Abstract
Clinical isolates of the opportunistic pathogen Mycobacterium avium complex (MAC) undergo a reversible switch between red and white colony morphotypes on agar plates containing the lipoprotein stain Congo red. Compared to their isogenic red counterparts, white morphotypic variants are more virulent and more resistant to multiple antibiotics. This report shows that the two-component regulatory system mtrAB is required for the red-to-white switch as well as for other morphotypic switches of MAC. A mutant with a transposon insertion in the histidine protein kinase gene mtrB was isolated from a morphotypically white parent clone. The mutant resembled a naturally occurring red morphotypic variant in that it stained with Congo red, was sensitive to multiple antibiotics, and was permeable by a fluorescent DNA stain. However, it differed from a red variant in that it could not switch to the white or transparent morphotype, and it could not survive intracellularly within macrophage-like cells. Transcomplementation with a cloned wild-type mtrB gene restored to the mutant the ability to form impermeable, drug-resistant white and transparent variants. Quantitative reverse transcriptase PCR showed that mtrB was required for the normal expression of cell surface Mce proteins, some of which are up-regulated in the red-to-white switch. The results indicate that mtrAB functions in regulating the composition and permeability of mycobacterial cell walls and plays a role in the reversible colony type switches of MAC.
The Mycobacterium avium complex (MAC) is the most significant of the environmental mycobacteria that opportunistically infect susceptible humans, especially AIDS patients. MAC infections are difficult to treat due to the intrinsic multidrug resistance of the organism. Drugs such as clarithromycin, azithromycin, rifabutin, ethambutol, amikacin, clofazamine, and fluoroquinolones, which are effective against primary isolates, frequently lose effectiveness unless administered in combination.
The intrinsic multidrug resistance of MAC is ascribed to cell wall impermeability, although additional factors are likely to contribute (1, 10, 11, 16, 24). Conditions that compromise cell wall integrity result in increased drug susceptibility (10, 17, 21, 26). Intrinsic multidrug resistance might involve nonspecific permeability barriers that decrease drug uptake, possibly working in synergy with more specific efflux mechanisms.
There is a correlation between drug susceptibility and colony type of MAC. Virtually all isolates of MAC form multiple colony morphotypes that vary with regard to infectivity and drug susceptibility. The conservation of this property within the species suggests that it confers selective advantages. The opaque-transparent switch is one such switch (1, 25, 33). Transparent variants are more resistant to multiple antibiotics than their opaque counterparts. They also predominate in patient samples and grow better in animal and macrophage models of infection. Opaque variants grow better on laboratory media and predominate after passage in vitro.
We described another morphotypic switch in MAC, termed red-white, that is visible when clinical isolates are grown on media containing the lipoprotein stain Congo red (CR) (2, 20). The red-white switch operates independently of the opaque-transparent switch, such that red opaque (RO), red transparent, white opaque (WO), and white transparent morphotypes can be distinguished by CR staining. White variants are more common than red variants in patient samples, and they grow better in mice and in human monocyte-derived macrophages. Morphotypic switching may help MAC populations to adapt to diverse environmental challenges.
New genetic tools facilitate the dissection of morphotypic switching and intrinsic drug resistance by MAC. Mutational analysis using a transposome mutagenesis system identified two genes, Maa2520 and pks12, that are required for the multidrug resistance associated with the white morphotype (24). These two genes encode a conserved hypothetical cell surface protein and a polyketide synthase, respectively, consistent with roles for cell surface structures in intrinsic drug resistance. Genome sequences of M. avium subsp. avium strain 104 (draft sequence) and M. avium subsp. paratuberculosis strain K10 (complete sequence) have been annotated (23, 28). Using these mutational and genomic tools, we found that the two-component regulatory system mtrAB is required for the red-to-white switch of MAC and for the multidrug resistance that accompanies it.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Stable, naturally occurring WO and RO clones of M. avium subsp. avium isolate HMC02 were described previously (2, 3). Bacteria were grown on Middlebrook 7H10 agar plates (Difco) supplemented with 10% oleic acid-albumin-dextrose-catalase (OADC) enrichment (BBL), 0.5% glycerol, and 100 μg/ml CR, or on Middlebrook 7H9 broth medium supplemented with 10% ADC enrichment and 0.2% glycerol. CR was stored and added to media as described previously (3). When appropriate, media were further supplemented with 100 μg/ml kanamycin, 30 μg/ml apramycin, or 0.2 μg/ml ciprofloxacin. Additional supplements used for drug susceptibility testing are described below. Clarithromycin was kindly provided for this study by Abbott Laboratories, North Chicago, IL. Other drugs were obtained from commercial vendors (ICN Biomedicals, Aurora, OH, and Sigma Chemical Co., St. Louis, MO). Bacteria were grown at 37°C under air.
Isolation of a mutant with a transposon insertion into mtrB.
We have used the EZ-TN 〈KAN-2〉 transposome from Epicentre (Madison, WI) to randomly mutagenize a clinical isolate of M. avium subsp. avium, strain HMC02 (12, 24). The 1.2-kb EZ-TN element, derived from Tn903, carries a kanamycin (Km) resistance gene flanked by an inverted repeat sequence. A library of ∼3,500 mutants of a WO variant of clinical isolate HMCO2 (HMC02-WO) was screened for mutants that exhibited decreased resistance to ciprofloxacin, as described previously (24). Briefly, WO cells of M. avium subsp. avium strain HMC02 mutagenized by EZ-TN were plated onto 7H10 agar plates with CR and 100 μg/ml kanamycin (7H10-CR-Km). Transformant colonies were picked into 96-well MasterBlock plates containing 7H9 broth with 100 μg/ml Km. After growth at 37°C for 1 week, a 96-prong replicator was used to replicate the clone grids onto 7H10-CR-Km plates containing 0.2 μg/ml ciprofloxacin and onto 7H10-CR-Km master plates. The remaining cell suspensions in MasterBlock wells were supplemented with 15% glycerol and stored frozen at −70°C. Mutants that were unable to grow on the ciprofloxacin plates were retrieved from the storage plates for further analysis. Transposon insertion sites were mapped by PCR (random primer or inverse) and automated sequencing as described previously (12).
