Abstract
The protease encoded by the human cytomegalovirus (HCMV) is an attractive target for antiviral drug development because of its essential function in viral replication. We describe here a cellular assay in the yeast Saccharomyces cerevisiae for the identification of small molecule inhibitors of HCMV protease by conditional growth in selective medium. In this system, the protease cleavage sequence is inserted into the N-(5′-phosphoribosyl)anthranilate isomerase (Trp1p), a yeast protein essential for cell proliferation in the absence of tryptophan. Coexpression of HCMV protease with the engineered Trp1p substrate in yeast cells results in site-specific cleavage and functional inactivation of the Trp1p enzyme, thereby leading to an arrest of cell proliferation. This growth arrest can be suppressed by the addition of validated HCMV protease inhibitors. The growth selection system presented here provides the basis for a high-throughput screen to identify HCMV protease inhibitors that are active in eukaryotic cells.
Herpesviruses are widely present in nature and afflict many species throughout the animal kingdom (14). The most frequent human infection is caused by the human cytomegalovirus (HCMV), affecting up to 80% of the general population. This highly prevalent member of the herpesvirus family is responsible for opportunistic infections in immunocompromised individuals, notably AIDS patients and organ transplant recipients (for reviews, see references 9 and 29). Antiviral agents currently licensed for the treatment of HCMV infections include ganciclovir and its orally bioavailable prodrug valganciclovir as well as foscarnet, cidofovir, and fomivirsen (9). All these drugs are nucleoside analogues that ultimately target, either directly or indirectly, the viral DNA polymerase. The only exception is fomivirsen, an antisense oligonucleotide approved for HCMV retinitis that blocks translation of HCMV immediate-early mRNA. Unfortunately for the patients, the clinical usefulness of these drugs is limited: they exhibit toxic side effects, including bone marrow toxicity and nephrotoxicity, and they need to be injected either intravenously or intraocularly (9). Moreover, HCMV strains with reduced susceptibility resulting from chronic antiviral treatment are becoming more and more frequent. Thus, improved alternative drugs with novel mechanisms of action are needed for treating HCMV infections.
Herpesviruses encode a serine protease that cleaves the assembly protein precursor, a major component of the intermediate capsid (reviewed in reference 23). Studies with herpes simplex virus type 1 protease, a close homologue of HCMV protease, showed that viral DNA cannot be packaged into virions without this cleavage, resulting in an empty nucleocapsid (17, 32). HCMV protease is expressed as a 708-amino-acid precursor encoded by the UL80 open reading frame. Autocatalytic cleavage is observed at two consensus sequences called maturational (M) and release (R) sites (23). M-site cleavage removes a 6-kDa C-terminal tail, which mediates interaction with the major capsid protein at the earliest stage of capsid assembly. Cleavage at the R-site releases the N-terminal 28-kDa proteolytic domain. Enzymatic in vitro assays with short peptide substrates demonstrated a 10-fold increase in the kcat/Km value for an M-site peptide substrate over that for an R-site substrate (37).
Four different groups elucidated the X-ray crystallographic structure of HCMV protease in 1996 (8, 33, 36, 38). These studies have unequivocally confirmed the status of this enzyme as a serine protease with a unique catalytic triad. Instead of the His-Ser-Asp/Glu residues present in the active site of classical serine proteases, HCMV protease contains an unusual His residue at the third position. Moreover, only the dimeric form is active, which sets it further apart from other serine proteases (28). The two active sites are well separated on opposite faces of the dimer and act in an independent manner (4). The fact that the protease is essential for the propagation of the virus and that it markedly differs from mammalian serine proteases makes this enzyme an attractive therapeutic target.
Though peptidomimetic drug design delivered a number of HCMV protease inhibitors with good in vitro potency in the submicromolar range, e.g., translactams (5) and 2-substituted benzoxazinones (1), their activity in cell culture remained limited. In addition, high-throughput screening (HTS) campaigns in the industry, based on enzymatic in vitro assays, did not lead to the identification of hits with good activity in cell culture assays. Remarkably, very few reports describe the investigation of HCMV protease activity in a cellular environment (26, 41), and to our knowledge, no cell-based HTS has been performed so far. More efforts should be invested in such cellular assays, since they not only allow for screening in a natural cellular environment but also directly exclude compounds that are unstable or toxic or that cannot penetrate biological membranes. Thus, appropriate cell-based assays can accelerate the drug discovery process (for reviews, see references 2 and 3).
In an effort to select for HCMV protease inhibitors in an in vivo environment, we have established a target-specific HTS system in Saccharomyces cerevisiae that detects protease activity by conditional growth in selective medium. In a proof-of-principle experiment, we show that the application of known HCMV protease inhibitors results in a concentration-dependent stimulation of cell proliferation.
MATERIALS AND METHODS
Yeast strains.
The genes encoding the three major ABC transporter proteins Pdr5p, Snq2p, and Yor1p were deleted in the S. cerevisiae JPY5 strain (MATα ura3-52 his3Δ200 leu2Δ1 trp1Δ63 lys2Δ385) to generate the RLY07 strain (MATα ura3-52 his3Δ200 leu2Δ1 trp1Δ63 lys2Δ385 pdr5Δ snq2Δ yor1Δ). All recombinant proteins described in this study were expressed in RLY07 cells.
Recombinant plasmids.
