Abstract
We have isolated and characterized in vitro mutants of the Lyme disease agent Borrelia burgdorferi that are resistant to spectinomycin, kanamycin, gentamicin, or streptomycin, antibiotics that target the small subunit of the ribosome. 16S rRNA mutations A1185G and C1186U, homologous to Escherichia coli nucleotides A1191 and C1192, conferred >2,200-fold and 1,300-fold resistance to spectinomycin, respectively. A 16S rRNA A1402G mutation, homologous to E. coli A1408, conferred >90-fold resistance to kanamycin and >240-fold resistance to gentamicin. Two mutations were identified in the gene for ribosomal protein S12, at a site homologous to E. coli residue Lys-87, in mutants selected in streptomycin. Substitutions at codon 88, K88R and K88E, conferred 7-fold resistance and 10-fold resistance, respectively, to streptomycin on B. burgdorferi. The 16S rRNA A1185G and C1186U mutations, associated with spectinomycin resistance, appeared in a population of B. burgdorferi parental strain B31 at a high frequency of 6 × 10−6. These spectinomycin-resistant mutants successfully competed with the wild-type strain during 100 generations of coculture in vitro. The aminoglycoside-resistant mutants appeared at a frequency of 3 × 10−9 to 1 ×10−7 in a population and were unable to compete with wild-type strain B31 after 100 generations. This is the first description of mutations in the B. burgdorferi ribosome that confer resistance to antibiotics. These results have implications for the evolution of antibiotic resistance, because the 16S rRNA mutations conferring spectinomycin resistance have no significant fitness cost in vitro, and for the development of new selectable markers.
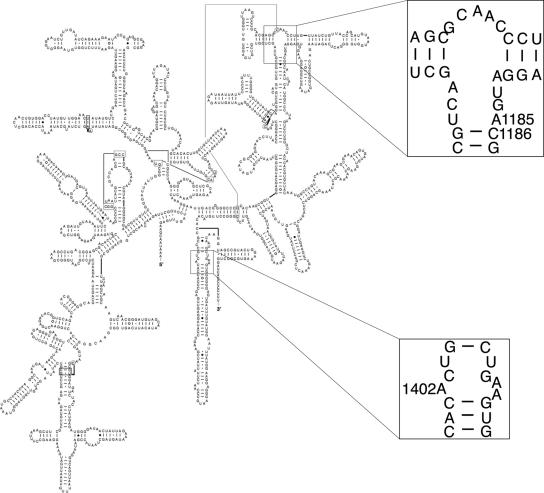
The spirochete Borrelia burgdorferi is a tick-borne pathogen that causes Lyme disease (62). Only a few antibiotics are available for clinical therapy of Lyme disease; the choice of new treatments depends on the pharmacodynamic interactions of the antimicrobial agent with the spirochete (30) and the frequency of mutations conferring resistance. In addition, only a few molecular tools are available for genetic manipulation of B. burgdorferi (53); the choice of new selectable markers, which is currently limited to antibiotic resistance (9, 16, 18, 56, 58), also depends on the mutation frequency. A high mutation rate conferring resistance to an antibiotic or a high frequency in the population of organisms resistant to the antibiotic interferes with selection. Further complicating the molecular genetics of B. burgdorferi is its unusual genome, which comprises a 910-kb linear chromosome as well as several linear and circular plasmids (2, 19). The chromosome contains two copies of the 23S and 5S rRNA genes and a single copy of the 16S rRNA gene (21, 22, 59). The secondary structure and sequence of the B. burgdorferi 16S rRNA (Fig. 1) are similar to those of other bacteria, including Escherichia coli and Thermus thermophilus, and to those of eukaryotic chloroplast 16S rRNA found in Chlamydomonas reinhardtii and Nicotiana tabacum (11). Because the 16S rRNA gene is present in only one copy in B. burgdorferi, a single mutational event in the 16S rRNA gene can confer resistance to any one of several antibiotics that target the small ribosomal subunit.
FIG. 1.
Borrelia burgdorferi 16S rRNA secondary-structure model. Enlarged regions show point mutations that confer resistance to spectinomycin (A1185 and C1186) and kanamycin or gentamicin (A1402). (Adapted from http://www.rna.icmb.utexas.edu/ [11] with permission.)
Many antibiotics that target ribosomes, such as spectinomycin and the aminoglycosides, either prevent translocation of tRNAs during translation or cause errors in selecting cognate tRNAs (3, 10, 12, 14, 50). Spectinomycin targets the 30S ribosomal subunit of many gram-negative bacteria, including E. coli (3, 8, 10, 67). Spectinomycin blocks elongation of the polypeptide during protein synthesis by binding to helix 34 of the 16S rRNA and apparently preventing the translocation of the peptidyl tRNA from the A-site to the P-site (3, 12). E. coli and other organisms have been shown to be resistant to spectinomycin when nucleotide C1192 or its cross-helix partner G1064, in helix 34 of the 16S rRNA, is mutated (5, 10, 40, 47, 60). Mutations at sites 1191 and 1193 in the chloroplast 16S rRNA of the eukaryotic organisms C. reinhardtii and N. tabacum confer resistance to spectinomycin as well (28, 65).
Kanamycin and gentamicin bind to helix 44 of the 16S rRNA and interfere with translational accuracy by increasing the affinity for noncognate tRNA selection (14, 15, 50, 51). Footprinting studies have demonstrated that resistance in E. coli results from decreased binding affinity of kanamycin or gentamicin to helix 44 upon mutation of site U1406 or A1408 in 16S rRNA (15, 43, 50, 51). Kanamycin and gentamicin resistance in Mycobacterium species have been mapped to mutations at homologous sites (49, 64).
Streptomycin also disrupts translational accuracy by increasing the affinity for noncognate tRNA (31, 32). This error-inducing activity is promoted by the formation of hydrogen bonds between streptomycin and several sites in 16S rRNA and ribosomal protein S12 (12, 61). Single mutations in the 16S rRNA or the S12 gene, rpsL, in E. coli have been shown to confer resistance to streptomycin (6, 20, 24, 39, 42, 46, 63). Mutations at homologous sites in the 16S rRNA and the S12 protein also confer resistance to streptomycin in many other organisms (17, 33, 41, 45, 61). An rpsL AAA-to-AGA mutation in codon 88, resulting in a K88R substitution in the S12 protein, confers resistance to streptomycin in the spirochete Leptospira biflexa (48).
There are only a few reports of antibiotic resistance in B. burgdorferi, including one describing resistance to erythromycin, which targets ribosomes in other bacteria (25, 57, 66). B. burgdorferi strain N40 was shown to be >3,000 times more resistant to the macrolide erythromycin than strain B31 (66). Sequencing of the 23S rRNA gene, a common site for mutations conferring erythromycin resistance in other bacteria, revealed no mutations in B. burgdorferi strain N40 (66). Although the mechanism of resistance has not been identified in strain N40, erythromycin-resistant clones showed changes in colony morphology on semisolid medium and lack of growth in liquid medium, indicating that resistance may be due to changes in metabolism (66).