In vitro drug susceptibility tests.
Resistance to ciprofloxacin, clarithromycin, and penicillin was quantified by a proportion method as described previously (24). A breakpoint of 1% was used with the following critical concentrations of antibiotics: clarithromycin, 0, 0.05, 0.25, 1.25, 6.25, 16, and 64 μg/ml; penicillin, 0, 0.5, 2.5, 12.5, and 62.5 μg/ml; and ciprofloxacin, 0, 0.025, 0.05, 0.25, 1.25, 6.25 μg/ml. AB Biodisk Etests (Remel, Lenexa, KS) were conducted as described previously (3, 24). To prevent artifactual results due to degradation of antibiotics, plates were read after no more than 2 weeks of incubation at 37°C. All drug susceptibility tests were conducted in triplicate.
Survival within THP1 cells.
The myelomonocytic cell line THP1 (ATCC T1B-202) was cultivated and infected with M. avium subsp. avium clones as described previously (24), except that infections were carried out in multiwell plates rather than on glass coverslips. Briefly, THP1 cells were cultivated and treated for adherence, as described previously (5, 7). Bacteria were grown with shaking at 37°C in 7H9 broth to an optical density at 600 nm of 0.3 to 0.5 and diluted in RPMI infection medium (10% fetal bovine serum and 2 mM l-glutamine) to uniform densities. Immediately prior to infection, two wells were used for the 0-h time point by lysing host cells with double-distilled H2O and plating them on 7H10-CR plates. Remaining wells were washed twice with RPMI to remove nonadherent THP1 cells and residual culture medium. Cells were then infected at a multiplicity of infection of 10. At time points of 2 h, 24 h, 48 h, 4 days, and 7 days postinfection, triplicate wells were washed three times with RPMI culture medium to remove extracellular bacteria. Cells were lysed in double-distilled H2O, serially diluted, and plated onto 7H10-CR plates. After incubation for 2 to 3 weeks at 37°C, results were quantified as the number of CFU per well. Means and standard deviations of each set of triplicate readings were plotted.
Permeation by SYTO16.
Actively growing bacteria in 7H9 broth (0.5 ml) were transferred to sterile microcentrifuge tubes, and SYTO16 (Molecular Probes, Eugene, OR) was added to 10 μM. After precisely 5 min of incubation at room temperature, 1.0 ml of ice-cold autoclaved distilled water was added, immediately followed by centrifugation for 3 min to pellet cells. After thorough removal of supernatants, cells were resuspended in ice-cold autoclaved distilled water and centrifuged again. After thorough removal of supernatant, cells were resuspended in 150 μl of water, and wet-mount slides were prepared. SYTO16 penetration was determined by fluorescence microscopy with a fluorescein isothiocyanate filter set. In order to minimize bleaching and assure uniformity, all images were photographed upon first exposure, viewed with uniform scale settings and minimal gamma, and saved as 16-bit TIFF files.
To quantify SYTO16 staining, images were analyzed using the MetaMorph Imaging System and software by Molecular Devices. For each image frame, visible and fluorescent images were taken with a 50-ms exposure time and saved in an 8-bit format. Cells were quantified using the cell count feature in the software. Pixel intensity thresholds for counting cells under visible light were set at 150 and 125, and the two values were averaged. The threshold was set at 100 for counting of cells in UV fluorescence images. Images with high background were excluded. A percentage was calculated based on the number of cells that fluoresced divided by the total number of visible cells in each frame. Each experiment included three replicate HMC02-WO (parent strain) control images, and values obtained in each experiment were normalized to the mean HMC02-WO value by dividing the percent fluorescence value in each image by the mean percent fluorescence value obtained for HMC02-WO.
Genetic complementation of the mtrB mutation.
A 2.9-kb fragment containing the contiguous intact mtrA and mtrB genes was PCR amplified from the genome of M. avium subsp. avium strain HMC02 using the primers MTRABfwd4 and MTRABrev1 (Table 1). These primers added terminal XbaI and HindIII restriction sites and spacers for cloning. The PCR product was purified from a 1% agarose gel using the QIAGEN QIAEX II gel extraction kit. The fragment was then digested with XbaI and HindIII and ligated into the mycobacterial shuttle vector pMH416. This extrachromosomally replicating plasmid, kindly provided by M. J. Hickey and D. R. Sherman, had been generated by replacing the kanamycin resistance cassette of pMH29 (6) with an apramycin resistance cassette. The apramycin cassette (22) does not confer cross-resistance to kanamycin, which was used to select EZ-TN insertions, or to azithromycin, which was used to test for intrinsic drug resistance. The mtrAB insert was oriented such that mtrAB transcription could not be driven by the MOP promoter on the plasmid but instead required a native promoter.
TABLE 1.
