Abstract
The formation, relative stability, and possible stoichiometries of two (self-)complementary peptide sequences (B and E) designed to form either a parallel homodimeric (B + B) or an antiparallel heterodimeric (B + E) coiled coil have been investigated. Peptide B shows a characteristic coiled coil pattern in circular dichroism spectra at pH 7.4, whereas peptide E is apparently random coiled under these conditions. The peptides are complementary to each other, with peptide E forming a coiled coil when mixed with peptide B. Molecular dynamics simulations show that combinations of B + B and B + E readily form a dimeric coiled coil, whereas E + E does not fall in line with the experimental data. However, the simulations strongly suggest the preferred orientation of the helices in the homodimeric coiled coil is antiparallel, with interactions at the interface quite different to that of the idealized model. In addition, molecular dynamics simulations suggest equilibrium between dimers, trimers, and tetramers of α-helices for peptide B.
INTRODUCTION
The prediction of protein structure from sequence is one of the grand challenges for the biomolecular sciences. One step toward this objective is to understand the formation of typical domains found in larger proteins. One of the simplest and most widespread motifs in nature is the so-called α-helical coiled coil (1), which consists of two or more α-helices wound around each other, forming a superhelix. Coiled coils commonly contain a heptad of residues labeled a–g repeated at least three or four times. The a and d positions are usually occupied by hydrophobic residues, which form the core of the structure (2,3). Because coiled coils frequently display common features, several computational programs (4–10) using different algorithms have been developed specifically to recognize this type of packing. By means of these tools, coiled coils involved in the stability of a large number of tertiary structures and oligomerization domains (2,11) as well as related structures such as α-sheets and α-cylinders (12) have been identified. It has been estimated (10) that between 5% and 10% of the sequences from various genome projects encode coiled coil regions. In particular coiled coils play an important role in the replication of DNA (13) and membrane fusion (14,15).
Synthetic short peptides or small domains from natural proteins that independently form helical bundles are also of interest as model systems to understand quaternary structure formation in proteins. With this aim several authors have studied the folding pathway of coiled coils. Most dimers of α-helices characterized so far suggest that folding and dimerization are coupled and that folding may best be described by a two-state model (3,16–18). However, the study of thermal unfolding by circular dichroism (CD) spectroscopy and differential scanning calorimetry (DSC) suggests that in certain cases small mutations may change the process from a two-state transition to one which is more complex and less cooperative (16,19). Also, recently it has been shown that the thermal unfolding of the coiled coil domain of yeast GCN4, a leucine zipper, exhibits at least three transitions. The first two involve transitions within the dimer itself, whereas the last one is associated with the process of dissociation (20).
The coiled coil motif is simple yet versatile. For example, dimers and trimers of α-helices have been designed to act as specific receptors for the molecular recognition of ligands (21) and also to deliver radionuclides to the surface of cancer cells (22). Coiled coils have been extensively investigated using a wide variety of approaches including NMR, x-ray diffraction, fluorescence and CD spectroscopy, protein engineering, isothermal titration, and DSC, as well as theoretical approaches including Monte Carlo and molecular dynamics (MD) simulation techniques (16,20–32). However, despite such intensive investigation, the precise nature of the folding pathway, the factors that determine the relative orientation of the helices, the oligomerization state, and the overall stability of coiled coils are unclear. A well-known example is the case of the Coil-Ser (33), an antiparallel trimer of peptides initially designed to fold into a parallel dimer. The antiparallel conformation was unexpected due to the repulsive electrostatic forces between the charged residues. A large number of mutations have been performed to help to understand the stability of this structure (34), and several hypotheses have been proposed (35,36). The parallel trimer Coil-VaLd, a mutant of Coil-Ser, suggests that the packing of the hydrophobic residues plays an important role. Nevertheless other contributions cannot be disregarded. One difficulty in studying coiled coils is that the most powerful tool to characterize the structure of proteins in solution, NMR, is often unable to distinguish relative orientation of peptides forming coiled coils because of the very similar primary structure. Computational approaches, such as MD simulation techniques, however, in principle offer a means to understand interactions in coiled coil systems in atomic detail.
