Abstract
Resistance of Plasmodium falciparum to drugs such as chloroquine and sulfadoxine-pyrimethamine is a major problem in malaria control. Artemisinin (ART) derivatives, particularly in combination with other drugs, are thus increasingly used to treat malaria, reducing the probability that parasites resistant to the components will emerge. Although stable resistance to artemisinin has yet to be reported from laboratory or field studies, its emergence would be disastrous because of the lack of alternative treatments. Here, we report for the first time, to our knowledge, genetically stable and transmissible ART and artesunate (ATN)-resistant malaria parasites. Each of two lines of the rodent malaria parasite Plosmodium chabaudi chabaudi, grown in the presence of increasing concentrations of ART or ATN, showed 15-fold and 6-fold increased resistance to ART and ATN, respectively. Resistance remained stable after cloning, freeze-thawing, after passage in the absence of drug, and transmission through mosquitoes. The nucleotide sequences of the possible genetic modulators of ART resistance (mdr1, cg10, tctp, and atp6) of sensitive and resistant parasites were compared. No mutations in these genes were identified. In addition we investigated whether changes in the copy number of these genes could account for resistance but found that resistant parasites retained the same number of copies as their sensitive progenitors. We believe that this is the first report of a malaria parasite with genetically stable and transmissible resistance to artemisinin or its derivatives.
The resistance of Plasmodium falciparum to more available, safe, and easily administered drugs, especially chloroquine and pyrimethamine-sulfadoxine (SP), has become a serious obstacle to the control of malaria. Artemisinin combination therapy (ACT), using carefully matched drugs, is now the recommended strategy both for clinical care and for the avoidance of drug resistance (36). The identification and monitoring of genes and mutations giving rise to resistance to artemisinin and its derivatives are essential for the evaluation of this strategy. While some of the genes involved in chloroquine and pyrimethamine-sulfadoxine resistance are known (9, 14), those determining the responses to artemisinin have yet to be identified. For instance, two genes, originally proposed to modulate sensitivity to chloroquine in P. falciparum, have also been investigated in the context of artemisinin resistance. These are pfmdr1 and pfcrt, encoding membrane transporter proteins, which are localized in the membrane of the parasite's food vacuole (6, 13). One particular mutation in the latter (pfcrt K76T) has been strongly associated with chloroquine resistance in a genetic cross, in allelic replacement transfections (13), and in natural populations of P. falciparum (9).
All artemisinin compounds induce a very rapid reduction of parasitemia, starting almost immediately after administration, killing all stages of the malaria parasite, including young gametocytes (1, 30). Artemisinin and its derivatives contain a stable endoperoxide bridge, which, it is suggested, is cleaved by intraparasitic heme. The cleaved endoperoxide becomes a carbon-centered free radical which then functions as an alkylating agent, reacting with both heme and parasite proteins (but not DNA) (1, 18, 30). A previous study with P. falciparum suggested that a sarcoplasmic and endoplasmic reticulum Ca2+ ATPase (SERCA)-type protein encoded by a gene denoted pfatp6 might be the major chemotherapeutic target of these drugs (11).
From natural populations of P. falciparum, a parasite's in vitro response to artemisinins may vary by manyfold, but even the highest 50% inhibitory concentrations (IC50s) documented thus far were not indicative of true resistance. When used as monotherapy, artemisinin derivatives are associated with low to moderate levels of recrudescence in vivo following single treatment (17), possibly due to their extremely short elimination half-lives. However, reports of high treatment failure and confirmed in vivo or in vitro resistance to these drugs have not yet been documented. Nevertheless, in light of previous experience with all other antimalarial drugs and because large parasite populations are increasingly being exposed to artemisinins, there is little doubt that resistance will develop (20a). Little is known about genes that may modulate sensitivity to these drugs, mainly because all attempts to select drug-resistant mutants of stable phenotype in the laboratory have failed. Nevertheless, a number of genes have been proposed as possible modulators of artemisinin susceptibility following experimental laboratory studies or field-based genotype-phenotype association surveys on P. falciparum isolates.
Artemisinin-resistant strains have been produced in the rodent malaria model Plasmodium yoelii (22, 27, 32), and shown to contain significantly more translationally controlled tumor protein (TCTP) than sensitive ones (2). However, the resistance phenotype was unstable and it was unclear whether sensitive and resistant strains were genetically related; hence a causal relationship between TCTP levels and artemisinin resistance remains to be established. For P. falciparum, allele replacement studies and the analysis of progeny from a genetic cross have shown that point polymorphisms in the pfmdr1 gene were correlated with small changes in artemisinin sensitivity (10, 26), although it is clear that they are not the major determinants of sensitivity to the drug. Additionally, increased copy number of pfmdr1 rather than single nucleotide polymorphisms was shown to be associated with reduced artesunate sensitivity in vitro in two previous studies on fresh isolates from Thailand (23, 24).
Ferrer-Rodriguez et al. (12) have recently reported that the mdr1 gene of P. yoelii was overexpressed in artemisinin-resistant lines, but in that case the resistant phenotype was only transiently stable, preventing the establishment of a causal association between artemisinin resistance and the mdr1 gene. A previous study investigating the haplotypes of the pfcrt gene from parasites of Asian, African, or South American origin suggested that some polymorphisms in the pfcrt gene may slightly reduce sensitivity to artemisinin (29).
Recent reports show that the inhibitory action of artemisinin on P. falciparum ATPase 6 activity in Xenopus laevis oocytes can be greatly influenced by site-directed mutagenesis of the gene (e.g., L263E). Also, the relative sensitivities (IC50) of Plasmodium vivax, P. falciparum, and Plasmodium berghei to artemisinin correlate with the relative inhibitions of the enzyme (Ki) to artemisinin in the X. laevis oocyte system (31).
While the determination of the genetic basis of pyrimethamine resistance and chloroquine resistance lagged far behind the emergence of drug resistance, the present work aims at using the rodent malaria model Plasmodium chabaudi chabaudi to identify candidate genes before resistance to artemisinin and its derivatives occurs.
The specific objectives of the work presented here were to select artemisinin and artesunate mutants in the rodent malaria parasite P. chabaudi chabaudi through prolonged exposure of drug-sensitive lines to low and increasing levels of the drug in mice and to study the possible involvement of the P. chabaudi chabaudi orthologues of pfatp6, pfcrt, pfmdr1, and pftctp in the selected mutant clones.
MATERIALS AND METHODS
Parasites and hosts.
P. chabaudi chabaudi clones were maintained in 4- to 6-week-old laboratory CD1 female mice and transmitted through Anopheles stephensi mosquitoes, as described by Walliker et al. (33). Mouse drinking water was supplemented with 0.05% paraminobenzoic acid.
