Abstract
A series of 3,5-dioxopyrazolidines was identified as novel inhibitors of UDP-N-acetylenolpyruvylglucosamine reductase (MurB). Compounds 1 to 3, which are 1,2-bis(4-chlorophenyl)-3,5-dioxopyrazolidine-4-carboxamides, inhibited Escherichia coli MurB, Staphyloccocus aureus MurB, and E. coli MurA with 50% inhibitory concentrations (IC50s) in the range of 4.1 to 6.8 μM, 4.3 to 10.3 μM, and 6.8 to 29.4 μM, respectively. Compound 4, a C-4-unsubstituted 1,2-bis(3,4-dichlorophenyl)-3,5-dioxopyrazolidine, showed moderate inhibitory activity against E. coli MurB, S. aureus MurB, and E. coli MurC (IC50s, 24.5 to 35 μM). A fluorescence-binding assay indicated tight binding of compound 3 with E. coli MurB, giving a dissociation constant of 260 nM. Structural characterization of E. coli MurB was undertaken, and the crystal structure of a complex with compound 4 was obtained at 2.4 Å resolution. The crystal structure indicated the binding of a compound at the active site of MurB and specific interactions with active-site residues and the bound flavin adenine dinucleotide cofactor. Peptidoglycan biosynthesis studies using a strain of Staphylococcus epidermidis revealed reduced peptidoglycan biosynthesis upon incubation with 3,5-dioxopyrazolidines, with IC50s of 0.39 to 11.1 μM. Antibacterial activity was observed for compounds 1 to 3 (MICs, 0.25 to 16 μg/ml) and 4 (MICs, 4 to 8 μg/ml) against gram-positive bacteria including methicillin-resistant S. aureus, vancomycin-resistant Enterococcus faecalis, and penicillin-resistant Streptococcus pneumoniae.
Rapidly developing resistance of pathogenic bacteria to available antibiotics is becoming a serious public health threat. For example, recently, two clinical strains of Staphyloccocus aureus resistant both to vancomycin and methicillin were isolated from patients in Pennsylvania and Michigan (16, 17). One of these strains demonstrated extremely high resistance to both antibiotics, with a MIC of 1,000 μg/ml (16). The appearance of such clinical strains is of great concern not only because of the isolates' high level of multidrug resistance but also because the highly anticipated transfer of vancomycin-resistant determinants (vanA gene complex) from Enteroccocus faecalis to methicillin-resistant S. aureus (MRSA) was found to occur in the wound of a patient coinfected with both bacteria (25). Therapeutic failure of vancomycin, which has been considered the antibiotic of choice for MRSA infections, and the development of multidrug resistance by other pathogenic bacteria have created an urgent need for the development of new antibiotics against novel targets. The enzymes involved in peptidoglycan biosynthesis are among the best-known targets in the search for new antibiotics.
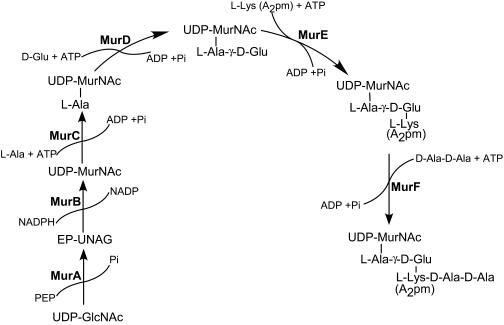
Bacterial peptidoglycan is an extensively cross-linked polymer unique to prokaryotic cells, and the enzymes involved in peptidoglycan biosynthesis are essential for bacterial cell survival. More than 10 synthetic transformations, each executed by a specific enzyme, are required to synthesize peptidoglycan (1, 14, 36). Figure 1 shows the enzymes involved in the biosynthesis of the UDP-MurNAc-pentapeptide precursor in the cytoplasm of both gram-positive and gram-negative bacteria. The MurA and MurB enzymes catalyze the synthesis of UDP-N-acetylmuramic acid (UDP-MurNAc) from UDP-N-acetylglucosamine (UDP-GlcNAc). Four different ligases—MurC, MurD, MurE, and MurF—sequentially add amino acids to UDP-MurNAc, producing the UDP-MurNAc-pentapeptide. The UDP-MurNAc-pentapeptide is then transferred to the plasma membrane, where MraY catalyzes the replacement of UDP by an undecaprenylpyrophosphate carrier lipid, producing Lipid I. This step is followed by the attachment of N-acetylglucosamine catalyzed by MurG to form Lipid II. After attachment of short peptide branches, Lipid II is translocated to the outer surface of the plasma membrane, where penicillin-binding proteins (PBPs) utilize it for cell wall assembly. Inhibition of any of these synthetic enzymes impacts peptidoglycan biosynthesis, resulting in disruption of the structural integrity of the cell wall, which leads to bacterial cell lysis. The most widely used antibiotics that disrupt cell wall assembly are β-lactam antibiotics, which bind to PBPs, and vancomycin, which binds to the Lipid II cell wall precursor. In spite of significant efforts to develop new antibiotics against the enzymes MurA through MurF (for a review, see reference 50), fosfomycin remains the only clinically used antibiotic targeting one of these enzymes, MurA (46).
FIG. 1.
Biosynthetic pathway of the peptidoglycan precursor UDP-MurNAc-pentapeptide in the cytoplasm of gram-positive and gram-negative bacteria. MurA and MurB synthesize UDP-MurNAc from UDP-GlcNAc. MurC, MurD, MurE, and MurF add l-Ala, d-Glu, l-Lys (or A2pm), and d-Ala-d-Ala, respectively, to form UDP-MurNAc-pentapeptide. The bond between l-Ala and d-Glu is formed with the γ-amino group of Glu. In most gram-negative bacteria the third position in the stem peptide is occupied by m-A2pm. In gram-positive bacteria, such as staphylococci, enterococci, and streptococci, this position is occupied by l-Lys.
MurB is essential for the viability of bacterial cells (36). The absence of a homologue in eukaryotic cells makes MurB an attractive target for small molecule inhibitors with the potential to have broad antibacterial activity. Biochemical characterization and X-ray structural analysis of MurB from Escherichia coli (3-5, 18, 45), S. aureus (6), and Streptococcus pneumoniae (49) have been published and have been used in structure-based approaches to design inhibitors (30). Based on a cocrystal structure study of E. coli MurB with enolpyruvyl-UDP-N-acetylglucosamine (EP-UNAG) (3, 4), it was found that the carboxylate of the substrate interacts with residues Arg159 and Glu325 and may be responsible for transition state stabilization, whereas the diphosphate moiety of the substrate interacts with residues Tyr190, Lys217, Asn233, and Glu288. This insight into diphosphate binding prompted the investigation of compounds containing diphosphate mimetics as potential MurB inhibitors. Trisubstituted thiazolidinones, presumably acting as diphosphate surrogates for the natural substrate, were identified as inhibitors of E. coli MurB (50% inhibitory concentrations [IC50s], 7 to 28 μM) (2). More recently, a related imidazolidinone that had activity against MurB (IC50, 12 μM) and was antibacterial was reported (11). Two inhibitors of S. aureus MurB, with Kd values in the submicromole range, were also reported (45). In addition, 2-phenyl-5,6-dihydro-2H-thieno[3,2-c]pyrazol-3-ols (33), phenyl thiazolyl ureas and carbamates (24), and 4-alkyl- and 4,4′-dialkyl 1,2-bis(4-chlorophenyl)pyrazolidine-3,5-diones (31) were identified as novel inhibitors of the enzymes MurA though MurD. These compounds also demonstrated activity against gram-positive bacteria.
In the present study, a set of novel 3,5-dioxopyrazolidines was identified as inhibitors of MurB. Detailed studies of the inhibitory activities of four compounds for E. coli MurB and S. aureus MurB are presented. A detailed biophysical characterization of this compound series was also pursued using a fluorescence-binding assay and X-ray crystallography employing E. coli MurB. The resulting profile of the 3,5-dioxopyrazolidines reveals potent and specific inhibitors that bind within the active site of MurB adjacent to the flavin adenine dinucleotide (FAD) cofactor and that display good activity against a range of gram-positive bacteria, including antibiotic-resistant strains.