We have identified five independent mutations in B. burgdorferi strain B31 conferring resistance to spectinomycin, kanamycin, gentamicin, or streptomycin. This study is the first to identify ribosomal mutations and one of only three, to our knowledge, in which genomic mutations have been associated with antibiotic resistance in B. burgdorferi (25, 57). The high frequency of spectinomycin-resistant mutants maintained in a population of strain B31-A and their ability to compete in vitro with wild-type cells imply that the antibiotic-resistant mutants not only are fit for the environment in which they were selected but also have maintained fitness in the absence of selection.
MATERIALS AND METHODS
Isolation and sequencing of antibiotic-resistant mutants.
Borrelia burgdorferi strain B31-A (9) was grown at 34°C in Barbour-Stoenner-Kelly (BSK)-H medium (Sigma). To determine the frequency of antibiotic-resistant cells in the B. burgdorferi population and to estimate the resistance levels of these strains, 107 or 108 cells (from a culture at 108 cells ml−1) were plated in semisolid BSK medium (54) with antibiotics (Sigma) at concentrations of 1.5, 6, or 30 μg ml−1 for spectinomycin, 40 μg ml−1 for kanamycin, 36 μg ml−1 for streptomycin, and 30 μg ml−1 for gentamicin. Kanamycin, gentamicin, and streptomycin plates with the same antibiotic concentrations were also inoculated with 109 cells obtained by concentrating cultures. The plates were incubated at 34°C for 11 to 14 days before colonies appeared. Seven spectinomycin-resistant colonies, three kanamycin-resistant colonies, four streptomycin-resistant colonies, and one gentamicin-resistant colony were isolated from the plates and grown in 10 ml of BSK-H medium containing the respective antibiotic; DNA was extracted as previously described (57).
DNA encoding the 3′ end of the 16S rRNA in the spectinomycin-, kanamycin-, and gentamicin-resistant strains was amplified by PCR using primers 16S 40F and 16S 659R (Table 1). The rpsE gene, which encodes the S5 protein, was also amplified in the spectinomycin-resistant mutants using primers rpsE U322F and rpsE 441R (Table 1). The rpsL gene, encoding protein S12, and the entire 16S rRNA gene were amplified in streptomycin-resistant strains using primers rpsL U106F and rpsG 58R and primers 16S U169F, 16S 40F, 16S 553F, 16S 659R, 16S 993R, 16S 1060F, 16S 1538R, BB425 3F, and BB426 288F, respectively (Table 1). The amplified products were purified using a QIAquick PCR purification kit (QIAGEN) and sequenced at the Murdock DNA Sequencing Facility (The University of Montana).
TABLE 1.
Oligonucleotides used for sequencing the genes encoding the 16S rRNA and the ribosomal proteins S5 and S12
| Oligonucleotide name | Gene | Sequence |
|---|---|---|
| rpsE U322F | rpsE (S5) | 5′-AATAAAGGACAAAATAGGG-3′ |
| rpsE 441R | rpsE (S5) | 5′-CAAAACTAAATCAAATGCC-3′ |
| 16S U169F | 16S rRNA | 5′-AATGACCTAAAATTAAGTCTAA-3′ |
| 16S 40F | 16S rRNA | 5′-TTGTTACGACTTCACCCCCCTC-3′ |
| 16S 553F | 16S rRNA | 5′-TCAAGCCCTGGTAAGGTTCC-3′ |
| 16S 659R | 16S rRNA | 5′-GAGTATGCTCGCAAGAGTG-3′ |
| 16S 993R | 16S rRNA | 5′-CGTTGTTCGGGATTATTG-3′ |
| 16S 1060F | 16S rRNA | 5′-TCATCACTTTGTCATTTC-3′ |
| 16S 1538R | 16S rRNA | 5′-AAATAACGAAGAGTTTGATCC-3′ |
| BB425 3F | 16S rRNA | 5′-GGAAGATGAGAGAAGGGAAG-3′ |
| BB426 288F | 16S rRNA | 5′-AGGATTCGCCTTTGCCAAG-3′ |
| rpsG 58R | rpsL (S12) | 5′-AATTATATCTGGTATCAACAAAAAC-3′ |
| rpsL U106F | rpsL (S12) | 5′-AAAATTAAAGTTAGTGAAAATATCG-3′ |
Susceptibility and growth assays.
Antibiotic susceptibility assays for parental strain B31-A and a representative strain for each of the five mutants were performed as previously described (18, 55). The concentration at which 50% of growth is inhibited (IC50) was determined for spectinomycin, streptomycin, kanamycin, and gentamicin.
Growth was assayed by spectrophotometry as previously described (55) with the following modifications. Cultures were inoculated with 106 cells of each strain into 50 ml of BSK-H, and 10 ml of each culture was removed every 24 h. The optical density at 600 nm was determined, and the doubling time was calculated during the exponential-growth phase. A Student t test was performed to evaluate the significance of the differences in doubling times between the wild type and four of the mutants.
Competition assay.
An in vitro competition assay between wild-type B31-A and the mutant antibiotic-resistant strains, similar to that described by Moine et al. (44), was performed. Cultures of B. burgdorferi strains B31-A, DCSPR3, DCSPR6, DCSmR4, and DCKAN3 were grown, without selection, in 10 ml of BSK-H to a density of 108 cells ml−1. Approximately 105 cells of each strain were added to 10 ml of BSK-H to formulate the following nine cultures: B31-A alone, DCSPR3 alone, DCSPR6 alone, DCSmR4 alone, DCKAN3 alone, B31-A plus DCSPR3, B31-A plus DCSPR6, B31-A plus DCSmR4, and B31-A plus DCKAN3. The cultures were incubated for 96 h until a density of about 108 cells ml−1 was reached, and 105 cells were passaged into 10 ml of BSK-H. After 5 or 10 passages (20 or 40 days), which correspond to about 50 or 100 generations, 103 cells from each of the nine different cultures were plated on semisolid BSK medium with and without the appropriate antibiotic (spectinomycin, streptomycin, or kanamycin) to screen for the ratio of antibiotic-resistant colonies to total colonies (antibiotic resistant plus wild type). In addition, 20 colonies were taken from the B31-A-plus-DCSPR3 plate with no selection and 20 colonies were taken from the B31-A-plus-DCSPR6 plate with no selection. Each of these colonies was added to one of the wells in a 24-well culture dish containing 2 ml of BSK-H with 30 μg ml−1 of spectinomycin. DCSPR3 or DCSPR6 mutants were added to three of the wells for positive controls, and one of the wells, to which no cells were added, served as a negative control. A color change in the BSK-H medium (from red to yellow; caused by the decrease in pH) was used to determine if the well was positive or negative for spectinomycin-resistant mutants.
