Abstract
Gentamicin continues to be a primary antibiotic against gram-negative infections. Unfortunately, associated nephro- and ototoxicity limit its use. Our previous mammalian studies showed that gentamicin is trafficked to the endoplasmic reticulum in a retrograde manner and subsequently released into the cytosol. To better dissect the mechanism through which gentamicin induces toxicity, we have chosen to study its toxicity using the simple eukaryote Saccharomyces cerevisiae. A recent screen of the yeast deletion library identified multiple gentamicin-sensitive strains, many of which participate in intracellular trafficking. Our approach was to evaluate gentamicin sensitivity under logarithmic growth conditions. By quantifying growth inhibition in the presence of gentamicin, we determined that several of the sensitive strains were part of the Golgi-associated retrograde protein (GARP) and homotypic fusion and vacuole protein sorting (HOPS) complexes. Further evaluation of their other components showed that the deletion of any GARP member resulted in gentamicin-hypersensitive strains, while the deletion of other HOPS members resulted in less gentamicin sensitivity. Other genes whose deletion resulted in gentamicin hypersensitivity included ZUO1, SAC1, and NHX1. Finally, we utilized a Texas Red gentamicin conjugate to characterize gentamicin uptake and localization in both gentamicin-sensitive and -insensitive strains. These studies were consistent with our mammalian studies, suggesting that gentamicin toxicity in yeast results from alterations to intracellular trafficking pathways. The identification of genes whose absence results in gentamicin toxicity will help target specific pathways and mechanisms that contribute to gentamicin toxicity.
Gentamicin is one of the primary aminoglycoside antibiotics used in the treatment of serious gram-negative infections. While gentamicin is highly efficacious, its use is limited because of associated nephro- and ototoxicity. In fact, aminoglycosides are a leading cause of acute renal failure in hospitalized patients (18, 34), with certain high-risk groups of patients experiencing an incidence rate of acute renal failure near 50% when treated (36). Thus, a better understanding of the causes of this toxicity may lead to therapeutic treatments that reduce or prevent renal toxicity while maintaining the aminoglycoside antibacterial activity. Since there are no new antibiotics for gram-negative bacteria in development, it is especially important that efforts be made to minimize the toxicity of existing gram-negative antibiotics (19, 28).
The use of model systems is being highly exploited for the analysis of complex biological problems. In particular, the baker's yeast Saccharomyces cerevisiae has proved to be a sophisticated model for such analysis. Indeed, although yeasts are relatively resistant to the toxic effects of antibiotics, Blackburn and Avery utilized a yeast strain library containing a deletion of each nonessential yeast gene individually within a single genetic background to screen for mutants that were hypersensitive to gentamicin (3). Their analysis uncovered 17 mutants that are more sensitive to gentamicin than the wild-type (WT) parent strain when grown on yeast extract-peptone-dextrose medium containing gentamicin.
Our work has concentrated on the intracellular trafficking pathways involved in cell toxicity in proximal tubule cells, the primary site of nephrotoxicity (30, 31). Following glomerular filtration, aminoglycosides rapidly bind to the apical membrane of proximal tubule cells and are endocytosed. Our data show that (32) a small percentage of the endocytosed gentamicin traffics via an endocytic retrograde pathway through the Golgi complex and into the endoplasmic reticulum (ER), where it is subsequently released into the cytosol. It is the cytosolic gentamicin that appears to trigger the associated intracellular pathologies (32).
To further delineate the intracellular mechanisms of aminoglycoside toxicity, we set out to utilize the yeast Saccharomyces cerevisiae as a model system to identify the intracellular pathways involved. We report here that deletions of the genes encoding the four known components of the Golgi-associated retrograde protein (GARP) complex all cause sensitivity to gentamicin. Furthermore, the absence of the GARP-associated proteins Vps45, Ypt6, and Tlg2 results in gentamicin sensitivity. Thus, the elimination of multiple proteins involved in vesicle targeting and fusion in addition to the nascent chain-associated complex (NAC) containing Zuo1, Ssb1/2, and Ssz1 results in gentamicin sensitivity. We have also utilized our Texas Red (TR)-gentamicin conjugate to begin characterizing its cellular transport in both wild-type and gentamicin-hypersensitive yeast strains.
MATERIALS AND METHODS
Evaluation of strains for gentamicin sensitivity, logarithmic growth assay.
The strains to be evaluated (Table 1) were inoculated from frozen stocks onto YPD (1% yeast extract, 2% peptone, 2% dextrose) plates. A freshly prepared overnight liquid culture in YPD at 30°C was diluted to 2.5 × 106 cells/ml, and eight 1-ml cultures were started, four untreated and four treated. The tubes were incubated continuously at 30°C for 4 to 5 h. At that time, the treated tubes received gentamicin and all tubes were incubated at 30°C for an additional 24 to 30 h. The growth of each sample was determined by measuring the optical density at 660 nm following sonication and dilution to enable measurement in the linear range. The percent inhibition of all treated samples was calculated by comparison to the same strain grown in the absence of gentamicin. Each strain was evaluated a minimum of eight times over 48 h, with the hypersensitive strains being measured up to 24 times. Data was analyzed using KaleidaGraph's (Synergy Software, Reading, PA) box plot. Each box encloses 50% of the data, with the median value displayed as the line in the box. The top and bottom of the box mark the limits of ±25% of the variable population. The lines extending from the top and bottom of each box mark the minimum and maximum values within the data set that fall within an acceptable range. Any value outside of this range is displayed as an individual point.
TABLE 1.