MATERIALS AND METHODS
Bacterial strains, plasmids, and reagents.
All reagents used in this study were purchased from Sigma Chemical Co. (St. Louis, MO) or Aldrich Chemical Co. (Milwaukeee, WI) unless otherwise specified. High-purity solvents (OmniSolv) were purchased from EM Science (Gibbstown, NJ). [14C]d-Ala-d-Ala, dl-[m-14C]diaminopimelic acid (A2pm) and [14C]N-acetylglucosamine were purchased from American Radiolabeled Chemicals, Inc. (St. Louis, MO). E. coli TOP10 and E. coli BL21(λDE3) were obtained from Invitrogen (San Diego, CA). Other bacterial strains were from the American Type Culture Collection (ATCC, Manassas, VA) or the Wyeth collection. E. coli K-12 imp BAS849 was kindly provided by S. A. Benson (44). EP-UNAG was synthesized and purified according to the published method (18). UDP-MurNAc, UDP-MurNAc-l-Ala, and UDP-MurNAc-l-Ala-d-Glu were isolated from S. aureus according to the method developed by Park (42) and were purified by high-performance liquid chromatography according to methods described previously (23).
Compound synthesis.
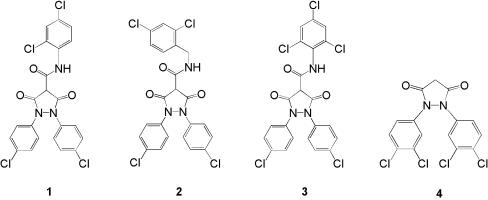
Compounds 1 to 4 (Fig. 2) were synthesized according to the following general procedures. The appropriately substituted anilines were converted to the diarylazo compounds using MnO2 in toluene (47), followed by reduction of the latter to the diaryl hydrazines with zinc dust and ammonium chloride (48). The diaryl hydrazine intermediates were condensed with diethyl malonate and sodium ethoxide in refluxing ethanol (43) to form the 1,2-bis(substituted phenyl)-3,5-dioxo-pyrazolidines. Compound 4, a 1,2-bis(3,4-dichlorophenyl)-3,5-dioxo-pyrazolidine, was prepared by this general procedure starting from 3,4-dichlorophenyl aniline. The 1,2-bis-(4-chlorophenyl)-3,5-dioxopyrazolidine-4-carboxamides (compounds 1 to 3) were obtained by reacting 1,2-bis(4-chlorophenyl)-3,5-dioxo-pyrazolidine with 2,4-dichlorophenyl, 2,4-dichlorobenzyl, or 2,4,6-trichlorophenyl isocyanates (39). Compound structures were confirmed by mass spectrometry.
FIG. 2.
Structures of 3,5-dioxopyrazolidines. Numbers refer to compounds 1 through 4.
Cloning of E. coli murB and S. aureus murB.
The murB-encoded UDP-N-acetylenolpyruvylglucosamine reductase was amplified from E. coli K-12 MG1655 genomic DNA using three PCRs. Reaction 1 was designed to amplify the full-length gene and introduce an EcoRI restriction site 5′ to the murB open reading frame, an NdeI endonuclease recognition sequence at the translation start site (boldfaced in the primer 1 sequence below), and a HindIII restriction site 3′ to the termination codon of the open reading frame. The oligonucleotides used to amplify the full-length gene were 5′-CCGGAATTCAGGCCTCAGGAAAGCACATATGAACCACTCCTTAAAACCCTG-3′ (primer 1) and 5′-CCCAAGCTTAGATCTGTGGCGAGCTCGCTCTTCTCATGAATTGTCTCC ACTGCGCTCACTAG-3′ (primer 2) (underlined sequences indicate EcoRI, NdeI, and HindIII restriction sites, respectively). In reaction 2, an NdeI recognition sequence located at nucleotides 634 to 639 within the murB open reading frame was inactivated using primer 3 (5′-GCGGTGTGTCACATGCGCACCACCAAACTCCCTGATCC-3′) in combination with primer 2, generating a 3′-terminal gene fragment. Primer 3 annealed to the region containing the internal NdeI site, replacing CATATG with CACATG (underlined in primer 3), encoding identical amino acids while removing the restriction site. The final PCR employed primer 1 as the sense primer and the DNA fragment created in reaction 2 as the antisense primer. Following cleavage by the endonucleases EcoRI and HindIII, the PCR product from reaction 3 was ligated into pUC119 restricted with EcoRI and HindIII, creating plasmid pUCEmurB. The murB sequence was confirmed by DNA sequence analysis. The murB coding region was cloned into a pET-29A expression vector (Novagen, Madison, WI) via restriction digestion and ligation of compatible NdeI and HindIII sites, creating plasmid pETEmurB. pETEmurB was transformed into E. coli BL21(λDE3) to enable expression of the E. coli MurB protein.
S. aureus murB was amplified from S. aureus Smith (ATCC 13709) genomic DNA using oligonucleotide primers 5′-CTGACATATGATAAATAAAGACATCTATCAAGC-3′ and 5′-GGATCCTTACGATTCCTTTGGATG-3′. The PCR product was cleaved with NdeI and BamHI and ligated into a similarly restricted protein expression vector, pET-29A, creating pETSmurB. The DNA sequence of the S. aureus murB contained within pETSmurB was confirmed by DNA sequence analysis, and the plasmid was transformed into E. coli BL21(λDE3) for expression of S. aureus MurB.
Cloning and preparation of E. coli MurA, MurC, MurD, MurE, and MurF.
The genes encoding the soluble cytoplasmic peptidoglycan-synthetic enzymes MurA (UDP-N-acetylenolpyruvylglucosamine transferase; NCBI accession no. AAC76221.1), MurC (UDP-MurNAc:l-alanine ligase; AAC73202.1), MurD (UDP-MurNAc-l-alanine:d-glutamate ligase; AAC73199.1), MurE (UDP-MurNAc-l-alanyl-d-glutamate:meso-diaminopimelic acid ligase; AAC73196.1), and MurF (UDP-MurNAc-l-alanyl-d-glutamyl-meso-diaminopimelic acid:d-alanyl-d-alanine ligase; AAC73197.1) were cloned. The genes were amplified from E. coli K-12 MG1655 genomic DNA. Appropriately restricted PCR products were ligated into pET28, a protein expression vector, and transformed into E. coli BL21(λDE3). Enzymes were purified from E. coli cells harboring the expression plasmid as described previously for MurA (12, 35), MurC (21, 34), MurD (7), MurE (38), and MurF (19). The purity of the enzyme preparations was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The enzymes were purified to 90% purity, with specific activity similar to that reported earlier (7, 12, 19, 34, 38) (data not shown).
Purification of MurB enzymes.
E. coli MurB and S. aureus MurB were purified according to published methods (5, 18) with modifications. E. coli BL21(λDE3) cells harboring the expression plasmid were grown in Luria-Bertani medium containing kanamycin (50 μg/ml) at 37°C. The expression of either E. coli MurB or S. aureus MurB was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM. The cultures were incubated for 2 h at 37°C, and cells were harvested, resuspended in 25 mM Tris-HCl buffer (pH 8.0)-1.0 mM dithiothreitol (DTT) (buffer A), and lysed by two passages through a French press at 15,000 lb/in2. The lysate was centrifuged at 100,000 × g for 30 min at 4°C. (NH4)2SO4 was added to the supernatant to a final concentration of 40%. Precipitated material was removed by centrifugation, and the supernatant was loaded onto a Phenyl-Sepharose 6 Fast Flow column (Amersham Biosciences Corp., Piscataway, NJ) equilibrated with buffer B containing 25 mM Tris-HCl (pH 8.0), 1.6 M (NH4)2SO4, and 5 mM DTT. MurB was eluted with a linear gradient of buffer B to buffer A. MurB was further purified using ion-exchange column chromatography on a MonoQ column (Amersham Biosciences Corp., Piscataway, NJ). The enzyme was eluted from the column using a gradient of KCl (0 to 1 M) in buffer A. MurB enzymatic activity was monitored throughout the purification. The purity of E. coli MurB and S. aureus MurB was analyzed using sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
MurB assay.