The parasite clones used to select artemisinin (ART) or artesunate (ATN) resistance were pyrimethamine- and/or chloroquine-resistant mutants (4) derived from a cloned isolate, P. chabaudi chabaudi clone AS-sens (Table 1). Briefly, the original AS isolate was cloned by limiting dilution (AS-sens) and then selected for resistance to pyrimethamine and recloned (4). This clone, designated AS-PYR, was then selected for resistance to chloroquine and a line resistant to six daily doses of this drug at 3 mg kg−1 obtained (28). This line was cloned and denoted AS-3CQ. AS-3CQ was then subjected to further stepwise increases of chloroquine, producing a line resistant to six daily doses of chloroquine at 15 mg kg−1 (21). This parasite was cloned (AS-15CQ) and subjected to further increments in chloroquine pressure to select parasites that survived six daily doses of chloroquine at 30 mg kg−1 (3, 21), which were cloned and termed AS-30CQ. Chloroquine resistance in all parasites was shown to be a stable phenotype after freeze-thawing, serial blood passages through mice in the absence of treatment, and transmission through mosquitoes (3, 4, 21, 28).
TABLE 1.
Clones of P. chabaudi chabaudi considered in this study
| Clone | Drug response |
|---|---|
| AS-sens | Drug sensitive |
| AS-PYR | Pyrimethamine resistant, chloroquine sensitive |
| AS-15CQ | Pyrimethamine resistant, intermediate chloroquine resistant |
| AS-30CQ | Pyrimethamine resistant, high chloroquine resistant |
Drug selection experiments and parasite cloning.
To select for artemisinin or artesunate resistance, P. chabaudi chabaudi clones AS-PYR, AS-15CQ, and AS-30CQ were exposed to gradually increasing concentrations of artemisinin or artesunate during several consecutive passages in mice.
We inoculated 107 infected red blood cells, diluted in 0.2 ml of citrate saline, intraperitoneally into groups of three mice. Artemisinin and artesunate at the chosen doses were freshly prepared in pure dimethyl sulfoxide and administered orally 3 h later, and at the same time on the following 4 days. Blood smears were taken every day from day 5 onwards and examined microscopically for parasites. Smears were fixed with methanol and stained in 20% Giemsa for 20 min, after which slides were washed and allowed to air dry. Parasitemia was calculated as the percentage of infected red blood cells/number of total red blood cells in 10 to 15 microscopic fields. Parasites which survived drug treatment were subinoculated into fresh mice and the treatment was repeated. In subsequent passages, drug concentrations were gradually increased. To discard the possibility that potential increases in drug tolerance could be to due to increased virulence attributed to multiple subinoculations, an untreated and unselected parasite line was maintained in parallel and passaged in untreated mice the same number of times as the drug selected line.
Drug tests and cloning of parasites.
In order to compare the drug responses of drug-selected parasites to those of unselected controls at given time points during the selection procedure, groups of 5 mice were each inoculated intraperitoneally with 107 parasites (day 0). Three mice were treated orally with drugs at various dosages, three hours after parasite inoculation on day 0, and at the same time on the following 4 days; the remaining two mice were given dimethyl sulfoxide only. Artemisinin or artesunate solutions were freshly prepared in dimethyl sulfoxide just prior to administration. Blood smears were taken every day from day 6 onwards, stained with Giemsa stain, and examined microscopically to determine the percent parasitemia in 10 microscopic fields.
Following these tests, drug-selected parasites showing a significant increase in drug tolerance in comparison with unselected control lines were cloned by the method of limiting dilution (28). Cloned parasites were retested for their responses to both artemisinin and artesunate.
Tests to evaluate the stability of drug resistance.
To assess whether artemisinin or artesunate resistance was a genetically stable feature, drug-resistant parasite clones were retested for their drug responses after each of three different procedures: freeze-thaw cycles in liquid nitrogen, at least 10 blood subinoculations in mice in the absence of drug treatment and transmission through Anopheles stephensi mosquitoes.
A measure of resistance in the drug-selected parasite clones was established in the following way. The minimum curative dose (MCD) of each drug was first assessed in drug-selected parasites and untreated control lines. The MCD was defined as the minimum dose of each drug that would prevent reappearance of parasites in all five mice within each treated group at any time during the first 10 days of the follow-up period. A resistance index was determined using the following equation: n-fold resistance = MCD drug-selected parasites/MCD drug-unselected parasites.
DNA and RNA extraction and cDNA synthesis.
Parasitized red cells were harvested from mice under general anesthesia, when trophozoite stages were most prevalent, into citrate saline (pH 7.2) and passed through a column of fibrous cellulose powder twice (CF11; Whatman) to remove mouse leukocytes (15). The resulting red blood cell pellet was washed twice in phosphate-buffered saline and parasite DNA was extracted by overnight incubation in lysis solution (10 mM Tris [pH 8.0], 50 mM EDTA, 0.1% sodium dodecyl sulfate, proteinase K [1 mg/ml]) at 42°C. After phenol extraction, DNA was precipitated using propan-2-ol and ammonium acetate (3 M) and dissolved in TE buffer (10 mM Tris-Cl, 1 mM EDTA, pH 8.0). DNA samples were stored at −20°C.
Parasite RNA was extracted using a commercial RNA isolation kit (AquaPure RNA isolation kit; Bio-Rad), following the manufacturer's instructions. RNA samples were stored at −70°C.
Parasite cDNA was synthesized using a RNA synthesis commercial kit (Fermentas) according to the manufacturer's instructions.
Identification of the P. chabaudi chabaudi tctp and atp6 genes.
The DNA sequences of the P. falciparum and P. yoelii tctp and atp6 genes were available online at the NCBI/NIH database (http://www.ncbi.nih.gov) with the following accession numbers: for P. chabaudi chabaudi tctp, NP_703454; P. yoelii tctp, AF124820; P. falciparum atp6, AB121053; and P. yoelii atp6, AABL01001880. To obtain the P. chabaudi chabaudi orthologues of these genes, these sequences were retrieved and used in BLAST searches against the available P. chabaudi chabaudi sequences (shotgun clones and genomic contigs), deposited at the P. chabaudi chabaudi genome database (http://www.sanger.ac.uk). The two sequences giving significant hits were retrieved and used to design P. chabaudi chabaudi-specific oligonucleotide primers to amplify overlapping DNA fragments spanning the coding region, introns, and both 5′ and 3′ noncoding sequences (Table 2). These were then used in PCR amplifications containing either P. chabaudi chabaudi genomic DNA or cDNA templates, as described below.
TABLE 2.