PCR and Q-RT-PCR primers
| Primer | Target gene | Sequence | Function |
|---|---|---|---|
| MTRABfwd4 | mtrAB | CTAGAAGCTTCGCCGCGACCACGAC | mtrAB cloning |
| MTRABrev1 | mtrAB | CTAGTCTAGAATCACAGCGACCTTTCGG | mtrAB cloning |
| OKH37 | aac(3)-IV | CCACTTGGACTGATCGAG | Detection of pMH416 in transformants |
| OKH38 | aac(3)-IV | CAGGAAGATCAACGGATC | Detection of pMH416 in transformants |
| Kan2FP1 | kan | ACCTACAACAAAGCTCTCATCAACC | Detection of EZ-TN insertion |
| MAA4214_2A | yrbE1B | ATCACCACGACCGTGCTCTACG | Q-RT-PCR (mce1 cluster) |
| MAA4214_2B | yrbE1B | ATGATGGCTTCCAGGAACGACC | Q-RT-PCR (mce1 cluster) |
| MAA4212_4A | mce1A | TCGGTCGATCGATGCGGTTCTC | Q-RT-PCR (mce1 cluster) |
| MAA4212_4B | mce1A | TTACACCGTGAGCGCGAAGTTG | Q-RT-PCR (mce1 cluster) |
| MAA1650_1A | yrbE3B | AATCCCGTTCAGGCACTTGTGG | Q-RT-PCR (mce3 cluster) |
| MAA1650_1B | yrbE3B | GAAGACCACGAAGATGTAGCTGC | Q-RT-PCR (mce3 cluster) |
| MAA1652_1A | mce3A | AATTCACGCATGGACACCATCG | Q-RT-PCR (mce3 cluster) |
| MAA1652_1B | mce3A | CGAGAATCGACAGGATGTGTTGC | Q-RT-PCR (mce3 cluster) |
| MAA0686_5A | yrbE4B | GCTGTTCTACGGCAAAGCGCT | Q-RT-PCR (mce4 cluster) |
| MAA0686_5B | yrbE4B | CGATCATCGCCAAAGTGCCTG | Q-RT-PCR (mce4 cluster) |
| MAA0687_5A | mce4A | CCGAAACTCAACGAGTCCCTGG | Q-RT-PCR (mce4 cluster) |
| MAA0687_5B | mce4A | GAACGAATCCAGGTCGCTGAGC | Q-RT-PCR (mce4 cluster) |
| SigA_3A | sigA | CACCAAGGGCTACAAGTTCTCC | Q-RT-PCR (sigA control) |
| SigA_3B | sigA | GCCTGACGGATCCACCAC | Q-RT-PCR (sigA control) |
The pMH416::mtrAB construct was transformed into Escherichia coli DH5α cells, and transformants were selected in LB broth containing 30 μg/ml apramycin. After extraction of the plasmid using the QIAprep spin miniprep kit, the structure and correct orientation of the insert were confirmed by restriction endonuclease digestion and agarose gel electrophoresis.
Repeated attempts to introduce pMH416::mtrB into the mtrB transposon mutant by our standard protocol for MAC (24) were unsuccessful, apparently because the mutant was killed by these manipulations. A modified protocol was used instead. Cells were grown at 37°C in a modified 7H9 broth that was supplemented with 0.5% glycerol and 10% OADC enrichment. After reaching mid-log phase, 0.1 volumes of 2 M glycine were added and the culture was incubated for another 24 h. The competent culture was centrifuged and washed three times in 10% glycerol and 0.5 M sucrose (electroporation solution). Cells were resuspended in electroporation solution to an optical density at 600 nm of 3.0. Plasmid DNA (1 μg) was added to 100 μl of cells for electroporation. Electroporation was carried out in three pulses at 1,000 Ω, 25 μF, 1.25 kV. Immediately after electroporation, cells were added to 900 μl of standard 7H9 broth and allowed to recover for 24 h. They were then plated on 7H10-CR agar plates with kanamycin (100 μg/ml) and apramycin (30 μg/ml) and allowed to grow for 2 weeks at 37°C.
Transformed clones were confirmed by using three PCRs. The first used primers OKH37 and OKH38 to generate a 600-bp product from the apramycin cassette of pMH416, confirming the presence of the plasmid. The second used the MTRABfwd4 and MTRABrev1 primers described above to confirm the presence of the cloned wild-type mtrAB allele. The third used the MTRABrev1 primer combined with the Kan2FP1 primer specific for the EZ-TN transposon (Table 1). These primers amplified a 717-bp region spanning the EZ-TN insertion, confirming that the original genomic insertion mutation was also intact in the heterodiploid transformants.
Q-RT-PCR.
Bacteria were grown in duplicate shaker flasks containing 50 ml of 7H9 broth supplemented with 1% Tween to disperse clumps and aid RNA extraction. On days 2, 3, and 4 postinoculation, 35-ml aliquots of each culture were harvested. Preliminary experiments indicated that the cultures were in exponential phase, late exponential phase, and early stationary phase at these time points, respectively. Cell pellets were washed in 0.5% Tween and 0.8% NaCl, and RNA was extracted as described previously (19). Briefly, the extraction procedure employed bead beating in a sodium acetate-sodium dodecyl sulfate-EDTA lysis buffer and acidified phenol. After centrifugation, the aqueous lysate was extracted with chloroform-isoamylalcohol. The RNA was isopropanol precipitated, washed, and then dissolved in RNase-free water. Residual DNA was digested on-column with DNase I (QIAGEN). RNA was quantified spectrophotometrically, and quality was checked by agarose gel electrophoresis. Gene expression levels were determined by quantitative reverse transcriptase PCR (Q-RT-PCR) with SYBR green, conducted on an Applied Biosystems Prism 7000 Sequence Detection System according to standard protocols used for mycobacteria (14). Expression of the housekeeping gene sigA was measured in every experiment, and the level of expression of each mce gene was divided by that of sigA to normalize for differences in total mRNA extracted (14, 19). Sequences of the primers used for Q-RT-PCR are listed in Table 1.
RESULTS
Isolation of an mtrB mutant.
The EZ-TN 〈KAN-2〉 transposon was used to randomly mutagenize a WO morphotypic clone of the clinical isolate M. avium subsp. avium HMC02, as described previously (24). A library of ∼3,500 mutants was screened for sensitivity to 0.2 μg/ml ciprofloxacin, to which the parent strain is intrinsically resistant. Approximately 43 such mutants were isolated, and EZ-TN insertions were mapped to 28 separate genetic loci. Six of these loci were mutagenized repeatedly, confirming their roles in intrinsic drug resistance. Two such genes, pks12 and Maa2520, were described previously (24). Additional multiply mutagenized genes were pstAB (coding for peptide synthases involved in glycopeptidolipid synthesis), ppdK (coding for a probable pyruvate-phosphate dikinase), Maa0577 (coding for a probable conserved exported protein), and Maa4466 (coding for a probable conserved transmembrane protein). The remaining 22 genes identified in this screen were mutagenized only once.
