentrations in a rodent model of malaria, model putative binding of these compounds in the enzyme active sites, and show in vitro inhibition of three of these inhibitors against two recombinant P. falciparum aspartic proteases, plasmepsin II (PM-II) and PM-IV.
MATERIALS AND METHODS
P. falciparum growth in vitro.
P. falciparum clone Dd2 and 3D7 parasites were cultured in blood group O+ human erythrocytes and serum (55). Cultures were maintained in a synchronous state by sorbitol treatment (30). In vitro growth inhibition of ring-stage-parasitized erythrocytes starting at 0.25% parasitemia and 2.5% hematocrit was determined by [3H]hypoxanthine incorporation using standard methods (2, 51). Ritonavir gel capsule formulation (Norvir; Abbott) was prepared as a 20 mM stock in dimethyl sulfoxide (DMSO). The gel capsule coformulation of ritonavir and lopinavir (Kaletra; Abbott) was prepared as a 20 mM stock based on the ritonavir concentration (92 mM lopinavir). Chloroquine (chloroquine diphosphate salt; Sigma) was prepared as a 10 mM stock in phosphate-buffered saline and included in each assay as a control. In all assays, the concentrations of DMSO and phosphate-buffered saline were maintained at 0.5%, concentrations that did not affect growth of control cultures (data not shown). The concentration of drug that inhibited parasite growth by 50% (EC50) was determined by linear interpolation of inhibition curves (26).
P. falciparum blood-stage development and hemoglobin digestion.
To determine the in vitro effects of drugs on P. falciparum-parasitized erythrocytes, Dd2 ring-stage-parasitized erythrocytes were incubated for 20 h with 10 μM E64, pepstatin A, ritonavir, saquinavir (Fortivase; Roche), or atazanavir (Reyataz; Bristol-Myers Squibb). In parallel controls, parasites were treated with an equivalent concentration of drug solvent (DMSO) or 1 μM chloroquine. Drug stocks were prepared as 20 mM solutions in DMSO (final concentration of DMSO in tests was 0.1%). After 20 h of growth in vitro, the morphology of drug-treated parasites was compared to that of controls by microscopic examination of Giemsa-stained thin blood smears. To determine the effect on hemoglobin digestion, parasites were processed as previously described (44) and extracts were electrophoresed via 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (5 × 106 parasites per lane), followed by staining with Coomassie blue.
Determination of drug concentrations in the plasma of mice.
Drugs were prepared from gel capsule formulations of saquinavir, ritonavir, and lopinavir-ritonavir in a suspension that resembles ritonavir vehicle (Cremaphor EL [65 mg/ml; Sigma], propylene glycol [0.156 mg/ml], peppermint oil [2.18 mg/ml], water-free citric acid [1.75 mg/ml], and 26.87% ethanol) (27). Groups of three 8-week-old female C57BL/6J mice each received a single 100-μl oral gavage in a dose ranging from 5 to 50 mg/kg body weight (average body weight, ∼20 g). After 4 hours (29), mice were euthanized by CO2 exposure and blood was collected by cardiac puncture. Plasma samples were stored at −20°C and transported overnight on ice before analysis. Lopinavir and ritonavir plasma concentrations were quantitated by high-performance liquid chromatography (HPLC) adapted from a previously reported method (41). Plasma (0.5 ml) and standards were extracted with 5.0 ml 1-chlorobutane. After mixing and centrifugation the organic solvent was evaporated and the residue was resuspended in mobile phase (0.5 ml acetonitrile:10 mM potassium phosphate buffer, pH 7.0; 50:50). The mobile phase was washed twice with 3 ml 99% hexane, and 50 μl was injected onto a phenyl hexyl HPLC column (250 × 4.6 mm; 5 μm) with UV detection at 205 nm. The calibration standards ranged from 50 to 10,000 μg/liter (r = 0.999), and the lower limit of detection was 25 μg/liter. Precision was better than 6% relative standard deviation, and accuracy was within 4% of the expected values for the assay.
Saquinavir plasma concentrations were quantitated by HPLC. Plasma (1.0 ml) and standards were alkalinized with 500 μl of 0.5 M sodium hydroxide and extracted with 7.0 ml diethyl ether (analytical reagent grade; Lab-Scan). After mixing and centrifugation and transfer to a new tube, the organic solvent was evaporated and the residue was resuspended in 6.0 ml 95% n-hexane with brief vortexing. Mobile phase (0.2 ml acetonitrile:25 mM sodium phosphate, pH 3.4; 42:58) was added to the 95% n-hexane and vortexed. After centrifugation the organic phase was discarded and 80 μl of the aqueous phase was transferred into an autosampler. Separation was performed on a cyano column (150 × 4.6 mm; 5 μm; Zorbax SB CN; Dupont) with UV detection at 239 nm. Calibration standards ranged from 10 to 1,000 μg/liter (r = 0.999). Precision was better than 10% relative standard deviation, and accuracy was within 11% of the expected values for the assay.
In vivo antimalarial studies.
The in vivo antimalarial activity of ARPIs was determined using the nonlethal murine malaria model of Plasmodium chabaudi AS (52) in 8-week-old C57BL/6J female mice. Mice were housed in a reverse light cycle cabinet (daylight, 10 p.m. to 10 a.m.), to ensure drug exposure during the trophozoite stages. Groups of six mice (average weight, ∼20 g) were infected intravenously in the tail vein with 105 parasitized erythrocytes from an infected donor mouse. Drugs were prepared from gel capsule formulations, as described in the previous section. Mice received drug in a 100-μl oral solution twice a day for 8 days, beginning 24 h postinfection (p.i.). Control groups received an equivalent volume of vehicle alone twice a day for 8 days, beginning 24 h p.i., or atovaquone-proguanil (Malarone; GlaxoSmithKline) at 0.2 mg/0.08 mg in 100 μl water orally once daily for 4 days (16), beginning 24 h p.i. Parasitemia was monitored from day 4 p.i. by daily microscopic examination of Giemsa-stained thin blood smears. Statistical analysis for differences in peak parasitemia between treatment groups was undertaken using the nonparametric Mann-Whitney U rank test. Animals were purchased from the Animal Resources Centre, Welleton, Western Australia, Australia, and were housed in the Queensland Institute of Medical Research (QIMR) animal facility under specific-pathogen-free conditions. Ethical approval for all animal work was obtained from the QIMR Animal Ethics Committee using protocols complying with guidelines set out by the National Health and Medical Research Council of Australia Animal Code of Practice.
Modeling studies.
The sequence of P. chabaudi plasmepsin (PcPM) was obtained from the SwissProt database, ID Q95V53. Structural coordinates of P. falciparum PM-II (PfPM-II) and PfPM-IV were obtained from the Protein Data Bank (PDB) database. Protein structures 1lf3 (PfPM-II) and 1ls5 (PfPM-IV) were used for docking studies. A homology model of PcPM and PfPM-I was developed using the modeler7v7 program and PfPM-II (1sme) as the template and further optimized in InsightII. PM proteins were compared by sequence and structural alignments using InsightII. Coordinates for amprenavir (1hpv), nelfinavir (1ohr), saquinavir (1hxb), ritonavir (1hxw), indinavir (1hsg), lopinavir (1mui), BR314 (1d4k), and BR124 (1d4l) bound to HIV type 1 (HIV-1) protease were obtained from the PDB. Pepstatin A coordinates were obtained from the complex with PfPM-II (1sme).
Docking studies were performed using Gold v2.1 on inhibitors containing statine and hydroxyethylamine isosteres to gain insight into possible interactions of these inhibitors with PfPM-II and PfPM-IV. Docking simulations were performed on known ligand-enzyme crystal structures (PDB, 1lf3 and 1ls5) to optimize structural constraints that generated the best docking ligand-enzyme models, as deduced through comparison between modeled ligand structures and protein-bound ligand-PM crystal structures. The optimal docking protocol was then used to derive the ligand-enzyme complexes for HIV-1 protease inhibitors. For each inhibitor, 10 independent docking experiments were performed. To accelerate calculations, docking was stopped when a root mean square deviation (RMSD) of 1.5 Å was reached for the three lowest-energy structures.