MurB activity was determined using a continuous assay (5) monitoring the oxidation of NADPH, as measured by reduction in the absorbance at 340 nm, using a SpectroMax 250-plate reader (Molecular Devices, Sunnyvale, CA) at room temperature. The reaction mixture contained 50 mM Tris-HCl (pH 8.0), 10 mM KCl, 100 μM NADPH, and 50 μM EP-UNAG in a total volume of 200 μl. The reaction was initiated by the addition of enzyme. The optical density at 340 nm (OD340) was monitored over 5 min. One unit of enzyme activity was defined as the amount of enzyme that catalyzed the oxidation of 1 μmol of NADPH/min. A molar absorption coefficient of 6,220 cm−1 · M−1 for NADPH absorption at 340 nm was used.
MurA, MurC, and MurD assays.
The activity of MurA, MurC, or MurD was measured indirectly by detecting the quantity of inorganic phosphate (32) released from the cleavage of phosphoenolpyruvate (PEP) for MurA or from ATP hydrolysis for MurC and MurD. The MurA assay followed the published method (35), and the reaction mixture contained 25 mM Tris-HCl buffer (pH 8.0), 150 nM MurA, 200 μM PEP, and 200 μM UDP-GlcNAc in a final volume of 25 μl. The MurC reaction mixture contained 83 mM Tris-HCl buffer (pH 8.5), 200 nM MurC, 50 mM MgCl2, 50 mM (NH4)2SO4, 2 mM ATP, 1.28 mM UDP-MurNAc, and 2 mM l-alanine in a final volume of 25 μl. The MurD reaction mixture (25 μl) contained 83 mM Tris-HCl buffer (pH 8.5), 68 nM MurD, 50 mM MgCl2, 50 mM (NH4)2SO4, 2 mM ATP, 0.38 mM UDP-MurNAc-l-Ala, and 2 mM d-glutamate. Each reaction was initiated by the addition of the appropriate UDP-linked substrate, and the reaction mixture was incubated at 37°C for 30 min. Reactions were terminated by the addition of 210 μl of malachite green reagent (0.03% malachite green, 1.4% ammonium molybdate, and 1.3 N HCl). In order to minimize the acid hydrolysis of ATP, 50 μl of 34% (wt/vol) sodium citrate was added to the MurC and MurD reaction products after addition of the malachite green reagent (32). One unit of enzyme activity was defined as the amount of enzyme that catalyzed the addition of 1 μmol of amino acid to a UDP-linked substrate/min at 37°C.
MurE assay.
MurE activity was determined using a thin-layer chromatography (TLC) method (1) employing substrates dl-[m-14C]A2pm and UDP-MurNAc-l-Ala-d-Glu. The reaction mixture (25 μl) contained 83 mM Tris-HCl buffer (pH 8.5), 1.6 μM MurE, 50 mM MgCl2, 50 mM (NH4)2SO4, 2 mM ATP, 0.2 mM UDP-MurNAc-l-Ala-d-Glu, and 1.6 mM (60 nCi) of dl-[m-14C]A2pm. The reaction was started by the addition of the UDP-linked substrate, and the reaction mixture was incubated at 37°C for 30 min. In the control reaction, the substrate was omitted. The reactions were terminated by heating at 100°C for 1 min. Two microliters of the reaction mixture was spotted onto a silica gel TLC plate (K6F, 60α, 250 μm, 10 by 20 cm; Whatman, Florham Park, NJ), and TLC was performed with isobutyric acid (1 M) and NH4OH (5:3, vol/vol) for 1.5 to 2 h. The TLC plate was dried and exposed to Kodak Biomax MR film with a BioMax MS intensifying screen at −80°C overnight. The amounts of radioactivity in the product UDP-MurNAc-l-Ala-d-Glu-[m-14C]A2pm and the substrate dl-[m-14C]A2pm were quantified with a Fluor-S MultiImager (Bio-Rad Laboratories, Hercules, CA).
MurE-MurF coupled assay.
Components and conditions were the same as for the MurE reaction except that nonradiactive dl-m-A2pm was used and [14C]d-Ala-d-Ala, at a final concentration of 360 μM (66 nCi), was added. The concentrations of MurE and MurF in the reaction were 1.6 μM and 0.1 μM, respectively.
MurB kinetic analysis.
MurB kinetic parameters for EP-UNAG were analyzed using substrate concentrations between 5 and 200 μM with a fixed concentration of NADPH, 50 μM. MurB kinetic parameters for NADPH were analyzed using concentrations from 2 to 100 μM at a fixed EP-UNAG concentration of 50 μM. Km and kcat values were derived using the Michaelis-Menten equation.
Inhibition of enzyme activity.
The inhibitory activities of the compounds against the enzymes were determined by enzymatic assays as described above with a 10-min preincubation of the enzyme and the compound. The preincubation time for MurB was 20 min. Preincubation was used in the assays because it has been demonstrated that preincubation is important for some inhibitors, particularly slow-binding inhibitors (20). Preincubation was shown to affect the IC50s of inhibitors against MurC. Compounds were dissolved in 100% dimethyl sulfoxide at 10 mg/ml. The final concentration of dimethyl sulfoxide in the reaction mixtures with or without compound was 1.25%. At least six concentrations of inhibitor (1.6, 3.2, 6.4, 12.8, 25, and 50 μM) were used for each compound tested. The percentage of MurF inhibition was estimated by subtracting the percent inhibition of the MurE reaction from the MurE-MurF reaction inhibition. IC50s were derived using the data analysis function of the Microsoft Excel program (sigmoid curve Hill analysis).
Crystallographic studies.
Purified E. coli MurB was concentrated to 17.3 mg/ml in 25 mM Tris-HCl (pH 7.4)-50 mM NaCl-10 mM DTT-10 mM MgCl2. The enzyme sample was combined with an equal molar amount of compound 4. Crystals were obtained at 20°C using the hanging drop vapor diffusion method with a 1:1 volume ratio of well solution (0.1 M Tris-HCl [pH 8.5], 20% polyethylene glycol 8000, 0.1 M calcium acetate) to protein solution. Crystals were transferred to a stabilizing solution, containing the well solution plus 25% glycerol, for 10 s before rapid cooling in liquid nitrogen. Data were collected at the Advanced Light Source, station 5.0.2, on a Quantum-4 detector (ADSC, Poway, CA) and processed with HKL software (41). The structure was determined by molecular replacement, using AMORE software (15), with the previously determined crystal structure (PDB code 1UXY) as a search model. This model was used to generate phases that were used with the CNS software package (13). These maps were of sufficient quality to allow subsequent rebuilding and refinement of the model.
Fluorescence-binding assay.
A binding assay based on the changes in the fluorescence emission spectra of FAD was developed to determine the binding affinity (Kd) and stoichiometry of binding between the inhibitor and E. coli MurB. Changes in the fluorescence of the isoalloxazine ring of the FAD cofactor were monitored. Binding experiments were conducted using an enzyme concentration of 1 μM. The concentration of the inhibitor was varied from 0 to 8 μM. The change in the fluorescence of FAD between 510 and 610 nm was determined after exciting the chromophore at 460 nm. The fluorescence emission spectra were acquired using a Fluoromax 3 spectrofluorometer (Jobin Yvon, Edison, NJ). The fluorescence emission of FAD was quenched due to specific interactions of the inhibitor with E. coli MurB. The changes in the fluorescence of FAD at the emission maximum of 520 nm were fit to a quadratic equation (28) to determine the binding affinity.
sPG biosynthesis assay.