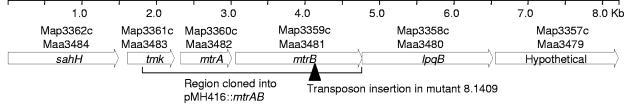
Among the 22 singly mutagenized genes was Maa3481, designated mtrB (28). The mutant WR8.1409 had an EZ-TN insertion at position 1083 within the 1,698-bp mtrB gene (Fig. 1). The predicted MtrB protein of M. avium subsp. avium was 87% identical to its ortholog in Mycobacterium tuberculosis (Rv3245c), a histidine protein kinase of a two-component regulatory system, MtrAB (32, 34). In the most recent release of the M. avium subsp. avium strain 104 genome sequence available at submission of this report (Institute for Genomic Research [www.tigr.org]), mtrB mapped 59 bp downstream of the response regulator gene mtrA (686 bp). The terminus of mtrB is the start codon of a downstream gene, lpqB (1,143 bp), which codes for a probable conserved lipoprotein. Genetic complementation results presented below show that the drug resistance defect of the mtrB insertion mutant was not due to polar effects on downstream genes. The genetic map of the region surrounding mtrAB is strongly conserved in the genomes of M. avium subsp. avium strain 104, M. avium subsp. paratuberculosis strain K10, M. tuberculosis strain H37Rv, and other Actinomycetales (18).
FIG. 1.
Map of the mtrAB region in the genome of M. avium subsp. avium strain 104. M. avium subsp. avium ORFs are numbered according to a recent annotation of the draft sequence (28). For reference, orthologous ORF numbers in the finished genome sequence of M. avium subsp. paratuberculosis strain K10 are also shown. The positions of the transposon insertion in mutant WR8.1409 and the cloned region are shown.
Growth characteristics of the mtrB mutant.
The growth rate of the mutant WR8.1409 on 7H10 agar plates was similar to that of the parent WO clone. The mutant formed smooth, opaque colonies that stained red and resembled naturally occurring RO colonies. Relative to the parent WO clone, the mutant was ≥5-fold more sensitive to ciprofloxacin, clarithromycin, and penicillin in vitro (Table 2). This level of multidrug sensitivity was similar to the Maa2520 and pks12 mutants described previously (24).
TABLE 2.
Drug susceptibilities of mutants derived from M. avium subsp. avium HMC02-WO
| Strain | MIC (μg/ml)a
|
||
|---|---|---|---|
| Clarithromycin | Ciprofloxacin | Penicillin | |
| HMC02-WO (parent) | 6.25 | 1.25 | 12.5 |
| WR8.1409 (mtrB) | 0.25* | 0.25* | 2.5* |
| WR6.389 (Maa2520) | 0.25* | 0.25* | 2.5* |
| WR6.440 (pks12) | 0.25 | 0.25 | 2.5 |
Values are means of three experiments. There was no variation between replicates. Asterisks indicate that no drug-resistant subvariants were observed. Other strains formed isolated drug-resistant colonies at frequencies of <1%.
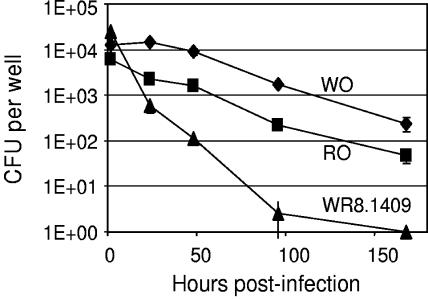
Although the mutant resembled naturally occurring RO variants in some regards, it differed from the latter in other regards. It was “phase locked” in the RO morphotype and could not form drug-resistant white or transparent subvariants even under antibiotic selective pressure (Table 2). This property was seen previously in Maa2520 mutants (24). Its growth in 7H9 broth was slower than that of the wild-type WO and RO clones. In addition, the mutant was unable to survive intracellularly within THP1 cells. When assessing intracellular survival and growth, a complicating factor was that mutagenesis had been carried out on an avirulent opaque variant, due to the difficulty of genetically transforming transparent variants. We followed a strategy used previously (24), in which THP1 cells were infected directly with opaque cells of parent and mutant strains and the ability of the bacteria to survive (if not grow) intracellularly was assessed. Viability of the HMC02-WO parent strain and the naturally occurring HMC02-RO variant declined slowly under these conditions but was not completely lost, presumably by virtue of subpopulations of more virulent variants. In contrast, mutant WR8.1409 declined to undetectable levels by 7 days postinfection (Fig. 2), consistent with an inability to form virulent variants.
FIG. 2.
Survival of WO, RO, and mtrB mutant clones within THP1 macrophage-like cells. The parent strain HMC02-WO (diamonds), a naturally occurring RO isolate from the same strain (squares), and the mtrB mutant WR8.1409 (triangles) were inoculated into THP1 cells and their intracellular survival was tracked as described in Materials and Methods. All data points are the means and standard deviations of triplicate measurements. All data points after 24 h are statistically different from each other (P < 0.01); however, the WO and RO curves have similar slopes and therefore may not represent truly different phenotypes under these conditions.
Permeability of the mtrB mutant.
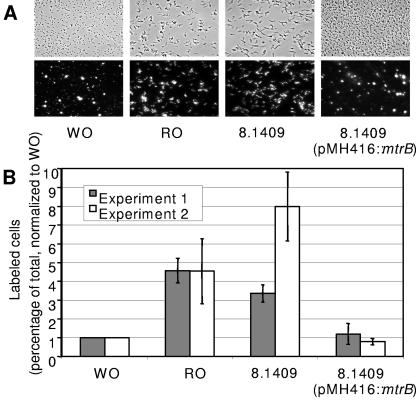
The effect of the mtrB mutation on cell wall permeability was assessed by measuring uptake of SYTO16, a cell-permeating green fluorescent nucleic acid stain. Because the fluorescence of SYTO16 is ∼40-fold enhanced upon binding to DNA, efficient staining requires penetration into the cytoplasm (9, 13). The stain labeled small clumps of wild-type WO cells, but most individual WO cells were not labeled. In contrast, most individual wild-type RO cells stained well and exhibited an elongated shape (Fig. 3A). The elongated shape is a consistent feature of RO cells and is seen throughout the growth cycle (unpublished observations). Quantitative image analysis confirmed that the percentage of RO cells labeled by SYTO16 was consistently greater than that of WO cells (Fig. 3B). Cells of the mtrB mutant were also elongated and stained consistently well with SYTO16 (Fig. 3A and B).