RESULTS
In a previous effort to develop a new selectable marker for manipulating the B. burgdorferi genome, we discovered a high frequency of background mutants in 1.5 μg ml−1 spectinomycin, which precluded using this antibiotic for selection (18). Several other bacteria with 16S rRNA mutations conferring spectinomycin resistance have been identified, so we hypothesized that B. burgdorferi strain B31-A may harbor a subpopulation with a 16S rRNA mutation. The susceptibilities of wild-type B31-A to spectinomycin, as well as to the aminoglycosides kanamycin, gentamicin, and streptomycin, were determined (Table 2) and found to be similar to our previously published values (18).
TABLE 2.
Antibiotic susceptibilities and growth rates of the B. burgdorferi wild-type strain B31-A and antibiotic-resistant mutants
| Strain | Mutation | IC50 (μg ml−1) of the following antibiotica:
|
Doubling time (h)b | |||
|---|---|---|---|---|---|---|
| SPT | STR | KAN | GEN | |||
| B31-A | None | 0.22 | 11 | 9 | 2.5 | 7.9 ± 0.5 |
| DCSPR3 | 16S rRNA C1186U | 300 | 6 | 17 | 2.5 | 8.1 ± 0.3 |
| DCSPR6 | 16S rRNA A1185G | >500 | 9 | 13 | 2.5 | 8.1 ± 0.1 |
| DCKAN3 | 16S rRNA A1402G | 0.12 | 6 | >800 | >600 | 8.8 ± 0.6 |
| DCSmR3 | S12 K88R | 0.10 | 80 | 13 | 1.5 | ND |
| DCSmR4 | S12 K88E | 0.08 | 110 | 16 | 1.5 | 8.0 ± 0.2 |
SPT, spectinomycin; STR, streptomycin; KAN, kanamycin; GEN, gentamicin.
Values are means ± standard errors of the means for three independent experiments with a total of five replicates. ND, not determined.
Frequency and identification of spectinomycin-resistant mutations.
The frequency of the spectinomycin-resistant mutants was determined by plating either 107 or 108 cells of B. burgdorferi strain B31-A in 1.5 μg ml−1, 6.0 μg ml−1, or 30 μg ml−1 of spectinomycin, concentrations 7-fold, 27-fold, or 136-fold higher than the IC50, respectively, and colonies typically appeared within 11 days. Each of these experiments yielded a frequency of spectinomycin mutants in the B. burgdorferi population of about 6 × 10−6 (Table 3).
TABLE 3.
Frequency of spectinomycin-resistant mutants in strain B31-A
| Spectinomycin concn (μg ml−1) | No. of cells plated | No. of colonies | Frequency (10−6) |
|---|---|---|---|
| 1.5 | 108 | 572 ± 83a | 5.7 |
| 107 | 69 ± 22a | 6.9 | |
| 6.0 | 108 | 694 ± 126a | 6.9 |
| 107 | 62 ± 21a | 6.2 | |
| 30 | 108 | 514 ± 118b | 5.1 |
| 107 | 62 ± 5a | 6.2 |
Values are means ± standard errors of the means for four independent experiments.
Value is the mean ± standard error of the mean for seven independent experiments.
Spectinomycin-resistant mutations have been mapped to the 16S rRNA in other bacteria (3, 5, 12, 26, 40, 47, 60). Therefore, we sequenced the 16S rRNA gene in seven spectinomycin-resistant mutants to determine if similar mutations were responsible for resistance in B. burgdorferi. Three mutants, DCSPR5, DCSPR6, and DCSPR7, had an A1185G mutation in 16S rRNA. The other four mutants, DCSPR1, DCSPR2, DCSPR3, and DCSPR4, had a C1186U mutation in 16S rRNA. The C1186U spectinomycin resistance mutation is homologous to the site identified in spectinomycin-resistant E. coli and Salmonella enterica (40, 47, 60), while the A1185G mutation is homologous to that found in chloroplasts of C. reinhardtii (28) and has only recently been identified in bacteria (5). There were no compensatory mutations at the cross-helix partners G1057 (1186) and U1058 (1185), which correspond to E. coli sites G1064 and U1065. Mutations in the rpsE gene, encoding ribosomal protein S5, are also known to confer resistance to spectinomycin (7, 23, 68). This gene was sequenced for our spectinomycin-resistant isolates, and no mutations were found.
Antibiotic susceptibilities of spectinomycin-resistant mutants.
Susceptibility assays with DCSPR3 (C1186U) and DCSPR6 (A1185G) revealed that both mutations confer a high level of resistance to spectinomycin. The IC50 of spectinomycin for wild-type B. burgdorferi strain B31-A is 0.2 μg ml−1. For the C1186U mutant, DCSPR3, the IC50 is ∼300 μg ml−1, representing at least a 1,000-fold increase in resistance to spectinomycin. The spectinomycin IC50 is >500 μg ml−1 for the A1185G mutant, DCSPR6, which is thus >2,000 times more resistant than the wild type (Table 2).
DCSPR3 and DCSPR6 did not have high levels of cross-resistance to the aminoglycosides streptomycin, kanamycin, and gentamicin (Table 2). The IC50s of streptomycin, kanamycin, and gentamicin for the wild-type B. burgdorferi strain B31-A were 11 μg ml−1, 9 μg ml−1, and 2.5 μg ml−1, respectively, nearly identical to those reported previously (18). Aminoglycoside susceptibilities of DCSPR3 and DCSPR6 were within twofold of those of the parental strain B31-A (Table 2).
Frequency and identification of aminoglycoside-resistant mutants.
The high frequency of spectinomycin-resistant mutants identified in our studies motivated us to examine the frequency and mechanism of resistance to closely related aminoglycoside antibiotics. The maintenance of spectinomycin-resistant mutants at high frequency indicates that there is a low fitness cost for these mutants. The structurally similar aminoglycosides were tested to determine if there is a corresponding fitness cost associated with these mutations. Identifying the frequency and levels of resistance of these mutants may also aid in choosing reliable selectable markers for genetic manipulation or clinical treatment of B. burgdorferi. Already, several markers have been constructed with aminoglycoside resistance cassettes (9, 16, 18, 58).
The frequency of kanamycin resistance was determined by plating 108 or 109 cells of B. burgdorferi strain B31-A on semisolid BSK medium containing 40 μg ml−1 kanamycin. Colonies appeared on kanamycin plates in 14 days, 3 days later than the appearance of spectinomycin-resistant colonies. The frequency of kanamycin-resistant mutants was between 1.3 × 10−8 and 1.4 × 10−7 (Table 4). Plating of 109 cells of B31-A on 30-μg ml−1 gentamicin plates yielded similar results: the frequency of gentamicin-resistant mutants was 1 × 10−8 (Table 4).
TABLE 4.
Frequency of aminoglycoside-resistant mutants in strain B31-A
| Antibiotic (μg ml−1) | No. of cells plated | No. of colonies | Frequency |
|---|---|---|---|
| Kanamycin (40) | 109 | 13.2 ± 4.3a | 1.3 × 10−8 |
| 108 | 14.0 ± 6.0b | 1.4 × 10−7 | |
| Streptomycin (36) | 109 | 3.4 ± 1.7a | 3.4 × 10−9 |
| 108 | 2.0 ± 1.0a | 2.0 × 10−8 | |
| Gentamicin (30) | 109 | 10.3 ± 1.2c | 1.0 × 10−8 |
Values are means ± standard errors of the means for five independent experiments.