The S. cerevisiae TRP1 gene was amplified by PCR from the plasmid YCplac22 (20). All TRP1-M constructs generated hereupon were subcloned via unique XbaI and SalI restriction sites in the CEN4-ARS1 plasmid pMH4. pMH4 carries a LEU2 auxotrophic marker and expresses subcloned TRP1-M constructs under the control of a 5′ truncated version of the ADH promoter and the GAL11 terminator. The HCMV protease cleavage sequence GGVVNA↓SCRLAGG, derived from the M site of the 75-kDa protease precursor, was flanked with unique NcoI and NotI sites and inserted by PCR directly to the C-terminal end of amino acid residues 49, 102, 132, 165, and 194 of the Trp1 protein. This generated a plasmid series encoding the recombinant proteins Trp149-M, Trp1102-M, Trp1132-M, Trp1165-M, and Trp1194-M. In order to generate Trp1194-Me (for Trp1194-M elongated), the 13-amino-acid cleavage sequence was replaced (via NcoI and NotI sites) by an extended M-type sequence consisting of 39 amino acids (see Fig. 3). For Western blotting, hemagglutinin (HA) epitope tags were added to both termini of the Trp1194-Me construct. The HCMV protease gene encoding amino acids 1 to 256 of the 708-amino-acid protease precursor was obtained by PCR from HCMV-infected MRC5 human cells and subcloned via unique XbaI and NotI sites in pMH51, a CEN4-ARS1 plasmid with a URA3 selection marker and a full-length (100%) GAL1 promoter. In the experiment for which results are shown in Fig. 4, the HCMV protease gene was subcloned in a plasmid series with distinct GAL1 promoters that express the protease with 71%, 46%, and 16% protein production levels relative to the original 100% GAL1 promoter. The GAL1 promoter contains four different pseudopalindromic binding sites for the transcription factor Gal4p. Liang et al. have shown that modifications in the number and type of Gal4p binding sites modulate transcription of a downstream cloned reporter gene (27). We have differentially deleted those binding modules and validated promoter strength with a downstream cloned lacZ reporter gene (above 71%, 46%, and 16% protein production levels).
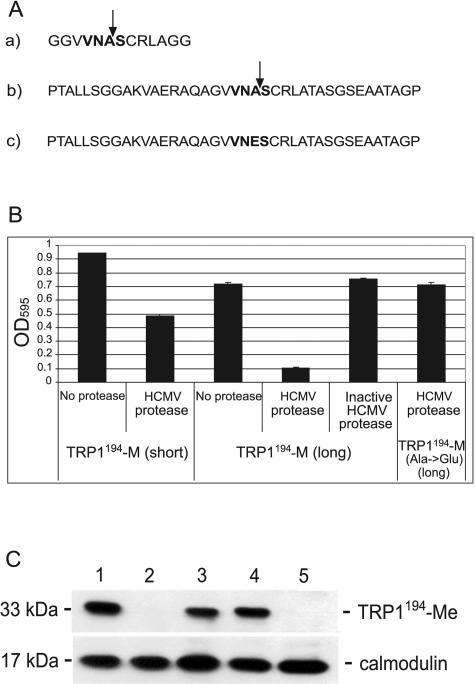
FIG. 3.
Sequence-specific cleavage of the Trp1194-Me substrate by the HCMV protease. (A) HCMV protease cleavage sequences inserted C terminal to G194 of Trp1p. a) 13 residues (short) with the M site; b) 39 residues (long) with the M site; c) long M-site sequence with the point mutation A→E at the scissile bond. (B) RLY07 cells were cotransformed with the different Trp1194-M substrates and a plasmid expressing either active or inactive (active-site point mutation S132A) HCMV protease, as indicated. Cells were grown in selective −Trp medium, and OD595 was measured after 38 h. Data are expressed as means from three independent experiments ± standard deviations. (C) Western blot analysis of Trp1194-Me cleavage by HCMV protease. The uncleaved 33-kDa Trp1194-Me substrate, which carries HA tags at both termini, is detected by immunoblotting with an anti-HA antibody. Protein extracts were made from transformed RLY07 cells expressing Trp1194-M (lane 1), Trp1194-M and active HCMV protease (lane 2), Trp1194-M and inactive HCMV protease (lane 3), Trp1194-Me(A→E) and active HCMV protease (lane 4), and empty vectors (negative control) (lane 5). The calmodulin antibody was used as an internal control for protein amount.
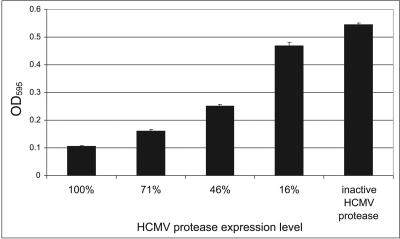
FIG. 4.
A gradual decrease of HCMV protease expression correlates with distinct stimulation of cell proliferation. In the first four columns, RLY07 cells were cotransformed with Trp1194-Me and with active HCMV protease expressed from a GAL1 promoter series (see Materials and Methods for detailed information). The last column shows cell growth mediated by the inactive protease. Cell densities were monitored after 30 h by measuring OD595. Data are expressed as means from three independent experiments ± standard deviations.
Yeast media and transformation.
All media were prepared according to Burke et al. (7). Transformation of yeast cells was performed following the lithium acetate method (19).
Spotting assay.
RLY07 cells transformed with the different TRP1-M constructs were inoculated in 3 ml synthetic dropout (SD) glucose medium lacking Leu (−Leu) and grown to an optical density at 595 nm (OD595) of 1 to reach an exponential growth phase. Cultures were hereupon washed with 5 ml H2O, resuspended in −Leu −Trp SD glucose medium and diluted to 106 cells/ml. Ten microliters of serially diluted culture was spotted on nonselective (−Leu) and selective (−Leu −Trp) glucose SD plates and incubated at 30°C for 3 days.
Liquid growth assays.