Oligonucleotide primers for amplification of the atp6 and tctp genes from P. chabaudi chabaudi
| Gene | Sense | Name | Sequence (5′-3′) | Nucleotides covered |
|---|---|---|---|---|
| atp6 | Sense | 5′UTR 1F | GAA ATA TTC ACA ACT CGT TTG GTC | −4064 to ATG |
| Antisense | 5′UTR 2R | GAA ACT TCG ATC TAC AAA TTG G | ||
| Sense | 5′UTR 3F | CTA CAA GAG TTT AAA TCG GAT TC | ||
| Antisense | 5′UTR 4R | CTT GTA GAT GAC AAA TGC AGG | ||
| Sense | 5′UTR 5F | CGG AGC ACT GTC CTA TAT C | ||
| Antisense | 5′UTR 6R | CGT AGA AAA TGA TAG CAC GG | ||
| Sense | 5′UTR 7F | GAC GTT TGC CAT TCT CGT G | ||
| Antisense | 5′UTR 8R | CCC TTT TCT CAC ACA CAT AAA G | ||
| Sense | 5′UTR 9F | ATT TAG CTT ATC CAA TTT CTT TAC | ||
| Antisense | 5′UTR 10R | GTG TCA GTT TTC TAC AAT GC | ||
| Sense | 5′UTR 11F | AAA CAC TAT ATG GTA TTT ACG C | ||
| Antisense | 5′UTR 12R | CTG TGG ATT TAC AAA ATA ATT GAT C | ||
| Sense | 5′UTR 13F | CAT AAT GCC AAA AAA TGC GTA G | ||
| Antisense | 5′UTR 14R | GAG TAG GTT AGC CCT TTG AT | ||
| Sense | 5′UTR 15F | GCT TTT CGT CGC AGC ATA TG | ||
| Antisense | 5′UTR 16R | GTA AAA GTA GAT GAA AAT CGG | ||
| Sense | 1F | GTA GAA GAT GTT TTG AGA GCA G | ATG to TAA | |
| Antisense | 2R | CTG CAT ATT TAT CAA CAG AGC | ||
| Sense | 3F | AAA GCA GAA CAA AGT ATG TTA AC | ||
| Antisense | 4R | CTC CTC TTT GAC ATA GTT GAT ATT C | ||
| Sense | 5F | ATC AAA TGA CAG CTA CTG TTT TTC | ||
| Antisense | 6R | GAA TCA GTT AAT GAC CGT ACA TC | ||
| Sense | 7F | ATT GTA AAG GTG CAC CAG AA | ||
| Antisense | 8R | GCA ACT TCT CCA ATA TTA CTA C | ||
| Sense | 9F | GAA GCT ATT AAA GAA GGA CGA TG | ||
| Antisense | 10R | CAT TAA GCG CAT TGA ACA TTT CG | ||
| Sense | 11F | AGG CAA GTA CCT TAT CAT TAT CC | ||
| Antisense | 12R | GCG TCT TCA ATT TTT GAC CAT AC | ||
| Sense | 13F | GCT AGA ATA TTT GGG GTT G | ||
| Antisense | 14R | CCC CCA AAT TAT GAA TAT GC | ||
| Sense | 3′UTR 1F | CAA AGA ACT CGG GTA TGG TC | TAA to 1114 | |
| Antisense | 5′UTR 2R | CAT CCC TAT GCA AAT GGC C | ||
| Sense | 3′UTR 3F | AAG GTG TGC GTA AAA TGA GG | ||
| Antisense | 3′UTR 4R | GAC ATA GGA AAG ACT GGT GAG | ||
| tctp | Sense | AAT GAT GAA GTA TGC TCT GAC TCA | ATG to TAA | |
| Antisense | TGA TAT GTA AAC AAA TCT AGG AG |
Amplification and sequencing of the mdr1, cg10, tctp, and atp6 genes of P. chabaudi chabaudi.
Genomic DNA or cDNA was used as the template in 50-μl PCRs, containing 0.2 μM of each oligonucleotide primer, 1× PCR buffer (Promega), 2.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, and 0.025 U/μl of Taq DNA polymerase. For amplification of the P. chabaudi chabaudi mdr1 and P. chabaudi chabaudi cg10 genes, oligonucleotide primers and PCR amplification conditions previously published were used (7, 16), based on DNA sequences characterized prior to this study (P. chabaudi chabaudi mdr1, AY123625; and P. chabaudi chabaudi cg10, AY304549).
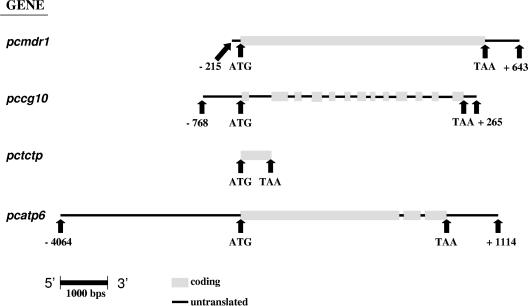
PCR products were purified using the QIAquick PCR purification kit from QIAGEN and sequenced using BigDye chain termination v3.1 (Applied Biosystems). The sequencing reactions were analyzed by Macrogen. The primers used in sequencing reactions were either those used for the initial amplification of the fragments, or M13 forward and reverse primers where amplified DNA fragments had been cloned into plasmid vectors. Gene and predicted amino acid sequences were manually compiled, and then compared between drug selected and unselected clones using an Internet-based interface denoted Multiple Sequence Alignment with hierarchical clustering (5), using default alignment parameters (http://prodes.toulouse.inra.fr/multalin/multalin.html). Figure 1 shows the extent of sequence generated with respect to the coding regions of the four genes.
FIG. 1.
Extent of DNA sequence obtained for each P. chabaudi chabaudi gene under study. The nucleotide sequences for each gene were obtained for all parasite clones under study, AS-15CQ, AS-30CQ, AS-ATN, and AS-ART. Numbers refer to nucleotides 5′ to the start codon ATG (negative) or 3′ to the termination codon (positive).
Estimation of copy numbers of the P. chabaudi chabaudi mdr1, tctp, and atp6 genes.
Real-time quantitative PCR (RTQ-PCR) was performed using a Roche LightCycler (34). Ten-microliter reactions in LightCycler capillaries (Roche) using FastStart DNA Master SYBR Green I kit reagents (Roche) were used according to the manufacturer's instructions. Magnesium chloride, primer concentration, denaturing, and annealing and elongation rates and times were varied to determine the conditions under which only the specific amplicon was produced. The average amplification efficiencies of the reactions were closely matched because small differences can result in large errors in quantification between samples. The average amplification efficiency, E, was determined from the slope of the standard curves produced in each experiment using the equation E = 10−1/slope.
PCRs where the amplicon doubles at every cycle have an optimal efficiency of 2.0 compared to reactions where no amplification occurs and efficiency is 1.0. The gene copy number was assessed by RTQ-PCR. We used sequences within a fragment of the merozoite surface protein of P. chabaudi chabaudi gene (MSP1) (accession no. L22982) (20) as an internal calibrator, since msp1 is a single-copy gene in P. chabaudi chabaudi. All samples were analyzed in replicate within each LightCycler run. The fluorescence signal produced from the amplicon was acquired at the end of the polymerization step at 72°C. The PCR conditions and primer sequences for gene quantifications are presented in Table 3.
TABLE 3.
PCRs for quantification of P. chabaudi chabaudi mdr1, tctp, and atp6 genes in comparison to msp1a
| Gene | Sense | Primer sequence (5′-3′) | MgCl2 concn/ primer concn |
|---|---|---|---|
| mdr | Sense | TCTCGACCAAATGTACCAATATA | 3 mM/0.8 μM |
| Antisense | GCATTAGTCATTTCTATATCATTTG | ||
| tctp | Sense | AATGATGAAGTATGCTCTGACTCA | 4.5 mM/0.8 μM |
| Antisense | CATTCCTTCTACTGCATCTTCAC | ||
| atp6 | Sense | AGGCAAGTACCTTATCATTATCC | 4.5 mM/0.8 μM |
| Antisense | GTGAAGAAGTAAAGATCCTATTG | ||
| msp1 | Sense | ACAGTAACACAAGAAGGAAC | 3.5 mM/0.4 μM |
| Antisense | GATACTTGTGTTGATGCTGG |
PCR amplification conditions: 40 cycles of 95°C for 600 min 95°C with a 0-min hold, cooling at 20°C/s to 63°C with a 7-min hold, heating at 20°C/s to 72°C with a 7-min hold. Heating at 20°C/s to 95°C with 0-min hold, cooling at 20°C/s at 65°C and heating at 0.2°C/s to 95°C in a continuous acquisition mode produced the melting curve.