In one modeling protocol, a distance constraint (2.5 to 3.5 Å) was used between the hydroxyl oxygen of the statine (or transition-state isostere) and the two catalytic aspartate carboxylate oxygens of PfPM-II and PfPM-IV. A second protocol was examined for comparison, with ligands also separately docked using two additional hydrogen bond restraints (P2 carbonyl group to OH of Ser79, P1′ carbonyl group to NH of Val78) on either side of the hydroxyl group of the statine in PfPM-II and one additional hydrogen bond constraint from the P2 carbonyl group to the NH of Gly78 in PfPM-IV. This second protocol gave results that more effectively matched the crystal structures, and so this was used for docking HIV-1 protease inhibitors in PfPMs. All modeling and docking were visualized in InsightII on Silicon Graphics R10000 and R12000 workstations. Docked structures were inspected manually to assess their binding mode(s), scored with GOLD, and then rescored using Ludi. The binding of substrates and substrate-based inhibitors of proteases is heavily influenced by hydrogen bonding networks. Ludi has been shown (9) to provide a well-developed analysis of hydrogen bonding as a contribution towards overall binding energy.
Recombinant plasmepsin II and IV chromogenic assays.
Recombinant plasmepsin II was prepared according to the method of Hill et al. (24), and recombinant plasmepsin IV was prepared according to the method of Wyatt and Berry (61). Fifty percent inhibitory concentration (IC50) values for the interaction of PM-II or PM-IV with protease inhibitors were calculated by monitoring the cleavage of the synthetic peptide substrate KERVF*ZALK (for PM-II) or KPIEF*ZRL (for PM-IV) (where * represents the scissile peptide bond and Z is nitrophenyl alanine) at a final enzyme concentration of 40 nM (PM-II) or 25 nM (PM-IV), in 100 mM sodium acetate buffer, pH 4.7 (with the ionic strength maintained at 100 mM by the addition of sodium chloride), at differing inhibitor concentrations. The final substrate concentration in the assay was at least threefold higher than the Km value, in order not to be rate-limiting. IC50 data were calculated from standardized, multiple experiments for each inhibitor concentration. The error on each data point was estimated to always be less than 15%. Pure lopinavir used in these assays was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
RESULTS
In vitro antimalarial activity.
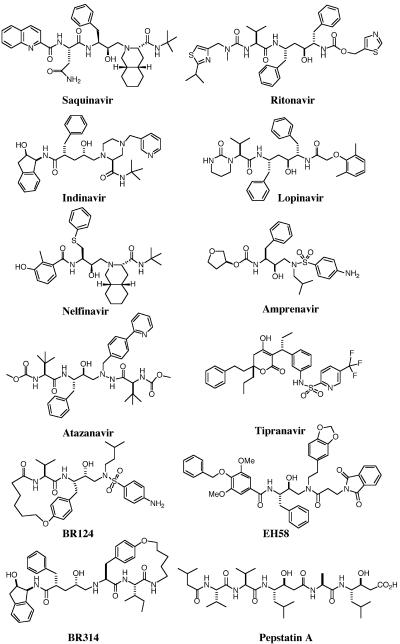
The in vitro antimalarial activities of nine structurally different HIV-1 protease inhibitors have been compared herein (Fig. 1; Table 1), seven of which are used clinically as antiretroviral drugs. As we have previously shown for the multidrug-resistant P. falciparum line Dd2 (51), the drug-sensitive line 3D7 is also sensitive to saquinavir and ritonavir, with an EC50 similar to that obtained with Dd2 (Table 1). Both Dd2 and 3D7 displayed in vitro EC50 values for atazanavir within clinically achievable drug plasma concentrations in humans (Table 1), while lopinavir displayed antimalarial activity well below achievable human plasma concentrations for both parasite lines (Table 1). For the coformulation of lopinavir and ritonavir, the EC50 with respect to the ritonavir component was the same as that obtained for ritonavir alone, whereas the EC50 of lopinavir, with respect to ritonavir, was approximately fourfold higher. Two other structurally quite distinct HIV-1 protease inhibitors, BR124 and BR314, which show potent antiretroviral activity in vitro (56), were also found to inhibit the growth of P. falciparum in vitro at comparable concentrations (Table 1).
FIG. 1.
Structures of aspartyl protease inhibitors.
TABLE 1.
In vitro efficacy of antiretroviral drugs against P. falciparum
| Compound | Antimalarial activitya (EC [μM])
|
Anti-HIV activity (EC50 [μM]) | HIV-1 protease inhibition (Ki [nM]) | Human cathepsin D IC50 (nM) | Log P (pH 7.0)b | Concn in human plasma (Cmin-max [μM]) | |||
|---|---|---|---|---|---|---|---|---|---|
| Dd2
|
3D7
|
||||||||
| EC50 | EC90 | EC50 | EC90 | ||||||
| Saquinavird | 0.6 (±0.2)m | 2.7 (±1.2) | 1.2 (±0.4) | 3.0 (±0.8) | 0.002 | 1.4 (±0.1) | >10,000 | 2.2 | 0.6-3.1c |
| Ritonavire | 0.8 (±0.4)m | 6.3 (±4.7) | 1.7 (±0.6) | 6.7 (±3.0) | 0.025 | 4.2 (±0.5) | 20 | 2.8 | 3.7-11.2c |
| Indinavirf | 2.4 (±1.4)m | >7 | NDn | ND | 0.056 | 2.9 (±0.4) | >10,000 | 1.6 | 0.21-11.1c |
| Lopinavir | 1.0 (±1.3) | 2.5 (±2.9) | 3.3 (±0.6) | 4.8 (±0.0) | 0.017 | 0.04 | ND | 4.9 | 8.8-15.3c |
| Kaletrag | |||||||||
| Ritonavir | 0.8 (±0.5) | 1.4 (±0.8) | 1.5 (±1.1) | 2.2 (±1.5) | 0.025 | 4.2 (±0.5) | 20 | 2.8 | 0.1-0.7c |
| Lopinavirh | 3.5 (±2.0) | 5.6 (4.3) | 6.9 (±5.2) | 10.3 (±6.7) | 0.017 | 0.04 | ND | 4.9 | 8.8-15.3c |
| Atazanaviri | 2.3 (±0.5) | 6.5 (±0.7) | 5.3 (±1.1) | 9.0 (±0.4) | 0.0014 | 2.7 (±0.3) | >10,000 | 4.3 | 0.3-6.7 |
| BR124j | 3.1 (±0.1) | >10 | ND | ND | 0.09 | 1.7 (±0.5) | 15,000 | 3.5 | ND |
| BR314j | 3.7 (±0.1) | >10 | ND | ND | 0.06 | 0.6 (±0.2) | 1,000 | 3.9 | ND |
| Amprenavirk | >10m | ND | ND | ND | 0.013 | 2.6 (±0.6) | 2.0 | 0.56-10.7 | |
| Nelfinavirl | >10m | ND | ND | ND | 0.014 | 3.3 (±0.4) | 435 | 4.3 | 1.7-7.0 |
This work.
Log P is predicted octanol solubility over water solubility and was calculated using the PrologD module within the computer program “Pallas for Windows version 2.1” (CompuDrug International Inc.). Log P 2 to 5 is considered necessary for good cell permeability(32).
Reference 42. Saquinavir free base (Fortovase), 1,200 mg three times daily, or saquinavir mesylate (Invirase), 600 mg three times daily.
Reference 28. Ritonavir (Norvir), 600 mg twice daily.
Reference 57. Indinavir (Crixivan), 800 mg three times daily.
Kaletra = lopinavir, 400 mg, together with ritonavir, 100 mg, twice daily, all with food.
Reference 49. EC50 relative to ritonavir concentration in drug formulation.
Reference 56.
Amprenavir (Agenerase), 1,200 mg twice daily.
Nelfinavir (Viracept), 750 mg three times daily.
Reference 51.
ND, not determined.