RLY07 cells transformed with the different TRP1-M constructs and the HCMV protease gene were inoculated in 3 ml −Leu −Ura galactose SD medium and grown at 30°C to an OD595 of 1. They were then washed with 5 ml H2O and resuspended in −Leu −Ura −Trp galactose SD medium supplemented with 10% glycerol (growth selection medium). It is likely to assume that glycerol promotes HCMV protease dimerization and subsequent proteolytic activity, as it has been shown in in vitro assays (28). In yeast, glycerol is taken up by a proton symport mechanism (13). Liquid growth assays were started at an inoculation OD595 of 0.01 in growth selection medium in 96-well microtiter plates in a volume of 150 μl per well. Cultures were incubated at 30°C without shaking. At defined time points (as specified in figure legends), plates were shaken at 1,000 rpm for 1 min to completely resuspend cells, and cell density was measured at 595 nm on a Tecan GENios reader (Männedorf, Switzerland). HCMV protease inhibitors BI31 and BI36 (Boeringher Ingelheim, Québec) were dissolved in dimethyl sulfoxide and added to the cultures directly at the beginning of the growth assay; final dimethyl sulfoxide concentration was 1%.
Western blot analysis.
Yeast whole-cell extracts were prepared as described by Burke et al. (7). Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and Western blot analysis was performed according to standard procedures. An anti-HA monoclonal antibody from Sigma (clone 3F10) was used at a concentration of 30 ng/ml to detect expression of Trp1194-Me.
RESULTS
A cell-based system to detect protease activity.
In the present work, we describe a cell growth selection system in the yeast S. cerevisiae that (i) can detect and characterize HCMV protease activity and (ii) is applicable to screen for HCMV protease inhibitors in a high-throughput format. To establish this system, we inserted an HCMV protease cleavage site into a yeast protein that is conditionally essential for cell proliferation. This protein was chosen from the auxotrophic growth markers, considering that most laboratory strains are already deleted for these genes and that therefore, such a system could be applied in almost any genetic background. Since the N-(5′-phosphoribosyl)anthranilate isomerase (Trp1p) enzyme has been intensively studied (10, 11, 22, 24) and its three-dimensional structure has been determined for different organisms, we have chosen this member of the tryptophan synthesis pathway as a substrate for HCMV protease.
Thus, in our setup, such an engineered Trp1 enzyme should be cleaved and subsequently inactivated by HCMV protease, resulting in a reduced growth in selective medium lacking tryptophan.
Insertion of the HCMV protease cleavage sequence at five different locations in Trp1p.
Two conditions are critical for appropriate operation of the above-described system. (i) Insertion of the HCMV protease cleavage sequence must preserve Trp1 function in the tryptophan biosynthesis pathway. (ii) On the other hand, cleavage at the inserted sequence must result in functional inactivation of the Trp1 enzyme.
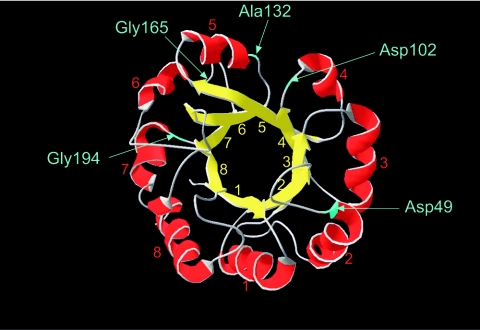
The Trp1 enzyme is a member of the prominent class of proteins that fold into a (β/α)8 barrel, which is the most commonly occurring fold among enzymes (12) (Fig. 1). The S. cerevisiae Trp1p structure has not yet been determined, but amino acid sequence alignments with the N-(5′-phosphoribosyl)anthranilate isomerase from Escherichia coli and Thermotoga maritima provide us with a reliable model. S. cerevisiae Trp1p shares 28% identical amino acids with E. coli Trp1p and 33% with T. maritima Trp1p. Alignment and modeling for S. cerevisiae Trp1p was performed with the SWISS-MODEL protein modeling server (http://swissmodel.expasy.org/repository/) (21, 35).
FIG. 1.
Modeled structure of the S. cerevisiae N-(5′-phosphoribosyl)anthranilate isomerase Trp1p. Residues preceding the inserted M-cleavage sequences are indicated in blue.
To determine suitable sites for insertion of the cleavage sequence, several constructs were designed. Since turn sequences are in general highly mutable in (β/α)8 barrels, five insertion sites were chosen that are located in such turns between an α-helix and a β-sheet, after amino acids Asp49, Asp102, Ala132, Gly165, and Gly194 (Fig. 1A). Respective constructs will therefore be referred to as Trp149-M, Trp1102-M, Trp1132-M, Trp1165-M, and Trp1194-M. The inserted sequence consists of 13 amino acids derived from the M site of the 75-kDa protease precursor (see Fig. 3A, sequence a).
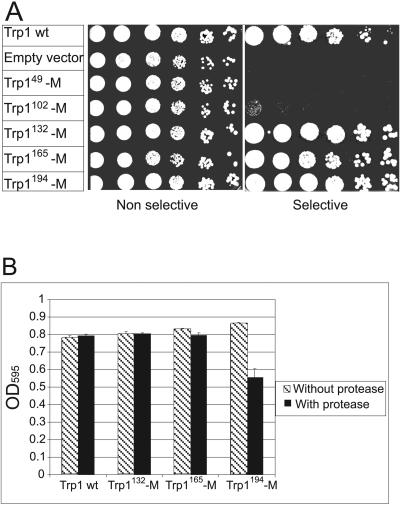
In order to evaluate the functionality of the Trp149-M, Trp1102-M, Trp1132-M, Trp1165-M, and Trp1194-M fusion proteins, a spotting assay was performed (Fig. 2A). RLY07 cells were transformed with these constructs, and their ability to proliferate was assessed on selective plates lacking tryptophan. For Trp1132-M-, Trp1165-M-, and Trp1194-M-expressing cells, growth was indistinguishable from that of cells expressing wild-type Trp1p, indicating that M-site insertion did not interfere with the functionality of the enzyme. In contrast, Trp149-M and Trp1102-M constructs produced nonfunctional enzymes, as demonstrated by likewise-transformed cells unable to grow on selective plates (Fig. 2A).