To date most of the MD simulations of coiled coils reported in the literature have been performed in vacuum (37–39), with specific constraints in the simulations (40) or using simplistic models (41). There have also been some studies of α-helical bundles in lipid bilayers (42,43), and Gorfe et al. (32) carried out MD simulations in coiled coil systems in explicit water but focused on pKa calculations rather than the properties of the coiled coils themselves. To our knowledge, no systematic MD study of the stability of coiled coils using explicit water and without structural restraints has yet been published.
In this work, we investigate the extent to which MD simulations can be used to study spontaneous formation and stability of coiled coils. The work is based on a series of model sequences developed in the group of Prof. Koksch (Berlin). A self-complementary sequence, peptide B, based on simple empirical rules was designed which shows a clear coiled coil pattern in the CD spectra. In addition a second sequence, peptide E, was also developed. Peptide E was designed to be nonself-complementary at pH 7.4 and showed no evidence of coiled coil formation at this pH but was intended to be complementary to peptide B, i.e., peptide E should form a coiled coil when mixed with peptide B. The peptides were designed to form either an ideal parallel homodimeric (B + B) or antiparallel heterodimeric (B + E) structure. Our aim in this study was to use MD simulations to investigate if it is possible to simulate the formation of coiled coils, to characterize the arrangement of residues in the interface for comparison to the original design principles, and to investigate if MD simulations can discriminate between different stoichiometries/orientations of coiled coils.
METHODS
Peptide synthesis
All peptides were synthesized by standard Fmoc chemistry on Fmoc-Leu-Wang resin (0.68 mmol/g) using a 431 A peptide synthesizer (Applied Biosystems, Foster City, CA). Purification was carried out by preparative reversed phase high-performance liquid chromatography (HPLC) on a Vydac C4 column (Grace Vydac, Hesperia, CA. The molecular weight of the products was determined by MALDI-TOF mass spectrometry using a Voyager MALDI-TOF Mass spectrometer (PerSeptive Biosystems, Framingham, MA), and their purity was determined by analytical HPLC.
CD spectroscopy
CD measurements of peptides in buffered solution (10 mM Tris, 10 mM NaCl, pH 7.4) were carried out on a J-715 spectrometer (Jasco Inc., Easton, MD) using a quartz cell of 1 mm path length. Spectra were recorded at 298 K from 190–240 nm at 0.5 nm resolution with a scan rate of 20 nm/min. Three scans were acquired and averaged for each sample. Raw data were noise reducted by smoothing and subtraction of buffer spectra. CD values were expressed as molar ellipticity.
Molecular dynamics
The MD simulations were performed using the GROMACS package version 3.0 (44–46) with the GROMOS96 (43a2) force field (47,48) and the simple point charge (SPC) water model (49). This force field treats aliphatic hydrogen atoms as united atoms, together with the carbon atom to which they are attached. Periodic boundary conditions with a rhombic dodecahedron box as the basic unit cell in the NPT ensemble were used. The pressure was maintained by weak coupling to a reference pressure of 1 bar, with a coupling time of 0.5 ps and an isothermal compressibility of 4.6 10−5 bar−1 (50). The number of water molecules used in each system varied between ∼7000 and ∼17,000 depending on the size of the simulation box. Water and peptides were coupled separately to a temperature bath (50) at 298 K using a coupling constant of 0.1 ps. Nonbonded interactions were evaluated using a twin range cutoff of 0.9 and 1.4 nm, interactions within the shorter and longer cutoffs being updated every step and every five steps, respectively. Beyond the 1.4 nm cutoff, a reaction field correction with a dielectric constant ɛ of 78.0 was used. The time step was 2 fs. The bond lengths and angle in water were constrained using the SETTLE algorithm (51), and the LINCS algorithm (52) was used to constrain bond lengths within the peptide. The equations of motion were integrated using the leapfrog method.