Value is the mean ± standard error of the mean for four independent experiments.
Value is the mean ± standard error of the mean for three independent experiments.
To determine the location of the mutation that conferred resistance to kanamycin and gentamicin, three colonies were isolated from kanamycin plates and one colony from a gentamicin plate. Sequencing of the 16S rRNA gene from all four of the clones revealed an A1402G mutation (Table 2), homologous to an A1408G mutation in E. coli (15, 50). The kanamycin-resistant clone DCKAN3 represents both the kanamycin-resistant mutants and the gentamicin-resistant mutant in further studies, because they have the same 16S rRNA mutation.
The frequency of streptomycin resistance is 2.0 × 10−8 to 3.4 × 10−9, based on the number of colonies that grew on BSK plates containing 36 μg ml−1 of streptomycin (Table 4). Colonies appeared within 11 days after plating. Sequence analysis of four streptomycin-resistant mutants revealed two different mutations in the rpsL gene, encoding the S12 protein. An A263G rpsL mutation in DCSmR3 resulted in a K88R substitution in the S12 protein, and an A262G mutation resulted in a K88E substitution in DCSmR4 (Table 2). Complete sequencing of the 16S rRNA gene on both strands revealed no 16S rRNA mutations in these streptomycin-resistant strains.
Antibiotic susceptibilities of aminoglycoside-resistant mutants.
The DCKAN3 mutant was assayed for susceptibility to kanamycin and the other antibiotics. The IC50 of kanamycin was >800 μg ml−1, more than 90 times the wild-type IC50 (Table 2). The kanamycin-resistant mutant was, as expected, cross-resistant to gentamicin at high levels. The IC50 of gentamicin for this A1402G mutant (DCKAN3) was >600 μg ml−1, indicating that it is >240-fold more resistant than wild-type B31-A (Table 2). The A1402G mutation provides no resistance to spectinomycin or streptomycin (Table 2).
The two streptomycin-resistant rpsL mutations conferred slightly different levels of resistance to streptomycin (Table 2). The IC50s were 80 μg ml−1 for the K88R mutant (DCSmR3) and 110 μg ml−1 for the K88E mutant (DCSmR4), which are thus 7-fold and 10-fold more resistant, respectively, than the wild type. Both of these streptomycin-resistant mutants had small increases in susceptibility to spectinomycin and gentamicin (Table 2).
Fitness of spectinomycin- and aminoglycoside-resistant mutants.
The frequency of spectinomycin-resistant mutants in the B31-A population was approximately 100-fold higher than the frequency of the aminoglycoside-resistant mutants. We hypothesized that this difference was a result of a lower fitness cost of the 16S rRNA C1186U (DCSPR3) and A1185G (DCSPR6) mutations conferring spectinomycin resistance. This lower fitness cost enables spectinomycin-resistant mutants to compete with wild-type B31-A and to be maintained at frequencies higher than the typical mutation frequency observed for most bacteria (1, 35-37). To test this hypothesis, growth and competition assays were performed with B31-A and four of the mutants.
There were no statistically significant differences in the growth rates of the mutants and wild-type B31-A (Table 2), although DCKAN3 grew modestly slower than the other strains. The doubling time for B31-A was calculated as 7.9 h, while the doubling times were 8.8 h for DCKAN3 (P = 0.30), 8.1 h for DCSPR3 (P = 0.53) and DCSPR6 (P = 0.77), and 8.0 h for DCSmR4 (P = 0.96).
At the start of each competition assay, approximately 103 cells from each culture were plated to estimate the percentage of antibiotic-resistant mutant cells and B31-A wild-type cells in the competition cocultures. The number of colonies on plates without antibiotic selection compared to plates with selection indicated that there were approximately equal numbers of mutant and wild-type cells (data not shown). Comparison of the percentage of colonies that grew with selection at the beginning of the assay and the percentage of colonies that grew with selection at the end of the assay indicated that streptomycin-resistant and kanamycin/gentamicin-resistant mutants (DCSmR4 and DCKAN3) were completely outgrown between 50 and 100 generations, while the spectinomycin-resistant mutants (DCSPR3 and DCSPR6) were still present at 100 generations (Table 5).
TABLE 5.
Competition of antibiotic-resistant strains with the wild type
| Strain | % of antibiotic-resistant colonies at:
|
|
|---|---|---|
| 50 generations | 100 generations | |
| B31-A + DCSPR3 | 41 ± 4.4a (DCSPR3) | 18 ± 4.4b (DCSPR3) |
| B31-A + DCSPR6 | 41 ± 2.1a (DCSPR6) | 8 ± 3.1b (DCSPR6) |
| B31-A + DCSmR4 | 6.0 ± 2.0c (DCSmR4) | 0 ± 0c (DCSmR4) |
| B31-A + DCKAN3 | 0.75 ± 0.25c (DCKAN3) | 0 ± 0c (DCKAN3) |
Values are means ± standard errors of the means for three independent experiments.
Values are means ± standard errors of the means for four independent experiments.
Values are means ± standard errors of the means for five independent experiments.
The spectinomycin-resistant mutants cultured with B31-A fared much better than the aminoglycoside-resistant mutants, surviving in the population for 100 generations. Comparison of the numbers of cells from the B31-A-plus-DCSPR3 cocultures plated with selection versus no selection indicated that 18% of the cells remaining after 100 generations were DCSPR3. Since there is no selection for wild-type B31-A or against spectinomycin-resistant cells on the plates without spectinomycin, a test was designed to assess the percentage of colonies that were spectinomycin resistant on the plates without antibiotic. Twenty colonies from a plate containing B31-A plus DCSPR3 with no selection were isolated and grown in liquid BSK-H with 30 μg ml−1 of spectinomycin. The results of this experiment were similar to that of the experiment comparing the number of colonies on semisolid medium with selection versus no selection: 4 of the 20 colonies (20%) were spectinomycin resistant. Similar, but lower, numbers were also obtained for the coculture of B31-A plus DCSPR6. Comparison of the number of colonies on spectinomycin plates versus the number of colonies on plates without spectinomycin indicated that 8% of the cells in the coculture were resistant to spectinomycin (DCSPR6) after 100 generations. Isolating 20 B31-A-plus-DCSPR6 colonies from plates with no selection and growing them in liquid BSK-H with 30 μg ml−1 of spectinomycin produced two spectinomycin-resistant wells, suggesting that 10% of the colonies from the coculture were still spectinomycin resistant (DCSPR6) after 100 generations. The experiment at 50 generations had more spectinomycin-resistant mutants present than that at 100 generations. Plating with and without selection showed that 41% of the cells were spectinomycin-resistant mutants in both the B31-A-plus-DCSPR3 and the B31-A-plus-DCSPR6 cocultures (Table 5).