FIG. 2.
Functional tests of the five engineered Trp1p constructs. (A) Serial dilutions of RLY07 cells transformed with wild-type (wt) Trp1p (positive control), empty vector (negative control), Trp149-M, Trp1102-M, Trp1132-M, Trp1165-M, or Trp1194-M were spotted on selective −Trp plates and incubated 3 days at 30°C. (B) Wild-type Trp1p, Trp1132-M, Trp1165-M, and Trp1194-M constructs, cotransformed in RLY07 either with an empty vector (dashed bars) or with active HCMV protease (black bars) were grown in selective −Trp medium for 35 h; cell density was measured at 595 nm. Data are expressed as means from three independent experiments ± standard deviations.
Sequence-specific cleavage of Trp1194-M by HCMV protease in yeast.
We next investigated whether the three functional Trp1-M proteins are cleaved and inactivated by HCMV protease. Trp1132-M, Trp1165-M, and Trp1194-M were coexpressed with the viral protease in the RLY07 strain, and cell proliferation was assayed by measuring OD595 of transformed cells cultured in growth selection medium (Fig. 2B). After 36 h, cells coexpressing Trp1194-M with the protease exhibited a growth reduction of 35% compared to control cells that contained an empty vector instead of the protease-expressing plasmid, indicating that cleavage between helix α7 and strand β8 reduces activity of the Trp1 enzyme. Interestingly, this region is situated close to the phosphate binding site of the anthranilate substrate. Crystallographic studies have shown that the two neighboring loops between β7 and α7 and between β8 and α8 are important for binding of the phosphate ion (40). Cleavage in this region might cause structural changes, thus preventing binding of the substrate. As opposed to Trp1194-M cells, Trp1132-M- as well as Trp1165-M-expressing cells did not show growth reduction upon coexpressing HCMV protease (Fig. 2B). Thus, the latter two engineered Trp1 substrates were either not cleaved or, alternatively, cleavage occurred but the separated fragments still formed an active enzyme.
To improve cleavage frequency at the M site of the Trp1194-M substrate, the 13-amino-acid target sequence was replaced by a longer sequence consisting of 39 amino acids (Fig. 3A, sequence b). Cells expressing the modified Trp1194-Me together with the active protease showed 85% reduction of cell proliferation compared to the 35% reduction with the original, shorter cleavage site (Fig. 3B). This indicates that the extended recognition site is more efficiently cleaved by the protease.
The HCMV protease has been shown to hydrolyze both the M site and R site of the protease precursor between an alanine and a serine (6). To demonstrate site-specific cleavage of the Trp1194-Me substrate, we replaced the alanine of the scissile bond with glutamic acid (Fig. 3A, sequence c), a mutation known to prevent cleavage (39). As expected, the proliferation of cells coexpressing the Trp1194-Me(A→E) mutant with HCMV protease was comparable to the proliferation of cells expressing Trp1194-Me alone, providing evidence that the Trp1194-Me substrate is cleaved in a sequence-specific manner at the scissile bond (Fig. 3B). A further control experiment was performed by using an enzymatically inactive version of HCMV protease harboring the S132A mutation at the active-site serine (8). As shown in Fig. 3B, this mutant was not able to cleave the Trp1194-Me substrate.
To demonstrate actual cleavage of the substrate in yeast, Trp1194-Me was tagged with an HA epitope and analyzed by Western blotting. The full-length substrate migrates at the level of 33-kDa proteins in a sodium dodecyl sulfate-polyacrylamide gel electrophoresis assay (Fig. 3C, lane 1). Coexpression of active HCMV protease (Fig. 3C, lane 2) causes the disappearance of the full-length substrate upon Western blot analysis of the respective yeast extracts. However, no cleaved fragments could be detected. We suppose that they are either degraded or below the detection threshold of the Western blot. Lane 3 of Fig. 3C shows that the inactive viral protease does not cleave Trp1194-Me since the full-length substrate band does not disappear. Lane 4 shows that the active protease has no effect on the point-mutated Trp1194-Me(A→E) substrate.
Taken together, the above-described experiments demonstrate that the Trp1194-Me substrate is cleaved in a sequence-specific manner by HCMV protease and that this cleavage results in a slow-growth phenotype.
Modulation of HCMV protease expression level correlates with distinct changes in cell proliferation.
A prerequisite for a successful HTS is that even only partial inhibition of the protease can be detected in the assay. We were therefore interested to find out whether different concentrations of intracellular active protease result in distinguishable and selectable levels of growth rate on the HTS read-out. In the experiments described above, HCMV protease was expressed from the full-length GAL1 promoter (100% activity). To express the protease at distinct lower levels, thus mimicking partial protease inhibition by a small molecule in a screening experiment, the protease was subcloned on a GAL1 vector series, with discrete protein production levels of 71%, 46%, and 16% compared to the original 100% promoter. These plasmids were cotransformed with the Trp1194-Me substrate in RLY07 cells, and cell growth was measured after 36 h at OD595. Figure 4 shows that a gradual decrease of promoter strength, and thus of intracellular protease activity, is inversely proportional to cell proliferation. For example, a reduction of 29% of protease expression (from the 100% promoter to the 71% promoter) resulted in a 53% stimulation of cell proliferation. Likewise, a reduction of 54% of protease expression caused a stimulation of 138%. Therefore, even weak inhibitors that cause only a partial reduction of HCMV protease activity should be detectable in our system.