Peptidoglycan biosynthesis was measured by determining the amount of soluble peptidoglycan (sPG) produced by a Staphyloccocus epidermidis strain. The original method (8) was modified to fit a 96-well microplate. Briefly, cells were grown in brain heart infusion broth to an OD600 of 0.6, harvested, washed twice with cold distilled H2O, and resuspended in enriched medium (8) at 10% of the original volume. The reaction mixture (250 μl) contained 200 μl of cell suspension, 50 μg/ml chloramphenicol, 100 μg/ml penicillin G, 2.9 μM (40nCi) [14C]N-acetylglucosamine, and the compound at a concentration of 0, 3.1, 6.2, 12.5, or 25 μg/ml. Reaction mixtures were incubated with agitation at 37°C for 1 h prior to centrifugation at 1,500 × g for 20 min. Two hundred microliters of the supernatant was transferred to a Millipore MultiScreen 1.2-μm glass fiber filter plate. Bovine serum albumin (BSA) and trichloroacetic acid were added to each well, giving final concentrations of 0.4% and 5%, respectively. After 30 min of incubation at 4°C, the plates were filtered and washed with 5% trichloroacetic acid. Radioactivity was measured on a Packard TopCount liquid scintillation counter using MicroScint scintillation fluid (Perkin-Elmer Life Sciences, Boston, MA).
Antimicrobial susceptibility tests.
The microorganisms used for testing antimicrobial activity included strains of methicillin-susceptible and methicillin-resistant S. aureus, penicillin-resistant S. pneumoniae, vancomycin-susceptible and vancomycin-resistant E. faecalis, E. coli, and a modified E. coli strain with increased outer membrane permeability (E. coli imp) (44). The MICs were determined by the microbroth dilution method using Mueller-Hinton II medium (Becton Dickinson Microbiology Systems, Cockeysville, MD) according to the recommendations of the Clinical and Laboratory Standards Institute (formerly the National Committee for Clinical Laboratory Standards) (40) after incubation at 35°C for 18 h. Bacterial inocula of 5 × 105 CFU/ml and a range of compound concentrations (0.06 to 128 μg/ml) were used.
Analytical methods.
Concentrations of EP-UNAG and UDP-linked substrates were determined by the Morgan-Elson reaction as described previously (29). Phosphate was quantified using malachite green reagent (32). Sterox detergent was omitted from the method. Protein concentrations were assayed using a protein assay kit from Bio-Rad Laboratories (Hercules, CA) according to the manufacturer's published method (9).
RESULTS
Characterization of MurB.
E. coli MurB and S. aureus MurB were overexpressed in E. coli and purified to >90% homogeneity (data not shown). The kinetic parameters for both enzymes are summarized in Table 1. The Km of EP-UNAG for E. coli MurB was 15 μM, two- to threefold lower than that for S. aureus MurB, 41 μM, consistent with the published values (6). The Km of NADPH for E. coli MurB (11 μM) was threefold higher than that for S. aureus MurB (3.7 μM). The kcat values of E. coli MurB for both substrates, 18 s−1 for EP-UNAG and 15 s−1 for NADPH, were two- to threefold higher than those of S. aureus MurB, 7.9 s−1, and 4.5 s−1, respectively. With substrate EP-UNAG, kcat/Km for E. coli MurB was fivefold higher (1,150 mM−1 s−1) than that for S. aureus MurB (192 mM−1 s−1). With substrate NADPH, the kcat/Km values were comparable for the two enzymes, 1,270 and 1,210 mM−1 s−1, respectively.
TABLE 1.
Comparisons of kinetic parameters for E. coli MurB and S. aureus MurB
| Enzyme | EP-UNAG
|
NADPH
|
||||
|---|---|---|---|---|---|---|
| Km (μM) | kcat (s−1) | kcat/Km (mM−1 s−1) | Km (μM) | kcat (s−1) | kcat/Km (mM−1 s−1) | |
| E. coli MurB | 15 ± 2.5 | 18 ± 3.2 | 1,150 | 11 ± 1.5 | 15 ± 2.3 | 1,270 |
| S. aureus MurB | 41 ± 11 | 7.9 ± 1.6 | 192 | 3.7 ± 0.93 | 4.5 ± 1.4 | 1,210 |
Inhibitory activity of the 3,5-dioxopyrazolidines against MurA, MurB, MurC, MurD, MurE, and MurF.
The inhibitory activities of compounds 1 to 4 against soluble peptidoglycan-biosynthetic enzymes are summarized in Table 2. Compounds 1 to 3 showed inhibitory activity against E. coli MurB, S. aureus MurB, and E. coli MurA. The IC50s of the 3,5-dioxopyrazolidine-4-carboxamides were in the range of 4.1 to 6.8 μM against E. coli MurB, 4.3 to 10.3 μM against S. aureus MurB, and 6.8 to 29.4 μM against E. coli MurA. Compound 2 also inhibited E. coli MurC, to a weaker extent. The C-4-unsubstituted 3,5-dioxopyrazolidine compound 4 showed moderate inhibitory activity against E. coli MurB, S. aureus MurB, and E. coli MurC (IC50s, 24.5 to 35 μM). Inhibition of MurD, MurE, or MurF by these compounds was negligible.
TABLE 2.
Inhibitory activities of 3,5-dioxopyrazolidines against peptidoglycan-synthetic enzymes
| Compound | IC50 (μM) against:
|
||||||
|---|---|---|---|---|---|---|---|
| E. coli MurA | E. coli MurB | S. aureus MurB | E. coli MurC | E. coli MurD | E. coli MurE | E. coli MurE-F | |
| 1 | 21.6 | 4.1 | 4.3 | >50 | >50 | >50 | >50 |
| 2 | 6.8 | 5.3 | 10.3 | 36.0 | >50 | >50 | >50 |
| 3 | 29.4 | 6.8 | 5.1 | >50 | >50 | >50 | >50 |
| 4 | >50 | 35.0 | 24.5 | 29.2 | >50 | >50 | >50 |
| Fosfomycin | 35.0 | >300 | >300 | >300 | >300 | >300 | >300 |
Fluorescence-binding assay.
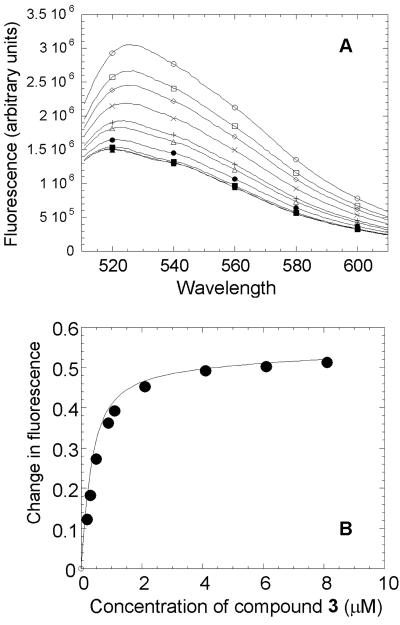
Data on the interactions of compound 3 with E. coli MurB are presented in Fig. 3. The fluorescence emission spectra of the isoalloxazine moiety of the FAD cofactor in both the absence and the presence of the compound are shown in Fig. 3A. Following excitation, the FAD cofactor emitted at a maximum wavelength of 525 nm in the absence of the inhibitor and at 520 nm in the presence of the inhibitor. Binding of the compound quenched the fluorescence of the FAD cofactor in a dose-dependent manner. The binding isotherm of the interactions between compound 3 and E. coli MurB is shown in Fig. 3B. A dissociation constant of 260 nM was estimated for the interactions of the inhibitor with the enzyme, with a stoichiometry consistent with 1:1 binding.
FIG. 3.