FIG. 3.
Permeation by SYTO16. Bacteria were exposed to SYTO16, and staining was quantified as described in Materials and Methods. Results obtained with the parent strain HMC02-WO, a naturally occurring RO isolate from the same strain, the mtrB mutant 8.1409, and a back-complemented clone of mutant 8.1409 are shown. (A) Images of typical fields under visible and fluorescence microscopy. (B) Percent labeling quantified and normalized to the WO control in each of two independent experiments. Values are means and standard deviations of three measurements made within each experiment.
The mtrB gene confers the ability to form drug-resistant morphotypes.
To test whether the mtrB mutation is directly associated with the phenotypes of the mutant, the contiguous wild-type mtrA and mtrB open reading frames (ORFs) were cloned into a mycobacterial shuttle vector, pMH416, and back-transformed into the mutant WR8.1409. The cloned region (Fig. 1) encompassed all of mtrA and mtrB but not the downstream lpqB gene. Introduction of the construct into the mutant was challenging because the mutant was killed by the electroporation procedures normally used to transform wild-type MAC strains (24). This fragility, which may have been related to the mutant's cell wall defects, required the development of the modified protocol outlined in Materials and Methods.
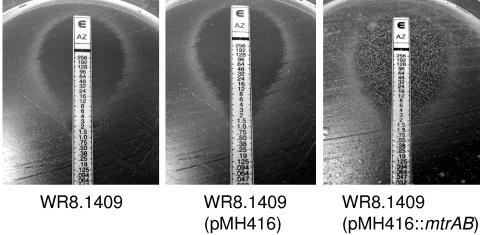
Two transformed clones carrying the wild-type mtrAB construct, designated WR8.1409 (pMH416::mtrAB), were characterized. Both clones were confirmed by PCR to carry the chromosomal mtrB::EZ-TN mutation as well as the plasmid-borne wild-type mtrAB allele. Initially they appeared identical to the mutant, forming red colonies on CR plates. However, when they were challenged by exposure to azithromycin or ciprofloxacin on Etest plates, as described previously (3), isolated white colonies appeared within the zones of inhibition formed by the major population of cells, indicating that the transformants had regained the ability to form drug-resistant morphotypic variants (Fig. 4). In contrast, the original mutant, and a vector-only control carrying pMH416 without the mtrAB insert, did not form such drug-resistant colonies, consistent with the phase-locked mutant phenotype.
FIG. 4.
Resistance to azithromycin in Etests. The mtrB mutant WR8.1409 (left), a vector-only control (center), and a clone carrying the wild-type mtrAB insert (right) were challenged with azithromycin Etest strips. A second clone of WR8.1409 (pMH416::mtrAB) yielded similar results, as did both clones when challenged with ciprofloxacin Etest strips.
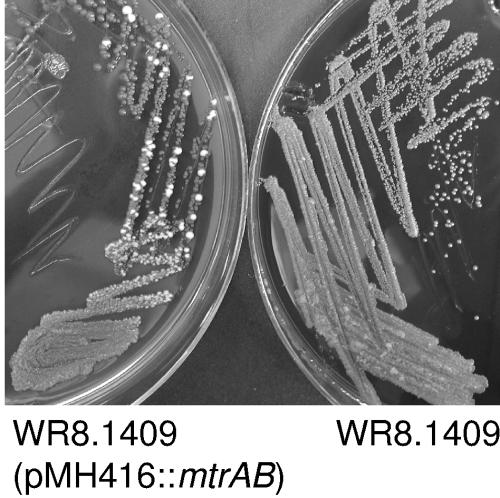
One clone of WR8.1409 (pMH416::mtrAB) also underwent spontaneous morphotypic switching to form isolated WO colonies in the absence of antibiotics other than the apramycin used to maintain the plasmid (Fig. 5). In contrast, the mutant WR8.1409, and the vector-only control WR8.1409 (pMH416), never formed such colonies over the course of several transfers. After isolation, WO colonies formed by WR8.1409 (pMH416::mtrB) exhibited a level of impermeability comparable to that of the wild-type WO strain (Fig. 3). These cells were also capable of differentiating further to form white transparent subvariants that were highly resistant to azithromycin (MIC > 256 μg/ml) and ciprofloxacin (MIC > 32 μg/ml) on Etest plates, identical to results seen when naturally occurring WO variants were challenged with Etests. Thus, the cloned wild-type mtrAB genes restored to the mutant the ability to switch to the impermeable white and transparent morphotypes, with the accompanying increases in multidrug resistance.
FIG. 5.
Spontaneous WO colonies formed by the back-complemented mtrB mutant. The white colonies formed by the mutant carrying pMH416::mtrAB are visible in the plate at left. Over the course of multiple transfers on the same medium, the original mutant (right) and the vector-only control (not shown) never formed such colonies.
Role for mtrB in mce gene expression.
Because mtrB is predicted to have a regulatory function, we asked whether it is required for the expression of genes that encode cell surface proteins, especially those that are differentially regulated in red and white variants of MAC. Preliminary analysis using a microarray of the M. avium subsp. avium strain 104 genome (28) identified approximately 130 genes that appeared to be down-regulated in naturally occurring RO cells relative to isogenic WO cells, of which 25 (19%) were within the mce1, mce3, and mce4 gene clusters (data not shown). Mce (for “mammalian cell entry”) proteins are located on the outer surfaces of mycobacterial cells and have been associated with virulence (4, 8, 27, 29) and with lipid translocation (S. A. Cantrell and L. W. Riley, Keystone Symposium on Tuberculosis: Integrating Host and Pathogen Biology, 2 to 7 April 2005, Whistler, BC). Q-RT-PCR was used to test the hypothesis that they are down-regulated in RO and mtrB mutant cells.