Strains evaluated in this study
| Strain | Genotype | Source |
|---|---|---|
| 4666 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 cax4Δ::KanMX4 | Open Biosystems |
| 4572 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 chc1Δ::KanMX4 | Open Biosystems |
| 2020 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 chs1Δ::KanMX4 | Open Biosystems |
| 3924 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 gcs1Δ::KanMX4 | Open Biosystems |
| 2778 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 mnn9Δ::KanMX4 | Open Biosystems |
| 4290 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 nhx1Δ::KanMX4 | Open Biosystems |
| 4105 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 pep3Δ::KanMX4 | Open Biosystems |
| 817 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 pep5Δ::KanMX4 | Open Biosystems |
| 3059 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 rib1Δ::KanMX4 | Open Biosystems |
| 4073 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 rub1Δ::KanMX4 | Open Biosystems |
| 5062 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 sac1Δ::KanMX4 | Open Biosystems |
| 4357 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 sps1Δ::KanMX4 | Open Biosystems |
| 6414 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ssz1Δ::KanMX4 | Open Biosystems |
| 1709 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 tlg2Δ::KanMX4 | Open Biosystems |
| 3774 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 vam6Δ::KanMX4 | Open Biosystems |
| 3236 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 vps15Δ::KanMX4 | Open Biosystems |
| 2783 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 vps16Δ::KanMX4 | Open Biosystems |
| 5305 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 vps33Δ::KanMX4 | Open Biosystems |
| 5149 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 vps34Δ::KanMX4 | Open Biosystems |
| 4015 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 vps41Δ::KanMX4 | Open Biosystems |
| 4462 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 vps45Δ::KanMX4 | Open Biosystems |
| 5091 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 vps51Δ::KanMX4 | Open Biosystems |
| 4318 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 vps52Δ::KanMX4 | Open Biosystems |
| FR4107 | MATα his3Δ1 leu2Δ0 ura3Δ lys2Δ0 vps53Δ::KanMX4 | Daniel Klionsky |
| 3966 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 vps54Δ::KanMX4 | Open Biosystems |
| 405 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 vps8Δ::KanMX4 | Open Biosystems |
| 4289 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ydr455cΔ::KanMX4 | Open Biosystems |
| 5171 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ypt6Δ::KanMX4 | Open Biosystems |
| 5937 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 zuo1Δ::KanMX4 | Open Biosystems |
Microscopy analysis of TR-gentamicin uptake.
TR was conjugated to gentamicin as previously described (32). Yeasts were incubated with TR-gentamicin or lucifer yellow (LY) for the specified period of time under the same growth conditions as those used for the growth assay. LY accumulates in the yeast vacuole via fluid-phase endocytosis (22). An aliquot of the yeast was removed and washed one time using fresh YPD medium and a 15-s spin in a microfuge. The yeasts were resuspended in 100 to 200 μl of YPD and placed onto a MatTek 35-mm coverslip dish that had been previously coated with concanavalin A. Yeasts were allowed to attach for 15 min and gently washed with YPD, and a 1-mm-thick 25% gelatin square was placed on top. Images were collected within 1 h by using a Zeiss UV LSM-510 confocal microscope system. TR-gentamicin was excited with the 543-nm line of the helium-neon laser (long-pass filter wavelength, 560 nm), and LY was excited with the 488-nm line of the argon laser (band-pass filter wavelength, 505 to 530 nm). A differential interference contrast (DIC) image was also collected using the 488-nm laser line. Image stacks were collected with a step size of 0.1 μm for three-dimensional reconstructions using Voxx software (8).
The staining of yeasts for the viability assay using methylene blue was performed according to established methods (38). Briefly, yeasts were incubated 1:1 with 0.01% methylene blue in 2% Na citrate solution. Viability was evaluated by counting a minimum of 300 cells on 4 different days for each strain.
Flow cytometry.
Yeasts in the logarithmic-growth phase were incubated for 5 h in the presence of 100 μg/ml Oregon Green (OG)-gentamicin. Yeasts were then pelleted and washed two times with YPD and resuspended into phosphate-buffered saline for propidium iodide (PI) staining and subsequent flow cytometry. A BD FACSCalibur was used, and 50,000 cells of each strain were analyzed for fluorescence using a 488-nm excitation wavelength and a 515-to-545-nm emission wavelength for OG-gentamicin and a 488-nm excitation wavelength and a 564-to-606-nm emission wavelength for PI. In data not shown, OG-gentamicin reacted with yeast in a manner indistinguishable from that of TR-gentamicin.
RESULTS
Evaluation of gentamicin-sensitive mutants under logarithmic-growth-phase conditions.
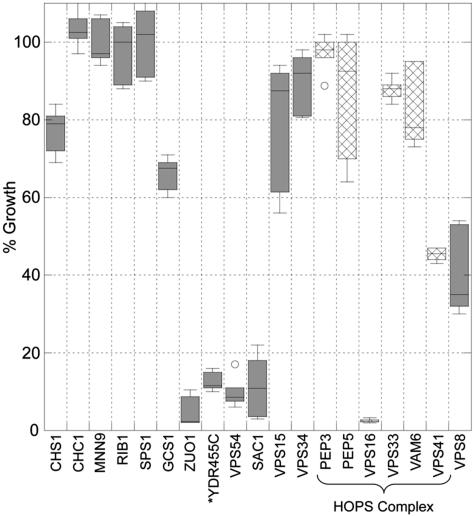
Our initial studies examined the effect on growth rate of a high concentration of gentamicin (500 μg/ml) added to both wild-type and mutant strains in the early logarithmic-growth phase. Figure 1 shows the percent growth of each respective strain in the presence of gentamicin compared to that for an additional culture of the same strain grown simultaneously in the absence of gentamicin. The strains that were previously identified to be most inhibited by gentamicin contained deletions of YDR455c, VPS16, SAC1, VPS54, and ZUO1. Each of these five strains is inhibited by over 80% and often by more than 90%. However, to our surprise, under these conditions, many of the strains that were previously identified as sensitive deletion strains (3) (Table 1; deleted for CHS1, CHC1, MNN9, RIB1, SPS1, PEP3, PEP5, VPS33, VPS15, OR VPS34) have, at most, 20% inhibition in the presence of 500 μg/ml gentamicin. Loss of Gcs1 resulted in about 40% inhibition, while loss of Cax4 (data not shown) is more variable but resulted in <20% inhibition with 100 μg/ml gentamicin.
FIG. 1.
Gentamicin addition to logarithmic-growth-phase cultures inhibits growth differentially. An overnight culture was used to start eight cultures of each strain at 2.5 × 106 cells/ml in YPD. After a 30°C incubation for 5 h, four cultures received 500 μg/ml gentamicin. Cultures were incubated for an additional 24 to 30 h. Cultures were then sonicated and diluted, and their optical densities at 660 nm were measured. All data were collected from a minimum of two overnight cultures and eight gentamicin-treated cultures. The data were graphed using a box plot in KaleidaGraph. The box itself contains the middle 50% of the data, with the vertical lines indicating the minimum and maximum data values. Any outliers are displayed as individual points. Note that Vps16 is the only HOPS member (crosshatches) with significant growth inhibition. YDR455c likely results in deletion of NHX1 which has similar gentamicin sensitivity in our assays. Error bars, one standard deviation.