Determination of binding affinity of compound 3 for E. coli MurB. (A) Fluorescence spectra of FAD bound to E. coli MurB in the absence and presence of compound 3. Each curve (described in order from top to bottom) represents one wavelength scan (excitation at 460 nm) of E. coli MurB at 1 μM alone or with 1, 2, 3, 4, 5, 6, 7, or 8 μM compound 3. (B) Binding isotherm for the interactions of compound 3 with E. coli MurB. Compound 3 binds E. coli MurB tightly with a Kd of 260 nM.
Crystallographic studies.
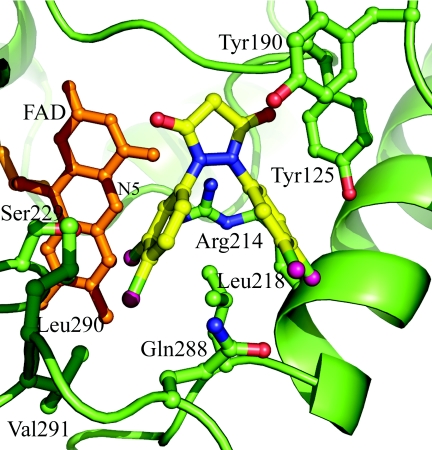
The crystal structure of the complex of E. coli MurB bound to compound 4 was obtained to 2.4 Å resolution following cocrystallization (Table 3; Fig. 4). Comparison of this complex with published crystal structures of E. coli MurB revealed that the overall protein conformation was closely related to that of the substrate-free form of E. coli MurB (PDB code 1MBT). Small differences were observed in loop regions distant from the active site. Within the active site, the most significant changes involved residues Tyr190, Tyr125, Lys217, and Met213. The extra density seen corresponding to compound 4 was proximal to these residues and the bound FAD cofactor. Comparison of the compound-binding site with the complex crystal structure of the N-acetylglucosamine substrate (PDB code 1MBB) indicated that the 3,5-dioxopyrazolidine core occupied the same region of the active site as the N-acetyl group of the substrate, parallel to the plane of the Tyr190 ring side chain. One of the two carbonyl atoms from the pyrazolidinedione formed a hydrogen bond to the backbone amide of Tyr125, mimicking the C-6-OH of the N-acetylglucosamine interaction. The second carbonyl was solvent exposed, and its closest interaction was more than 4 Å from the side chain of Asn233. The two dichloro-substituted phenyl groups of compound 4 both had intimate interactions with the side chain of Leu218, making van der Waals interactions with opposite faces of the branched alkyl atoms of the leucine side chain. This defining interaction resulted in one of these groups being positioned such that it was proximal and perpendicular to the edge of the plane of the FAD molecule, with the closest interaction being 3.9 Å from the N-5 of the isoalloxazine ring. The second dichloro-substituted phenyl group of compound 4 was partially exposed to solvent and interacted with the side chains of Tyr125, Lys217, and Met213.
TABLE 3.
Statistics from the crystallographic analysis of E. coli MurB
| Parametera | Value |
|---|---|
| Resolution (Å) | 2.0-2.4 |
| Space group | P21 |
| Cell dimensions | a = 41.99 Å, b = 89.27 Å, c = 50.93 Å, β = 111.12° |
| Unique observations | 13,723 |
| Completeness (%) | 99.0 |
| Rsym | 4.2 |
| Refinement | |
| Rwork (%) | 21.4 |
| Rfree (%) | 26.4 |
| RMSD bonds (Å) | 0.006 |
| RMSD angles (°) | 1.277 |
Rsym = ∑|I − <I>|/∑I, where I is the observed intensity and <I> is the average intensity of multiple observations of symmetry-related reflections. Rwork = ∑||Fobs| − |Fcalc|/∑|Fobs|. Rfree is equivalent to Rwork, but calculated for a randomly chosen 5% of reflections omitted from the refinement process. Fobs is the observed structure factor amplitude derived from the mean measured intensities, and Fcalc is the calculated protein structure factor derived from the atomic model. RMSD, root mean square deviation from ideal geometry and in the B factor of bonded atoms.
FIG. 4.
Crystal structure of E. coli MurB in complex with compound 4. Compound 4 is bound within the active site of E. coli MurB. Compound 4 (shown as a ball-and-stick model, with bonds colored yellow and chlorine atoms colored pink) binds close to the FAD cofactor (shown as a ball-and-stick model colored orange) (closest distance is 3.9 Å) and the atoms of the protein side chains (shown as a ball-and-stick model, colored green for carbon, red for oxygen, and blue for nitrogen). The electron density for the side chain of Tyr190 is weak. Conformational changes compared to substrate-bound E. coli MurB (PDB code 2MBR) are localized within the active-site region.
Effects of the 3,5-dioxopyrazolidines on peptidoglycan biosynthesis in a whole-cell assay.
The effects of the 3,5-dioxopyrazolidines on peptidoglycan biosynthesis were evaluated in an sPG biosynthesis assay utilizing S. epidermidis. The assay measured the synthesis of the non-cross-linked soluble peptidoglycan, released into the growth medium. Penicillin G (100 μg/ml) was added to the cells to inhibit the PBPs, preventing cross-linking of sPG into the cell wall matrix, thereby forming insoluble peptidoglycan. Inhibition of protein synthesis by chloramphenicol (50 μg/ml) prevented sPG degradation by newly synthesized endogenous autolytic enzymes. The results (Table 4) indicated that exposure of the bacteria to the compounds resulted in reductions in soluble peptidoglycan biosynthesis. The 3,5-dioxopyrazolidines inhibited sPG synthesis with IC50s in the range of <0.39 to 11.1 μg/ml. The comparison of sPG inhibition with MICs against S. epidermidis revealed a trend for compounds with lower MICs to demonstrate lower IC50s.
TABLE 4.
Inhibitory activities of 3,5-dioxopyrazolidines on soluble peptidoglycan biosynthesis in S. epidermidis
| Compound | IC50 (μg/ml) | MIC (μg/ml) |
|---|---|---|
| 1 | 1.5 | 0.50 |
| 2 | 5.9 | 16 |
| 3 | 0.39 | 2 |
| 4 | 11.1 | 32 |
| Fosfomycin | 4.1 | 32 |
Antibacterial activities of 3,5-dioxopyrazolidines.
The antibacterial activities of the 3,5-dioxopyrazolidines are summarized in Table 5. The compounds showed activity against strains of S. aureus including MRSA, E. faecalis including vancomycin-resistant enterococci (VRE), and penicillin-resistant S. pneumoniae. The growth of these strains was inhibited at compound concentrations in the range of 0.25 to 16 μg/ml. However, these compounds showed no activity against E. coli. In order to evaluate the effect of outer membrane permeability on the antibacterial activities of the compounds, E. coli imp, an E. coli strain with improved outer membrane permeability, was employed (10, 44). Vancomycin was used in the assay to validate the permeability of the E. coli imp strain. Vancomycin, a large (1.5-kDa) antimicrobial agent, is inactive against gram-negative bacteria, because it cannot cross the outer membrane. As expected, vancomycin showed no activity against wild type E coli but was active against the E. coli imp strain, with a MIC of 0.25 μg/ml. These data suggest that the outer membrane of the E. coli imp strain is permeable to vancomycin. Interestingly, only one of the 3,5-dioxopyrazolidines, compound 1, showed activity against the E. coli imp strain, with a MIC of 16 μg/ml. Fosfomycin demonstrated activity against both E. coli strains, with slightly better activity (1 dilution) against E. coli imp.
TABLE 5.