No colonies from the coculture of B31-A plus DCKAN3 grew on plates with kanamycin, and no colonies from the coculture of B31-A plus DCSmR4 appeared on plates with streptomycin after 100 generations. However, colonies from the same cultures grew on plates with no selection, indicating that wild-type B31-A was the only cell type in the culture. Plating of the B31-A-plus-DCSmR4 coculture at 50 generations did produce colonies on the streptomycin plates. Independent experiments produced similar results, indicating that 6% of the cells in the B31-A-plus-DCSmR4 coculture were DCSmR4 at 50 generations of the competition assay. Less than 1% of the cells in the B31-A-plus-DCKAN3 coculture were DCKAN3 mutants at 50 generations (Table 5).
Single cultures used as controls for the competition assays revealed that each of the mutant strains, DCSPR3, DCSPR6, DCSmR4, and DCKAN3, grew on plates with and without antibiotic selection after 100 generations, indicating that these cells were still viable and antibiotic resistant (data not shown).
DISCUSSION
The high frequency of antibiotic-resistant colonies appearing upon spectinomycin selection (18) prompted an investigation to identify mutations responsible for resistance. We hypothesized that mutations in the 16S rRNA or small ribosomal subunit proteins were the cause of spectinomycin resistance (3, 7, 10, 23, 26, 40, 47, 60) and that the high frequency of these mutants in the B. burgdorferi B31-A population was due either to a high mutation rate or to a low fitness cost (1, 37). We have identified five different mutations conferring resistance to spectinomycin, streptomycin, kanamycin, and gentamicin; the spectinomycin-resistant mutants have a lower fitness cost than the aminoglycoside-resistant mutants.
Spectinomycin resistance.
Mutations at several sites including 1191 to 1193 in the 16S rRNA of E. coli, S. enterica, C. reinhardtii, and Chlamydia psittaci have been shown to confer resistance to spectinomycin (5, 10, 28, 40, 47, 60). We have identified an A1185G mutation and a C1186U mutation in Borrelia burgdorferi 16S rRNA (nucleotide positions homologous to E. coli A1191 and C1192) that confer high levels of resistance to spectinomycin (Table 2). The 1,300-fold increase in resistance to spectinomycin due to the C1186U mutation is similar to the resistance levels reported for a homologous mutant of S. enterica (47). Perhaps of more interest is the A1185G mutation, which confers over 2,200-fold resistance to spectinomycin, since it has only recently been described in a bacterium (5). A homologous mutation, which confers a high level of resistance to spectinomycin, has been identified in the chloroplast of the eukaryote C. reinhardtii (A1123G) (28).
Sequencing of the 16S rRNA gene revealed no other mutations including the complementary nucleotides 1057 and 1058 (E. coli nucleotides 1063 and 1064). In addition, no mutations were found in the rpsE gene encoding the S5 protein, a site of spectinomycin resistance mutations in other bacteria (7, 23, 68). Secondary mutations in E. coli have been shown to confer spectinomycin dependence when more than one mutation occurs in the small ribosomal subunit involving S4 or S5 proteins (13). Bacillus subtilis also appears to be spectinomycin dependent when double mutations occur in the ribosomal genes (29). This lack of dependence was confirmed by the ability of DCSPR3 and DCSPR6 to grow in the absence of spectinomycin (data not shown).
Mutants DCSPR6 (A1185G) and DCSPR3 (C1186U) had the same susceptibility to gentamicin as wild-type B31-A. However, both of these spectinomycin-resistant mutants were slightly more susceptible to streptomycin and slightly more resistant to kanamycin (Table 2). These minor differences in aminoglycoside susceptibilities may be a result of small conformational changes along the 16S rRNA that affect the binding of streptomycin and kanamycin to the 16S rRNA.
The frequency of the spectinomycin-resistant phenotype includes both the C1186U and A1185G mutations; the frequency of individual mutations was not determined. Additional point mutations in the 16S rRNA gene, which have been identified in other organisms as conferring spectinomycin resistance (5, 10, 26, 28, 65), may contribute to the phenotypic mutation rate as well but were not represented in the seven mutants sequenced. The concentration of spectinomycin (1.5 to 30 μg ml−1) and the number of B31-A cells plated (107 or 108) did not affect the frequency of mutants (Table 3). The frequency of spectinomycin-resistant mutants is considerably higher than the frequencies of the aminoglycoside-resistant B. burgdorferi mutants also reported in this study (Table 4) and the spectinomycin-resistant mutation frequencies reported for most other bacteria. Spectinomycin-resistant mutants appear at frequencies of 2 × 10−9 in E. coli (35), 3 × 10−8 in Salmonella enterica serovar Enteritidis (35), and ∼10−8 in Mycoplasma mycoides (36), although the frequency for C. psittaci is higher, at 5 × 10−5 (5).
DCSPR3 and DCSPR6 were able to compete with B31-A better than either of the aminoglycoside-resistant mutants DCSmR4 or DCKAN3. The high frequency of the spectinomycin-resistant mutants (6 × 10−6) compared to the aminoglycoside-resistant mutants in the B31-A population is likely the result of a lower fitness cost associated with these mutations (1, 37). This lower fitness cost probably indicates that the spectinomycin-resistant ribosomes function with efficiency comparable to wild-type ribosomes. Sartakova et al. reported that B. burgdorferi was considerably more susceptible to spectinomycin in liquid medium than in semisolid medium (58). Our data suggest that this difference was due to the greater number of spirochetes assayed on semisolid medium (1 × 108 compared to 2 × 105), which revealed the subpopulation of spectinomycin-resistant mutants.
Kanamycin and gentamicin resistance.
Resistance to the aminoglycosides kanamycin and gentamicin in E. coli is conferred as a result of U1406A or A1408G mutations in 16S rRNA that reduce the binding affinity of the antibiotic for helix 44 (50, 51). We have identified an A1402G mutation in B. burgdorferi strain B31-A conferring high levels of resistance to kanamycin and gentamicin (Table 2). High levels of resistance to kanamycin and gentamicin are also found in E. coli with the homologous A1408G mutation (15, 43, 50) and Mycobacterium tuberculosis with the homologous A1400G mutation (64). Susceptibility to the antibiotics spectinomycin and streptomycin was slightly increased with the A1402G mutation (Table 2).
The frequencies of kanamycin-resistant mutants and streptomycin-resistant mutants depended on the number of cells plated (Table 4), which was not the case with spectinomycin-resistant mutants (Table 3). We do not know the reason for this but assume that it is an artifact of the plating technique, which requires concentrating a culture to yield 109 cells.
Streptomycin resistance.
Mutations at several sites in the 16S rRNA gene and the rpsL gene encoding S12 have been identified as conferring resistance to streptomycin in many organisms. We have identified two mutations in the B. burgdorferi rpsL gene conferring resistance to streptomycin. These mutations at codon 88 encode K88R and K88E amino acid substitutions in the ribosomal S12 protein. The frequency of the streptomycin-resistant mutations (Table 4), which includes both the K88R (DCSmR3) and K88E (DCSmR4) mutations, is in agreement with previous reports for other bacteria. The mutation frequency for the rpsL gene is ∼10−8 in Mycobacterium smegmatis (61), ∼4 × 10−10 in E. coli (38), and ∼10−7 in T. thermophilus (27). The frequency of the K88R and K88E mutants is similar to that of the kanamycin-resistant mutants and 100-fold less than that of the spectinomycin-resistant mutants in B. burgdorferi.