Spontaneous dimerization
The amino acid sequence of peptide B is given in Fig. 1 a. In Fig. 1, b and c, models of antiparallel and parallel coiled coils consisting of two B peptides are presented. To attempt to study the spontaneous formation of parallel and antiparallel coiled coils, different initial configurations, in which the relative positions of two B peptides were varied, were constructed using tools from the GROMACS package and the program visual molecular dynamics (VMD) (53). The arrangement of the peptides used as initial conformations was based on a hypothetical ideal parallel coiled coil in which the leucines of both peptides are oriented to the core of the structure. Starting from this reference structure, the distance, the relative orientation of the leucines, and the angle between the axis of the helices as well as combinations of these three variables were varied (see Fig. 2), generating 12 different initial structures. In all cases, the secondary structure of the individual peptide chains was an ideal α-helix, generated using the program WHATIF (54). In these initial trials, the starting conformation for the two peptides was chosen to be an ideal α-helix so as to facilitate the formation of a coiled coil. The protonation state of the residues was chosen appropriate for approximately pH 7. The N-termini and lysines were protonated, whereas the C-termini and glutamates were deprotonated. Each of these structures was placed in a rhombic dodecahedron box in which the distance between the geometrical center of the adjacent unit cells was between 6.5 and 7 nm. The boxes were solvated with preequilibrated SPC water molecules. No counterions were added. The total charge in each simulation box was zero since the basis sequence has no net charge. The initial velocities of the atoms were taken from a Maxwell distribution at 298 K. Different random number seeds were used for each simulation. Before carrying out the MD simulations, a steepest descent minimization was performed. Three independent simulations were carried out for certain of the initial configurations, giving 24 simulations in total. In Table 1 the distances and orientations of the helices together with the number of water molecules used for each simulation are listed. Configurations were stored every 20 ps during 10 ns for analysis. For each configuration, the distance to the nearest periodic image of each peptide was calculated and found to be in most cases >2 nm and never <1.6 nm, i.e., always larger than the cutoff used for the long-range interactions.
FIGURE 1.
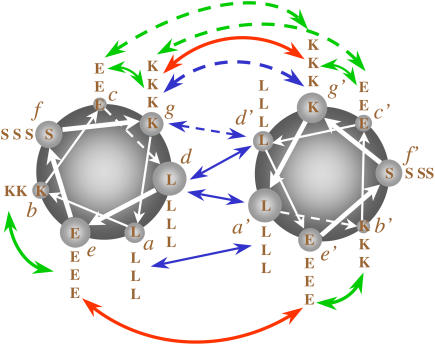
Amino acid sequences of the B and E peptides (a). Idealized models of homodimeric antiparallel (b) and parallel (c) coiled coils consisting of two copies of peptide B. Idealized models of heterodimeric antiparallel (d) and parallel (e) coiled coils consisting of one peptide B and one peptide E. An idealized model of a homotrimeric coiled coil consisting of three copies of peptide B (f). Expected attractive and repulsive electrostatic interactions are represented by green and red arrows, respectively. Blue arrows denote the hydrophobic interactions at the interface of the peptides. The italic letters indicate the position of each amino acid in the heptad. The small capital letters represent the name of the residue. The name of the peptides is in capital letters in the center of each article.
FIGURE 2.
Representation of the parameters used to define the initial conformation of the peptides in the dimer. (d) The distance between the centers of both helices. (α) The relative orientation of the helices; 0° corresponds to a parallel orientation. (β) The relative orientation of the leucine residues; 0° means that the leucines of both helices are oriented toward the interface of the dimer.
TABLE 1.