P. falciparum development and hemoglobin digestion.
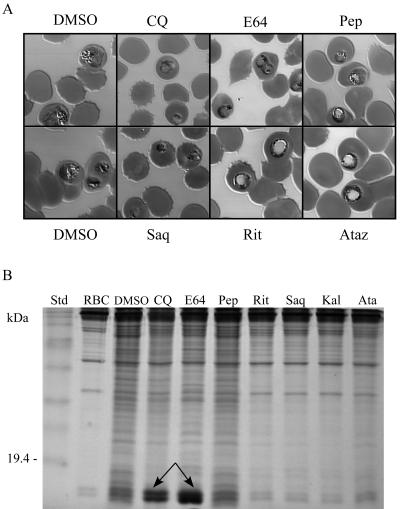
To gain a better understanding of the effect of HIV aspartyl protease inhibitors on the development of P. falciparum, we examined the morphology and hemoglobin digestion pattern of ring-stage parasites treated for 20 h with 10 μM saquinavir, ritonavir, atazanavir, E46, or pepstatin A or 1 μM chloroquine. While control parasites treated with DMSO all matured to the trophozoite stage and contained regular arrays of hemozoin (Fig. 2A), parasites treated with chloroquine appeared stunted and morphologically abnormal. As expected (44), hemoglobin digestion was inhibited in chloroquine-treated parasites (Fig. 2B). Parasites treated with the cysteine protease inhibitor E64 or the aspartic protease inhibitor pepstatin A showed morphologies altered and distinct both from each other and from control parasites (Fig. 2A). E64-treated parasites in particular showed food vacuole swelling, as has been previously reported (5, 44). Consistent with previous findings (44), E64 inhibited hemoglobin digestion in in vitro-cultured parasites (Fig. 2B) while the hemoglobin digestion profile for pepstatin A-treated parasites was similar to that for the DMSO control (Fig. 2B). The morphology of ritonavir- and atazanavir-treated parasites is clearly distinct from that of DMSO-treated control parasites and from parasites treated with E64 and pepstatin. Although it appears that these drugs cause enlargement of the digestive vacuole, these effect cannot be clearly distinguished from nonspecific vacuolization of dying parasites, particularly given the results of hemoglobin digestion experiments where a significant reduction in total protein content in parasitized red blood cells exposed to these drugs is apparent compared to DMSO-, chloroquine-, E64-, and pepstatin A-treated cultures.
FIG. 2.
Effect of protease inhibitors on P. falciparum development and hemoglobin digestion. Ring-stage in vitro-cultured Dd2-parasitized erythrocytes were incubated for 20 h with either 1 μM chloroquine or 10 μM E64, pepstatin A (Pep), ritonavir (Rit), saquinavir (Saq), atazanavir (Ataz), or ritonavir-lopinavir (Kaletra [Kal]). In parallel controls, parasites were treated with an equivalent concentration of DMSO drug solvent. (A) Micrographs of representative Giemsa-stained parasitized erythrocytes. (B) Effect of protease inhibitors on P. falciparum hemoglobin hydrolysis (15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis). Arrows indicate inhibition of hemoglobin digestion by chloroquine (CQ; partial) and E64. RBC, uninfected red blood cells.
While similar hemoglobin digestion profiles were obtained for both saquinavir and ritonavir-lopinavir (Fig. 2B), the effect of these two inhibitors on parasite morphology appeared to be different from that of either ritonavir or atazanavir. Treatment with 10 μM saquinavir for 20 h resulted in parasites that appeared pyknotic, a morphological appearance previously associated with exposure to aspartic proteinase inhibitors (5, 46). Ritonavir-lopinavir killed all parasites (not shown), an expected finding given that parasites treated with this combination were exposed to 10 μM ritonavir plus 46 μM lopinavir.
Plasma concentrations of ARPIs in mice.
Prior to investigation of the in vivo activity of ARPIs in a murine model of malaria, the plasma concentrations of these drugs were measured in mice following the oral administration of different doses of saquinavir, ritonavir, a combination of saquinavir and ritonavir, or coformulated lopinavir-ritonavir (Table 2). Saquinavir was not detected in the plasma of mice after administration of a 5-mg/kg dose. However, when saquinavir was given at a dose of 10 mg/kg with the potent cytochrome P450 inhibitor ritonavir (5, 10, or 50 mg/kg), the plasma concentration of saquinavir exceeded the minimum target required for suppression of HIV-1 replication in humans (Table 1) (4).
TABLE 2.
Plasma concentrations of orally administered HIV-1 protease inhibitors in mice
| Drug | Dose (mg/kg) | Plasma concn (μM)a
|
||
|---|---|---|---|---|
| Saquinavir | Ritonavir | Lopinavir | ||
| Saquinavir | 5 | 0 | ||
| Ritonavir | 10 | 0 | ||
| 50 | 10.1 | |||
| Saquinavir-ritonavir | 5/50 | 0.5 | 12.7 | |
| 10/5 | 0.1 | 0 | ||
| 10/10 | 1.7 | 2.6 | ||
| 10/50 | 1.3 | 7.6 | ||
| Lopinavir-ritonavirb | 10/2.5 | 0 | 0 | |
| 20/5 | 0 | 0.5 | ||
| 40/10 | 0 | 5.3 | ||
Ritonavir, saquinavir, and lopinavir concentrations were measured from the pooled plasma of three mice 4 hours after oral administration.
Administered as lopinavir-ritonavir (Kaletra).
When saquinavir was administered at 10 mg/kg with 50-mg/kg ritonavir, the plasma concentration of saquinavir was lower than that obtained when administered with 10 mg/kg ritonavir (Table 2; 1.3 versus 1.7 μM). A similar effect has been observed previously in rats and has been attributed to saturation of the first-pass metabolism of saquinavir by excess ritonavir (29). The plasma concentration of ritonavir increased in a dose-dependent manner when coadministered at 5, 10, and 50 mg/kg with 10-mg/kg saquinavir. However, higher ritonavir concentrations were observed when the drug was administered with less saquinavir (5-mg/kg saquinavir, 50-mg/kg ritonavir). Interestingly, when ritonavir was administered at doses of 10 mg/kg or below, either alone or in combination with lopinavir, no measurable ritonavir could be detected in the plasma of mice. In contrast, when ritonavir was administered at a dose of 10 mg/kg in combination with 10-mg/kg saquinavir, a ritonavir plasma concentration of 2.6 μM was observed. A dose-dependent increase in lopinavir concentration was observed in mice administered the lopinavir-ritonavir coformulation (Table 2).
Antimalarial activity in vivo.
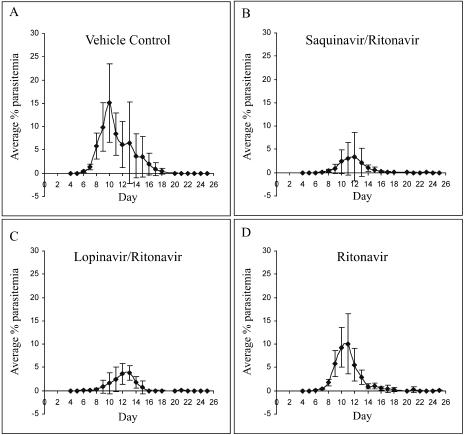
The in vivo activity of the HIV-1 aspartyl protease inhibitors saquinavir, ritonavir, ritonavir-saquinavir, and ritonavir-lopinavir was determined in a nonlethal murine malaria model. Female C57BL/6J mice were infected with 105 P. chabaudi AS-parasitized erythrocytes, and the antiretroviral drugs, or vehicle control, were administered orally twice daily for 8 consecutive days, starting 24 h postinfection. Drug doses were selected based on plasma concentrations achieved in mice (Table 2) and were as follows: 10 mg/kg ritonavir, 10 mg/kg ritonavir-40 mg/kg lopinavir, 10 mg/kg each ritonavir and saquinavir, and 10 mg/kg saquinavir.