Neither mutant exhibited any of the characteristics associated with streptomycin dependence. Complete sequencing of the rpsL and 16S rRNA genes revealed no secondary mutations associated with streptomycin dependence. Another characteristic of this phenotype is increased resistance to kanamycin and gentamicin. Although the level of resistance to kanamycin did increase slightly, the DCSmR4 mutant was able to grow on semisolid medium without streptomycin, and resistance to gentamicin decreased slightly, which is not consistent with the streptomycin-dependent phenotype.
The resistance levels for DCSmR3 (7-fold) and DCSmR4 (10-fold) are much lower than those for DCSPR3, DCSPR6, and DCKAN3, which are all >100-fold more resistant to the respective antibiotics used to select them (Table 2). The levels of resistance in B. burgdorferi are typical of those of other streptomycin-resistant mutants. Similar susceptibilities have been identified in M. smegmatis mutants that have the homologous mutations in rpsL as well as in 16S rRNA (61). The slight differences in streptomycin resistance between DCSmR3 (the conservative K88R substitution) and DCSmR4 (K88E) may be the result of the change in the charge at this position in the S12 protein.
DCSmR4 was unable to compete with wild-type B31-A, indicating a significant fitness cost for this mutation (Table 5). S12 mutations have been shown to affect the selection of cognate tRNAs and the rates of polypeptide chain elongation in E. coli and T. thermophilus, which may be the source of this fitness cost (34, 51). Mutations in the E. coli rpsL gene slow the rates of polypeptide chain elongation by nearly 20% (34). A lower rate of translation could explain the inability of DCSmR4 to compete with wild-type B31-A. Conversely, spectinomycin-resistant mutations have been shown to have little if any effect on ribosome function even in the absence of antibiotic (10, 40), although recently some synergistic and antagonistic effects were demonstrated when additional mutations were combined with the C1192U spectinomycin resistance mutation in E. coli (52). Recently described spectinomycin-resistant mutants of C. psittaci demonstrated significant decreases in fitness (4). Notably, the C. psittaci C1192U mutant was more fit than the A1191G mutant, which is similar to our data with B. burgdorferi (Tables 2 and 5).
Antibiotic resistance as a selectable marker.
Although none of the antibiotics in this study are used clinically to treat Lyme disease, resistance to aminoglycosides has been employed as a selectable marker for genetic manipulation of the spirochete (9, 16, 18, 58). We have found that a large subpopulation of spectinomycin-resistant B. burgdorferi strain B31-A prevents functional selection of the recombinant aadA cassette (18) with spectinomycin. More selectable markers are needed for increasingly sophisticated genetic experiments with the Lyme disease agent (53); the caveat regarding the frequency of background mutants resistant to an antibiotic should be considered prior to developing candidate genetic markers.
Acknowledgments
We thank Mike Gilbert, Scott Hennelly, and Ira Schwartz for thoughtful and critical reading of the manuscript; Patty McIntire (Murdock DNA Sequencing Facility) for DNA sequencing; Jill Dion, Barbara Wright, Sharyl Bundle, Meghan Lybecker, Dave Patterson, Mike Minnick, Kate Pflughoeft, Laura Smitherman, Kristi Frank, Amanda Ng, Michael Travisano, and Frank Rosenzweig for useful discussions or assistance with experiments; Patti Rosa and Tom Schwan for strains; and the anonymous reviewers for constructive suggestions.
This work was supported by grants from the National Science Foundation (MCB-9722408) and the National Institutes of Health (AI053195 and AI051486) to D.S.S.
REFERENCES
- 1.Andersson, D. I. 2003. Persistence of antibiotic resistant bacteria. Curr. Opin. Microbiol. 6:452-456. [DOI] [PubMed] [Google Scholar]
- 2.Barbour, A. G., and C. F. Garon. 1987. Linear plasmids of the bacterium Borrelia burgdorferi have covalently closed ends. Science 237:409-411. [DOI] [PubMed] [Google Scholar]
- 3.Bilgin, N., A. A. Richter, M. Ehrenberg, A. E. Dahlberg, and C. G. Kurland. 1990. Ribosomal RNA and protein mutants resistant to spectinomycin. EMBO J. 9:735-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Binet, R., and A. T. Maurelli. 2005. Fitness cost due to mutations in the 16S rRNA associated with spectinomycin resistance in Chlamydia psittaci 6BC. Antimicrob. Agents Chemother. 49:4455-4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Binet, R., and A. T. Maurelli. 2005. Frequency of spontaneous mutations that confer antibiotic resistance in Chlamydia spp. Antimicrob. Agents Chemother. 49:2865-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bohman, K., T. Ruusala, P. C. Jelenc, and C. G. Kurland. 1984. Kinetic impairment of restrictive streptomycin-resistant ribosomes. Mol. Gen. Genet. 198:90-99. [DOI] [PubMed] [Google Scholar]
- 7.Bollen, A., J. Davies, M. Ozaki, and S. Mizushima. 1969. Ribosomal protein conferring sensitivity to the antibiotic spectinomycin in Escherichia coli. Science 165:85-86. [DOI] [PubMed] [Google Scholar]
- 8.Bollen, A., T. Helser, T. Yamada, and J. Davies. 1969. Altered ribosomes in antibiotic-resistant mutants of E. coli. Cold Spring Harbor Symp. Quant. Biol. 34:95-100. [DOI] [PubMed] [Google Scholar]
- 9.Bono, J. L., A. F. Elias, J. J. Kupko III, B. Stevenson, K. Tilly, and P. Rosa. 2000. Efficient targeted mutagenesis in Borrelia burgdorferi. J. Bacteriol. 182:2445-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brink, M. F., G. Brink, M. P. Verbeet, and H. A. de Boer. 1994. Spectinomycin interacts specifically with the residues G1064 and C1192 in 16S rRNA, thereby potentially freezing this molecule into an inactive conformation. Nucleic Acids Res. 22:325-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cannone, J. J., S. Subramanian, M. N. Schnare, J. R. Collett, L. M. D'Souza, Y. Du, B. Feng, N. Lin, L. V. Madabusi, K. M. Muller, N. Pande, Z. Shang, N. Yu, and R. R. Gutell. 2002. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3:2. (Correction, 3:15, 2002.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carter, A. P., W. M. Clemons, D. E. Brodersen, R. J. Morgan-Warren, B. T. Wimberly, and V. Ramakrishnan. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407:340-348. [DOI] [PubMed] [Google Scholar]
- 13.Dabbs, E. R. 1977. A spectinomycin dependent mutant of Escherichia coli. Mol. Gen. Genet. 151:261-267. [DOI] [PubMed] [Google Scholar]
