Identifying submicroscopic corrals in the plasma membrane has been of interest since the classic Sheetz barnyard model of confinement of proteins by the erythrocyte membrane skeleton (1). In this issue, Wawrezinieck et al. (2) present a novel fluorescence correlation spectroscopy (FCS) method to identify corrals.
What is a corral? From the standpoint of cell biology, there are several possible corrals: a), the spectrin-actin membrane skeleton, a lattice of triangles of side ∼75 nm (1); b), actin filaments near the membrane, possibly with transiently or permanently bound transmembrane proteins forming the pickets of Kusumi's picket fence model, side 32–110 nm (3); c), a lipid domain, either raft or nonraft, with the raft size 0–700 nm (4); d), the extracellular matrix (5); and e), tethers connecting the mobile species to an immobile structure. To identify a corral from diffusion measurements, a corral must be taken to be an area with boundaries impenetrable enough that a mobile particle is confined to the area for a time much longer than the diffusion time across the area. One must be able to distinguish real confinement from the apparent confinement that is an inherent fluctuation in an unconfined random walk. This statistical problem is discussed in the context of single-particle tracking by Saxton and Jacobson (6) and in the supplemental material of Kusumi et al. (3).
How can a corral be detected? In a fluorescence photobleaching recovery (FPR) experiment, it is necessary to replenish the entire bleached spot to obtain a complete recovery curve. Diffusion is therefore over an area on the order of 10 times the spot size, where the spot size is diffraction-limited or larger. For the membrane skeleton or actin corrals, the measurement thus averages over many corrals and the diffusion coefficient is proportional to the corral area/mean residence time. Lipid domains, however, may be large enough to be detectable. In FPR measurements of lipid and protein diffusion in fibroblasts, Yechiel and Edidin (7) varied the spot size over a range of 0.35–5.0 μm, and found that the mobile fraction decreased monotonically with spot size. They interpreted their results as showing protein-rich domains of radius 0.4–1.5 μm surrounded by a protein-poor continuum.
For years, most measurements of lateral diffusion were made by FPR, but a combination of technical advances, reviewed by Webb (8), brought about a renaissance in FCS. It is now highly popular, as a walk through the appropriate poster sessions at the Biophysical Society annual meeting shows. FCS has single-molecule sensitivity, and averaging is carried out to obtain the autocorrelation function, so data analysis is less difficult than in single-particle tracking (reviews: 9,10,11).
How can this highly sensitive method be used to identify corrals? The experimental approach, developed independently by Masuda et al. (5) and Wawrezinieck et al. (2), is to vary the beam size. The major advance made by Wawrezinieck et al. (2) is in the method of data analysis. In their FCS measurements and simulations, they varied the beam size w, and measured τ, the width of the autocorrelation function at half-maximum. For free diffusion, τ is linear in the beam area,
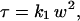 |
with k1 proportional to 1/D, where D is the diffusion coefficient. But for diffusion in the plasma membrane, they found experimentally, and confirmed by extensive simulations of confined diffusion, that the function is not linear but affine,
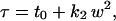 |
with t0 and k2 constants. Furthermore, the y-intercept t0 is positive for trapping by an isolated corral but negative for trapping in a network of barriers. Plots from the simulations show three regions:
Beam area ≪ corral area: free diffusion; linear dependence.
Beam area ∼ corral area: transitional region.
Beam area ≫ corral area: affine dependence.
The beam radius is 190–400 nm, but as the authors point out, the autocorrelation function can be affected by processes on smaller length scales. (Trivially, consider a point high-affinity binding site for the diffusing species.) Their estimate of the minimum detectable microdomain radius is 60 nm. Single-particle tracking gives higher resolution, with particle positions resolved to tens of nanometers (6), but the ability to detect 60-nm corrals by FCS is an important advance.
The work of Wawrezinieck et al. (2) is a promising beginning. The theory remains to be done, and a test of the method and the resolution in a well-defined (nanofabricated?) model system. It would be useful for the authors to revisit the experiments of Yechiel and Edidin (7) to see whether the new FCS approach can tell us more about these influential results. Likewise it would be interesting to examine by FCS the larger corrals found in the single-particle tracking experiments of the Kusumi laboratory (3). The most informative experiments would use the same cell types, diffusing species, and labels as the earlier measurements.
References
- 1.Sheetz, M. P. 1983. Membrane skeletal dynamics: Role in modulation of red cell deformability, mobility of transmembrane proteins, and shape. Semin. Hematol. 20:175–188. [PubMed] [Google Scholar]
- 2.Wawrezinieck, L., H. Rigneault, D. Marguet, and P.-F. Lenne. 2005. Fluorescence correlation spectroscopy diffusion laws to probe the submicron cell membrane organization. Biophys. J. 89:4029–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kusumi, A., C. Nakada, K. Ritchie, K. Murase, K. Suzuki, H. Murakoshi, R. S. Kasai, J. Kondo, and T. Fujiwara. 2005. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: High-speed single-molecule tracking of membrane molecules. Annu. Rev. Biophys. Biomol. Struct. 34:351–378. [DOI] [PubMed] [Google Scholar]
- 4.Anderson, R. G. W., and K. Jacobson. 2002. A role for lipid shells in targeting proteins to caveolae, rafts, and other lipid domains. Science. 296:1821–1825. [DOI] [PubMed] [Google Scholar]
- 5.Masuda, A., K. Ushida, and T. Okamoto. 2005. New fluorescence correlation spectroscopy enabling direct observation of spatiotemporal dependence of diffusion constants as an evidence of anomalous transport in extracellular matrices. Biophys. J. 88:3584–3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saxton, M. J., and K. Jacobson. 1997. Single-particle tracking: Applications to membrane dynamics. Annu. Rev. Biophys. Biomol. Struct. 26:373–399. [DOI] [PubMed] [Google Scholar]
- 7.Yechiel, E., and M. Edidin. 1987. Micrometer-scale domains in fibroblast plasma membranes. J. Cell Biol. 105:755–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Webb, W. W. 2001. Fluorescence correlation spectroscopy: inception, biophysical experimentations, and prospectus. Appl. Opt. 40:3969–3983. [DOI] [PubMed] [Google Scholar]
- 9.Haustein, E., and P. Schwille. 2003. Ultrasensitive investigations of biological systems by fluorescence correlation spectroscopy. Methods. 29:153–166. [DOI] [PubMed] [Google Scholar]
- 10.Lagerholm, B. C., G. E. Weinreb, K. Jacobson, and N. L. Thompson. 2005. Detecting microdomains in intact cell membranes. Annu. Rev. Phys. Chem. 56:309–336. [DOI] [PubMed] [Google Scholar]
- 11.Rigler, R., and E. Elson. 2001. Fluorescence Correlation Spectroscopy: Theory and Applications. Springer, Berlin.