The genomic DNA samples used were the same as those used for gene amplification and sequencing. The purity and quantity of DNA were determined by fluorimetry, UV spectrophotometry, and electrophoresis of serially diluted ethidium bromide-stained samples on agarose gels. Standards consisting of 10-fold serial dilutions of genomic DNA were freshly prepared from concentrated stocks for each RTQ-PCR experiment.
Samples of a 10-fold serial dilution of AS-15CQ, AS-30CQ, AS-ART (derived from AS-30CQ selected for artemisinin resistance), and AS-ATN (derived from AS-15CQ selected for artesunate resistance) genomic DNA in the range of 60 to 0.006 ng were used as quantification standards for the LightCycler calibration curve. Maximum recovery filter tips (AXYGEN Scientific) and No Stick microtubes (Alpha Laboratories) were used in these experiments.
The analysis of relative gene expression data was performed using the 2−ΔΔCt method described in detail by Livak and Schmittgen (19); the control gene was P. chabaudi chabaudi msp-1 and the target genes were the P. chabaudi chabaudi mdr1, tctp, and atp6 genes. The average cycle threshold (Ct) was calculated for both control and target genes and the ΔCt (Ct target gene − Ct msp-1) was determined. ΔΔCt was calculated for the relative quantification of the target gene as follows: ΔΔCt = (Ct target gene − Ct MSP-1)α − (Ct target gene − Ct MSP-1)β, where α is the ART- or ATN-resistant sample and β is the AS-30CQ or AS-15CQ sample. After validation of the method, results for each sample were expressed in n-fold changes in α target gene expression, normalized to msp-1 relative to the expression of β, according to the equation amount of target = 2−ΔΔCt.
RESULTS
Selection and cloning of artemisinin- and artesunate-resistant parasites.
Three related cloned lines of P. chabaudi chabaudi, derived sequentially from a drug-sensitive cloned isolate denoted AS-sens, were used for the artemisinin or artesunate resistance selection experiments (Table 1). These were (i) a chloroquine-sensitive, pyrimethamine-resistant clone denoted AS-PYR, (ii) a clone exhibiting intermediate-level chloroquine resistance denoted AS-15CQ, and (iii) a highly chloroquine-resistant clone denoted AS-30CQ.
The drug selection procedures involved administration of subcurative treatment to infected mice with daily drug doses given over a total period of 5 days. Attempts to establish stable artemisinin or artesunate resistance in AS-PYR were unsuccessful (data not shown). The clones AS-30CQ and AS-15CQ were thus used to select for artemisinin and artesunate resistance, respectively.
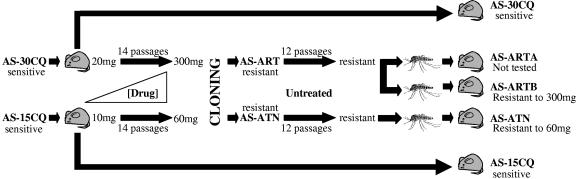
Initial drug testing revealed that the maximum dose of artemisinin tolerated by the AS-30CQ parasites was 20 mg/kg/day. Parasites that survived that initial dose were subinoculated and subjected to successive increments in drug doses, as described in Materials and Methods. AS-30CQ responded well to this artemisinin selection procedure, and a line that tolerated a daily dose of 300 mg/kg/day was selected after 15 passages (Fig. 2). Similarly, the AS-15CQ clone, which initially survived 10 mg/kg/day artesunate, tolerated gradual increases in artesunate treatment well, and after 14 subinoculations under drug pressure a population of parasites was selected which survived treatment of 60 mg/kg/day (Fig. 2). At this stage, it was considered that a significant level of resistance had been achieved and thus the artemisinin and artesunate selection procedures were suspended.
FIG. 2.
Artemisinin and artesunate selection procedure. Parasite clones (AS-30CQ and AS-15CQ) were passaged in the absence and presence of gradually increasing doses of drug (artemisinin and artesunate, respectively). Initial drug sensitivities (20 or 10 mg/kg/day) decreased (to 300 or 60 mg/kg/day) after 15 and 14 passages, respectively. The drug responses after cloning, passage in the absence of drug (untreated), and transmission through mosquitoes remained unchanged. Control selection procedures in the absence of drug are also shown.
Parasite lines containing populations of artemisinin- and artesunate-resistant parasites were then subjected to cloning by limiting dilution. Seven and five clones were successfully established from the artemisinin- and artesunate-resistant parasite populations, respectively. Of these, one clone from each line was chosen on the basis of its faster growth rate. The artemisinin- and artesunate-resistant clones were designated P. chabaudi chabaudi AS-ART and AS-ATN, respectively (Fig. 2). AS-ART and AS-ATN were tested for their responses to artemisinin and artesunate, respectively, immediately after cloning, and shown to retain the same phenotype as that of the drug-resistant population from which they had been derived (data not shown). These parasites were then used in subsequent studies to investigate further whether the observed drug resistance was stable.
Stability of artesunate or artemisinin resistance.
In order to confirm whether an heritable genetic mutation underlies the responses of AS-ART and AS-ATN to artemisinin and artesunate, respectively, the stabilities of the drug resistance in these clones were evaluated by measuring drug responses after (i) multiple blood passages in the absence of the drug, (ii) freeze-thawing of parasites in liquid nitrogen, and (iii) transmission through a mosquito host, Anopheles stephensi.
AS-ART and AS-ATN parasite lines were therefore subjected to 12 further passages in untreated mice, after which they were tested for their drug responses. They both retained tolerance to the corresponding drug (data not shown). Thus, artemisinin and artesunate resistance in AS-ART and AS-ATN, respectively, were shown to be stable phenotypes in the absence of drugs.
AS-ART and AS-ATN parasite lines were deep-frozen in liquid nitrogen, thawed after 1 week, and inoculated back into mice. Again, the drug phenotypes remained unchanged in both cases (data not shown).
Mice infected with AS-ART or AS-ATN were used to feed A. stephensi mosquitoes. AS-ART was successfully transmitted through mosquitoes on two separate occasions, and the resulting blood forms that developed in mice were renamed AS-ARTA and AS-ARTB (Fig. 2). The MCDs of both AS-ARTA and AS-ARTB were then assessed in parallel to the untreated control line, AS-30CQ. AS-ARTB showed a 15-fold increase in MCD to artemisinin relative to AS-30CQ (Table 4). Thus, artemisinin resistance remained stable after transmission of the resistant parasites through mosquitoes. In a similar fashion, AS-ATN was also subjected to mosquito transmission and showed a sixfold increase in MCD to artesunate relative to AS-15CQ (Table 4). These assays showed that the sensitivity of AS-ATN to artesunate was significantly lower than that of its sensitive progenitor AS-15CQ; therefore we also consider artesunate resistance to be stable after the sporogonic cycle.
TABLE 4.