Within the previously identified mutants were cells lacking components of several protein complexes, including the NAC, the HOPS (homotypic fusion and vacuole protein sorting) complex, and the GARP complex (13, 40). Therefore, we postulated that mutants lacking other components of these complexes would exhibit similar sensitivities to gentamicin. To test this hypothesis, we determined the gentamicin sensitivity of cells with mutations for other components of these complexes. Zuo1 is part of the NAC complex, and in agreement with Craig's laboratory, we found that a strain lacking an associated protein, Ssz1, is also inhibited by over 90% (14). Another protein complex that was implicated by the previous data was the HOPS complex, whose members include Pep3, Pep5, Vam6, Vps16, Vps33, and Vps41 (40). This complex has been localized to specific domains on the vacuole where fusion occurs (41). Specificity of fusion for both target membrane and vesicle depend also on the interacting t- and v-SNARE proteins. Evaluation of the HOPS members and Vps8, which coprecipitates with Pep3 and Vps16, shows only a mutant lacking Vps16 to be strongly inhibited by gentamicin, while mutants lacking Vps41 and Vps8 had their growth inhibited by approximately 50% in the presence of gentamicin. The deletion of YDR455c disrupts the coding region for NHX1, which encodes a Na+/H+ exchanger that was recently implicated in regulating vesicle trafficking out of the late endosome compartment (4). SAC1 encodes a lipid phosphoinositide phosphatase which may function along with the phosphatidylinositol 4-kinase, Stt4, to regulate which membranes contain PI4P, which in turn contributes to endocytic and vacuolar trafficking (37).
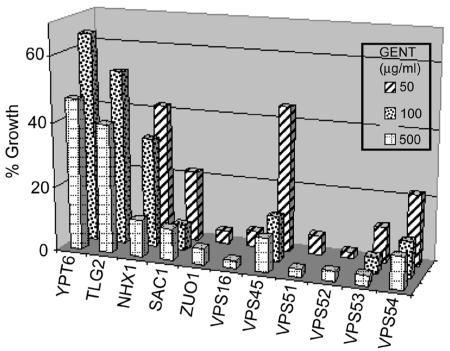
In Fig. 2, an evaluation of the most sensitive strains at several concentrations is shown. Mutants lacking Nhx1 show sensitivities similar to those lacking Ydr455c at 500 μg/ml. This supports the conclusion that the YDR455c deletion actually disrupts NHX1. Loss of Nhx1 causes significantly less inhibition when the gentamicin concentration is reduced to 100 or 50 μg/ml. In contrast, mutants lacking Sac1, Zuo1, and Vps16 are inhibited strongly by a concentration of 100 μg/ml, shown for the Sac1 mutant, while a concentration of 50 μg/ml inhibited only mutants lacking Zuo1 and Vps16 strongly. Vps54 is a member of the GARP complex and forms a 1:1:1 complex with Vps52 and Vps53 within the GARP complex (9). The GARP complex is another membrane-tethering complex that participates in membrane fusion, which in this case, involves endosomes arriving at the trans-Golgi network (40). Mutants deleted for VPS52, VPS53, or VPS54 are inhibited strongly by gentamicin. This is in agreement with data from Conibear and Stevens showing that, if one of these proteins is missing, the other two proteins are unstable (10). Vps51 assembles with Vps52, Vps53, and Vps54 and also binds the t-SNARE protein Tlg1, which is in close proximity to the t-SNARE protein Tlg2. Another GARP-associated factor, the Rab-GTPase Ypt6, resides on the vesicle and binds to the GARP complex when GTP is bound. Vps45 is a member of the Sec1p/Munc18 (SM) family whose members have essential roles in regulating multiple membrane transport pathways (39). The absence of Vps45 results in strong gentamicin sensitivity, while TLG2 and YPT6 deletion strains are inhibited approximately 50%.
FIG. 2.
Effect of lower gentamicin (GENT) concentrations on growth in sensitive strains and examination of other GARP members and their associated proteins. The growth assay was performed as for Fig. 1. The mean for each concentration and strain is presented. Note the hypersensitivity of cell mutants for all GARP members and ZUO1 and VPS16.
Comparison of TR-gentamicin and lucifer yellow uptake in mutant strains.
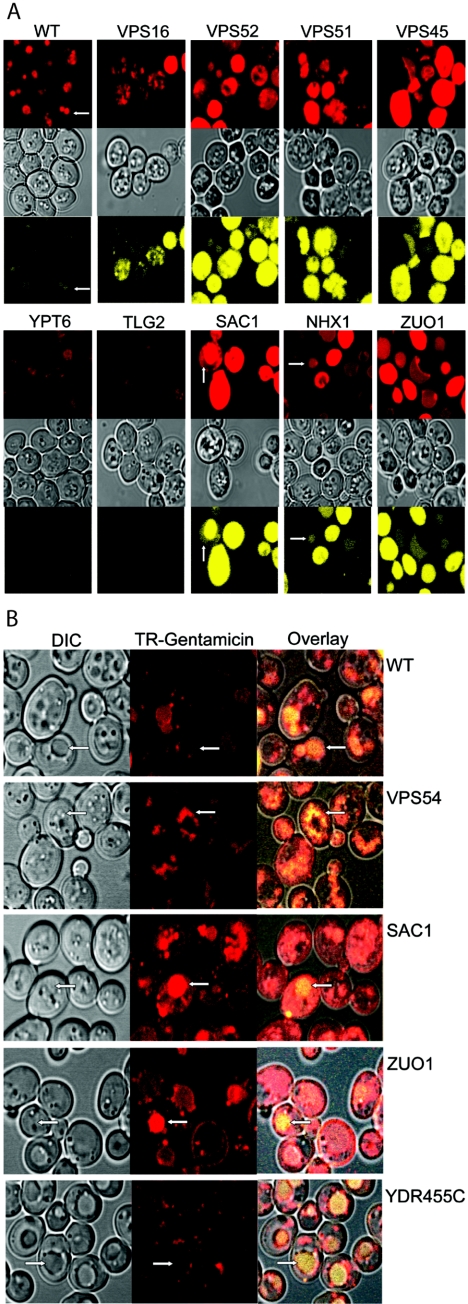
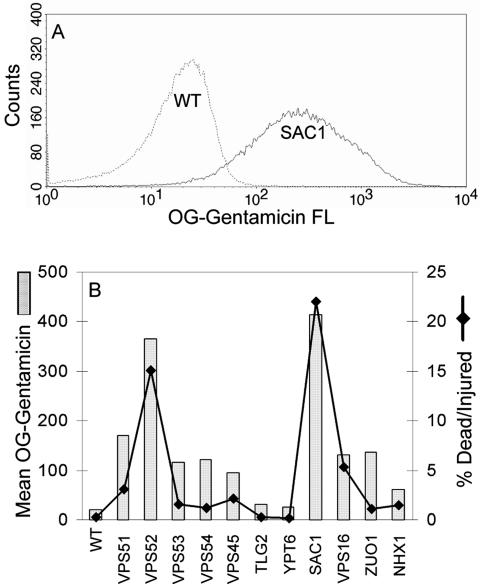