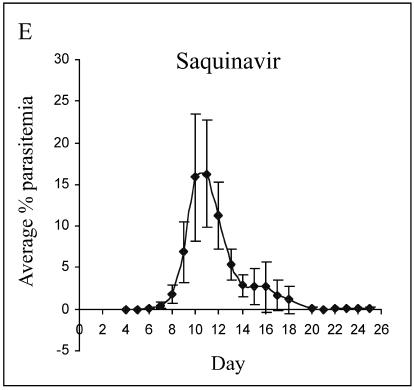
The antiretroviral drug regimens most effective against P. chabaudi AS infection were combinations of ritonavir-saquinavir and ritonavir-lopinavir. Ritonavir-saquinavir, administered at 10-mg/kg doses, resulted in a reduction in the median peak parasitemia (Fig. 3B; median peak parasitemia of 2% compared to 20% in the vehicle control group; P = 0.01), and a 2-day delay in the onset of parasitemia compared to the vehicle control group was observed. Combined ritonavir (10 mg/kg) and lopinavir (40 mg/kg) (administered as Kaletra) resulted in a median peak parasitemia of 4% compared to the vehicle control group median of 20% (P < 0.004), as well as a 2-day delay in the onset of parasitemia compared to the vehicle control group (Fig. 3A and C). Mice given ritonavir alone at a dose of 10 mg/kg (Fig. 3D) also showed a significant reduction in median peak parasitemia compared to the vehicle control group (9% compared to 20% in the vehicle control group; P = 0.037). The course of parasitemia in those mice administered saquinavir at a dose of 10 mg/kg did not differ from that observed in vehicle-treated controls (Fig. 3A and E).
FIG. 3.
In vivo efficacy of HIV-1 protease inhibitors against murine malaria. Female C57BL/6J mice infected with 105 P. chabaudi AS-parasitized erythrocytes were treated orally, twice daily, for 8 consecutive days starting 24 h p.i. with either (A) vehicle, (B) 10 mg/kg each ritonavir and saquinavir, (C) 10 mg/kg ritonavir and 40 mg/kg lopinavir, (D) 10 mg/kg ritonavir, or (E) 10 mg/kg saquinavir. Parasitemia was monitored for 25 days from day 4 p.i. No parasitemia developed in Malarone-treated mice (not shown).
Comparison between active sites of P. falciparum and P. chabaudi plasmepsins.
Since the in vitro studies above were conducted using the human-infecting P. falciparum parasite while the in vivo studies were performed using the mouse-infecting P. chabaudi parasite, we compared the structures of their respective food vacuole aspartyl protease enzymes to determine whether there is likely to be a significant difference in drug binding between this rodent malaria digestive vacuole plasmepsin and those of the human pathogen P. falciparum.
We found some important differences between the active sites of the crystal structure (1lf3) of P. falciparum PfPM-II and our homology model of P. chabaudi plasmepsin (PcPM) (Fig. 4). In PfPM-II the S1, S2, and S1′ subsites were generally smaller than their equivalent pockets in PcPM (Fig. 4). At S1, PfPM-II has a Phe residue at position 111, whereas in PcPM the equivalent amino acid is leucine. Similarly, PfPM-II has a Leu at residue 292 forming the S2 subsite, whereas PcPM has valine. At S1′ PfPM-II has positioned a Phe (residue number 294), thus creating a smaller subsite than in PcPM, which has an Ile in the same location. In addition, the polarity of the S3 and S3′ subsites also differs between the two enzymes. At the S3′ subsite, Asn 76 in PfPM-II is substituted for a Val in PcPM and at S3 in PfPM-II (residue 114) a Thr is replaced by an Ile.
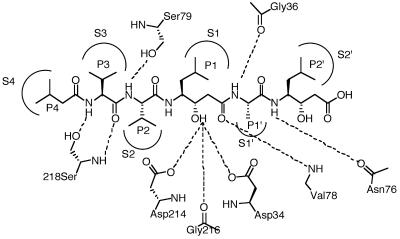
FIG. 4.
Comparison of active sites of P. falciparum plasmepsin II (PDB, 1LF3) and P. chabaudi plasmepsin (homology model) showing side chain variations. Pepstatin A residues (P) are shown bound to P. falciparum plasmepsin II binding sites (S). Prime and nonprime designations distinguish C-terminal from N-terminal sides of the cleavage site. Subsite pocket residue differences in P. falciparum versus P. chabaudi (residues in italics) are as follows: S4 (I290T), S3 (T114I), S2 (L292V), S1 (F111L), S1′ (V78G; N76V), S2′ (N76V) (all residues numbered according to PfPM-II numbering).
Comparison between active sites of P. falciparum plasmepsins I, II, and IV.
Overall, the active sites of PfPM-I and PfPM-II are similar, differing mainly in the S1′ subsite, which is much larger in PfPM-I, the Phe 294 in PfPM-II being larger than the corresponding Leu in PfPM-I. The active site substrate-binding grooves of PfPM-I and PfPM-II are, however, significantly different from that in PfPM-IV. For example, PfPM-II and PfPM-IV have significant changes in polarity in the S3 and S3′ pockets caused by the alterations of Thr 114 in PfPM-II to Ile in PfPM-IV at the S3 pocket and the contributions of Met 75 and Asn 76 (PfPM-II) compared to Ser and Ile in PfPM-IV at S3′. The S2 and S1 subsites are also smaller in PfPM-II than in PfPM-IV. The difference in the S2 subsite is due to the substitution of Leu 292 in PfPM-II for Val in PfPM-IV, and the change in the S1 pocket is due to the substitution of Phe at residue 111 (PfPM-II) for Leu in PfPM-IV. Changes are also found in the S1′ subsite as the replacement of Phe 294 and Val 78 with Ile and Gly, respectively, in PfPM-IV results in this pocket being smaller in PfPM-II than in PfPM-IV. These variations suggest that PfPM-II has smaller subsites and is more limiting in its capacity to accommodate ligands with bulky substituents, and this can be seen experimentally using synthetic inhibitors (61). Ligand docking studies below have focused principally on the active sites of PfPM-II and PfPM-IV, for which there are crystal structures.
Comparison between ligand conformations bound to plasmepsin II and those bound to HIV-1 protease.
Before docking HIV protease inhibitors into plasmepsins, we compared and analyzed known crystal structures of the HIV-1 protease in complex with the ligands in Table 1 and compared them with crystal structures of PfPM-II in complex with known ligands (Fig. 5). In each case there was high conservation of an extended beta-strand conformation for ligand backbones (RMSD, 0.78 Å and 0.39 Å, respectively) and highly conserved ligand side chain locations. This striking superimposition of these two sets of protease-bound ligand conformations (Fig. 5, bottom) reveals high similarity in the aspartyl protease binding of the ligands and supports the idea that HIV-1 protease ligands may have affinity for PfPM-II.
FIG. 5.
Comparison of superimposed crystal structures of ligand-protease complexes (proteases not shown) for HIV-1 protease and P. falciparum plasmepsin II. (Top) Superimposition of crystal structures for HIV protease inhibitor complexes showing protease-bound ligand conformations: lopinavir (PDB code, 1mui), indinavir (1hsg), ritonavir (1hxw), amprenavir (1hpv), saquinavir (1hxb), nelfinavir (1ohr), BR124 (1d4l), BR314 (1d4k). Overlay was performed using backbone atoms N, Cα, and C (or equivalent) from P2-P1′ using the Search and Compare module within InsightII. (Middle) Superimposition of crystal structures for P. falciparum PM-II inhibitor complexes showing protease-bound conformations: pepstatin A (1m43), pepstatin A (1sme), statine-based ligand (1me6), rs367 (1lee), rs370 (1lf2), EH58 (1lf3). Overlay was performed using backbone atoms N, Cα, and C (or equivalent) from P3-P1′. (Bottom) Superimposition of above enzyme-bound HIV-1 protease inhibitors (black) on PfPM-II inhibitors (gray) using backbone atoms N, Cα, and C (or equivalent) from P2-P1′.
Inhibition of PfPM-II and PfPM-IV enzymes.
To validate the hypothesis that plasmepsins may be targets of the ARPIs, three inhibitors (saquinavir, ritonavir, and lopinavir) were assayed against recombinant PfPM-II and PfPM-IV enzymes (Table 3). Only one compound (saquinavir) showed submicromolar inhibitory potency against PfPM-II and PfPM-IV. The other two compounds had low and similar IC50 values, reflecting their similar structures. All of their IC50 concentrations are below the levels achieved in individuals treated with these drugs for HIV infection (Table 1), suggesting that they would be active at the concentrations reached in vivo during antiretroviral therapy.