Resistance of P. chabaudi chabaudi AS-ATN and AS-ARTBa
| P. chabaudi chabaudi | MCD ATN (mg/kg/day) | MCD ART (mg/kg/day) | n-fold ATN | n-fold ART |
|---|---|---|---|---|
| AS-15CQ | 10 | 6 | ||
| AS-ATN | 60 | 160 | 6 | 26 |
| AS-30CQ | 8 | 20 | ||
| AS-ARTB | 40 | 300 | 5 | 15 |
The absolute and relative drug sensitivities of AS-ATN and AS-ART after blood passage in the absence of treatment, freeze-thawing, and mosquito transmission are given.
In summary, the artemisinin and artesunate resistance phenotypes of the drug-selected parasite clones were unaltered after passage in the absence of drug pressure, after freezing and thawing, and after transmission through laboratory mosquitoes. We believe, therefore, that artemisinin and artesunate resistance in the clones AS-ART and AS-ATN is genetically encoded.
Cross-resistance to artemisinin and artesunate.
In order to evaluate whether the mechanisms of resistance to artemisinin and to artesunate share similar features, the responses of AS-ART to artesunate and AS-ATN to artemisinin were tested. AS-ART showed a fivefold increase in the MCD to artesunate relative to AS-30CQ, while AS-ATN showed a greater than 10-fold increase in the MCD to artemisinin relative to AS-15CQ (Table 4). These tests therefore revealed that both clones showed cross-resistance and that in both parasites there were greater increases in artemisinin resistance (15- to 26-fold) than for artesunate resistance (five- to sixfold). This suggests that resistance to artemisinin and artesunate does share similar features, at least in the parasite clones described here.
Identification and characterization of TCTP and ATP6 genes.
We wished to investigate whether the P. chabaudi chabaudi orthologues of P. falciparum TCTP and ATP6 play a role in the resistance to artemisinin derivatives in P. chabaudi chabaudi. P. chabaudi chabaudi-specific database sequences were used to design oligonucleotide primers for amplifying both genes (Table 5).
TABLE 5.
Comparison between predicted amino acid sequences of the P. falciparum and P. chabaudi chabaudi tctp and atp6 orthologues
| Gene | P. falciparum chromosome | Length (residues)
|
% Identity
|
||
|---|---|---|---|---|---|
| P. falciparum | P. chabaudi chabaudi | Nucleotide | Amino acid | ||
| tctp | 5 | 516 | 516 | 84 | 90 |
| atp6 | 1 | 3,687 | 3,357 | 72 | 72 |
PCR amplifications were initially carried out using genomic or cDNA from the drug-sensitive parasite clone AS-sens to amplify and sequence an array of several overlapping PCR fragments spanning the desired genes. Following alignment of these fragments, full sequences from genomic and cDNA were assembled and aligned in order to predict the exon-intron structure. Coding sequences were translated using internet-based bioinformatics tools at http://www.bioinformatics.org/sms/, and predicted peptides aligned with their P. falciparum protein homologues.
The P. chabaudi chabaudi tctp gene was first characterized in the drug-sensitive parasite AS-sens (GenBank accession number AY304545). In AS-sens, the gene is encoded by a continuous open reading frame containing 516 bp, which translates into a predicted peptide of 171 amino acids. The predicted AS-sens and P. falciparum amino acid sequences for TCTP showed 90% identity between the two species, confirming that the P. chabaudi chabaudi orthologue of the gene had been successfully identified (Table 5). Subsequent comparisons of the predicted amino acid sequence of P. chabaudi chabaudi TCTP with that of P. yoelli and P. berghei orthologues show amino acid identities of 97 and 98%, respectively.
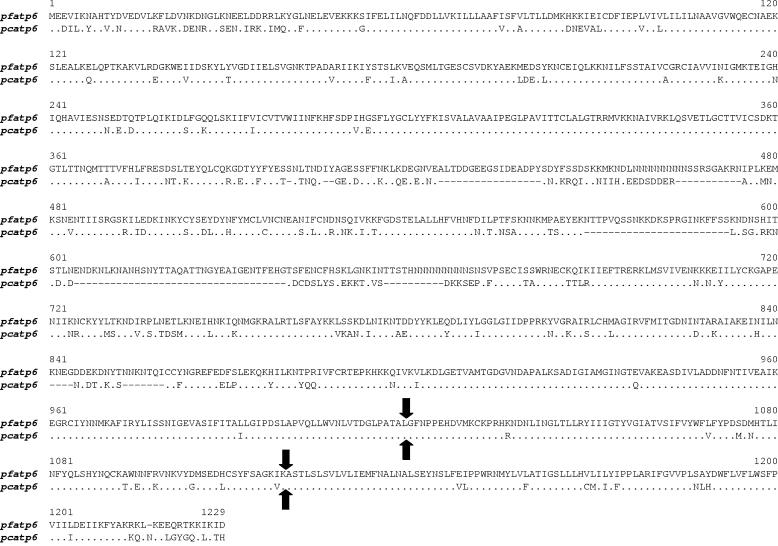
The atp6 gene from AS-sens was similarly characterized (GenBank accession number DQ171938) and was also found to be similar to that of P. falciparum. The P. chabaudi chabaudi atp6 gene has two introns (intron 1, 92 bp; intron 2, 90 bp) located near the 3′ end of the gene and in identical positions to the predicted introns in the P. falciparum gene (Fig. 3). Translation of the coding sequence of the P. chabaudi gene resulted in a predicted protein of 1,118 amino acids. This sequence shares 72% identity with that of the P. falciparum orthologue (Table 5). The NCBI Conserved Domain Search revealed that this sequence contained a segment of 242 residues (107 to 348) typical of a putative conserved domain of E1- and E2-type ATPases. Additionally, PROSITE scanning (http://us.expasy.org/prosite/) revealed a characteristic E1-E2 ATPase phosphorylation signature, DKTGTLT, between residues 358 and 364. E1-E2. ATPases are enzymes that form an aspartyl phosphate intermediate in the course of ATP hydrolysis and can be divided into four major groups, one of which constitutes the Ca2+-transporting ATPases and of which P. falciparum and P. chabaudi chabaudi atp6 are members.
FIG. 3.
Alignment between predicted amino acid sequences of ATPase 6 proteins of P. falciparum (pf) and P. chabaudi (pc). Arrows indicate predicted positions of two introns in each gene.
Amplification and sequencing of P. chabaudi chabaudi mdr1, cg10, tctp, and atp6 in sensitive and drug-resistant parasites.
In order to identify possible point mutations in the cg10, mdr1, tctp, or atp6 gene, in artemisinin- and artesunate-resistant parasites, AS-ARTB and AS-ATN, respectively, relative to AS-30CQ and AS-15CQ, respectively, we proceeded to determine the nucleotide sequences of these genes in sensitive and resistant parasites (Fig. 1). Comparisons of coding regions and introns between all parasites revealed that the sequences of all genes in the mutants were found to be identical to those of the sensitive parasites from which they had been derived. Therefore, point mutations in mdr1, cg10, tctp, and atp6 were not involved in the generation of artemisinin or artesunate resistance at any stage in these lineages.
We also wished to determine whether mutations in noncoding sequences lying proximal to the gene had been selected, in view of the increasing evidence that P. falciparum atp6 may play a role in the mechanism of action of artemisinin and in possible changes of sensitivity to the drug. Such mutations may affect the level of transcription. However, no differences in nucleotide sequence between the resistant clones and their sensitive progenitors were found in 4 kb of nucleotide sequence upstream or in 1 kb downstream of the gene.