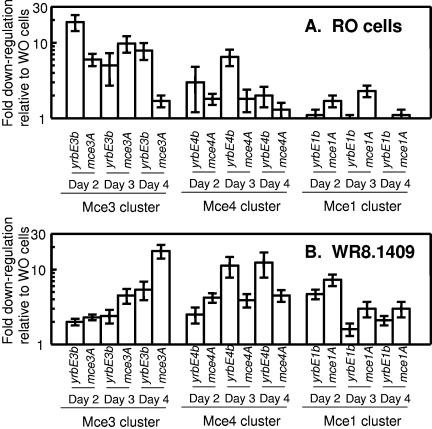
Expression of two representative genes within each mce cluster (yrbE-B and mce-A) was measured in WO and RO cells harvested 2, 3, and 4 days postinoculation. Expression of mce genes was normalized to that of sigA, a housekeeping gene that is commonly used for this purpose in analysis of mycobacterial gene expression (14, 19). Consistent down-regulation of genes in the mce3 cluster was observed in RO cells relative to WO cells; however, down-regulation of the other two mce clusters was more modest (Fig. 6A). The mce3 cluster was also down-regulated in the mtrB mutant WR8.1409 relative to the WO parent strain, although this down-regulation became apparent later in the growth curve. Down-regulation of the other two mce clusters was somewhat more pronounced in the mutant than in naturally occurring RO cells (Fig. 6B). Replicate experiments yielded substantially similar results, with the three mce clusters broadly down-regulated in the mtrB mutant and somewhat more narrowly down-regulated in the RO clone. The data suggest that mtrB is required for the expression of cell surface-associated Mce proteins in WO cells, consistent with a trans-acting regulatory function.
FIG. 6.
Down-regulation of mce mRNA in RO and mtrB mutant cells relative to WO cells. Q-RT-PCR analysis was conducted as described in the text. HMC02-WO cells were grown and harvested under the same conditions as HMC02-RO cells (A) and the mtrB mutant (B). Values are down-regulation (n-fold) of each locus relative to HMC02-WO cells. Each data point is the mean and standard deviation of triplicate Q-RT-PCRs. For each comparison, RNA extracts were diluted 1:1, 1:10, and 1:100, and the dilution that yielded the smallest standard deviation is shown.
Q-RT-PCR also detected weak (average, twofold) down-regulation of mtrA but not mtrB in RO cells relative to WO cells. Similarly, mtrA but not mtrB was two- to threefold down-regulated in the mtrB mutant WR8.1409 relative to the parent strain (data not shown). These observations are consistent with a self-regulatory function for mtrAB. They also suggest that transcription of mtrA and mtrB may not be coupled; however, additional analysis is needed to confirm this transcriptional structure.
DISCUSSION
The mtrB mutant was phase locked in the RO morphotype and could not form drug-resistant white or transparent subvariants even under antibiotic selective pressure. Introduction of a cloned wild-type mtrB allele restored to the mutant the ability to undergo these morphotypic switches. We hypothesize that mtrAB regulates the expression of multiple cell surface gene products, some of which are required for the cell envelope impermeability and multidrug resistance associated with the white and transparent morphotypes.
The intrinsic multidrug resistance of MAC has long been ascribed to cell envelope impermeability. Mutational data presented recently (24) and in this report are consistent with this model. It is easy to conceive of a nonspecific permeability barrier that excludes structurally diverse antibiotics from intracellular sites of activity. However, it is difficult to imagine how such a barrier could function without also inhibiting the uptake of beneficial compounds. For MAC, the solution may be its morphotypic switches, which enable the organism to toggle between permeable and impermeable forms. Such switches could help this environmental organism to adapt to diverse extracellular and intracellular challenges.
Interestingly, restoration of cloned wild-type mtrAB genes did not automatically convert the mutant to a visible, uniform white morphotype. Instead, it restored the ability of cells within the population to undergo the switch. This ability became apparent when the transformants were challenged with antibiotics on Etest plates or simply were transferred several times on CR plates. mtrAB may function directly in one or more morphotypic switches that occur in a subpopulation of cells in response to unknown signals. Alternatively, it may function in independent regulatory pathways that overlap the morphotypic switches. In this capacity it could be required for the expression of genes that are also required for the expression of drug-resistant morphotypes. In-depth genomic array analysis of morphotypic variants and mutants will help to discern these possibilities.
Although colony type switching and intrinsic multidrug resistance are peculiar to MAC, mtrAB is present in the genomes of multiple Mycobacterium species (18). mtrA, but not mtrB, is required for growth of M. tuberculosis in vitro (34). The response regulator protein MtrA is itself regulated by SigC, which is required for growth of M. tuberculosis in mice (31). It is an immunodominant antigen expressed in rabbits and humans during preclinical tuberculosis (30), and it has been found in the cell wall fraction of Mycobacterium leprae (15). These observations suggest that MtrAB functions during intracellular as well as extracellular growth, possibly at the cell surface.
More recent reports support this model. Deletion of mtrAB in Corynebacterium glutamicum resulted in increased sensitivity to penicillin and vancomycin and elongated cell shape, consistent with our observations in MAC (18). The C. glutamicum mutant exhibited altered expression of genes, including mepA, which was correlated with the cell shape phenotype, and several genes associated with osmoregulation. The mutant did not exhibit altered osmosensitivity; however, a role for mtrAB in osmoregulation remains a possibility (18). More recently, a shotgun transposon mutagenesis similar to ours, in which mutagenized clones of were screened for altered colony morphology, was conducted with M. tuberculosis. A single mutant was isolated with an insertion in the 3′ half of mtrB. The mutant exhibited altered colony morphology, altered cell wall lipids, and reduced virulence in mice, consistent with a role for mtrB in regulating the expression of cell surface structures (J. Lynnette and R. W. Stokes, Keystone Symposium on Tuberculosis: Integrating Host and Pathogen Biology, 2 to 7 April 2005, Whistler, BC). Most Mycobacterium species do not exhibit the striking colony type switches seen in MAC; however, they may undergo less conspicuous developmental processes that involve their cell walls. The conservation of mtrAB within the genus suggests that these processes are important to the biology of the organisms.