TABLE 3.
Enzyme inhibitory potencies and predicted relative enzyme affinities (Ludi) for HIV-1 protease inhibitors binding to PM-II and PM-IV
| Plasmepsin and drug | IC50 (nM)a | Ludi scoreb | Hbndc | Lipod |
|---|---|---|---|---|
| PM-II | ||||
| EH58 | 100e | 844 | 138 | 896 |
| Saquinavir | 750 | 748 | 110 | 796 |
| Lopinavir | 1,600 | 655 | 73 | 740 |
| Ritonavir | 1,250 | 587 | 50 | 792 |
| PM-IV | ||||
| Pepstatin A | ∼1 | 521 | 157 | 763 |
| Saquinavir | 375 | 714 | 182 | 690 |
| Lopinavir | >2,000 | 672 | 88 | 790 |
| Ritonavir | 1,800 | 653 | 181 | 711 |
In vitro enzyme inhibition.
Sum Ludi score = 100 log Kd.
Hbnd, hydrogen bond contribution to total binding energy.
Lipo, hydrophobic interaction energy contribution. Rotational penalty scores not shown.
Reference 3.
Docking of HIV protease inhibitors in P. falciparum plasmepsins II and IV.
Initial validation of the ligand docking protocol was first performed (using the GOLD program) on the known PfPM-II inhibitor EH58 (PDB code 1lf3) into PfPM-II and pepstatin A into PfPM-IV (PDB code 1ls5). Docking results were compared with the crystal structures and agreed best (PfPM-II: RMSD, 0.63; PfPM-IV: RMSD, 0.73) when the modeling protocol incorporating all three (PfPM-II) or two (PfPM-IV) H-bond restraints was used.
Using this validated modeling protocol, the three HIV protease inhibitors for which we have plasmepsin inhibition data were docked into PfPM-II and PfPM-IV. Docked structures for saquinavir, ritonavir (Fig. 6), and lopinavir were analyzed for hydrogen bonding interactions, interaction energies, and hydrophobic contacts using Ludi. The structural results suggest that the hydrophobic and hydrogen bonding interactions between the drugs and HIV-1 protease are also possible with PfPM-II and PfPM-IV.
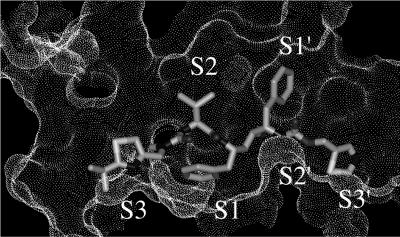
FIG. 6.
Connolly surface of active site of PfPM-II (1lf3) in complex with docked ritonavir showing enzyme subsites S3-S2′.
EH58 had the highest predicted affinity for PfPM-II, far greater than for saquinavir, while ritonavir and lopinavir had about half the affinity of saquinavir. Lopinavir and ritonavir had predicted affinities of approximately 1 μM based on Ludi values of about 600 for PfPM-II. These predicted affinities are in reasonable agreement with experimentally determined inhibitory potencies from the enzyme assays. Both EH58 and saquinavir have much higher hydrogen bonding scores than ritonavir and lopinavir, the higher ranking for EH58 being attributed to a better hydrophobic fit. Both EH58 and saquinavir are distinguished from the other two inhibitors in having steric bulk at P3 and P1′, which are complementary for the S3 and S1 pockets of PfPM-II.
For PfPM-IV, the single pepstatin A-bound crystal structure (1ls5) has larger active-site S2, S1, and S1′ pockets than the corresponding pepstatin A-bound PfPM-II (1sme). The docking results for PfPM-IV (Table 3) show similar trends and rankings as for PfPM-II, with saquinavir having higher affinity than ritonavir or lopinavir. Pepstatin A ranked poorly due to small side chains at P4, P3, P2, and P1′ not optimally filling the corresponding S4, S3, S2, and S1′ pockets of PfPM-IV.
DISCUSSION
In this paper we have compared antimalarial activities of nine HIV-1 protease inhibitors (Table 1). All compounds are potent in vitro antiviral agents against HIV-infected human cells (EC50, <100 nM) and exhibit reasonable cell permeability (log P, 1.6 to 4.9) (Table 1). Seven of the compounds exhibited antimalarial activity between 0.5 and 4.0 μM, while two drugs (amprenavir and nelfinavir) showed minimal activity even at 10 μM. The EC50 values obtained for the multidrug-resistant line Dd2 (reference 51 and this study) are lower than those recently reported (40); however, like the 3D7 EC50 values reported here, they are either within or below levels achieved in clinical use of these drugs (Table 1). The 90% effective concentration values obtained for saquinavir, ritonavir, and lopinavir are also within the target plasma range for these drugs in humans receiving antiretroviral therapy. This is important as subtherapeutic plasma levels of these drugs with respect to malaria may lead to the selection of drug-resistant parasites. Investigation of field isolates of P. falciparum from patients taking these protease inhibitors as part of their antiretroviral therapy should therefore be a priority in future studies. While further work will be required to determine drug availability within parasitized erythrocytes, another aspartyl protease inhibitor displaying potent antimalarial activity (the pentapeptide pepstatin A) has been shown to enter the parasite via a parasite-induced permeation pathway (47). Of those inhibitors that showed antimalarial activity in vitro, two were structurally quite different macrocyclic compounds (26) that are highly selective for HIV-1 protease over human aspartic proteases, three were older ARPIs (saquinavir, ritonavir, and indinavir) now in clinical use in humans for the treatment of HIV/AIDS, and two were newer ARPIs (lopinavir-ritonavir [Kaletra] and atazanavir) also in clinical use.
To investigate the potential clinical activity of ARPIs against malaria, three of the HIV-1 protease inhibitors (saquinavir, ritonavir, and lopinavir) were also evaluated in vivo for antimalarial efficacy in C57BL/6J mice infected with P. chabaudi AS, a nonlethal murine model of malaria. P. chabaudi parasites are known to have at least seven plasmepsin aspartic proteases (35), one of which has similarity to the putative targets of HIV protease inhibitors in P. falciparum, the digestive vacuole plasmepsins. Ritonavir alone and combined with saquinavir or lopinavir significantly attenuated parasitemia, with the most active regimen being a combination of ritonavir and saquinavir or ritonavir and lopinavir (Fig. 3B and C). Whether this decrease in parasitemia is the result of a direct effect on parasite growth or an indirect effect resulting from stimulation of the hosts' immune system remains to be determined. Saquinavir given alone was ineffective, an expected finding given its poor bioavailability when administered alone. Saquinavir is now generally administered along with ritonavir, a potent cytochrome P450 inhibitor (12), for so-called “boosting” to increase the bioavailability of saquinavir. This benefit was also reflected in the drug concentrations that we observed in the plasma of mice after oral administration of a single dose of saquinavir compared to when it was administered in combination with ritonavir (Table 2).
HIV-1 protease inhibitors also had significant effects on the morphology of P. falciparum parasites and their hemoglobin digestion. Treatment of ring-stage Dd2 parasites with 10 μM saquinavir resulted in pyknotic or dead parasites, an observation that has been made previously in parasites treated with P. falciparum aspartic protease inhibitors such as pepstatin (5) and SC-50083 (19). In contrast, after 20 h of treatment of ring-stage parasites with ritonavir or atazanavir (10 μM) a distinct alteration, namely, vacuolar enlargement, was observed, an appearance similar to that seen with the cysteine protease inhibitor E64 (Fig. 2). It is not yet known whether this is due to these drugs acting on digestive vacuole processes or nonspecific vacuolization of dying parasites. Although we cannot account for the differences in parasite morphology after treatment with saquinavir versus ritonavir or atazanavir, it may be that saquinavir exerts its effect more rapidly or perhaps acts on a different parasite target. Similar morphological effects have recently been observed for a geographically distinct drug-sensitive line (HB3) treated with saquinavir (40).