Two validated HCMV protease inhibitors stimulate cell proliferation in a screening assay format.
The assay described above was developed to identify cell-permeable small molecule inhibitors of HCMV protease in high-throughput screening experiments. We have already shown that the system is able to distinguish different levels of intracellular protease activity. In order to assess sensitivity of the selection system towards small molecules, the two validated HCMV protease inhibitors BI31 and BI36 of Boehringer Ingelheim were applied (42). BI31 and BI36 show 50% inhibitory concentration values of 1.7 and 0.5 μM in an enzymatic in vitro assay and inhibit viral replication in cell culture with 50% effective concentration (EC50) values of 95 and 78 μM, respectively (42). Both compounds are based on a 4-thioalkyl β-lactam scaffold. Lactam derivatives have initially been published as inhibitors of classical serine proteases, such as human leukocyte elastase (15). Further development of such scaffolds by rational design delivered inhibitors specific for HCMV protease (42).
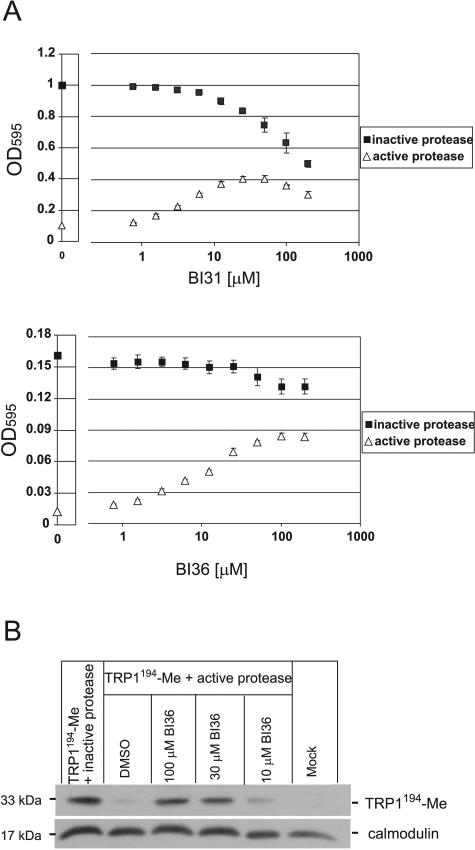
RLY07 cells coexpressing the Trp1194-Me substrate and HCMV protease were incubated with various concentrations of BI31 and BI36 in a 96-well microtiter plate and cultivated under selective conditions at 30°C for 44 and 28 h, respectively. Figure 5A shows that increasing concentrations of both BI31 and BI36 in cells expressing the active protease correlate with increasing cell proliferation. The calculated EC50 of 31 μM for BI36 suggests that the yeast-based assay exhibits the same sensitivity as that of a mammalian cellular assay, at least for this class of compounds (42). At BI36 concentrations of >100 μM, HCMV protease was strongly inhibited, as evidenced by proliferation rates similar to those of cells expressing the inactive protease. Though BI31 also restored cell growth in a concentration-dependent manner, stimulation was not as pronounced as with BI36 due to cellular toxicity of this compound. This toxicity is also shown by the fact that increasing concentrations of BI31 in RLY07 cells expressing the inactive (active-site mutated S132A) HCMV protease caused a gradual decrease of cell proliferation. Importantly, despite this toxicity, BI31 still stimulated growth of cells expressing the active protease. For example, at 50 μM cell density is multiplied by a factor 4 despite 25% toxicity. This suggests that in an HTS, compounds will be scored as positives even if they exert some intrinsic toxicity.
FIG. 5.
Validated HCMV protease inhibitors BI31 (upper panel) and BI36 (lower panel) stimulate cell proliferation in a concentration-dependent manner. (A) RLY07 cells expressing Trp1194-Me and active HCMV protease (triangles) or Trp1194-Me and inactive protease (squares) were cultivated either in the absence (0) or presence of increasing concentrations of BI31 and BI36. Cells were grown in 150-μl cultures in a 96-well microplate for 44 h (BI31) and 28 h (BI36) before OD595 was measured. Data are expressed as means from three independent experiments ± standard deviations. (B) Western blot analysis with HA antibody showing that application of BI36 prevents cleavage of the Trp1194-Me substrate by HCMV protease. RLY07 cells were transformed with Trp1194-Me and inactive protease (positive control), Trp1194-Me and active protease, or Trp1194-Me with active protease in the presence of distinct concentrations of BI36, as indicated. RLY07 cells containing two empty plasmids were used as a negative control. The calmodulin antibody served as an internal control for protein amount. DMSO, dimethyl sulfoxide.
Inhibition of HCMV protease activity in yeast by BI36 was also addressed by Western blot analysis (Fig. 5B). As already shown in Fig. 3C, a 33-kDa band corresponding to the uncleaved full-length Trp1194-Me substrate can be detected in cells coexpressing the Trp substrate with the inactive (S132A) protease. In cells cotransformed with the active protease instead of the inactive version, disappearance of the 33-kDa band can be observed. Importantly, the addition of 100 μM BI36 to the growth selection medium of those cells almost completely prevented cleavage of the substrate (Fig. 5B). When using lower concentrations of BI36 (10 μM), the full-length substrate band disappears again, suggesting that at concentrations well below the calculated EC50 of 31 μM in yeast, only a small percentage of protease activity is inhibited.