The mtrB mutant of MAC exhibited reduced expression of genetically scattered mce genes, consistent with a trans-acting regulatory function for mtrB. The mce1, mce3, and mce4 clusters were chosen for this analysis because (i) preliminary microarray data suggested that they were differentially regulated in naturally occurring red and white variants, (ii) their products are known to function at the cell surface, and (iii) they have been associated with mycobacterial virulence. The down-regulation of mce genes in the mutant was not attributable to a general decrease in transcriptional activity, because expression was normalized to expression of sigA. It is not yet known whether MtrAB acts directly on mce gene expression or indirectly through other regulatory systems.
The mce3 genes were also seen to be down-regulated in naturally occurring red variants of MAC. Because mce genes have been associated with mycobacterial virulence (4, 27, 29), it is tempting to speculate that up-regulation of these genes in white variants contributes to the virulence of the white morphotype. However, the function of Mce proteins in mycobacterial virulence remains unclear. There are nine mce gene clusters in the M. avium subsp. avium 104 genome (28), and these genes are present in multiple copies in pathogenic as well as nonpathogenic mycobacteria. Therefore, their functions probably are not confined to virulence. A recent report offers a new model for mce function that is consistent with our observations. Based on the presence of fatty acid dehydrogenase (fad) genes within the mce1 cluster of M. tuberculosis, it was hypothesized that mce genes might function in lipid synthesis or translocation. Consistent with this, thin-layer chromatography and mass spectroscopy revealed decreased amounts of certain mycolic acids in the cell walls of mce1 mutants of M. tuberculosis (S. A. Cantrell and L. W. Riley, Keystone Symposium on Tuberculosis: Integrating Host and Pathogen Biology). In MAC, each of the three mce clusters examined in the present study has at least one fad gene mapping nearby (within 17 ORFs of the mceA gene in each cluster). If mce genes are involved in the synthesis or translocation of cell wall lipids, then their down-regulation in the RO morphotype and in the mtrB mutant could contribute to the cell wall permeability and drug susceptibility exhibited by these forms.
With additional studies it should be possible to identify the genes that are regulated by mtrAB in the Actinomycetales. There are likely to be variations between organisms, as indicated by the fact that C. glutamicum, which carries a functional mtrAB system (18), does not have mce genes, while M. avium subsp. avium lacks strong homologs to mepA, betP, and other genes that are regulated by mtrAB in C. glutamicum (18, 28). However, in all organisms examined, mtrAB is seen to function in regulating the composition and function of the cell envelope. The existence of such a regulatory system suggests that the complex cell walls of mycobacteria and related organisms are dynamic structures that can change in response to various physiological needs.
Acknowledgments
We are grateful to Jennifer Lynnette, Richard Stokes, Sally Cantrell, Arnold Smith, and Tige Rustad for helpful discussions.
This work was supported by grants RO1-AI25767 and R21-AI061006 from the National Institutes of Health and by a grant from the Akibene Foundation. Equipment grants from the Firland Foundation and the M. J. Murdock Charitable Trust are also gratefully acknowledged. Preliminary M. avium genome sequence data were obtained from the Institute for Genomic Research (TIGR) website at http://www.tigr.org. Sequencing of the M. avium genome is being carried out by TIGR with support from the National Institutes of Health.
REFERENCES
- 1.Belisle, J. T., and P. J. Brennan. 1994. Molecular basis of colony morphology in Mycobacterium avium. Res. Microbiol. 145:237-242. [DOI] [PubMed] [Google Scholar]
- 2.Cangelosi, G. A., C. O. Palermo, and L. E. Bermudez. 2001. Phenotypic consequences of red-white colony type variation in Mycobacterium avium. Microbiology 147:527-533. [DOI] [PubMed] [Google Scholar]
- 3.Cangelosi, G. A., C. O. Palermo, J. P. Laurent, A. M. Hamlin, and W. H. Brabant. 1999. Colony morphotypes on Congo red agar segregate along species and drug susceptibility lines in the Mycobacterium avium-intracellulare complex. Microbiology 145:1317-1324. [DOI] [PubMed] [Google Scholar]
- 4.Chitale, S., S. Ehrt, I. Kawamura, T. Fujimura, N. Shimono, N. Anand, S. Lu, L. Cohen-Gould, and L. W. Riley. 2001. Recombinant Mycobacterium tuberculosis protein associated with mammalian cell entry. Cell. Microbiol. 3:247-254. [DOI] [PubMed] [Google Scholar]
- 5.Garcia, R. C., E. Banfi, and M. G. Pittis. 2000. Infection of macrophage-like THP-1 cells with Mycobacterium avium results in a decrease in their ability to phosphorylate nucleolin. Infect. Immun. 68:3121-3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.George, K. M., Y. Yuan, D. R. Sherman, and C. E. Barry. 1995. The biosynthesis of cyclopropanated mycolic acids in Mycobacterium tuberculosis. J. Biol. Chem. 270:27292-27298. [DOI] [PubMed] [Google Scholar]
- 7.Hayashi, T., A. Catanzaro, and S. P. Rao. 1997. Apoptosis of human monocytes and macrophages by Mycobacterium avium sonicate. Infect. Immun. 65:5262-5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hou, J. Y., J. E. Graham, and J. E. Clark-Curtiss. 2002. Mycobacterium avium genes expressed during growth in human macrophages detected by selective capture of transcribed sequences (SCOTS). Infect. Immun. 70:3714-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ibrahim, P., A. S. Whiteley, and M. R. Barer. 1997. SYTO16 labelling and flow cytometry of Mycobacterium avium. Lett. Appl. Microbiol. 25:437-441. [DOI] [PubMed] [Google Scholar]
- 10.Jarlier, V., and H. Nikaido. 1994. Mycobacterial cell wall: structure and role in natural resistance to antibiotics. FEMS Microbiol. Lett. 123:11-18. [DOI] [PubMed] [Google Scholar]