The mechanism by which gentamicin induces toxicity is likely to be related to the cellular components it encounters. We have previously characterized and used TR-gentamicin to follow the uptake of gentamicin and retrograde trafficking in LLCPK cells (29-32). To compare the uptake of TR-gentamicin in the different deletion strains, we added 500 μg/ml TR-gentamicin and 1 mg/ml LY (vacuole marker) to logarithmically growing cultures. After 12 to 15 h, each strain was imaged by confocal microscopy. To permit an accurate comparison of uptake between strains, all images in Fig. 3A were collected using the same laser power and gain. For each strain, three images were acquired simultaneously to show the location of TR-gentamicin and LY as well as the morphology of the cells by DIC. Under these conditions, it was clear that the most sensitive gentamicin strains contained more TR-gentamicin and more LY than did less sensitive strains. This result was observed for all strains, including the less sensitive strains not shown in Fig. 3A. Note also that, in most cases, the TR-gentamicin appears to colocalize with the LY in the vacuole (arrows). Figure 3B shows that 2-h incubation of cells with TR-gentamicin and LY also results in their colocalization in most mutants examined. Many of the Vps mutants (lacking Vps16, Vps52, Vps53, or Vps54) are known to have altered vacuolar morphology which is quite evident in cells lacking Vps16 (Fig. 3A) or Vps54 (Fig. 3B). Note also the distinct punctate staining in many mutant strains, which likely corresponds to a vesicular intermediate and/or dispersed vacuolar component. In addition, cells with SAC1 and ZUO1 mutations have clear membrane labeling in addition to vacuolar staining with TR-gentamicin. Interestingly, at this early time point, cells deleted for YDR455c appear to have less TR-gentamicin concentrated in the vacuole, with more present at the vacuolar membrane, often at punctate sites. To quantify gentamicin uptake, flow cytometry was used. Yeasts in logarithmic-growth phase were incubated with 100 μg/ml OG-gentamicin for 5 h. In Fig. 4A, the raw data from the WT and SAC1 deletion strain are presented. Note the significant shift to increased OG-gentamicin uptake for the SAC1 deletion strain. In Fig. 4B, the mean value of OG-gentamicin is plotted for each strain analyzed along with the percent dead/injured values calculated from PI staining. Note the strong linear correlation (R value 0.95, r2 = 0.90) between increased gentamicin uptake and death/injury.
FIG.3.
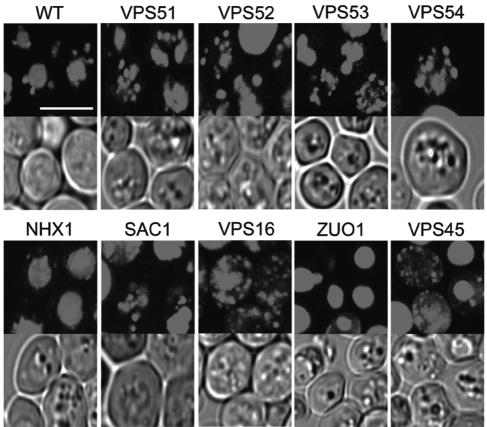
(A) Differential uptake of TR-gentamicin and lucifer yellow in cells with the indicated genes mutated. A total of 500 μg/ml TR-gentamicin and 1 mg/ml lucifer yellow was added to cultures in the logarithmic-growth phase. Incubation at 30°C continued for 12 to 15 h. Yeasts were imaged using confocal microscopy with identical microscopic settings to enable direct comparison between strains. TR-gentamicin is shown in the top panel in red, DIC is shown in the middle panel, and lucifer yellow is shown in the bottom panel in yellow for each strain. Arrows identify the colocalization of TR-gentamicin and lucifer yellow in the vacuole. (B) Short-term uptake of TR-gentamicin and lucifer yellow. Overnight cultures were incubated with 500 μg/ml TR-gentamicin and 1 mg/ml lucifer yellow for 2 h. Imaging was conducted as for panel A. DIC image is shown in the left panel, with the TR-gentamicin shown in red in the middle panel. The overlay of all three channels was performed in Zeiss LSM Image Browser and imported into Adobe Photoshop, where color enhancement was performed. This process enabled the dimmer lucifer yellow to be visible when overlaid with the TR-gentamicin. Note the concentration of TR-gentamicin with lucifer yellow in the vacuole but also at distinct punctate locations in the cytoplasm.
FIG. 4.
Quantitation of gentamicin uptake. A total of 100 μg/ml OG-gentamicin was added to yeasts in logarithmic-growth phase for 5 h. Each strain was then washed and stained for PI prior to flow (FL) analysis. The raw data for OG-gentamicin uptake for the WT and the SAC1 deletion strains are presented in panel A, while the mean uptake values for all strains are shown in panel B along with the percent dead/injured cells based on PI staining.
TR-gentamicin location in sensitive strains visualized by Voxx.