In summary, we have shown that two well-validated active-site inhibitors of HCMV protease cause a concentration-dependent stimulation of cell growth in our yeast assay. Importantly, sensitivity of the yeast system towards these two compounds is comparable to the sensitivity of a mammalian cell-based assay. The calculated Z′ factor values of 0.85 for 50 μM BI36 and 0.75 for 50 μM BI31 clearly demonstrate the suitability of this bioassay for high-throughput applications.
DISCUSSION
The frequent emergence of cytomegalovirus strains resistant against classical antiviral agents, which all directly or indirectly target viral DNA polymerase, spurred the search for alternative therapeutic targets of the virus (reviewed in reference 29). The cytomegalovirus protease, or assemblin, has gained much attention as a target due to the unique serine protease fold and catalytic triad that is expected to facilitate the design of virus-specific inhibitors. Though past screening campaigns have culminated in the identification of inhibitors with nanomolar activities in enzymatic in vitro assays, their activity in cell-based assays was limited in most cases. Examples include benzoxazinones (16), lactams (5, 18), and the highly toxic enedione derivatives (30). To our knowledge, no cell-based HTS on herpesviral proteases has so far been performed successfully. Compared to in vitro assays, cellular systems allow for the screening of the therapeutic target in its natural configuration within the cell. In addition, they select for compounds that are stable within a metabolic environment, that are able to penetrate biological membranes, and that show no or only limited toxic effects on the cell (for reviews, see references 2 and 3). Therefore, appropriate cellular systems potentially accelerate the drug discovery process.
We have established a cellular assay in the yeast S. cerevisiae that detects HCMV protease activity and that enables screening for small molecule protease inhibitors with a positive growth read-out. Yeast provides a eukaryotic environment in a single cell with a high degree of conserved basic molecular mechanisms with mammalian cells. In addition, yeast is inexpensive to propagate and can be easily genetically engineered.
For establishing the growth selection system described here, a 39-amino-acid HCMV protease cleavage sequence derived from the M site of the 75-kDa protease precursor was inserted into the phosphoribosylanthranilate isomerase, a yeast enzyme involved in the biosynthesis of tryptophan and therefore essential for cell proliferation in the absence of tryptophan. Coexpression of HCMV protease with this engineered substrate caused sequence-specific cleavage at the M site, resulting in growth arrest (Fig. 3B). Importantly, the addition of validated cell-active β-lactam inhibitors restored cell proliferation in a concentration-dependent manner (Fig. 5B). Since yeast encodes a pleiotrophic drug resistance network with a variety of ABC transporters that efficiently clear small molecules within the cell, we sensitized the strain by deleting the three major drug efflux pumps Pdr5p, Snq2p, and Yor1p. In such a setup, the yeast system shows a sensitivity towards small molecules similar to that shown in a mammalian cellular assay, at least for the β-lactam inhibitors tested, with a calculated EC50 of 31 μM in the yeast assay versus 78 μM in a mammalian plaque reduction assay (42).
HCMV protease has been shown to be active as a dimer (28). Remarkably, enzymatic in vitro assays require high concentrations of dimerization-promoting agents, such as antichaotropic salts and glycerol, that dramatically increase the kcat/Km value to obtain reasonable turnover rates of the enzyme (25, 28). The fact that in our cellular assay a gradual increase of glycerol concentration correlates inversely with cell proliferation (data not shown) underscores the hypothesis that HCMV protease is also a dimer in yeast. This is confirmed by the observation that the point mutation L221A completely inactivates HCMV protease in yeast (data not shown). It has been demonstrated for Kaposi's sarcoma-associated herpesvirus protease that mutating the homologous residue L196 to an alanine abrogates protease activity by preventing dimerization (31).
It is a subject of debate whether the 29-kDa assemblin or the 75-kDa protease precursor is the physiologically relevant protease responsible for capsid maturation and therefore the appropriate therapeutic target. While all enzymatic HTS has been performed with the 29-kDa domain, Robertson et al. suggested the 75-kDa precursor to be the “real” target. Indeed, transcomplementation studies with mutant herpes simplex virus type 1 viruses have shown that the proteolytic activity essential for capsid maturation is provided by the full-length precursor protein (34). We coexpressed Trp1194-Me with the 75-kDa protease precursor and observed a minor but statistically significant reduction in cell growth (data not shown). We are currently investigating whether this proteolytic activity is derived from the protease precursor per se or whether the 29-kDa assemblin, generated upon autocatalytic cleavage, is responsible for cleavage of the Trp1 substrate.
Applicability of the above-described Trp1p-based yeast assay is not limited to HCMV protease and can be exploited for other proteases. We have replaced the M site of the Trp1194-Me substrate with a 13-amino-acid cleavage sequence of the coxsackievirus 3C cysteine protease. Coexpression of the likewise-engineered Trp1 substrate with the 3C protease resulted in an arrest of cell proliferation (data not shown). In a proof-of-concept study, we have used the Trp1-M system as a validation tool for hits identified in a high-throughput screen of a 15,000-chemical-compound library. With this approach, we have identified a number of compounds that also show activity in a mammalian plaque reduction assay in the low-micromolar range (unpublished data).
Acknowledgments
We thank Boehringer Ingelheim (Québec, Canada) for kindly providing the BI31 and BI36 compounds.