- 11.Khoo, K. H., J. B. Tang, and D. Chatterjee. 2001. Variation in mannose-capped terminal arabinan motifs of lipoarabinomannans from clinical isolates of Mycobacterium tuberculosis and Mycobacterium avium complex. J. Biol. Chem. 276:3863-3871. [DOI] [PubMed] [Google Scholar]
- 12.Laurent, J. P., K. A. Hauge, K. Burnside, and G. A. Cangelosi. 2003. Mutational analysis of cell wall biosynthesis in Mycobacterium avium. J. Bacteriol. 185:5003-5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mailaender, C., N. Reiling, H. Engelhardt, S. Bossmann, S. Ehlers, and M. Niederweis. 2004. The MspA porin promotes growth and increases antibiotic susceptibility of both Mycobacterium bovis BCG and Mycobacterium tuberculosis. Microbiology 150:853-864. [DOI] [PubMed] [Google Scholar]
- 14.Manganelli, R., E. Dubnau, S. Tyagi, F. R. Kramer, and I. Smith. 1999. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol. Microbiol. 31:715-724. [DOI] [PubMed] [Google Scholar]
- 15.Marques, M. A. M., S. Chitale, P. J. Brennan, and M. C. V. Pessolani. 1998. Mapping and identification of the major cell wall-associated components of Mycobacterium leprae. Infect. Immun. 66:2625-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mdluli, K., J. Swanson, E. Fischer, R. E. Lee, and C. E. Barry. 1998. Mechanisms involved in the intrinsic isoniazid resistance of Mycobacterium avium. Mol. Microbiol. 27:1223-1233. [DOI] [PubMed] [Google Scholar]
- 17.Mizuguchi, Y., T. Uodo, and T. Yamada. 1983. Mechanism of antibiotic resistance in Mycobacterium intracellulare. Microbiol. Immunol. 27:425-431. [DOI] [PubMed] [Google Scholar]
- 18.Moker, N., M. Brocker, S. Schaffer, R. Kramer, S. Morbach, and M. Bott. 2004. Deletion of the genes encoding the MtrA-MtrB two-component system of Corynebacterium glutamicum has a strong influence on cell morphology, antibiotic susceptibility and expression of genes involved in osmoprotection. Mol. Microbiol. 54:420-438. [DOI] [PubMed] [Google Scholar]
- 19.Mostowy, S., C. Cleto, D. R. Sherman, and M. A. Behr. 2004. The Mycobacterium tuberculosis complex transcriptome of attenuation. Tuberculosis 84:197-204. [DOI] [PubMed] [Google Scholar]
- 20.Mukherjee, S., M. Petrofsky, K. Yaraei, L. E. Bermudez, and G. A. Cangelosi. 2001. The white morphotype of Mycobacterium avium-intracellulare is common in infected humans and virulent in infection models. J. Infect. Dis. 184:1480-1484. [DOI] [PubMed] [Google Scholar]
- 21.Nikaido, H., and V. Jarlier. 1991. Permeability of the mycobacterial cell wall. Res. Microbiol. 142:437-443. [DOI] [PubMed] [Google Scholar]
- 22.Paget, E., and J. Davies. 1996. Apramycin resistance as a selective marker for gene transfer in mycobacteria. J. Bacteriol. 178:6357-6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paustian, M. L., V. Kapur, and J. P. Bannantine. 2005. Comparative genomic hybridizations reveal genetic regions within the Mycobacterium avium complex that are divergent from Mycobacterium avium subsp. paratuberculosis isolates. J. Bacteriol. 187:2406-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Philalay, J. S., C. O. Palermo, K. A. Hauge, T. R. Rustad, and G. A. Cangelosi. 2004. Genes required for intrinsic multidrug resistance in Mycobacterium avium. Antimicrob. Agents Chemother. 48:3412-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prinzis, S., B. Rivoire, and P. J. Brennan. 1994. Search for the molecular basis of morphological variation in Mycobacterium avium. Infect. Immun. 62:1946-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rastogi, N., K. S. Goh, and S. Clavel-Seres. 1997. Stazyme, a mycobacteriolytic preparation from a Staphylococcus strain, is able to break the permeability barrier in multiple drug resistant Mycobacterium avium. FEMS Immunol. Med. Microbiol. 19:297-305. [DOI] [PubMed] [Google Scholar]
- 27.Sassetti, C. M., and E. J. Rubin. 2003. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. USA 100:12989-12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Semret, M., G. Zhai, S. Mostowy, D. Alexander, G. A. Cangelosi, D. Cousins, D. van Soolingen, and M. Behr. 2004. Extensive genomic polymorphism within Mycobacterium avium. J. Bacteriol. 186:6332-6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimono, N., L. Morici, N. Casali, S. Cantrell, B. Sidders, S. Ehrt, and L. W. Riley. 2003. Hypervirulent mutant of Mycobacterium tuberculosis resulting from disruption of the mce1 operon. Proc. Natl. Acad. Sci. USA 100:15918-15923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh, K. K., X. Zhang, A. S. Patibandla, P. Chien, Jr., and S. Laal. 2001. Antigens of Mycobacterium tuberculosis expressed during preclinical tuberculosis: serological immunodominance of proteins with repetitive amino acid sequences. Infect. Immun. 69:4185-4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun, R., P. J. Converse, C. Ko, S. Tyagi, N. E. Morrison, and W. R. Bishai. 2004. Mycobacterium tuberculosis ECF sigma factor sigC is required for lethality in mice and for the conditional expression of a defined gene set. Mol. Microbiol. 52:25-38. [DOI] [PubMed] [Google Scholar]
- 32.Via, L. E., R. Curcic, M. H. Mudd, S. Dhandayuthapani, R. J. Ulmer, and V. Deretic. 1996. Elements of signal transduction in Mycobacterium tuberculosis: in vitro phosphorylation and in vivo expression of the response regulator MtrA. J. Bacteriol. 178:3314-3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woodley, C. L., and H. L. David. 1976. Effect of temperature on the rate of transparent to opaque colony type transition in Mycobacterium avium. Antimicrob. Agents Chemother. 9:113-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zahrt, T. C., and V. Deretic. 2000. An essential two-component signal transduction system in Mycobacterium tuberculosis. J. Bacteriol. 182:3832-3838. [DOI] [PMC free article] [PubMed] [Google Scholar]