Assessment of copy numbers of the mdr1, tctp, and atp6 genes of P. chabaudi chabaudi.
After identification of oligonucleotide primers (Table 3) and optimization of the real-time PCR conditions, we investigated if ART and ATN resistance could be related to changes in the gene copy numbers of the mdr1, tctp, and atp6 genes. Differences in the relative number of copies of each target gene were thus compared between the artemisinin- and artesunate-resistant parasites AS-ARTB and AS-ATN relative to their sensitive progenitors, AS-30CQ and AS-15CQ, respectively.
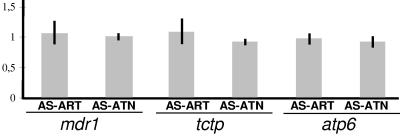
Three independent experiments were carried out; the mean n-fold amplifications of genes in these experiments are presented in Fig. 4. There were no observed changes in the gene copy numbers of the mdr1, tctp, and atp6 genes during the selection for artemisinin and artesunate resistance, showing that the phenotype of artemisinin and artesunate resistance is not associated with an increase in the gene copy number of mdr1, tctp, or atp6.
FIG. 4.
Relative differences (n-fold) in gene copy number between artemisinin- (AS-ART) and artesunate- (AS-ATN) resistant parasites and their sensitive progenitors, AS-30CQ and AS-15CQ, respectively. The vertical axis represents the mean n-fold gene number (gray bars) and standard deviation (vertical lines) of each of the genes under study normalized against that of MSP-1, generated after three independent assays.
DISCUSSION
Although there have been considerable scientific advances over the past hundred years, the overall burden of malaria is currently increasing, especially in sub-Saharan Africa. In the absence of a fully protective antimalarial vaccine, the control of malaria relies principally on the use of drugs for treatment or prophylaxis. Much of the increasing burden of malaria is due to the spread of resistance of the major human malaria pathogen, P. falciparum, to most drugs presently available. Consequently, the World Health Organization and health authorities in malaria-endemic countries are recommending new therapies, based on the use of artemisinin derivatives, combined with other drugs, the so-called artemisinin combination therapy (35). Reports of resistance of natural populations of P. falciparum to these drugs have not been forthcoming thus far, but most agree that resistant parasites will occur in the future, based upon previous experience with other antimalarial drugs. In the light of this, understanding the mechanisms underlying artemisinin resistance, before treatment failures are commonly observed, will provide tools to evaluate prevent or delay its appearance and spread.
The genetic basis of drug resistance in malaria may be investigated in different ways. One of the most appropriate is to use genetically stable resistant mutants selected through drug pressure from cloned sensitive parasite lines. In this way, drug-sensitive and drug-resistant parasites are genetically identical (isogenic) except for any mutations involved in resistance; such mutations can then be pinpointed using different approaches. Efforts to select genetically stable artemisinin resistant mutants of P. falciparum, P. berghei, and P. yoelii have yet to be successful.
We report here for the first time to our knowledge, malaria parasites that have genetically stable and transmissible resistance to artemisinin and artesunate. The successful selection of these parasites and the fact that they can be transmitted through Anopheles sp. mosquitoes is a significant achievement for two main reasons. Firstly, these observations demonstrate that malaria parasites are not only genetically and biologically capable of sustaining stable resistance to artemisinins, but also that this resistance can be selected through drug pressure. Consequently it is conceivable that in human malaria, artemisinin resistance may arise in the future due to extensive and/or inappropriate drug usage; once it does, it may spread in the parasite population and become established. Second, the resistant P. chabaudi chabaudi parasites reported here can now be used to investigate the genetic determinants of resistance to these drugs.
We note that we were able to generate ART or ATN resistance in clones (AS-15CQ and AS-30CQ) which were already resistant to chloroquine, while efforts to generate ART or ATN resistance from the chloroquine-sensitive clone (AS-PYR) were abandoned when 10 passages in the presence of drug failed to lead to observable decreases in parasite sensitivity. This may be a result of a genetic potentiation of the parasites' ability to generate mutations in response to drug treatment (called the accelerated resistance to multiple drugs phenotype) (25) which might have occurred during the generation of the chloroquine resistant lines. Alternatively, it is possible that the ART resistance phenotype is expressed only in chloroquine-resistant clones. This would tend to suggest that there is some functional interaction between the pathways underlying chloroquine and artemisinin resistance. This interpretation would be supported by showing that allelic replacement of the wild-type allele of the gene (underlying ART resistance) with the mutant allele decreases ART sensitivity in chloroquine-resistant clones only. These questions have significant relevance to the practical lifetime of a drug in areas where resistance to other drugs (or to chloroquine specifically) is prevalent.
Previous to the present study, artemisinin resistance was achieved in vivo in Plasmodium yoelii (22, 27, 32) but the resistance phenotype in these parasites was lost upon removal of drug pressure, suggesting one of two hypotheses: that resistance was due to genetic mutations that are not sustainable by the parasites and therefore lost in the absence of treatment, or that continuous exposure of these parasites to artemisinin, triggered physiological or epigenetic adaptations that increased the parasite's tolerance to the drug, but were manifested only in a transient manner and were consequently lost after stopping drug treatment. In these studies, two different genes, P. chabaudi chabaudi mdr1 (12) and tctp (2), were suggested to be potential modulators of resistance to artemisinin, as discussed in the introduction.
This study has shown that there are no changes in nucleotide sequence or copy number for P. chabaudi chabaudi mdr1 in the artemisinin- or artesunate-resistant parasites compared to their sensitive progenitors. For tctp, we found that the P. chabaudi chabaudi gene is very similar to its orthologue in the other rodent malaria parasites as well as to the P. falciparum tctp gene, in terms of both structure and amino acid sequence, indicating a high degree of conservation among malaria species. We then confirmed that there were no differences in coding sequence or copy number of tctp in artemisinin- or artesunate-sensitive and resistant parasites. These data indicate that resistance to artemisinin derivatives in P. chabaudi chabaudi is not determined by changes in either the sequence or the gene copy number of either the tctp or mdr1 gene.
In P. chabaudi chabaudi, we have previously demonstrated that chloroquine resistance is independent of the P. chabaudi crt orthologue cg10 (16). However, in light of the results with the human malaria parasite (29), we wished to investigate whether artemisinin or artesunate could have selected for mutations in this gene in AS-ATN and AS-ARTB. We have shown that the DNA sequences and copy numbers of P. chabaudi chabaudi cg10 in artemisinin- and artesunate-resistant parasites are unchanged relative to their sensitive progenitors and we conclude that the P. chabaudi chabaudi orthologue of P. falciparum crt is not involved in the generation of artemisinin or artesunate resistance in these parasites.
It has recently been suggested that a major chemotherapeutic target of artemisinin in P. falciparum is a calcium-translocating P-type ATPase of the SERCA type (11), encoded by a gene denoted P. falciparum atp6. Thus, artemisinin derivatives inhibit P. falciparum ATPase 6 activity in Xenopus oocytes and this pattern of inhibition is altered by site-directed mutagenesis of residues predicted to lie close to the artemisinin-binding site (31). Importantly, specific amino acid substitutions at residue 263 (263L, L263S, and L263A) in P. falciparum ATPase 6 influence the Ki with respect to artemisinin inhibition in a manner which parallels the IC50s of P. falciparum, P. berghei, and P. vivax, respectively, from which these substitutions derive.