REFERENCES
- 1.Abood, N. A., L. A. Schretzman, D. L. Flynn, K. A. Houseman, A. J. Wittwer, V. M. Dilworth, P. J. Hippenmeyer, and B. C. Holwerda. 1997. Inhibition of human cytomegalovirus protease by benzoxazinones and evidence of antiviral activity in cell culture. Bioorg. Med. Chem. Lett. 7:2105-2108. [Google Scholar]
- 2.Barberis, A. 2002. Cell-based high-throughput screens for drug discovery. Eur. BioPharm. Rev. 2002:93-96.
- 3.Barberis, A., T. Gunde, C. Berset, S. Audetat, and U. Lüthi. 2005. Yeast as a screening tool. Drug Discov. Today 2:187-192. [DOI] [PubMed] [Google Scholar]
- 4.Batra, R. 2001. Molecular mechanism for dimerization to regulate the catalytic activity of human cytomegalovirus protease. Nat. Struct. Biol. 8:810-817. [DOI] [PubMed] [Google Scholar]
- 5.Borthwick, A. D. 2005. Design of translactam HCMV protease inhibitors as potent antivirals. Med. Res. Rev. 25:427-452. [DOI] [PubMed] [Google Scholar]
- 6.Burck, P. J., D. H. Berg, T. P. Luk, L. M. Sassmannshausen, M. Wakulchik, D. P. Smith, H. M. Hsiung, G. W. Becker, W. Gibson, and E. C. Villarreal. 1994. Human cytomegalovirus maturational proteinase: expression in Escherichia coli, purification, and enzymatic characterization by using peptide substrate mimics of natural cleavage sites. J. Virol. 68:2937-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burke, D., D. Dawson, and T. Stearns. 2000. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 8.Chen, P., H. Tsuge, R. J. Almassy, C. L. Gribskov, S. Katoh, D. L. Vanderpool, S. A. Margosiak, C. Pinko, D. A. Matthews, and C. C. Kan. 1996. Structure of the human cytomegalovirus protease catalytic domain reveals a novel serine protease fold and catalytic triad. Cell 86:835-843. [DOI] [PubMed] [Google Scholar]
- 9.De Clercq, E. 2003. New inhibitors of human cytomegalovirus (HCMV) on the horizon. J. Antimicrob. Chemother. 51:1079-1083. [DOI] [PubMed] [Google Scholar]
- 10.Eder, J., and K. Kirschner. 1992. Stable substructures of eightfold beta alpha-barrel proteins: fragment complementation of phosphoribosylanthranilate isomerase. Biochemistry 31:3617-3625. [DOI] [PubMed] [Google Scholar]
- 11.Eder, J., and M. Wilmanns. 1992. Protein engineering of a disulfide bond in a beta/alpha-barrel protein. Biochemistry 31:4437-4444. [DOI] [PubMed] [Google Scholar]
- 12.Farber, G. K., and G. A. Petsko. 1990. The evolution of alpha/beta barrel enzymes. Trends Biochem. Sci. 15:228-234. [DOI] [PubMed] [Google Scholar]
- 13.Ferreira, C., F. van Voorst, A. Martins, L. Neves, R. Oliveira, M. C. Kielland-Brandt, C. Lucas, and A. Brandt. 2005. A member of the sugar transporter family, Stl1p is the glycerol/H+ symporter in Saccharomyces cerevisiae. Mol. Biol. Cell 16:2068-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fields, B. N., D. M. Knipe, and P. M. Howley. 1996. Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 15.Finke, P. E., S. K. Shah, D. S. Fletcher, B. M. Ashe, K. A. Brause, G. O. Chandler, P. S. Dellea, K. M. Hand, A. L. Maycock, D. G. Osinga, et al. 1995. Orally active beta-lactam inhibitors of human leukocyte elastase. 3. Stereospecific synthesis and structure-activity relationships for 3,3-dialkylazetidin-2-ones. J. Med. Chem. 38:2449-2462. [DOI] [PubMed] [Google Scholar]
- 16.Flynn, D. L., N. A. Abood, and B. C. Holwerda. 1997. Recent advances in antiviral research: identification of inhibitors of the herpesvirus proteases. Curr. Opin. Chem. Biol. 1:190-196. [DOI] [PubMed] [Google Scholar]
- 17.Gao, M., L. Matusick-Kumar, W. Hurlburt, S. F. DiTusa, W. W. Newcomb, J. C. Brown, P. J. McCann III, I. Deckman, and R. J. Colonno. 1994. The protease of herpes simplex virus type 1 is essential for functional capsid formation and viral growth. J. Virol. 68:3702-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerona-Navarro, G., M. J. Perez de Vega, M. T. Garcia-Lopez, G. Andrei, R. Snoeck, J. Balzarini, E. De Clercq, and R. Gonzalez-Muniz. 2004. Synthesis and anti-HCMV activity of 1-acyl-beta-lactams and 1-acylazetidines derived from phenylalanine. Bioorg. Med. Chem. Lett. 14:2253-2256. [DOI] [PubMed] [Google Scholar]
- 19.Gietz, D., A. St. Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 21.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 22.Hennig, M., R. Sterner, K. Kirschner, and J. N. Jansonius. 1997. Crystal structure at 2.0 Å resolution of phosphoribosyl anthranilate isomerase from the hyperthermophile Thermotoga maritima: possible determinants of protein stability. Biochemistry 36:6009-6016. [DOI] [PubMed] [Google Scholar]
- 23.Holwerda, B. C. 1997. Herpesvirus proteases: targets for novel antiviral drugs. Antivir. Res. 35:1-21. [DOI] [PubMed] [Google Scholar]