TR-gentamicin was added to logarithmic-growth-phase cultures at a concentration of 50 μg/ml, with an additional 50 μg/ml unlabeled gentamicin added to wild-type cells as well as cells with VPS54, VPS45, NHX1, and SAC1 mutations. The objective was to have gentamicin at a concentration that inhibited growth (Fig. 1 and 2). Cells were imaged between 12 to 15 h after adding TR-gentamicin. In order to better evaluate the location of TR-gentamicin in the sensitive strains, a three-dimensional, voxel-based rendering program, Voxx, was used (8). Image stacks were collected with a 0.1-μm z-step enabling three-dimensionally rendered images to be constructed (Fig. 5). Images are rendered at maximum intensity so comparisons can be made only on location, not quantity, of TR-gentamicin. Based on TR-gentamicin's colocalization with LY, it is evident that most TR-gentamicin is delivered to the vacuole. Fragmentation of the vacuole is known to occur in GARP deletion strains with VPS51, VPS52, VPS53, and VPS54 mutations, and TR-gentamicin is found in multiple spherical-shaped structures, which is consistent with vacuole localization in these deletion strains. The VPS16 deletion strain contains no normal vacuolar structure, while VPS45 mutants have one large central vacuole (16). In the VPS16 mutant strain, TR-gentamicin is distributed throughout the cell in both diffuse and distinct staining patterns. For the VPS45 mutant, we observe some cells with one large vacuole but also see smaller punctate structures, often at the cell periphery. Both NHX1 and ZUO1 mutants appear to have concentrated TR-gentamicin in their vacuole. In the gentamicin-sensitive strains, TR-gentamicin is often observed to be present in a diffuse pattern that may represent cytosolic release from a membrane-bound organelle, though at this time, the appearance of TR-gentamicin in highly fragmented vacuoles cannot be excluded.
FIG. 5.
Three-dimensional reconstruction of TR-gentamicin in sensitive strains. Logarithmic-phase cultures were incubated overnight with 50 μg/ml TR-gentamicin. Confocal stacks were collected using a 0.1-μm z-axis step size. Images were opened in Voxx, and maximum intensity rendering was performed. For each strain, the rendered TR-gentamicin image is shown above the corresponding single plane DIC image. The white bar shown in the wild-type image is 5 μm, and all images are presented at the same magnification. The supplemental material contains movies that rotate the full field in both x and y axes directions to better show the cellular distribution of the TR-gentamicin.
Evaluation of yeast viability following gentamicin treatment.
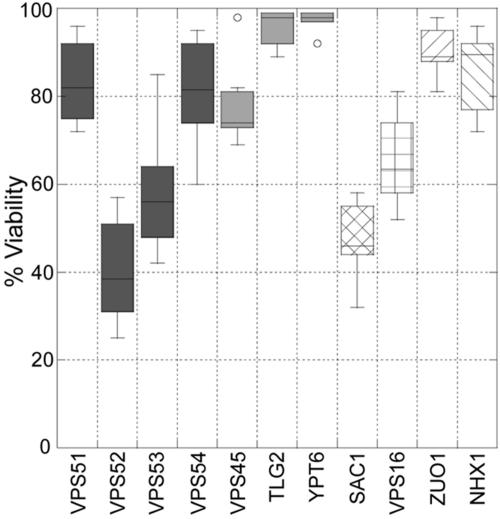
The gentamicin-induced inhibition in the growth assay could be caused by a fungistatic or fungicidal action of the aminoglycoside. To address the viability of the gentamicin-sensitive strains, we utilized a standard methylene blue assay. Gentamicin was added to logarithmically growing cultures and incubated for an additional 20 to 30 h, at which point viability was assessed. We chose to evaluate viability using the lowest concentration of gentamicin that resulted in significant growth inhibition for the respective strain. Note that, while growth inhibition is severely reduced at similar gentamicin concentrations, viability assessed with methylene blue was quite variable, even among strains lacking components of specific protein complexes (Fig. 6). This difference may simply reflect the fact that the growth assay is a more sensitive indicator of cell inhibition than viability assays.
FIG. 6.
Measurement of yeast viability following gentamicin treatment. Yeast strains were incubated overnight with gentamicin concentrations (500 μg/ml for TLG2 and YPT6 mutants, 100 μg/ml for VPS45, VPS53, VPS54, SAC1, and NHX1 mutants, and 50 μg/ml for VPS51, VPS52, VPS16, and ZUO1 mutants) by using the same conditions as those detailed in the legend for Fig. 1. The strains were than stained with methylene blue and counted. Data are presented using a box plot.
DISCUSSION
Cellular injury caused by gentamicin is most pronounced in kidney proximal tubule cells, which accumulate the highest concentrations of this antibiotic. Following unimpaired filtration of gentamicin across the glomerulus, cellular uptake involves both fluid-phase and receptor-mediated, megalin-associated endocytosis (21, 35). The majority of internalized gentamicin is delivered to the lysosome. The mechanism by which gentamicin produces cell injury is still unresolved, though recent studies from our laboratory support the importance of retrograde movement of gentamicin through the endosomal system. Utilizing TR-gentamicin in proximal tubule cells, we showed that 5 to 10% of internalized gentamicin is initially trafficked from endosomes to the Golgi (29, 31). Further analysis revealed gentamicin delivery to the endoplasmic reticulum, with subsequent cytosolic release and subcellular compartment association, including the nucleus and mitochondria (32). Furthermore, we observed a reduction in mitochondrial potential in gentamicin-treated cells which was at times consistent with cytosolic delivery of gentamicin, suggesting one possible mechanism of toxicity. The movement of gentamicin through multiple membrane-bound organelles and its eventual cytosolic delivery would potentially enable this polycationic antibiotic to encounter and disrupt multiple biochemical processes occurring in both membrane and cytosolic locations. The mechanism(s) by which gentamicin undergoes retrograde trafficking to the ER may involve pathways utilized by other cellular toxins (20, 33). Therefore, investigating the trafficking and interactions of gentamicin in a eukaryotic model system that is more amenable to genetic, biochemical, and molecular dissection could lead to a better understanding of the pathophysiology of gentamicin and other toxins.
The recent screening of a yeast deletion library for antibiotic-sensitive strains by Blackburn and Avery (3) uncovered many gentamicin-sensitive strains possessing mutations in proteins involved in intracellular trafficking pathways. These observations provided findings for the eukaryotic model system that were consistent with our mammalian studies (29-32). Therefore, we set out to further characterize the specific pathways and protein complexes, which if deleted, would lead to increased gentamicin sensitivity. We also utilized TR-gentamicin as a tool to begin characterizing its transport in yeast and evaluating differences between the gentamicin-sensitive strains as a tool to further understand cellular toxicity.
We did note many similarities and some differences between our findings and those of Blackburn and Avery (3). These differences most likely resulted from differences in the sensitivities of our screens. Their screen was performed under a single condition (YPD with 256 μg/ml gentamicin and growth evaluated after 2 days at 30°C). While false positives were unlikely to be overlooked due to the careful analysis of mutants after the preliminary screen, the uneven growth rate of the strains within the mutant collection as well as the uneven number of cells present in each initial culture could have caused difficulties in determining antibiotic sensitivities with a fraction of the mutants.