Since these results may indicate a possible role for ATPase 6 in the mode of action of artemisinin and in possible resistance mechanisms, we characterized the SERCA homologue of P. chabaudi chabaudi in artemisinin-sensitive and -resistant parasites in order to investigate whether mutations or copy number could determine the resistance phenotypes. The atp6 gene of P. chabaudi chabaudi was shown here to be structurally very similar to its P. falciparum orthologue, a coding sequence interrupted by two relatively small introns located near the 5′ end of the gene. In terms of the degree of conservation among predicted protein sequence, the P. chabaudi chabaudi SERCA showed 72 and 86% identity to its P. falciparum and P. yoelii homologues, respectively.
Our observations demonstrated that the P. chabaudi chabaudi atp6 sequence and copy number had remained unaltered following selection with artesunate or artemisinin, implying that artemisinin treatment does not select mutations in the gene encoding its putative chemotherapeutic target, or mutations which affect its 5′ and 3′ predicted promoter region. We note that the amino acid residue at position 263, like that in P. berghei, is serine. Although we are unable to measure the IC50 of artemisinin because of the difficulties of propagating P. chabaudi chabaudi parasites in culture, we have shown that parasites AS-15CQ and AS-30CQ do not appear to be very sensitive to artemisinin in vivo, as is the case with P. berghei. The resistance phenotype which we have investigated may therefore represent high-level artemisinin resistance, which depends upon mutations in genes other than atp6.
There are, however, other reasons which may explain why mutations in the P. chabaudi chabaudi atp6 gene did not mediate the resistant phenotype in the rodent malaria system. Firstly of course, ATPase 6 may not be the target in either P. chabaudi chabaudi or P. falciparum. The evidence supporting a role for P. falciparum ATPase 6 in artemisinin sensitivity in P. falciparum arises from an ex vivo heterologous system (Xenopus oocytes). Inhibition of activity in such a system should not imply antimalarial therapeutic activity in the mammalian host. It is relevant to point out that the Ki for artemisinin inhibition of P. falciparum ATPase 6 may be over an order of magnitude greater than the IC50. This may suggest either concentration of artemisinin in the host or the presence of other targets. Second, the resistance mechanism may involve components distinct from the target molecules. However, the correspondence between the inhibition of mutated P. falciparum ATPase 6 and the relative sensitivities of P. falciparum, P. berghei, and P. vivax does begin to suggest a role for this protein in the resistance mechanism. Third, if P. falciparum ATPase 6 turns out to be the major modulator of artemisinin responses in P. falciparum, it is possible that the rodent malaria parasite P. chabaudi chabaudi may reveal alternative mechanisms of resistance to those of P. falciparum, as is the case with chloroquine resistance (16).
This study is the first to report malaria parasites that present resistance to artemisinin and artesunate, that are genetically stable in the absence of drug pressure and transmissible through malaria vectors. These parasites can now be used to identify the genes underlying resistance to these important classes of drugs, using high-throughput comparative genomic studies based on genome-wide approaches. In this context, the recently reported novel technique of linkage group selection (8) is now considered the method of choice to identify the mutations which underlie artemisinin resistance in this rodent model of malaria. If relevant, the resulting information may be used to prevent or monitor artemisinin resistance in human malaria.
Acknowledgments
Artemisinin and artesunate were kind gifts from African Artemisia and Dafra Pharma (Eerse, Belgium), respectively.
We acknowledge the financial support of CMDT/IHMT and FCT of Portugal (project ref. POCTI/ESP/42666/2001) and the European Union (project RESMALCHIP, ref QLK2-CT-2002-1503). A.A. is funded by FCT of Portugal (SFRH/BD/8913/2002). P.H. was supported by the Medical Research Council (United Kingdom). S.C. was supported by the Wellcome Trust (United Kingdom).
REFERENCES
- 1.Akompong, T., S. Eksi, K. Williamson, and K. Haldar. 2000. Gametocytocidal activity and synergistic interactions of riboflavin with standard antimalarial drugs against growth of Plasmodium falciparum in vitro. Antimicrob. Agents Chemother. 44:3107-3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhisutthibhan, J., X. Q. Pan, P. A. Hossler, D. J. Walker, C. A. Yowell, J. Carlton, J. B. Dame, and S. R. Meshnick. 1998. The Plasmodium falciparum translationally controlled tumour protein homologue and its reaction with the antimalarial drug artemisinin. J. Biol. Chem. 273:16192-16198. [DOI] [PubMed] [Google Scholar]
- 3.Carlton, J., M. Mackinnon, and D. Walliker. 1998. A chloroquine resistance locus in the rodent malaria parasite Plasmodium chabaudi. Mol. Biochem. Parasitol. 93:57-72. [DOI] [PubMed] [Google Scholar]
- 4.Carter, R., and D. Walliker. 1975. New observations on the malaria parasites of rodents of the Central African Republic-Plasmodium vinckei petteri subsp. nov. and Plasmodium chabaudi Landau, 1965. Ann. Trop. Med. Parasitol. 69:187-196. [DOI] [PubMed] [Google Scholar]
- 5.Corpet, F., J. Gouzy, and D. Kahn. 1998. The ProDom database of protein domain families. Nucleic Acids Res. 26:323-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cowman, A. F., S. Karcz, D. Galatis, and J. G. Culvenor. 1991. A P-glycoprotein homologue of Plasmodium falciparum is localized on the digestive vacuole. J. Cell Biol. 113:1033-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cravo, P. V., J. M. Carlton, P. Hunt, L. Bisoni, R. A. Padua, and D. Walliker. 2003. Genetics of mefloquine resistance in the rodent malaria parasite Plasmodium chabaudi. Antimicrob. Agents Chemother. 47:709-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Culleton, R., A. Martinelli, P. Hunt, and R. Carter. 2005. Linkage group selection: rapid gene discovery in malaria parasites. Genome Res. 15:92-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Djimde, A., O. K. Doumbo, J. F. Cortese, K. Kayentao, S. Doumbo, Y. Diourte, A. Dicko, X. Z. Su, T. Nomura, D. A. Fidock, T. E. Wellems, C. V. Plowe, and D. Coulibaly. 2001. A molecular marker for chloroquine-resistant falciparum malaria. N. Engl. J. Med. 344:257-263. [DOI] [PubMed] [Google Scholar]