- 24.Hommel, U., M. Eberhard, and K. Kirschner. 1995. Phosphoribosyl anthranilate isomerase catalyzes a reversible amadori reaction. Biochemistry 34:5429-5439. [DOI] [PubMed] [Google Scholar]
- 25.Khayat, R., R. Batra, G. A. Bebernitz, M. W. Olson, and L. Tong. 2004. Characterization of the monomer-dimer equilibrium of human cytomegalovirus protease by kinetic methods. Biochemistry 43:316-322. [DOI] [PubMed] [Google Scholar]
- 26.Lawler, J. F., Jr., and S. H. Snyder. 1999. Viral protease assay based on GAL4 inactivation is applicable to high-throughput screening in mammalian cells. Anal. Biochem. 269:133-138. [DOI] [PubMed] [Google Scholar]
- 27.Liang, S. D., R. Marmorstein, S. C. Harrison, and M. Ptashine. 1996. DNA sequence preferences of GAL4 and PPR1: how a subset of Zn2 Cys6 binuclear cluster proteins recognizes DNA. Mol. Cell. Biol. 16:3773-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Margosiak, S. A., D. L. Vanderpool, W. Sisson, C. Pinko, and C. C. Kan. 1996. Dimerization of the human cytomegalovirus protease: kinetic and biochemical characterization of the catalytic homodimer. Biochemistry 35:5300-5307. [DOI] [PubMed] [Google Scholar]
- 29.Martinez, A., A. Castro, C. Gil, and C. Perez. 2001. Recent strategies in the development of new human cytomegalovirus inhibitors. Med. Res. Rev. 21:227-244. [DOI] [PubMed] [Google Scholar]
- 30.Pinto, I. L., R. L. Jarvest, B. Clarke, C. E. Dabrowski, A. Fenwick, M. M. Gorczyca, L. J. Jennings, P. Lavery, E. J. Sternberg, D. G. Tew, and A. West. 1999. Inhibition of human cytomegalovirus protease by enedione derivatives of thieno[2,3-d]oxazinones through a novel dual acylation/alkylation mechanism. Bioorg. Med. Chem. Lett. 9:449-452. [DOI] [PubMed] [Google Scholar]
- 31.Pray, T. R., K. K. Reiling, B. G. Demirjian, and C. S. Craik. 2002. Conformational change coupling the dimerization and activation of KSHV protease. Biochemistry 41:1474-1482. [DOI] [PubMed] [Google Scholar]
- 32.Preston, V. G., J. A. Coates, and F. J. Rixon. 1983. Identification and characterization of a herpes simplex virus gene product required for encapsidation of virus DNA. J. Virol. 45:1056-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiu, X., J. S. Culp, A. G. DiLella, B. Hellmig, S. S. Hoog, C. A. Janson, W. W. Smith, and S. S. Abdel-Meguid. 1996. Unique fold and active site in cytomegalovirus protease. Nature 383:275-279. [DOI] [PubMed] [Google Scholar]
- 34.Robertson, B. J., P. J. McCann III, L. Matusick-Kumar, W. W. Newcomb, J. C. Brown, R. J. Colonno, and M. Gao. 1996. Separate functional domains of the herpes simplex virus type 1 protease: evidence for cleavage inside capsids. J. Virol. 70:4317-4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shieh, H. S., R. G. Kurumbail, A. M. Stevens, R. A. Stegeman, E. J. Sturman, J. Y. Pak, A. J. Wittwer, M. O. Palmier, R. C. Wiegand, B. C. Holwerda, and W. C. Stallings. 1996. Three-dimensional structure of human cytomegalovirus protease. Nature 383:279-282. [DOI] [PubMed] [Google Scholar]
- 37.Stevens, J. T., C. Mapelli, J. Tsao, M. Hail, D. O'Boyle, II, S. P. Weinheimer, and C. L. Diianni. 1994. In vitro proteolytic activity and active-site identification of the human cytomegalovirus protease. Eur. J. Biochem. 226:361-367. [DOI] [PubMed] [Google Scholar]
- 38.Tong, L., C. Qian, M. J. Massariol, P. R. Bonneau, M. G. Cordingley, and L. Lagace. 1996. A new serine-protease fold revealed by the crystal structure of human cytomegalovirus protease. Nature 383:272-275. [DOI] [PubMed] [Google Scholar]
- 39.Welch, A. R., L. M. McNally, M. R. Hall, and W. Gibson. 1993. Herpesvirus proteinase: site-directed mutagenesis used to study maturational, release, and inactivation cleavage sites of precursor and to identify a possible catalytic site serine and histidine. J. Virol. 67:7360-7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilmanns, M., C. C. Hyde, D. R. Davies, K. Kirschner, and J. N. Jansonius. 1991. Structural conservation in parallel beta/alpha-barrel enzymes that catalyze three sequential reactions in the pathway of tryptophan biosynthesis. Biochemistry 30:9161-9169. [DOI] [PubMed] [Google Scholar]
- 41.Wittwer, A. J., C. L. Funckes-Shippy, and P. J. Hippenmeyer. 2002. Recombinant full-length human cytomegalovirus protease has lower activity than recombinant processed protease domain in purified enzyme and cell-based assays. Antivir. Res. 55:291-306. [DOI] [PubMed] [Google Scholar]
- 42.Yoakim, C., W. W. Ogilvie, D. R. Cameron, C. Chabot, C. Grand-Maitre, I. Guse, B. Hache, S. Kawai, J. Naud, J. A. O'Meara, R. Plante, and R. Deziel. 1998. Potent beta-lactam inhibitors of human cytomegalovirus protease. Antivir. Chem. Chemother. 9:379-387. [DOI] [PubMed] [Google Scholar]