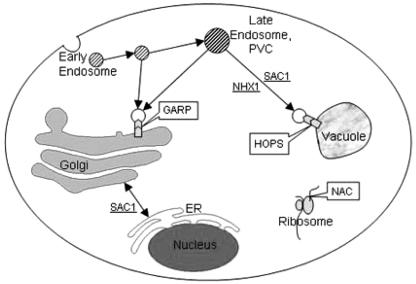
Our results, as shown in Fig. 7, suggest that the deletion of a protein from any of several intracellular vesicular pathways and or complexes resulted in sensitivity to gentamicin. One such complex that appeared critical for tolerating gentamicin is the NAC containing Zuo1 (13, 14). Zuo1 is a ribosome-associated J protein and a partner of Ssb1 and Ssb2 of Hsp70 and the Hsp70-related protein Ssz1. Together, they act as a molecular chaperone to facilitate protein folding of nascent proteins. Their absence clearly caused gentamicin sensitivity, and a recent study suggests the mechanism may be due to altered plasma membrane function that results in ion transport changes (14, 15). Kim and Craig also suggested this chaperone complex may participate in the folding of WD40 proteins, many of which are involved in the secretory pathway (15). Their studies emphasized the importance of understanding both a protein's interaction(s) and the cellular pathway(s) in which it participates.
FIG. 7.
Diagram showing the location of several complexes (HOPS, GARP, and NAC) and individual genes whose absence results in gentamicin-hypersensitive strains. The delivery of gentamicin to the Golgi, ER, and vacuole and cytoplasmic interaction with ribosomal-associated proteins is consistent with our mammalian studies. Additional studies using these and other mutants will help to establish the mechanism of delivery and toxicity to these various cellular locations.
Two other protein complexes, HOPS and GARP, have multiple members whose absence resulted in significant gentamicin sensitivity. Blackburn and Avery identified four members of the HOPS complex (Pep3, Pep5, Vps33, and Vps16) as affecting gentamicin resistance (3). In our growth assay, cells with VPS16 mutations were gentamicin sensitive while cells lacking the other HOPS components were minimally affected. We also examined cells with two other HOPS-member mutations (Vam6 and Vps41) and found that the loss of Vps41 resulted in an approximately 50% growth inhibition in the presence of gentamicin, while the loss of Vam6 caused little growth retardation. Interestingly, the loss of Vps8, which has an association with Pep3, Pep5, Vps16, and Vps33 based on affinity precipitation, resulted in a growth inhibition of greater than 50%. The HOPS complex plays an important role in vacuole fusion events (40). The variable effects we observed in the HOPS deletion mutants suggest either that the vacuole fusion event is not important in inducing gentamicin sensitivity or that some HOPS members participate in other pathways with a greater effect on gentamicin sensitivity. In support of the latter, Vps41 has been shown to be involved in both vesicle budding and fusion during vacuole biogenesis and Vps16, whose loss results in one of the most gentamicin-sensitive strains, was shown to have more than a 50% reduction in Dcp1-dependent mRNA decapping activity (23-25, 27, 42, 43). Reduction in decapping activity would remove a key signal needed to increase translation, which is potentially important for adaptation following gentamicin exposure.
Vps54 (Luv1) was identified by Blackburn and Avery as important for gentamicin resistance though they did not discuss its role as a component of the GARP complex (3). This complex contains at least four proteins (Vps51, Vps52, Vps53, and Vps54) and has an associated Rab, Ypt6, in addition to interacting with various t- and v-SNAREs, and the SM protein, Vps45 (39, 40). This complex has a critical role in the docking and fusion of endosome-derived vesicles with the trans-Golgi network. The absence of any of the GARP members resulted in significant and similar gentamicin sensitivities. This is not surprising for Vps52, Vps53, or Vps54 since each of these proteins is necessary for the stability of the remaining components of the complex, and the absence of any one of them results in the rapid degradation of the other two (10). In contrast, loss of Vps51 does not eliminate the other GARP members (26). Consequently, the similar gentamicin sensitivities we observed in cells with Vps51 mutations or the other GARP members support our conclusion that the loss of the GARP complex is a key contributor to inducing gentamicin toxicity. In further support of this, the deletion of YPT6, TLG2, or VPS45 resulted in significant gentamicin sensitivity. The loss of Vps45 results in down-regulation of Tlg2 but not Tlg1, and the truncation of Tlg2 inhibits the association of Vps45 with the t-SNAREs (7). The increased gentamicin sensitivity caused by loss of Vps45 compared to loss of Tlg2 suggests that the loss of Vps45 affects another pathway in addition to GARP. Recent studies have shown the importance of this SM protein in both yeast and mammalian cells. There are only four SM proteins in yeasts and seven in mammals, while many more SNAREs exist (39). The SM proteins are essential for intracellular membrane fusion events and appear to work closely with specific SNAREs to provide specificity in the membrane fusion process (11). Vps45 and Tlg2 have also been shown to have a role in the constitutive Cvt pathway (1). Future analysis of strains containing specific point mutations of the gentamicin-sensitive deletion proteins and double mutants will help to define what pathways and interactions, when absent, result in toxicity.
The final two strains that exhibited significant gentamicin toxicity had SAC1 or NHX1 mutations. NHX1 is present on the opposite DNA strand but overlaps YDR455c (4). We found similar results when analyzing either YDR455c or NHX1 mutants. Nhx1 is one of the oldest members of the Na+/H+ exchanger family (5). Members of this family have important roles in multiple functions including salt tolerance, transepithelial Na+ transport, vesicle trafficking, and vesicle biogenesis. Several studies have shown the importance of Nhx1 in endosomal trafficking, where it appears to regulate the pH of endosomes (2, 6). NHX1 mutants missort CPY and have a general defect in vesicle trafficking out of the endosome that appears to be a pH-dependent event. SAC1 codes for a lipid phosphatase that has been localized to ER and Golgi membranes (12, 17). Its enzymatic activity acts on phosphatidylinositol 4-phosphate [PtdIns(4)P]. SAC1 mutants accumulate PtdIns(4)P at ER and vacuolar membranes, which in turn results in altered late endocytic and vacuolar trafficking (37). A common attribute of many of the gentamicin-sensitive mutants is altered trafficking involving endosomes. This information, when taken together, enables better predictions to be made concerning how and why specific gene deletions result in increased gentamicin toxicity.