We, and others, have previously hypothesized that the target of ARPIs in the malaria parasite may be a family of P. falciparum aspartyl proteases called PMs (40, 48, 51). It has been reported that the PMs, together with cysteine proteinases (falcipains), are involved in hemoglobin degradation by the parasite, a process necessary for normal parasite growth and development. The amino acids liberated by hemoglobin degradation, which occurs in the parasite digestive vacuole, are incorporated into parasite proteins with the released toxic heme product polymerizing into inert, coordinated hematin dimers, or hemozoin (15, 39, 53). It has also been suggested that removal of hemoglobin provides space for the growing parasite within the red blood cell (20).
P. falciparum has 10 PMs, two of which (PfPM-I and PfPM-II) have been shown to be involved in the in vitro and in vivo initiation of hemoglobin degradation in blood stages (6, 11, 18, 21-23), along with two further digestive vacuole plasmepsins (histoaspartic proteinase [PfHAP] and PfPM-IV) that are also thought to be involved in hemoglobin degradation (6, 7, 14, 61). Only one of the seven plasmepsins from P. chabaudi murine malaria, believed to be the hemoglobin-degrading aspartic protease, appears to be equivalent to the P. falciparum digestive vacuole enzymes with sequence identities to PM-I (48%), PM-II (49%), and PM-IV (53%) (35). The enzymes involved in this metabolic pathway, including PMs, are recognized targets for new drug therapies (13, 45). However, given that individual digestive vacuole plasmepsins of P. falciparum are not essential for parasite survival (33, 38), it is unlikely that only a single P. falciparum plasmepsin is the target for ARPIs (50).
Several experimental PIs have been shown to inhibit recombinant plasmepsin activity and parasite growth, including pepstatin A (reviewed in reference 17). Other synthetic inhibitors, such as Ro40-5576, produce Ki values of 6 nM, 250 nM, and 15 nM against PfPM-I, PfPM-II, and PfPM-IV, respectively (61). Here we found that ritonavir, lopinavir, and saquinavir were considerably less potent inhibitors of both PfPM-II and PfPM-IV (Table 3) and also less potent than the nonspecific tightly binding plasmepsin inhibitor pepstatin A (PfPM-II and PfPM-IV Ki between 0.1 and 0.7 nM) (61). Our findings for PfPM-II are consistent with another study showing that ritonavir and lopinavir inhibit recombinant PfPM-II activity in vitro (ritonavir IC50, 3.1 μM; lopinavir IC50, 2.7 μM) (40).
As members of different families of the aspartic proteinases (dimeric and monomeric), the HIV-1 protease and the plasmepsins share some similarities (54). Here we have performed extensive modeling experiments with two digestive vacuole P. falciparum plasmepsins and the equivalent P. chabaudi plasmepsin to assess possible binding of HIV protease inhibitors to plasmepsins involved in hemoglobin digestion by malaria parasites. The modeling experiments indicate that the dimensions of the active site of P. falciparum PM-II are smaller and more limiting than either PfPM-IV or the P. chaubaudi PM. This may explain our findings that the ARPI combination of lopinavir-ritonavir was more active in vivo than ritonavir alone against P. chabaudi, while no difference in activity of ritonavir versus lopinavir-ritonavir was observed in vitro against P. falciparum under the assay conditions used. Previous work where cysteine protease inhibitors were evaluated as drugs targeting degradation of hemoglobin in the digestive vacuole also suggests that structural differences in target enzymes between rodent and human malarias may be critical considerations when evaluating drug leads (31).
Our in silico experiments suggest that inhibitors of HIV-1 protease can fit the active sites of PfPM-II and PfPM-IV, adopting very similar ligand backbone conformations, while also positioning side chains in very similar locations. The active sites of PfPM-II and PfPM-IV appear sufficiently flexible and large enough to capture the HIV protease inhibitors investigated herein, as well as more flexible and smaller inhibitors like pepstatin A. It is therefore conceivable that their antimalarial activities are due at least in part to inhibition of PfPMs. While the HIV PIs do not fit as well as potent PM inhibitors such as EH58 (Fig. 1), the predicted binding is consistent with the observed micromolar activity in vitro against P. falciparum in cell culture. Both PfPM-II and PfPM-IV clearly have a great deal more space available at S3, S1, and S3′ than can be occupied by either the HIV protease inhibitors or most reported PM inhibitors. As well, PfPM-IV has a significantly larger S1′ subsite than PfPM-II. Compounds with much bulkier substituents would therefore be predicted to have higher affinities for PMs. Our in vitro antimalarial studies and measured versus predicted ligand affinities for PfPM-II and PfPM-IV support the possible involvement of plasmepsins in the action of these drugs.
The observed inhibitory activity of ARPIs against malaria parasites, P. falciparum in vitro and P. chabaudi in vivo, raises the prospect of their potential antimalarial benefit in HIV- and malaria-coinfected individuals. They have the advantage over new experimental antimalarial agents in development as they are already clinically accepted drugs that are widely prescribed for treatment of HIV/AIDS. Although they are expensive for people in the developing world, multinational agreements in recent years have provided much greater access to these drugs in regions where HIV and P. falciparum are both endemic. However, to better determine the antimalarial potential of these drugs, further studies are required, including clinical trials, parasite resistance studies, and pharmacodynamic investigations on interactions with existing antimalarial therapies.
Acknowledgments
We thank the Australian Research Council (grant no. DP00210598), the National Health and Medical Research Council of Australia (grant nos. 102582 and 290208), Roche Australia, and the Mark Nicholson and Alice Hill and the Tudor Foundation for partial support. K.T.A. and L.A.M. were supported by the Australian Centre for International and Tropical Health and Nutrition.
We thank the Brisbane Red Cross Blood Service for kindly providing blood and sera for P. falciparum culture.