List of MD simulations performed for dimers
| Name of trajectory*
|
Starting structure†
|
No. water molecules
|
|||
|---|---|---|---|---|---|
| d (nm) | α (deg) | β (deg) | Coiled coil‡ | ||
| aB1–aB3 | 1.43 | 180 | 0 | 6145 | √ (3) |
| aB4–aB6 | 1.69 | 180 | 180 | 7063 | |
| aB7 | 2.02 | 180 | 0 | 7752 | |
| aB8 | 1.98 | 180 | 180 | 7735 | |
| pB1–pB3 | 1.42 | 0 | 0 | 6137 | √ (1) |
| pB4–pB6 | 1.68 | 0 | 180 | 6738 | |
| pB7 | 2.02 | 0 | 0 | 7745 | |
| pB8 | 1.98 | 0 | 180 | 7735 | |
| xB1–xB3 | 1.42 | 90 | 0 | 6138 | |
| xB4– xB6 | 1.68 | 90 | 180 | 6147 | |
| xB7 | 2.02 | 90 | 0 | 7754 | |
| xB8 | 1.98 | 90 | 180 | 7744 | |
| aE1 | 1.98 | 180 | 0 | 16751 | |
| aE2 | 1.11 | 180 | 0 | 16765 | |
| aE3 | 1.22 | 180 | 0 | 16748 | |
| pE1 | 1.95 | 0 | 0 | 16749 | |
| pE2 | 1.06 | 0 | 0 | 16767 | |
| pE3 | 1.43 | 0 | 0 | 16746 | |
| aBE1 | 1.97 | 180 | 0 | 16754 | |
| aBE2 | 1.07 | 180 | 0 | 11739 | √ |
| aBE3 | 0.81 | 180 | 0 | 7776 | √ |
| pBE1 | 1.98 | 0 | 0 | 16754 | |
| pBE2 | 1.06 | 0 | 0 | 11744 | |
| pBE3 | 0.80 | 0 | 0 | 7761 | |
Lines between rows separate groups of simulations of the same peptides starting from the same relative orientation of the helices.
a, p, and x denote antiparallel, parallel, and 90°, respectively, between the axis of the helices in the initial conformation. B and E indicate homodimers consisting of peptide B or E, and BE indicates a heterodimer consisting of one peptide B and another peptide E. The last number in the name of the trajectories is an index to distinguish different trajectories. The simulation time was 10 ns for all the trajectories except for aB2, aB4, pB1, and xB8, for which it was 110 ns.
α is the relative orientation of the helices, 0° corresponds to a parallel orientation, and β is the relative orientation of the leucine residues; 0° means that the leucines of both helices are oriented toward the interface of the dimer. d is the distance between the centers of both helices. The meaning of α, β, and d are also illustrated in Fig. 2.
Marks only appear when coiled coils were observed after the corresponding simulation time, and the number on their right is the number of simulations providing coiled coils given one starting structure.
Stability of dimers
The simulations of the B peptides that successfully formed coiled coils were used to investigate the interactions stabilizing the dimers. In addition, to study the effect of changes in the electrostatic and/or hydrophobic interactions on the stability of a coiled coil, a second peptide (peptide E) was also investigated. The amino acid sequence of peptide E is also listed in Fig. 1 a. Three simulations of dimers formed by combining E + E and E + B starting from an antiparallel and a parallel orientation were performed (12 simulations in total). Fig. 1, d and e, shows models of the antiparallel and parallel heterodimeric coiled coils. For the simulations of E + E, a distance of 9 nm between centers of adjacent unit cells was used because peptide E was not expected to be stable as an α-helix due to repulsion between charged groups. For the E + B dimers, boxes with three different sizes (7, 8, and 9 nm) were used. The distances, the orientations of the helices, and the number of water molecules used to solvate the peptides are listed in Table 1. The initial structure of peptide E in all simulations was an ideal helix generated with the program WHATIF.
The protonation state of the residues and N- and C-termini was the same as for peptide B. The net charge of peptide E was −4 e. The question of whether counterions should be added to balance the overall charge in simulations is a matter of continuing debate. In this regard it should be noted that due to its high dielectric constant, explicit water very effectively screens charge-charge interactions at distance. Furthermore, due to the small box size, adding ions to balance the charge in this system would lead to very high effective salt concentration, which could affect the stability of the coiled coils. Thus, so that a direct comparison could be made with the simulations of peptide B, again no counterions were added. This also avoided difficulty associated with the very slow equilibration of the ion distribution.