Our previous studies in mammalian cells documented the utility of TR-gentamicin as a probe to monitor the intracellular location of gentamicin. In the present studies, the level of TR-gentamicin uptake correlated qualitatively with growth inhibition and LY uptake (Fig. 3). To quantitatively address uptake, we utilized OG-gentamicin and flow analysis (Fig. 4). While there is a clear positive correlation between gentamicin uptake and dead/injured cells, additional studies are needed to establish for each strain whether localization or uptake is the most critical parameter. Delivery of gentamicin to a specific cellular location may be critical for inducing cell toxicity. Localization of TR-gentamicin clearly showed that delivery to LY staining vacuole structures was the predominant pathway. This is consistent with mammalian studies in which the majority of intracellular gentamicin was localized to the lysosome. A critical question is whether gentamicin is trafficked to other locations and/or whether it causes missorting in the trafficking pathways. The present studies support the importance of the GARP, HOPS, and NAC protein complexes whose absences resulted in gentamicin-sensitive strains. Visual analysis of these strains using TR-gentamicin suggested localization to smaller punctate structures, possibly endosomes, and fragmented vacuole pieces for the GARP mutants. Distinguishing fragmented vacuole localization of TR-gentamicin from endosomal or cytosolic locations will require further investigation. However, the known effect of gentamicin on membrane fusion and documented cytosolic release in mammalian cells suggests that cytosolic release is a likely mechanism by which gentamicin induces toxicity in both yeast and mammalian cells.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants P50-DK-61594 and PO1-DK-53465, a Veterans Affairs Merit Review (to B. A. Molitoris), NSF grant MCB-0091317 (to M. G. Goebl), and an Indiana Genomics Initiative grant (INGEN) from the Lilly Endowment to Indiana University School of Medicine.
We would like to thank Pat Smith of the IU Pulmonary Flow Facility for her assistance in performing the flow cytometry experiments and Stacy Bennett for technical assistance.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Abeliovich, H., T. Darsow, and S. D. Emr. 1999. Cytoplasm to vacuole trafficking of aminopeptidase I requires a t-SNARE-Sec1p complex composed of Tlg2p and Vps45p. EMBO J. 18:6005-6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ali, R., C. L. Brett, S. Mukherjee, and R. Rao. 2004. Inhibition of sodium/proton exchange by a Rab-GTPase-activating protein regulates endosomal traffic in yeast. J. Biol. Chem. 279:4498-4506. [DOI] [PubMed] [Google Scholar]
- 3.Blackburn, A. S., and S. V. Avery. 2003. Genome-wide screening of Saccharomyces cerevisiae to identify genes required for antibiotic insusceptibility of eukaryotes. Antimicrob. Agents Chemother. 47:676-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowers, K., B. P. Levi, F. I. Patel, and T. H. Stevens. 2000. The sodium/proton exchanger Nhx1p is required for endosomal protein trafficking in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 11:4277-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brett, C. L., M. Donowitz, and R. Rao. 2005. Evolutionary origins of eukaryotic sodium/proton exchangers. Am. J. Physiol. Cell Physiol. 288:C223-239. [DOI] [PubMed] [Google Scholar]
- 6.Brett, C. L., Y. Wei, M. Donowitz, and R. Rao. 2002. Human Na+/H+ exchanger isoform 6 is found in recycling endosomes of cells, not in mitochondria. Am. J. Physiol. Cell Physiol. 282:C1031-C1041. [DOI] [PubMed] [Google Scholar]
- 7.Bryant, N. J., and D. E. James. 2001. Vps45p stabilizes the syntaxin homologue Tlg2p and positively regulates SNARE complex formation. EMBO J. 20:3380-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clendenon, J. L., C. L. Phillips, R. M. Sandoval, S. Fang, and K. W. Dunn. 2002. Voxx: a PC-based, near real-time volume rendering system for biological microscopy. Am. J. Physiol. Cell Physiol. 282:C213-218. [DOI] [PubMed] [Google Scholar]
- 9.Conibear, E., J. N. Cleck, and T. H. Stevens. 2003. Vps51p mediates the association of the GARP (Vps52/53/54) complex with the late Golgi t-SNARE Tlg1p. Mol. Biol. Cell 14:1610-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conibear, E., and T. H. Stevens. 2000. Vps52p, Vps53p, and Vps54p form a novel multisubunit complex required for protein sorting at the yeast late Golgi. Mol. Biol. Cell 11:305-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dulubova, I., T. Yamaguchi, Y. Gao, S.-W. Min, I. Huryeva, T. C. Sudhof, and J. Rizo. 2002. How Tlg2p/syntaxin 16 ‘snares’ Vps45. EMBO J. 21:3620-3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foti, M., A. Audhya, and S. D. Emr. 2001. Sac1 lipid phosphatase and Stt4 phosphatidylinositol 4-kinase regulate a pool of phosphatidylinositol 4-phosphate that functions in the control of the actin cytoskeleton and vacuole morphology. Mol. Biol. Cell 12:2396-2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gautschi, M., A. Mun, S. Ross, and S. Rospert. 2002. A functional chaperone triad on the yeast ribosome. Proc. Natl. Acad. Sci. USA 99:4209-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang, P., M. Gautschi, W. Walter, S. Rospert, and E. A. Craig. 2005. The Hsp70 Ssz1 modulates the function of the ribosome-associated J-protein Zuo1. Nat. Struct. Mol. Biol. 12:497-504. [DOI] [PubMed] [Google Scholar]
- 15.Kim, S.-Y., and E. A. Craig. 2005. Broad sensitivity of Saccharomyces cerevisiae lacking ribosome-associated chaperone Ssb or Zuo1 to cations, including aminoglycosides. Eukaryot. Cell 4:82-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koning, A. J., L. L. Larson, E. J. Cadera, M. L. Parrish, and R. L. Wright. 2002. Mutations that affect vacuole biogenesis inhibit proliferation of the endoplasmic reticulum in Saccharomyces cerevisiae. Genetics 160:1335-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Konrad, G., T. Schlecker, F. Faulhammer, and P. Mayinger. 2002. Retention of the yeast Sac1p phosphatase in the endoplasmic reticulum causes distinct changes in cellular phosphoinositide levels and stimulates microsomal ATP transport. J. Biol. Chem. 277:10547-10554. [DOI] [PubMed] [Google Scholar]