REFERENCES
- 1.Abbenante, G., and D. P. Fairlie. 2005. Protease inhibitors in the clinic. Med. Chem. 1:71-104. [DOI] [PubMed] [Google Scholar]
- 2.Andrews, K. T., A. Walduck, M. J. Kelso, D. P. Fairlie, A. Saul, and P. G. Parsons. 2000. Anti-malarial effect of histone deacetylation inhibitors and mammalian tumour cytodifferentiating agents. Int. J. Parasitol. 30:761-768. [DOI] [PubMed] [Google Scholar]
- 3.Asojo, O. A., S. V. Gulnik, E. Afonina, B. Yu, J. A. Ellman, T. S. Haque, and A. M. Silva. 2003. Novel uncomplexed and complexed structures of plasmepsin II, an aspartic protease from Plasmodium falciparum. J. Mol. Biol. 327:173-181. [DOI] [PubMed] [Google Scholar]
- 4.Back, D. J., S. H. Khoo, S. E. Gibbons, and C. Merry. 2001. The role of therapeutic drug monitoring in treatment of HIV infection. Br. J. Clin. Pharmacol. 52:89S-96S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailey, E., R. Jambou, J. Savel, and G. Jaureguiberry. 1992. Plasmodium falciparum: differential sensitivity in vitro to E-64 (cysteine protease inhibitor) and pepstatin A (aspartyl protease inhibitor). J. Protozool. 39:593-599. [DOI] [PubMed] [Google Scholar]
- 6.Banerjee, R., J. Liu, W. Beatty, L. Pelosof, M. Klemba, and D. E. Goldberg. 2002. Four plasmepsins are active in the Plasmodium falciparum food vacuole, including a protease with an active-site histidine. Proc. of the Natl. Acad. Sci. USA 99:990-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berry, C., M. J. Humphreys, P. Matharu, R. Granger, P. Horrocks, R. P. Moon, U. Certa, R. G. Ridley, D. Bur, and J. Kay. 1999. A distinct member of the aspartic proteinase gene family from the human malaria parasite Plasmodium falciparum. FEBS Lett. 447:149-154. [DOI] [PubMed] [Google Scholar]
- 8.Boffito, M., D. Back, M. Stainsby-Tron, A. Hill, G. Di Perri, G. Moyle, M. Nelson, J. Tomkins, B. Gazzard, and A. Pozniak. 2005. Pharmacokinetics of saquinavir hard gel/ritonavir (1000/100 mg twice daily) when administered with tenofovir diproxil fumarate in HIV-1-infected subjects. Br. J. Clin. Pharmacol. 59:38-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bohm, H. J. 1998. Prediction of binding constants of protein ligands: a fast method for the prioritization of hits obtained from de novo design or 3D database search programs. J. Comput. Aided Mol. Des. 12:309-323. [DOI] [PubMed] [Google Scholar]
- 10.Bold, G., A. Fassler, H. G. Capraro, R. Cozens, T. Klimkait, J. Lazdins, J. Mestan, B. Poncioni, J. Rosel, D. Stover, M. Tintelnot-Blomley, F. Acemoglu, W. Beck, E. Boss, M. Eschbach, T. Hurlimann, E. Masso, S. Roussel, K. Ucci-Stoll, D. Wyss, and M. Lang. 1998. New aza-dipeptide analogues as potent and orally absorbed HIV-1 protease inhibitors: candidates for clinical development. J. Med. Chem. 41:3387-3401. [DOI] [PubMed] [Google Scholar]
- 11.Bray, P. G., O. Janneh, K. J. Raynes, M. Mungthin, H. Ginsburg, and S. A. Ward. 1999. Cellular uptake of chloroquine is dependent on binding to ferriprotoporphyrin IX and is independent of NHE activity in Plasmodium falciparum. J. Cell Biol. 145:363-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buss, N., P. Snell, J. Bock, A. Hsu, and K. Jorga. 2001. Saquinavir and ritonavir pharmacokinetics following combined ritonavir and saquinavir (soft gelatin capsules) administration. Br. J. Clin. Pharmacol. 52:255-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coombs, G. H., and J. C. Mottram. 1997. Parasite proteinases and amino acid metabolism: possibilities for chemotherapeutic exploitation. Parasitology 114(Suppl.):S61-S80. [PubMed] [Google Scholar]
- 14.Dame, J. B., C. A. Yowell, L. Omara-Opyene, J. M. Carlton, R. A. Cooper, and T. Li. 2003. Plasmepsin 4, the food vacuole aspartic proteinase found in all Plasmodium spp. infecting man. Mol. Biochem. Parasitol. 130:1-12. [DOI] [PubMed] [Google Scholar]
- 15.Egan, T. J., J. M. Combrinck, J. Egan, G. R. Hearne, H. M. Marques, S. Ntenteni, B. T. Sewell, P. J. Smith, D. Taylor, D. A. van Schalkwyk, and J. C. Walden. 2002. Fate of haem iron in the malaria parasite Plasmodium falciparum. Biochem. J. 365:343-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elliott, S. R., R. D. Kuns, and M. F. Good. 2005. Heterologous immunity in the absence of variant-specific antibodies after exposure to subpatent infection with blood-stage malaria. Infect. Immun. 73:2478-2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fidock, D. A., P. J. Rosenthal, S. L. Croft, R. Brun, and S. Nwaka. 2004. Antimalarial drug discovery: efficacy models for compound screening. Nat. Rev. Drug Discov. 3:509-520. [DOI] [PubMed] [Google Scholar]
- 18.Francis, S. E., I. Y. Gluzman, A. Oksman, D. Banerjee, and D. E. Goldberg. 1996. Characterization of native falcipain, an enzyme involved in Plasmodium falciparum hemoglobin degradation. Mol. Biochem. Parasitol. 83:189-200. [DOI] [PubMed] [Google Scholar]
- 19.Francis, S. E., I. Y. Gluzman, A. Oksman, A. Knickerbocker, R. Mueller, M. L. Bryant, D. R. Sherman, D. G. Russell, and D. E. Goldberg. 1994. Molecular characterization and inhibition of a Plasmodium falciparum aspartic hemoglobinase. EMBO J. 13:306-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ginsburg, H. 1990. Some reflections concerning host erythrocyte-malarial parasite interrelationships. Blood Cells 16:225-235. [PubMed] [Google Scholar]
- 21.Gluzman, I. Y., S. E. Francis, A. Oksman, C. E. Smith, K. L. Duffin, and D. E. Goldberg. 1994. Order and specificity of the Plasmodium falciparum hemoglobin degradation pathway. J. Clin. Investig. 93:1602-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldberg, D. E., A. F. Slater, R. Beavis, B. Chait, A. Cerami, and G. B. Henderson. 1991. Hemoglobin degradation in the human malaria pathogen Plasmodium falciparum: a catabolic pathway initiated by a specific aspartic protease. J. Exp. Med. 173:961-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldberg, D. E., A. F. Slater, A. Cerami, and G. B. Henderson. 1990. Hemoglobin degradation in the malaria parasite Plasmodium falciparum: an ordered process in a unique organelle. Proc. Natl. Acad. Sci. USA 87:2931-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hill, J., L. Tyas, L. H. Phylip, J. Kay, B. M. Dunn, and C. Berry. 1994. High level expression and characterisation of plasmepsin II, an aspartic proteinase from Plasmodium falciparum. FEBS Lett. 352:155-158. [DOI] [PubMed] [Google Scholar]
- 25.Hsu, A., J. Isaacson, S. Brun, B. Bernstein, W. Lam, R. Bertz, C. Foit, K. Rynkiewicz, B. Richards, M. King, R. Rode, D. J. Kempf, G. R. Granneman, and E. Sun. 2003. Pharmacokinetic-pharmacodynamic analysis of lopinavir-ritonavir in combination with efavirenz and two nucleoside reverse transcriptase inhibitors in extensively pretreated human immunodeficiency virus-infected patients. Antimicrob. Agents Chemother. 47:350-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huber, W., and J. C. Koella. 1993. A comparison of three methods of estimating EC50 in studies of drug resistance of malaria parasites. Acta Trop. 55:257-261. [DOI] [PubMed] [Google Scholar]
- 27.Huisman, M. T., J. W. Smit, H. R. Wiltshire, R. M. Hoetelmans, J. H. Beijnen, and A. H. Schinkel. 2001. P-glycoprotein limits oral availability, brain, and fetal penetration of saquinavir even with high doses of ritonavir. Mol. Pharmacol. 59:806-813. [DOI] [PubMed] [Google Scholar]