- 14.Davies, J., L. Gorini, and B. D. Davis. 1965. Misreading of RNA codewords induced by aminoglycoside antibiotics. Mol. Pharmacol. 1:93-106. [PubMed] [Google Scholar]
- 15.De Stasio, E. A., D. Moazed, H. F. Noller, and A. E. Dahlberg. 1989. Mutations in 16S ribosomal RNA disrupt antibiotic-RNA interactions. EMBO J. 8:1213-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elias, A. F., P. E. Stewart, D. Grimm, M. J. Caimano, C. H. Eggers, K. Tilly, J. L. Bono, D. R. Akins, J. D. Radolf, T. G. Schwan, and P. Rosa. 2002. Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect. Immun. 70:2139-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finken, M., P. Kirschner, A. Meier, A. Wrede, and E. C. Böttger. 1993. Molecular basis of streptomycin resistance in Mycobacterium tuberculosis: alterations of the ribosomal protein S12 gene and point mutations within a functional 16S ribosomal RNA pseudoknot. Mol. Microbiol. 9:1239-1246. [DOI] [PubMed] [Google Scholar]
- 18.Frank, K. L., S. F. Bundle, M. E. Kresge, C. H. Eggers, and D. S. Samuels. 2003. aadA confers streptomycin-resistance in Borrelia burgdorferi. J. Bacteriol. 185:6723-6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J.-F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quakenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. K. Adams, J. Gocayne, J. Weidman, T. Utterback, L. Watthey, L. McDonald, P. Artiach, C. Bowman, S. Garland, C. Fujii, M. D. Cotton, K. Horst, K. Roberts, B. Hatch, H. O. Smith, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 20.Frattali, A. L., M. K. Flynn, E. A. De Stasio, and A. E. Dahlberg. 1990. Effects of mutagenesis of C912 in the streptomycin binding region of Escherichia coli 16S ribosomal RNA. Biochim. Biophys. Acta 1050:27-33. [DOI] [PubMed] [Google Scholar]
- 21.Fukunaga, M., and M. Sohnaka. 1992. Tandem repeat of the 23S and 5S ribosomal RNA genes in Borrelia burgdorferi, the etiological agent of Lyme disease. Biochem. Biophys. Res. Commun. 183:952-957. [DOI] [PubMed] [Google Scholar]
- 22.Fukunaga, M., Y. Yanagihara, and M. Sohnaka. 1992. The 23S/5S ribosomal RNA genes (rrl/rrf) are separate from the 16S ribosomal RNA gene (rrs) in Borrelia burgdorferi, the aetiological agent of Lyme disease. J. Gen. Microbiol. 138:871-877. [DOI] [PubMed] [Google Scholar]
- 23.Funatsu, G., E. Schiltz, and H. G. Wittmann. 1972. Ribosomal proteins. XXVII. Localization of the amino acid exchanges in protein S5 from two Escherichia coli mutants resistant to spectinomycin. Mol. Gen. Genet. 114:106-111. [DOI] [PubMed] [Google Scholar]
- 24.Funatsu, G., and H. G. Wittmann. 1972. Ribosomal proteins. XXXIII. Location of amino-acid replacements in protein S12 isolated from Escherichia coli mutants resistant to streptomycin. J. Mol. Biol. 68:547-550. [DOI] [PubMed] [Google Scholar]
- 25.Galbraith, K. M., A. C. Ng, B. J. Eggers, C. R. Kuchel, C. H. Eggers, and D. S. Samuels. 2005. parC mutations in fluoroquinolone-resistant Borrelia burgdorferi. Antimicrob. Agents Chemother. 49:4354-4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galimand, M., G. Gerbaud, and P. Courvalin. 2000. Spectinomycin resistance in Neisseria spp. due to mutations in 16S rRNA. Antimicrob. Agents Chemother. 44:1365-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gregory, S. T., J. H. Cate, and A. E. Dahlberg. 2001. Streptomycin-resistant and streptomycin-dependent mutants of the extreme thermophile Thermus thermophilus. J. Mol. Biol. 309:333-338. [DOI] [PubMed] [Google Scholar]
- 28.Harris, E. H., B. D. Burkhart, N. W. Gillham, and J. E. Boynton. 1989. Antibiotic resistance mutations in the chloroplast 16S and 23S rRNA genes of Chlamydomonas reinhardtii: correlation of genetic and physical maps of the chloroplast genome. Genetics 123:281-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henkin, T. M., K. M. Campbell, and G. H. Chambliss. 1979. Spectinomycin dependence in Bacillus subtilis. J. Bacteriol. 137:1452-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunfeld, K.-P., P. Kraiczy, E. Kekoukh, V. Schafer, and V. Brade. 2002. Standardised in vitro susceptibility testing of Borrelia burgdorferi against well-known and newly developed antimicrobial agents—possible implications for new therapeutic approaches to Lyme disease. Int. J. Med. Microbiol. 291(Suppl. 33):125-137. [DOI] [PubMed] [Google Scholar]
- 31.Karimi, R., and M. Ehrenberg. 1994. Dissociation rate of cognate peptidyl-tRNA from the A-site of hyper-accurate and error-prone ribosomes. Eur. J. Biochem. 226:355-360. [DOI] [PubMed] [Google Scholar]
- 32.Karimi, R., and M. Ehrenberg. 1996. Dissociation rates of peptidyl-tRNA from the P-site of E. coli ribosomes. EMBO J. 15:1149-1154. [PMC free article] [PubMed] [Google Scholar]
- 33.Kavanagh, T. A., K. M. O'Driscoll, P. F. McCabe, and P. J. Dix. 1994. Mutations conferring lincomycin, spectinomycin, and streptomycin resistance in Solanum nigrum are located in three different chloroplast genes. Mol. Gen. Genet. 242:675-680. [DOI] [PubMed] [Google Scholar]
- 34.Kurland, C., D. Hughes, and M. Ehrenberg. 1996. Limitations of translational accuracy, p. 979-1004. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, B. K. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 35.LeClerc, J. E., B. Li, W. L. Payne, and T. A. Cebula. 1996. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 274:1208-1211. [DOI] [PubMed] [Google Scholar]
- 36.Lee, D. H., R. J. Miles, and J. R. M. Inal. 1987. Antibiotic sensitivity and mutation rates to antibiotic resistance in Mycoplasma mycoides ssp. mycoides. Epidemiol. Infect. 98:361-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lenski, R. E. 1998. Bacterial evolution and the cost of antibiotic resistance. Int. Microbiol. 1:265-270. [PubMed] [Google Scholar]
- 38.Levin, B. R., V. Perrot, and N. Walker. 2000. Compensatory mutations, antibiotic resistance and the population genetics of adaptive evolution in bacteria. Genetics 154:985-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lodmell, J. S., R. R. Gutell, and A. E. Dahlberg. 1995. Genetic and comparative analyses reveal an alternative secondary structure in the region of nt 912 of Escherichia coli 16S rRNA. Proc. Natl. Acad. Sci. USA 92:10555-10559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Makosky, P. C., and A. E. Dahlberg. 1987. Spectinomycin resistance at site 1192 in 16S ribosomal RNA of E. coli: an analysis of three mutants. Biochimie 69:885-889. [DOI] [PubMed] [Google Scholar]