Antimicrobial activities of 3,5-dioxopyrazolidines
| Compound | MIC (μg/ml) for:
|
||||||
|---|---|---|---|---|---|---|---|
| S. aureusa | E. faecalisb | MSCNSc GC6464 | S. pneumoniae GC1894d | S. pneumoniae GC1894 + BSAf | E. coli GC4559 | E. coli (imp)e BAS849 | |
| 1 | 1-2 | 0.25 | 0.5 | 0.50 | >128 | >128 | 16 |
| 2 | 2-16 | 0.25 | 16 | 0.25 | >128 | >128 | >128 |
| 3 | 1-2 | 0.25/0.5 | 2 | 0.25 | >128 | >128 | >128 |
| 4 | 8 | 4/8 | 4 | 4 | >128 | >128 | >128 |
| Fosfomycin | 4-8 | 64 | ND | 32 | 32 | 32 | 16 |
| Vancomycin | 0.5-2 | <0.12/>128 | 1 | <0.12 | <0.12 | >128 | 0.25 |
Values are ranges for three S. aureus strains including MRSA.
Values are ranges for two E. faecalis strains including VRE.
MSCNS, methicillin-susceptible coagulase-negative staphylococcus. ND, not determined.
Penicillin-resistant S. pneumoniae.
Strain with improved outer membrane permeability.
In the presence of 4% BSA.
Protein-binding properties of 3,5-dioxopyrazolidines.
In order to assess the protein-binding properties of the compounds, the MICs of the test compounds were determined using an S. pneumoniae strain in the presence or absence of 4% BSA in the growth medium. No antimicrobial activity for 3,5-dioxopyrazolidines was detected in the presence of 4% BSA, while vancomycin and fosfomycin MICs were not affected by the addition of BSA (Table 5).
DISCUSSION
The strategy of in vitro screening to identify small molecule inhibitors of the six cytoplasmic peptidoglycan-synthetic enzymes (MurA through MurF) has led to the identification of a number of small molecule inhibitors (50). Recently, inhibitors of MurA (22), MurB (11, 24, 31), MurC (20), MurD (27), and MurF (26) were reported. MurB is an attractive target, not only because of its critical and unique role in bacterial cell wall synthesis but also because the MurB enzymes from several bacterial species have been characterized both biochemically and structurally (3-6, 18, 45, 49).
In this study, the genes encoding MurB from both E. coli and S. aureus were cloned and expressed in E. coli, and the MurB enzymes were purified under identical conditions. A side-by-side comparison of the enzyme kinetics of E. coli MurB and S. aureus MurB indicated that the enzymes catalyzed the reduction of EP-UNAG with only minor differences. E. coli MurB showed higher affinity for EP-UNAG than S. aureus MurB, while S. aureus MurB showed slightly greater affinity for NADPH. The kinetic parameters of E. coli MurB (Km values for EP-UNAG and NADPH) were comparable to those previously reported (12). The Km of S. aureus MurB for NADPH (3.7 μM) was lower than that reported in the literature (15 μM) (6). There was a single-amino-acid difference between these two S. aureus MurB enzymes: residue 168 was a threonine in the enzyme used for the studies reported here and an alanine in the enzyme in the previous report (6).
The enzyme-inhibitory activities observed for compounds in the 3,5-dioxopyrazolidine series tested against E. coli MurB and S. aureus MurB were generally similar. The most significant difference was observed for compound 2, with IC50s of 5.3 μM against E. coli MurB and 10.3 μM against S. aureus MurB. This suggests that the structural and sequence differences between E. coli MurB and S. aureus MurB, i.e., the absence of the two loops in the S. aureus MurB structure, and amino acid variations within the active sites of the two enzymes, had a minimal effect on the susceptibilities of the enzymes to inhibition by the 3,5-dioxopyrazolidines.
To further the understanding of these compounds, biophysical experiments were conducted. Interactions of the 3,5-dioxopyrazolidines with E. coli MurB were characterized by using a fluorescence assay to determine the binding affinity and the solvent environment of the binding site, as well as by X-ray crystallography studies. MurB contains one noncovalently bound molecule of FAD, a cofactor acting as a hydride transfer mediator between NADPH and the substrate EP-UNAG. The intrinsic fluorescence properties of this flavin molecule were employed to serve as an intrinsic probe of the enzyme's active site. In the presence of the inhibitor, the FAD fluorescence is shifted from 525 nm to a lower wavelength, 520 nm, consistent with a less solvent accessible environment of the chromophore relative to the free enzyme. The estimated dissociation constant of 260 nM for compound 3 with E. coli MurB indicated that the inhibitor was tightly bound to the enzyme, with a stoichiometry consistent with 1:1 binding.
The crystal structure of compound 4 with E. coli MurB was resolved to 2.4 Å (Table 1; Fig. 4). The structure revealed a diverse series of interactions between the compound and the active site of the enzyme as well as the flavin ring system of the bound FAD. This is consistent with the results of the fluorescence assay. The changes in protein conformation to accommodate binding of the compound were small in comparison to those seen when the EP-UNAG substrate is bound (6). The location of the compound in the active site would preclude binding of the substrate and provides a clear mechanism for the inhibition. In considering the differences between S. aureus MurB and E. coli MurB, a superposition of crystal structures indicated that the important interactions of compound 4 with the FAD flavin ring system and Tyr125 are conserved. The side chain of Leu218 bifurcates the two chlorophenyl groups of compound 4 and appears to provide a structural platform resulting in the compound's proximity to FAD and its hydrogen bond to the main chain of Tyr125. A comparison of published crystal structures revealed an equivalent side chain residue in S. aureus MurB, Gln229. This observation, combined with significant structural differences in the loop from β13 to α3 (6), implies that a different set of molecular interactions would exist between the 3,5-dioxopyrazolidines and gram-negative versus gram-positive MurB enzymes.
To understand the effects of the 3,5-dioxopyrazolidines on peptidoglycan biosynthesis, the inhibitory activities of the compounds were evaluated in an sPG biosynthesis assay utilizing S. epidermidis. Reduced production of sPG following exposure of bacteria to a compound suggests that inhibition of a peptidoglycan-synthetic enzyme (MurB by compounds 1 and 3, MurA and MurB by compound 2, and MurB and MurC by compound 4) affected peptidoglycan biosynthesis, leading to the inhibition of cell growth.
As shown in Table 2, compounds 1 to 4 were all inhibitors of MurB. In addition, several of these compounds also demonstrated inhibitory activity against MurA and/or MurC. This is not unexpected, since the product of MurA is the substrate for MurB and the product of MurB is the substrate for MurC. It is therefore reasonable that an inhibitor of MurB could fit into the active site of MurA and/or MurC and function as an inhibitor of these enzymes as well. The observation that the compounds are not inhibitors of the later enzymes in the pathway, MurD through MurF, fits with the need for these enzymes to recognize the growing amino acid side chain in order to ensure the addition of the amino acids in the correct sequence. Thus, an inhibitor of MurB may not effectively fit the active site of MurD, MurE, or MurF, making insufficient critical contacts to provide the affinity that would enable it to function as an inhibitor of any of these three enzymes.
The MICs shown in Table 5 indicate good antibacterial activities for the 3,5-dioxopyrazolidines tested against strains of S. aureus including MRSA, E. faecalis strains including VRE, and penicillin-resistant S. pneumoniae. The compounds tested showed no activity against wild-type E. coli. Only compound 1 demonstrated antimicrobial activity against E. coli imp, a strain with improved outer membrane permeability. The failure of the compounds to inhibit the E. coli imp strain was unexpected, given that this strain is susceptible to vancomycin (MIC, 0.25 μg/ml), a large antibiotic with no activity against wild-type E. coli. We do not have an explanation for this observation at this time. We speculate that it may be due either to the compounds' physicochemical properties, preventing penetration of the gram-negative cell membranes, or to binding to components specific to gram-negative bacterial cells, for example, lipopolysaccharides.