- 28.Kempf, D. J., K. C. Marsh, J. F. Denissen, E. McDonald, S. Vasavanonda, C. A. Flentge, B. E. Green, L. Fino, C. H. Park, X. P. Kong, et al. 1995. ABT-538 is a potent inhibitor of human immunodeficiency virus protease and has high oral bioavailability in humans. Proc. Natl. Acad. Sci. USA 92:2484-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kempf, D. J., K. C. Marsh, G. Kumar, A. D. Rodrigues, J. F. Denissen, E. McDonald, M. J. Kukulka, A. Hsu, G. R. Granneman, P. A. Baroldi, E. Sun, D. Pizzuti, J. J. Plattner, D. W. Norbeck, and J. M. Leonard. 1997. Pharmacokinetic enhancement of inhibitors of the human immunodeficiency virus protease by coadministration with ritonavir. Antimicrob. Agents Chemother. 41:654-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lambros, C., and J. P. Vanderberg. 1979. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J. Parasitol. 65:418-420. [PubMed] [Google Scholar]
- 31.Lee, B. J., A. Singh, P. Chiang, S. J. Kemp, E. A. Goldman, M. I. Weinhouse, G. P. Vlasuk, and P. J. Rosenthal. 2003. Antimalarial activities of novel synthetic cysteine protease inhibitors. Antimicrob. Agents Chemother. 47:3810-3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lipinski, C. A., F. Lombardo, B. W. Dominy, and P. J. Feeney. 1997. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 23:3-25. [DOI] [PubMed] [Google Scholar]
- 33.Liu, J., I. Y. Gluzman, M. E. Drew, and D. E. Goldberg. 2005. The role of Plasmodium falciparum food vacuole plasmepsins. J. Biol. Chem. 280:1432-1437. [DOI] [PubMed] [Google Scholar]
- 34.Marsh, K. 1998. Malaria disaster in Africa. Lancet 352:924. [DOI] [PubMed] [Google Scholar]
- 35.Martins, T. M., C. Novo, V. E. do Rosario, and A. Domingos. 2003. Aspartic proteases from Plasmodium chabaudi: a rodent model for human malaria. Acta Trop. 89:1-12. [DOI] [PubMed] [Google Scholar]
- 36.Musial, B. L., J. K. Chojnacki, and C. I. Coleman. 2004. Atazanavir: a new protease inhibitor to treat HIV infection. Am. J. Health Syst. Pharmacol. 61:1365-1374. [DOI] [PubMed] [Google Scholar]
- 37.Nathoo, S., L. Serghides, and K. C. Kain. 2003. Effect of HIV-1 antiretroviral drugs on cytoadherence and phagocytic clearance of Plasmodium falciparum-parasitised erythrocytes. Lancet 362:1039-1041. [DOI] [PubMed] [Google Scholar]
- 38.Omara-Opyene, A. L., P. A. Moura, C. R. Sulsona, J. A. Bonilla, C. A. Yowell, H. Fujioka, D. A. Fidock, and J. B. Dame. 2004. Genetic disruption of the Plasmodium falciparum digestive vacuole plasmepsins demonstrates their functional redundancy. J. Biol. Chem. 279:54088-54096. [DOI] [PubMed] [Google Scholar]
- 39.Pagola, S., P. W. Stephens, D. S. Bohle, A. D. Kosar, and S. K. Madsen. 2000. The structure of malaria pigment beta-haematin. Nature 404:307-310. [DOI] [PubMed] [Google Scholar]
- 40.Parikh, S., J. Gut, E. Istvan, D. E. Goldberg, D. V. Havlir, and P. J. Rosenthal. 2005. Antimalarial activity of human immunodeficiency virus type 1 protease inhibitors. Antimicrob. Agents Chemother. 49:2983-2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ray, J., E. Pang, and D. Carey. 2002. Simultaneous determination of indinavir, ritonavir and lopinavir (ABT 378) in human plasma by high-performance liquid chromatography. J. Chromatogr. 775:225-230. [DOI] [PubMed] [Google Scholar]
- 42.Roberts, N. A., J. A. Martin, D. Kinchington, A. V. Broadhurst, J. C. Craig, I. B. Duncan, S. A. Galpin, B. K. Handa, J. Kay, A. Krohn, et al. 1990. Rational design of peptide-based HIV proteinase inhibitors. Science 248:358-361. [DOI] [PubMed] [Google Scholar]
- 43.Robinson, B. S., K. A. Riccardi, Y. F. Gong, Q. Guo, D. A. Stock, W. S. Blair, B. J. Terry, C. A. Deminie, F. Djang, R. J. Colonno, and P. F. Lin. 2000. BMS-232632, a highly potent human immunodeficiency virus protease inhibitor that can be used in combination with other available antiretroviral agents. Antimicrob. Agents Chemother. 44:2093-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosenthal, P. J. 1995. Plasmodium falciparum: effects of proteinase inhibitors on globin hydrolysis by cultured malaria parasites. Exp. Parasitol. 80:272-281. [DOI] [PubMed] [Google Scholar]
- 45.Rosenthal, P. J. 1999. Proteases of protozoan parasites. Adv. Parasitol. 43:105-159. [DOI] [PubMed] [Google Scholar]
- 46.Rosenthal, P. J., J. H. McKerrow, M. Aikawa, H. Nagasawa, and J. H. Leech. 1988. A malarial cysteine proteinase is necessary for hemoglobin degradation by Plasmodium falciparum. J. Clin. Investig. 82:1560-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saliba, K. J., and K. Kirk. 1998. Uptake of an antiplasmodial protease inhibitor into Plasmodium falciparum-infected human erythrocytes via a parasite-induced pathway. Mol. Biochem. Parasitol. 94:297-301. [DOI] [PubMed] [Google Scholar]
- 48.Savarino, A., R. Cauda, and A. Cassone. 2005. Aspartic proteases of Plasmodium falciparum as the target of HIV-1 protease inhibitors. J. Infect. Dis. 191:1381-1382. [DOI] [PubMed] [Google Scholar]
- 49.Sham, H. L., D. J. Kempf, A. Molla, K. C. Marsh, G. N. Kumar, C. M. Chen, W. Kati, K. Stewart, R. Lal, A. Hsu, D. Betebenner, M. Korneyeva, S. Vasavanonda, E. McDonald, A. Saldivar, N. Wideburg, X. Chen, P. Niu, C. Park, V. Jayanti, B. Grabowski, G. R. Granneman, E. Sun, A. J. Japour, D. W. Norbeck, et al. 1998. ABT-378, a highly potent inhibitor of the human immunodeficiency virus protease. Antimicrob. Agents Chemother. 42:3218-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skinner-Adams, T. S., L. W. Guddat, D. L. Gardiner, J. S. McCarthy, and K. T. Andrews. 2005. Are Plasmodium falciparum plasmepsins the target of HIV protease inhibitors? J. Infect. Dis. 191:1382-1383. [Google Scholar]
- 51.Skinner-Adams, T. S., J. S. McCarthy, D. L. Gardiner, P. M. Hilton, and K. T. Andrews. 2004. Antiretrovirals as antimalarial agents. J. Infect. Dis. 190:1998-2000. [DOI] [PubMed] [Google Scholar]
- 52.Stevenson, M. M., J. J. Lyanga, and E. Skamene. 1982. Murine malaria: genetic control of resistance to Plasmodium chabaudi. Infect. Immun. 38:80-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sullivan, D. J., Jr., I. Y. Gluzman, and D. E. Goldberg. 1996. Plasmodium hemozoin formation mediated by histidine-rich proteins. Science 271:219-222. [DOI] [PubMed] [Google Scholar]
- 54.Tacconelli, E., A. Savarino, F. De Bernardis, R. Cauda, and A. Cassone. 2004. Candidiasis and HIV-protease inhibitors: the expected and the unexpected. Curr. Med. Chem. Immunol. Endocr. Metab. Agents 4:49-59. [Google Scholar]
- 55.Trager, W., and J. B. Jensen. 1976. Human malaria parasites in continuous culture. Science 193:673-675. [DOI] [PubMed] [Google Scholar]
- 56.Tyndall, J. D., R. C. Reid, D. P. Tyssen, D. K. Jardine, B. Todd, M. Passmore, D. R. March, L. K. Pattenden, D. A. Bergman, D. Alewood, S. H. Hu, P. F. Alewood, C. J. Birch, J. L. Martin, and D. P. Fairlie. 2000. Synthesis, stability, antiviral activity, and protease-bound structures of substrate-mimicking constrained macrocyclic inhibitors of HIV-1 protease. J. Med. Chem. 43:3495-3504. [DOI] [PubMed] [Google Scholar]
- 57.Vacca, J. P., B. D. Dorsey, R. B. Levin, S. L. McDaniel, J. P. Vacca, J. P. Guare, P. L. Darke, J. A. Zugay, E. A. Emini, W. A. Schleif, J. C. Quintero, et al. 1994. L-735,524: the design of a potent and orally bioavailable HIV protease inhibitor. J. Med. Chem. 37:3443-3451. [DOI] [PubMed] [Google Scholar]
- 58.Vernick, K. D., and A. P. Waters. 2004. Genomics and malaria control. N. Engl. J. Med. 351:1901-1904. [DOI] [PubMed] [Google Scholar]
- 59.West, M. L., and D. P. Fairlie. 1995. Targeting HIV-1 protease: a test of drug-design methodologies. Trends Pharmacol. Sci. 16:67-75. [DOI] [PubMed] [Google Scholar]
- 60.World Health Organization. 2002, posting date. Statistical annex. In The world health report: reducing risks, promoting healthy life. World Health Organization, Geneva, Switzerland. [Online.]
- 61.Wyatt, D. M., and C. Berry. 2002. Activity and inhibition of plasmepsin IV, a new aspartic proteinase from the malaria parasite, Plasmodium falciparum. FEBS Lett. 513:159-162. [DOI] [PubMed] [Google Scholar]