- 41.Meier, A., P. Kirschner, F. C. Bange, U. Vogel, and E. C. Böttger. 1994. Genetic alterations in streptomycin-resistant Mycobacterium tuberculosis: mapping of mutations conferring resistance. Antimicrob. Agents Chemother. 38:228-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Melançon, P., C. Lemieux, and L. Brakier-Gingras. 1988. A mutation in the 530 loop of Escherichia coli 16S ribosomal RNA causes resistance to streptomycin. Nucleic Acids Res. 16:9631-9639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moazed, D., and H. F. Noller. 1987. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature 327:389-394. [DOI] [PubMed] [Google Scholar]
- 44.Moine, H., C. L. Squires, B. Ehresmann, and C. Ehresmann. 2000. In vivo selection of functional ribosomes with variations in the rRNA-binding site of Escherichia coli ribosomal protein S8: evolutionary implications. Proc. Natl. Acad. Sci. USA 97:605-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Montandon, P. E., P. Nicolas, P. Schürmann, and E. Stutz. 1985. Streptomycin-resistance of Euglena gracilis chloroplasts: identification of a point mutation in the 16S rRNA gene in an invariant position. Nucleic Acids Res. 13:4299-4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Montandon, P. E., R. Wagner, and E. Stutz. 1986. E. coli ribosomes with a C912 to U base change in the 16S rRNA are streptomycin resistant. EMBO J. 5:3705-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O'Connor, M., and A. E. Dahlberg. 2002. Isolation of spectinomycin resistance mutations in the 16S rRNA of Salmonella enterica serovar Typhimurium and expression in Escherichia coli and Salmonella. Curr. Microbiol. 45:429-433. [DOI] [PubMed] [Google Scholar]
- 48.Picardeau, M., A. Brenot, and I. Saint Girons. 2001. First evidence for gene replacement in Leptospira spp. Inactivation of L. biflexa flaB results in non-motile mutants deficient in endoflagella. Mol. Microbiol. 40:189-199. [DOI] [PubMed] [Google Scholar]
- 49.Prammananan, T., P. Sander, B. A. Brown, K. Frischkorn, G. O. Onyi, Y. Zhang, E. C. Böttger, and R. J. Wallace, Jr. 1998. A single 16S ribosomal RNA substitution is responsible for resistance to amikacin and other 2-deoxystreptamine aminoglycosides in Mycobacterium abscessus and Mycobacterium chelonae. J. Infect. Dis. 177:1573-1581. [DOI] [PubMed] [Google Scholar]
- 50.Recht, M. I., S. Douthwaite, and J. D. Puglisi. 1999. Basis for prokaryotic specificity of action of aminoglycoside antibiotics. EMBO J. 18:3133-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Recht, M. I., and J. D. Puglisi. 2001. Aminoglycoside resistance with homogeneous and heterogeneous populations of antibiotic-resistant ribosomes. Antimicrob. Agents Chemother. 45:2414-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodriguez-Correa, D., and A. E. Dahlberg. 2004. Genetic evidence against the 16S ribosomal RNA helix 27 conformational switch model. RNA 10:28-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosa, P. A., K. Tilly, and P. E. Stewart. 2005. The burgeoning molecular genetics of the Lyme disease spirochaete. Nat. Rev. Microbiol. 3:129-143. [DOI] [PubMed] [Google Scholar]
- 54.Samuels, D. S. 1995. Electrotransformation of the spirochete Borrelia burgdorferi. Methods Mol. Biol. 47:253-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Samuels, D. S., and C. F. Garon. 1993. Coumermycin A1 inhibits growth and induces relaxation of supercoiled plasmids in Borrelia burgdorferi, the Lyme disease agent. Antimicrob. Agents Chemother. 37:46-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Samuels, D. S., K. E. Mach, and C. F. Garon. 1994. Genetic transformation of the Lyme disease agent Borrelia burgdorferi with coumarin-resistant gyrB. J. Bacteriol. 176:6045-6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Samuels, D. S., R. T. Marconi, W. M. Huang, and C. F. Garon. 1994. gyrB mutations in coumermycin A1-resistant Borrelia burgdorferi. J. Bacteriol. 176:3072-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sartakova, M. L., E. Y. Dobrikova, D. A. Terekhova, R. Devis, J. V. Bugrysheva, O. V. Morozova, H. P. Godfrey, and F. C. Cabello. 2003. Novel antibiotic-resistance markers in pGK12-derived vectors for Borrelia burgdorferi. Gene 303:131-137. [DOI] [PubMed] [Google Scholar]
- 59.Schwartz, J. J., A. Gazumyan, and I. Schwartz. 1992. rRNA gene organization in the Lyme disease spirochete, Borrelia burgdorferi. J. Bacteriol. 174:3757-3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sigmund, C. D., M. Ettayebi, and E. A. Morgan. 1984. Antibiotic resistance mutations in 16S and 23S ribosomal RNA genes of Escherichia coli. Nucleic Acids Res. 12:4653-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Springer, B., Y. G. Kidan, T. Prammananan, K. Ellrott, E. C. Böttger, and P. Sander. 2001. Mechanisms of streptomycin resistance: selection of mutations in the 16S rRNA gene conferring resistance. Antimicrob. Agents Chemother. 45:2877-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Steere, A. C. 2001. Lyme disease. N. Engl. J. Med. 345:115-125. [DOI] [PubMed] [Google Scholar]
- 63.Strigini, P., and L. Gorini. 1970. Ribosomal mutations affecting efficiency of amber suppression. J. Mol. Biol. 47:517-530. [DOI] [PubMed] [Google Scholar]
- 64.Suzuki, Y., C. Katsukawa, A. Tamaru, C. Abe, M. Makino, Y. Mizuguchi, and H. Taniguchi. 1998. Detection of kanamycin-resistant Mycobacterium tuberculosis by identifying mutations in the 16S rRNA gene. J. Clin. Microbiol. 36:1220-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Svab, Z., and P. Maliga. 1991. Mutation proximal to the tRNA binding region of the Nicotiana plastid 16S rRNA confers resistance to spectinomycin. Mol. Gen. Genet. 228:316-319. [DOI] [PubMed] [Google Scholar]
- 66.Terekhova, D., M. L. Sartakova, G. P. Wormser, I. Schwartz, and F. C. Cabello. 2002. Erythromycin resistance in Borrelia burgdorferi. Antimicrob. Agents Chemother. 46:3637-3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wallace, B. J., P.-C. Tai, and B. D. Davis. 1974. Selective inhibition of initiating ribosomes by spectinomycin. Proc. Natl. Acad. Sci. USA 71:1634-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wilcox, S. K., G. S. Cavey, and J. D. Pearson. 2001. Single ribosomal protein mutations in antibiotic-resistant bacteria analyzed by mass spectrometry. Antimicrob. Agents Chemother. 45:3046-3055. [DOI] [PMC free article] [PubMed] [Google Scholar]