- 18.Leehey, D., B. Braun, D. Tholl, L. Chung, C. Gross, J. Roback, and J. Lentino. 1993. Can pharmacokinetic dosing decrease nephrotoxicity associated with aminoglycoside therapy? J. Am. Soc. Nephrol. 4:81-90. [DOI] [PubMed] [Google Scholar]
- 19.Levy, S. B., and B. Marshall. 2004. Antibacterial resistance worldwide: causes, challenges and responses. Nat. Med. 10:S122-S129. [DOI] [PubMed] [Google Scholar]
- 20.Lord, J. M., and L. M. Roberts. 1998. Toxin entry: retrograde transport through the secretory pathway. J. Cell Biol. 140:733-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moestrup, S., S. Cui, H. Vorum, C. Bregengard, S. Bjorn, K. Norris, J. Gliemann, and E. Christensen. 1995. Evidence that epithelial glycoprotein 330/megalin mediates uptake of polybasic drugs. J. Clin. Investig. 96:1404-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munn, A. L., A. Heese-Peck, B. J. Stevenson, H. Pichler, and H. Riezman. 1999. Specific sterols required for the internalization step of endocytosis in yeast. Mol. Biol. Cell 10:3943-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peterson, M. R., and S. D. Emr. 2001. The class C Vps complex functions at multiple stages of the vacuolar transport pathway. Traffic 2:476-486. [DOI] [PubMed] [Google Scholar]
- 24.Piper, R. C., N. J. Bryant, and T. H. Stevens. 1997. The membrane protein alkaline phosphatase is delivered to the vacuole by a route that is distinct from the VPS-dependent pathway. J. Cell Biol. 138:531-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Price, A., D. Seals, W. Wickner, and C. Ungermann. 2000. The docking stage of yeast vacuole fusion requires the transfer of proteins from a cis-SNARE complex to a Rab/Ypt protein. J. Cell Biol. 148:1231-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reggiori, F., C.-W. Wang, P. E. Stromhaug, T. Shintani, and D. J. Klionsky. 2003. Vps51 is part of the yeast Vps fifty-three tethering complex essential for retrograde traffic from the early endosome and Cvt vesicle completion. J. Biol. Chem. 278:5009-5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rehling, P., T. Darsow, D. J. Katzmann, and S. D. Emr. 1999. Formation of AP-3 transport intermediates requires Vps41 function. Nat. Cell Biol. 1:346-353. [DOI] [PubMed] [Google Scholar]
- 28.Ridley, R. G. 2004. Research on infectious diseases requires better coordination. Nat. Med. 10:S137-S140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sandoval, R., J. Leiser, and B. Molitoris. 1998. Aminoglycoside antibiotics traffic to the Golgi complex in LLC-PK1 cells. J. Am. Soc. Nephrol. 9:167-174. [DOI] [PubMed] [Google Scholar]
- 30.Sandoval, R. M., R. L. Bacallao, K. W. Dunn, J. D. Leiser, and B. A. Molitoris. 2002. Nucleotide depletion increases trafficking of gentamicin to the Golgi complex in LLC-PK1 cells. Am. J. Physiol. Renal Physiol. 283:F1422-F1429. [DOI] [PubMed] [Google Scholar]
- 31.Sandoval, R. M., K. W. Dunn, and B. A. Molitoris. 2000. Gentamicin traffics rapidly and directly to the Golgi complex in LLC-PK1 cells. Am. J. Physiol. Renal Physiol. 279:F884-F890. [DOI] [PubMed] [Google Scholar]
- 32.Sandoval, R. M., and B. A. Molitoris. 2004. Gentamicin traffics retrograde through the secretory pathway and is released in the cytosol via the endoplasmic reticulum. Am. J. Physiol. Renal Physiol. 286:F617-F624. [DOI] [PubMed] [Google Scholar]
- 33.Sandvig, K., M. Ryd, O. Garred, E. Schweda, P. Holm, and B. van Deurs. 1994. Retrograde transport from the Golgi complex to the ER of both Shiga toxin and the nontoxic Shiga B-fragment is regulated by butyric acid and cAMP. J. Cell Biol. 126:53-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schentag, J., M. Plaut, and F. Cerra. 1981. Comparative nephrotoxicity of gentamicin and tobramycin: pharmacokinetic and clinical studies in 201 patients. Antimicrob. Agents Chemother. 19:859-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmitz, C., J. Hilpert, C. Jacobsen, C. Boensch, E. I. Christensen, F. C. Luft, and T. E. Willnow. 2002. Megalin deficiency offers protection from renal aminoglycoside accumulation. J. Biol. Chem. 277:618-622. [DOI] [PubMed] [Google Scholar]
- 36.Smith, C. R., R. D. Moore, and P. S. Lietman. 1986. Studies of risk factors for aminoglycoside nephrotoxicity. Am. J. Kidney Dis. 8:308-313. [DOI] [PubMed] [Google Scholar]
- 37.Tahirovic, S., M. Schorr, and P. Mayinger. 2005. Regulation of intracellular phosphatidylinositol-4-phosphate by the Sac1 lipid phosphatase. Traffic 6:116-130. [DOI] [PubMed] [Google Scholar]
- 38.Teparic, R., I. Stuparevic, and V. Mrsa. 2004. Increased mortality of Saccharomyces cerevisiae cell wall protein mutants. Microbiology 150:3145-3150. [DOI] [PubMed] [Google Scholar]
- 39.Toonen, R. F., and M. Verhage. 2003. Vesicle trafficking: pleasure and pain from SM genes. Trends Cell Biol. 13:177-186. [DOI] [PubMed] [Google Scholar]
- 40.Whyte, J. R. C., and S. Munro. 2002. Vesicle tethering complexes in membrane traffic. J. Cell Sci. 115:2627-2637. [DOI] [PubMed] [Google Scholar]
- 41.Wickner, W. 2002. Yeast vacuoles and membrane fusion pathways. EMBO J. 21:1241-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wurmser, A. E., T. K. Sato, and S. D. Emr. 2000. New component of the vacuolar class C-Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion. J. Cell Biol. 151:551-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang, S., C. J. Williams, K. Hagan, and S. W. Peltz. 1999. Mutations in VPS16 and MRT1 stabilize mRNAs by activating an inhibitor of the decapping enzyme. Mol. Cell. Biol. 19:7568-7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.