- 10.Duraisingh, M. T., C. Roper, D. Walliker, and D. C. Warhurst. 2000. Increased sensitivity to the antimalarials mefloquine and artemisinin is conferred by mutations in the pfmdr1 gene of Plasmodium falciparum. Mol. Microbiol. 36:955-961. [DOI] [PubMed] [Google Scholar]
- 11.Eckstein-Ludwig, U., R. J. Webb, I. D. Van Goethem, J. M. East, A. G. Lee, M. Kimura, P. M. O'Neill, P. G. Bray, S. A. Ward, and S. Krishna. 2003. Artemisinins target the SERCA of Plasmodium falciparum. Nature. 424:957-961. [DOI] [PubMed] [Google Scholar]
- 12.Ferrer-Rodriguez, I., J. Perez-Rosado, G. W. Gervais, W. Peters, B. L. Robinson, and A. E. Serrano. 2004. Plasmodium yoelii: identification and partial characterization of an MDR1 gene in an artemisinin-resistant line. J. Parasitol. 90:152-160. [DOI] [PubMed] [Google Scholar]
- 13.Fidock, D. A., T. Nomura, A. K. Talley, R. A. Cooper, S. M. Dzekunov, M. T. Ferdig, L. M. Ursos, A. B. Sidhu, B. Naude, K. W. Deitsch, X. Z. Su, J. C. Wootton, P. D. Roepe, and T. E. Wellems. 2000. Mutations in the P. falciparum digestive vacuole transmembrane protein Pfcrt and evidence for their role in chloroquine resistance. Mol. Cell 6:861-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayton, K., and X. Z. Su. 2004. Genetic and biochemical aspects of drug resistance in malaria parasites. Curr. Drug Targets Infect. Disord. 4:1-10. [DOI] [PubMed] [Google Scholar]
- 15.Homewood, C. A., and K. D. Neame. 1976. A comparison of methods used for the removal of white cells from malaria-infected blood. Ann. Trop. Med. Parasitol. 70:249-251. [DOI] [PubMed] [Google Scholar]
- 16.Hunt, P., P. V. Cravo, P. Donleavy, J. M. Carlton, and D. Walliker. 2004. Chloroquine resistance in Plasmodium chabaudi: are chloroquine resistance transporter (crt) and multidrug resistance (mdr1) orthologues involved? Mol. Biochem. Parasitol. 133:27-35. [DOI] [PubMed] [Google Scholar]
- 17.Ittarat, W., A. L. Pickard, P. Rattanasinganchan, P. Wilairatana, S. Looareesuwan, K. Emery, J. Low, R. Udomsangpetch, and S. R. Meshnick. 2003. Recrudescence in artesunate-treated patients with falciparum malaria is dependent on parasite burden not on parasite factors. Am. J. Trop. Med. Hyg. 68:147-152. [PubMed] [Google Scholar]
- 18.Kamchonwongpaisan, S., and S. R. Meshnick. 1996. The mode of action of the antimalarial artemisinin and its derivatives. Gen. Pharmacol. 27:587-592. [DOI] [PubMed] [Google Scholar]
- 19.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 20.McKean, P. G., K. O'Dea, and K. N. Brown. 1993. Nucleotide sequence analysis and epitope mapping of the merozoite surface protein 1 from Plasmodium chabaudi chabaudi AS. Mol. Biochem. Parasitol. 62:199-209. [DOI] [PubMed] [Google Scholar]
- 20a.Meshnick, S. R. 2002. Artemisinin mechanisms of action, resistance and toxicity. Int. J. Parasitol. 32:1655-1660. [DOI] [PubMed] [Google Scholar]
- 21.Padua, R. A. 1981. Plasmodium chabaudi: genetics of resistance to chloroquine. Exp. Parasitol. 52:419-426. [DOI] [PubMed] [Google Scholar]
- 22.Peters, W., and B. L. Robinson. 2000. The chemotherapy of rodent malaria. LVIII. Drug combinations to impede the selection of drug resistance. Part. 2: the new generation-artemisinin or artesunate with long acting blood schizontocides. Ann. Trop. Med. Parasitol. 94:23-35. [DOI] [PubMed] [Google Scholar]
- 23.Pickard, A. L., C. Wongsrichanalai, A. Purfield, D. Kamwendo, K. Emery, C. Zalewski, F. Kawamoto, R. Scott Miller, and S. R. Meshnick. 2003. Resistance to antimalarials in Southeast Asia and genetic polymorphisms in pfmdr1. Antimicrob. Agents Chemother. 47:2418-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price, R. N., A. C. Uhlemann, A. Brockman, R. McGready, E. Ashley, L. Phaipun, K. Patel Laing, S. Looareesuwan, N. J. White, F. Nosten, and S. Krishna. 2004. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet 364:438-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rathod, P. K., T. McErlean, and P. C. Lee. 1997. Variations in frequencies of drug resistance in Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 94:9389-9393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reed, M. B., K. J. Saliba, S. R. Caruana, K. Kirk, and A. F. Cowman. 2000. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature 403:906-909. [DOI] [PubMed] [Google Scholar]
- 27.Robinson, B. L., and W. Peters. 1999. The chemotherapy of rodent malaria. LVI. Studies on the development of resistance to natural and synthetic endoperoxides. Ann. Trop. Med. Parasitol. 93:325-329. [DOI] [PubMed] [Google Scholar]
- 28.Rosario, V. E. 1976. Genetics of chloroquine resistance in malaria parasites. Nature 261:585-586. [DOI] [PubMed] [Google Scholar]
- 29.Sidhu, A. B., D. Verdier-Pinard, and D. A. Fidock. 2002. Chloroquine resistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutations. Science 298:210-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ubalee, R., D. Songthammawat, K. Na-Bangchang, P. Tan-ariya, and J. Karbwang. 1999. Ex vivo blood schizontocidal activities of artemisinin derivatives against Plasmodium falciparum. Southeast Asian J. Trop. Med. Public Health 30:225-231. [PubMed] [Google Scholar]
- 31.Uhlemann, A. C., A. Cameron, U. Eckstein-Ludwig, J. Fischbarg, P. Iserovich, F. A. Zuniga, M. East, A. Lee, L. Brady, R. K. Haynes, and S. Krishna. 2005. A single amino acid residue can determine the sensitivity of SERCAs to artemisinins. Nat. Struct. Mol. Biol. 12:628-629. [DOI] [PubMed] [Google Scholar]
- 32.Walker, D. J., J. L. Pitsch, M. M. Peng, B. L. Robinson, W. Peters, J. Bhisutthibhan, and S. R. Meshnick. 2000. Mechanisms of artemisinin resistance in the rodent malaria pathogen Plasmodium yoelii. Antimicrob. Agents Chemother. 44:344-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walliker, D., R. Carter, and A. Sanderson. 1975. Genetic studies on Plasmodium chabaudi: recombination between enzyme markers. Parasitology 70:19-24. [DOI] [PubMed] [Google Scholar]
- 34.Wittwer, C. T., K. M. Ririe, R. V. Andrew, D. A. David, R. A. Gundry, and U. J. Balis. 1997. The LightCycler: a microvolume multisample fluorimeter with rapid temperature control. BioTechniques. 22:176-181. [DOI] [PubMed] [Google Scholar]
- 35.Wongsrichanalai, C., A. L. Pickard, W. H. Wernsdorfer, and S. R. Meshnick. 2002. Epidemiology of drug-resistant malaria. Lancet Infect. Dis. ii:209-218. [DOI] [PubMed] [Google Scholar]
- 36.Yeung, S., W. Pongtavornpinyo, I. M. Hastings, A. J. Mills, and N. J. White. 2004. Antimalarial drug resistance, artemisinin-based combination therapy, and the contribution of modeling to elucidating policy choices. Am. J. Trop. Med. Hyg. 71:179-186. [PubMed] [Google Scholar]