The decline in activity for all compounds against S. pneumoniae from a MIC in the range of 0.5 to 4 μg/ml (without BSA) to >128 μg/ml in the presence of 4% BSA is consistent with high protein-binding properties of these compounds. In contrast, the presence of BSA did not change the MIC of vancomycin or fosfomycin. The effects of serum and serum albumin on the MICs of a wide variety of antibiotics have been extensively studied (37). A close correlation was demonstrated between the amount of free, unbound compound and a compound's antibacterial activity, suggesting that protein-bound antibiotics are not available to interact with their bacterial target and therefore have reduced activity. Our data indicate that this is likely the case for the 3,5-dioxopyrazolidines in the presence of 4% BSA. This observation prevents further testing of these compounds for efficacy in animal models. Similar data—high albumin binding—were reported earlier for a MurC inhibitor (20) and MurB inhibitors such as 2-phenyl-5,6-dihydro-2H-thieno[3,2-c]pyrazol-3-ols (33), phenyl thiazolyl ureas, carbamate derivatives (24), and 4-alkyl and 4,4-dialkyl 1,2-bis(4-chlorophenyl)pyrazolidine-3,5-dione derivatives (31).
In summary, the 3,5-dioxopyrazolidines studied are potent inhibitors targeting MurB. This class of new inhibitors establishes a strong relationship between enzyme inhibition, decreased soluble peptidoglycan synthesis, and antibacterial activity. These findings are encouraging, because they indicate the possibility of developing novel antibacterial agents targeting an early step in peptidoglycan biosynthesis.
Acknowledgments
We thank Craig Caufield, George Ellestad, and John Primeau for useful suggestions and discussions.
REFERENCES
- 1.Anderson, J. S., M. Matsuhashi, M. A. Haskin, and J. L. Strominger. 1967. Biosynthesis of the peptidoglycan of bacterial cell walls. II. Phospholipid carriers in the reaction sequence. J. Biol. Chem. 242:3180-3190. [PubMed] [Google Scholar]
- 2.Andres, C. J., J. J. Bronson, S. V. D'Andrea, M. S. Deshpande, P. J. Falk, K. A. Grant-Young, W. E. Harte, H. T. Ho, P. F. Misco, J. G. Robertson, D. Stock, Y. Sun, and A. W. Walsh. 2000. 4-Thiazolidinones: novel inhibitors of the bacterial enzyme MurB. Bioorg. Med. Chem. Lett. 10:715-717. [DOI] [PubMed] [Google Scholar]
- 3.Benson, T. E., C. T. Walsh, and J. M. Hogle. 1997. X-ray crystal structures of the S229A mutant and wild-type MurB in the presence of the substrate enolpyruvyl-UDP-N-acetylglucosamine at 1.8-Å resolution. Biochemistry 36:806-811. [DOI] [PubMed] [Google Scholar]
- 4.Benson, T. E., C. T. Walsh, and V. Massey. 1997. Kinetic characterization of wild-type and S229A mutant MurB: evidence for the role of Ser 229 as a general acid. Biochemistry 36:796-805. [DOI] [PubMed] [Google Scholar]
- 5.Benson, T. E., J. L. Marquardt, A. C. Marquardt, F. A. Etzkorn, and C. T. Walsh. 1993. Overexpression, purification, and mechanistic study of UDP-N-acetylenolpyruvylglucosamine reductase. Biochemistry 32:2024-2030. [DOI] [PubMed] [Google Scholar]
- 6.Benson, T. E., M. S. Harris, G. H. Choi, J. I. Cialdella, J. T. Herberg, J. P. Martin, Jr., and E. T. Baldwin. 2001. A structural variation for MurB: X-ray crystal structure of Staphylococcus aureus UDP-N-acetylenolpyruvylglucosamine reductase (MurB). Biochemistry 40:2340-2350. [DOI] [PubMed] [Google Scholar]
- 7.Bertrand, J. A., G. Auger, E. Fanchon, L. Martin, D. Blanot, J. van Heijenoort, and O. Dideberg. 1997. Crystal structure of UDP-N-acetylmuramoyl-l-alanine:d-glutamate ligase from Escherichia coli. EMBO J. 16:3416-3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boothby, D., L. Daneo-Moore, and G. D. Shockman. 1971. A rapid, quantitative, and selective estimation of radioactively labeled peptidoglycan in gram-positive bacteria. Anal. Biochem. 44:645-653. [DOI] [PubMed] [Google Scholar]
- 9.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 10.Braun, M., and T. J. Silhavy. 2002. Imp/OstA is required for cell envelope biogenesis in Escherichia coli. Mol. Microbiol. 45:1289-1302. [DOI] [PubMed] [Google Scholar]
- 11.Bronson, J. J., K. L. DenBleyker, P. J. Falk, R. A. Mate, H.-T. Tso, M. J. Pucci, and B. Snyder. 2003. Discovery of the first antibacterial small molecule inhibitors of MurB. Bioorg. Med. Chem. Lett. 13:873-875. [DOI] [PubMed] [Google Scholar]
- 12.Brown, E. D., J. L. Marquardt, J. P. Lee, C. T. Walsh, and K. S. Anderson. 1994. Detection and characterization of a phospholactoyl-enzyme adduct in the reaction catalyzed by UDP-N-acetylglucosamine enolpyruvoyl transferase, MurZ. Biochemistry 33:10638-10645. [DOI] [PubMed] [Google Scholar]
- 13.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 14.Bugg, T. D. H., and C. T. Walsh. 1992. Intracellular steps of bacterial cell wall peptidoglycan biosynthesis: enzymology, antibiotics, and antibiotic resistance. Nat. Prod. Rep. 9:199-215. [DOI] [PubMed] [Google Scholar]
- 15.CCP4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 16.Centers for Disease Control and Prevention. 2002. Vancomycin-resistant Staphylococcus aureus—Pennsylvania, 2002. Morb. Mortal. Wkly. Rep. 51:902. [PubMed] [Google Scholar]
- 17.Chang, S., D. M. Sievert, J. C. Hageman, M. L. Boulton, F. C. Tenover, F. P. Downes, S. Shah, J. T. Rudrik, G. R. Pupp, W. J. Brown, D. Cardo, S. K. Fridkin, and the Vancomycin-Resistant Staphylococcus aureus Investigative Team. 2003. Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N. Engl. J. Med. 348:1342-1347. [DOI] [PubMed] [Google Scholar]
- 18.Dhalla, A. M., J. Yanchunas, H.-T. Ho, P. J. Falk, J. J. Villafranca, and J. G. Robertson. 1995. Steady-state kinetic mechanism of Escherichia coli UDP-N-acetylenolpyruvylglucosamine reductase. Biochemistry 34:5390-5402. [DOI] [PubMed] [Google Scholar]
- 19.Duncan, K., J. van Heijenoort, and C. T. Walsh. 1990. Purification and characterization of the d-alanyl-d-alanine-adding enzyme from Escherichia coli. Biochemistry 29:2379-2386. [DOI] [PubMed] [Google Scholar]
- 20.Ehmann, D. E., J. E. Demeritt, K. G. Hull, and S. L. Fisher. 2004. Biochemical characterization of an inhibitor of Escherichia coli UDP-N-acetylmuramyl-l-alanine ligase. Biochim. Biophys. Acta 1698:167-174. [DOI] [PubMed] [Google Scholar]
- 21.Emanuele, J. J., Jr., H. Jin, B. L. Jacobson, C. Y. Chang, H. M. Einspahr, and J. J. Villafranca. 1996. Kinetic and crystallographic studies of Escherichia coli UDP-N-acetylmuramate:l-alanine ligase. Protein Sci. 5:2566-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eschenburg, S., M. A. Priestman, F. A. Abdul-Latif, C. Delachaume, F. Fassy, and E. Schönbrunn. 2005. A novel inhibitor that suspends the induced-fit mechanism of UDP-N-acetylglucosamine enolpyruvyl transferase (MurA). J. Biol. Chem. 280:14070-14075. [DOI] [PubMed] [Google Scholar]
- 23.Flouret, B., D. Mengin-Lecreulx, and J. van Heijenoort. 1981. Reverse-phase high-pressure liquid chromatography of uridine diphosphate N-acetylmuramyl peptide precursors of bacterial cell wall peptidoglycan. Anal. Biochem. 114:59-63. [DOI] [PubMed] [Google Scholar]
- 24.Francisco, G. D., Z. Li, J. D. Albright, N. H. Eudy, A. H. Katz, P. J. Petersen, P. Labthavikul, G. Singh, Y. Yang, B. A. Rasmussen, Y. I. Lin, and T. S. Mansour. 2004. Phenyl thiazolyl urea and carbamate derivatives as new inhibitors of bacterial cell-wall biosynthesis. Bioorg. Med. Chem. Lett. 14:235-238. [DOI] [PubMed] [Google Scholar]
- 25.González-Zorn, B., and P. Courvalin. 2003. vanA-mediated high level glycopeptide resistance in MRSA. Lancet Infect. Dis. 3:67-68. [DOI] [PubMed] [Google Scholar]
- 26.Gu, Y. G., A. S. Florjancic, R. F. Clark, T. Zhang, C. S. Cooper, D. D. Anderson, C. G. Lerner, J. O. McCall, Y. Cai, C. L. Black-Schaefer, G. F. Stamper, P. J. Hajduk, and B. A. Beutel. 2004. Structure-activity relationships of novel potent MurF inhibitors. Bioorg. Med. Chem. Lett. 14:267-270. [DOI] [PubMed] [Google Scholar]
- 27.Horton, J. R., J. M. Bostock, I. Chopra, L. Hesse, S. E. V. Phillips, D. J. Adams, A. P. Johnson, and C. W. G. Fishwick. 2003. Macrocyclic inhibitors of the bacterial cell wall biosynthesis enzyme MurD. Bioorg. Med. Chem. Lett. 13:1557-1560. [DOI] [PubMed] [Google Scholar]
- 28.Jamieson, E. R., M. P. Jacobson, C. M. Barnes, C. S. Chow, and S. J. Lippard. 1999. Structural and kinetic studies of a cisplatin-modified DNA icosamer binding to HMG1 domain B. J. Biol. Chem. 274:12346-12354. [DOI] [PubMed] [Google Scholar]
- 29.Johnson, A. R. 1971. Improved method of hexosamine determination. Anal. Biochem. 44:628-635. [DOI] [PubMed] [Google Scholar]
- 30.Katz, A. H., and C. E. Caufield. 2003. Structure-based design approaches to cell wall biosynthesis inhibitors. Curr. Pharm. Des. 9:857-866. [DOI] [PubMed] [Google Scholar]
- 31.Kutterer, K. M. K., J. M. Davis, G. Singh, Y. Yang, W. Hu, A. Severin, B. A. Rasmussen, G. Krishnamurthy, A. Failli, and A. H. Katz. 2005. 4-Alkyl and 4,4′-dialkyl 1,2-bis(chlorophenyl)pyrazolidine-3,5-dione derivatives as new inhibitors of bacterial cell wall biosynthesis. Bioorg. Med. Chem. Lett. 15:2527-2531. [DOI] [PubMed] [Google Scholar]
- 32.Lanzetta, P. A., J. L. Alvarez, P. S. Reinach, and O. A. Candia. 1979. An improved assay for nanomole amounts of inorganic phosphate. Anal. Biochem. 100:95-97. [DOI] [PubMed] [Google Scholar]
- 33.Li, Z., G. D. Francisco, W. Hu, P. Labthavikul, P. J. Petersen, A. Severin, G. Singh, Y. Yang, B. A. Rasmussen, Y. I. Lin, J. S. Skotnicki, and T. S. Mansour. 2003. 2-Phenyl-5,6-dihydro-2H-thieno[3,2-c]pyrazol-3-ol derivatives as new inhibitors of bacterial cell wall biosynthesis. Bioorg. Med. Chem. Lett. 13:2591-2594. [DOI] [PubMed] [Google Scholar]
- 34.Liger, D., A. Masson, D. Blanot, J. van Heijenoort, and C. Parquet. 1996. Study of the overproduced uridine-diphosphate-N-acetylmuramate:l-alanine ligase from Escherichia coli. Microb. Drug Resist. 2:25-27. [DOI] [PubMed] [Google Scholar]
- 35.Marquardt, J. L., D. A. Siegele, R. Kolter, and C. T. Walsh. 1992. Cloning and sequencing of Escherichia coli murZ and purification of its product, a UDP-N-acetylglucosamine enolpyruvyl transferase. J. Bacteriol. 174:5748-5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mengin-Lecreulx, D., B. Flouret, and J. van Heijenoort. 1982. Cytoplasmic steps of peptidoglycan synthesis in Escherichia coli. J. Bacteriol. 151:1109-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merrikin, D. J., J. Briant, and G. N. Rolinson. 1983. Effect of protein binding on antibiotic activity in vivo. J. Antimicrob. Chemother. 11:233-238. [DOI] [PubMed] [Google Scholar]
- 38.Michaus, C., D. Mengin-Lecreulx, J. van Heijenoort, and D. Blanot. 1990. Overproduction, purification and properties of the UDP-N-acetylmuramoyl-l-alanyl-d-glutamate:meso-diaminopimelic acid ligase from Escherichia coli. Eur. J. Biochem. 194:853-861. [DOI] [PubMed] [Google Scholar]
- 39.Mueller, N., P. Andrews, and W. Stendel. 27May1987. Preparation of pyrazolidinediones as endo- and exoparasiticides. German patent application DE 3540934 A1.
- 40.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically: Approved standard M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 41.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 42.Park, J. T. 1966. Membrane associated reaction involved in bacterial cell wall muropeptide synthesis. Methods Enzymol. 8:466-472. [Google Scholar]
- 43.Pesin, V. G., A. M. Khaletskii, and Z.-S. Den. 1958. Chemistry of pyrazolidine. II. Halogenation and thiocyanation of 1,2-diphenyl-3,5-dioxopyrazolidine and its derivatives. Zh. Obshch. Khim. 28:2816-2820. [Google Scholar]
- 44.Sampson, B. A., R. Misra, and S. A. Benson. 1989. Identification and characterization of a new gene of Escherichia coli K-12 involved in outer membrane permeability. Genetics 122:491-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarver, R. W., J. M. Rogers, and D. E. Epps. 2002. Determination of ligand-MurB interactions by isothermal denaturation: application as a secondary assay to complement high throughput screening. J. Biomol. Screen. 7:21-28. [DOI] [PubMed] [Google Scholar]
- 46.Schito, G. C. 2003. Why fosfomycin trometamol is first line therapy for uncomplicated UTI? Int. J. Antimicrob. Agents 22(Suppl. 2):S79-S83. [DOI] [PubMed] [Google Scholar]
- 47.Shine, H. J., and E. S. Rhee. 1984. The preparation of 4,4′-dichloroazobenzene-15N,15N′ and -4,4′-13C2 and 4-chloroazobenzene-15N,15N′. J. Label. Comp. Radiopharm. 21:569-573. [Google Scholar]
- 48.Shine, H. J., L. Kupczyk-Subotkowska, and W. Subotkowski. 1985. Heavy-atom kinetic isotope effects in the acid-catalyzed rearrangement of N-2-naphthyl-N′-phenylhydrazine. Rearrangement is shown to be a concerted process. J. Am. Chem. Soc. 107:6674-6678. [Google Scholar]
- 49.Sylvester, D. R., E. Alvarez, A. Patel, K. Ratnam, H. Kallender, and N. G. Wallis. 2001. Identification and characterization of UDP-N-acetylenolpyruvylglucosamine reductase (MurB) from the gram-positive pathogen Streptococcus pneumoniae. Biochem. J. 355:431-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zoeiby, A. E., M. Beaumont, E. Dubuc, F. Sanschagrin, N. Voyer, and R. C. Levesque. 2003. Combinatorial enzymatic assay for the screening of a new class of bacterial cell wall inhibitors. Bioorg. Med. Chem. 11:1583-1592. [DOI] [PubMed] [Google Scholar]