Oligomerization state
The stability of trimers and tetramers consisting of monomers of peptide B was studied in a similar manner. A total of six simulations, three with antiparallel and three with parallel conformations, were performed to study the trimers and eight simulations, four antiparallel and four parallel, to investigate the stability of different tetramers. In all cases, the initial structure was a model coiled coil consisting of three or four α-helices with the leucine residues oriented to the core of the structure. The antiparallel trimeric models consisted of two parallel helices and one antiparallel helix, whereas the antiparallel tetrameric structure consisted of a bundle of four α-helices, each one antiparallel to the two adjacent helices and parallel to the diagonal helix. In Fig. 1 f a representation of an antiparallel trimeric coiled coil is shown. Note, each of the initial configurations differed slightly in respect to the distance or the orientation of the leucines to reduce systematic bias. The distance between the geometrical centers of adjacent unit cells was 8 or 9 nm and the boxes solvated with water molecules. No template structure was used to generate the initial configurations. The individual peptides were simply placed in close proximity in the proposed orientation with the leucine residues oriented toward the core of the oligomer.
The total length of the simulations presented in this work is 0.9 μs. Analysis of the trajectories was performed with tools from the GROMACS package, using VMD version 1.8 and RASMOL version 2.7 (55).
RESULTS AND DISCUSSION
Spontaneous dimerization
A classic coiled coil structure is formed from two peptides that are predominately α-helical and interact in a parallel or antiparallel orientation by means of a closely packed hydrophobic interface. Longer peptides also show a degree of supercoiling. According to this definition, only 4 of the 24 simulations of peptide B show the formation of a coiled coil dimer from two separated monomers. This is despite the fact that coiled coil formation is believed to be highly cooperative and the initial configurations consisted of two preformed ideal α-helices in close proximity, a configuration specifically chosen to facilitate the formation of coiled coils. From this it is already clear that to simulate the formation of coiled coils from peptides placed randomly in an arbitrary conformation would not be possible on the timescales investigated in this study.
The relative orientation of the peptides forming coiled coils was antiparallel in three trajectories (aB1–aB3) and parallel in the fourth (pB1). The peptides are too short for supercoiling to be observed. In each of these cases the initial separation between the two monomers was small. The initial conformation of the peptides in trajectories aB1–aB3 is shown in Fig. 3 a with the structures after 10 ns from trajectories aB2 and aB3 shown in Fig. 3, b and c. The initial conformation for the trajectory pB1, Fig. 3 d, was similar to aB1–aB3 except the helices were initially arranged parallel. The conformation at 10 ns for this trajectory is shown in Fig. 3 e. Starting from the same initial conformation two other simulations (pB2 and pB3) did not form an antiparallel coiled coil. The 20 simulations which did not form coiled coils resulted in structures consisting of partially folded α-helices, either separated or interacting with each other in an irregular fashion. An example is shown in Fig. 3 f, the conformation at 10 ns of trajectory pB2.
FIGURE 3.
Initial configuration of the peptides in trajectories aB1–aB3 (a). Configurations at 10 ns obtained from trajectories aB2 (b) and aB3 (c). The initial configuration of the peptides in trajectories pB1-pB3 (d). Configurations at 10 ns obtained from trajectories pB1 (e) and pB2 (f). In a and d, the side chains of the leucine residues are highlighted in red. In b, c, e, and f, the leucines at positions a, d, a′, and d′ are highlighted in green, yellow, orange, and red, respectively, to show the packing of the leucines in different antiparallel and parallel coiled coils. Figures generated using VMD.
The variation of the solvent accessible surface (SAS) of the hydrophobic residues for these 24 trajectories was calculated (56). For the trajectories that formed classic coiled coil dimers, a sharp reduction in the SAS was observed during the first nanosecond, which then remained almost constant during the rest of the simulation. A slow, more continuous decrease was observed in the trajectories which did not lead to coiled coils. The root mean square positional deviation (RMSD) for the same 24 simulations was also calculated taking as reference the corresponding initial conformation in each case. The trajectories leading to coiled coils are the most stable. They also remain closer to the initial structure. The results suggest that on this timescale the final structure is highly dependent on the initial conformation. Coiled coils could only be obtained starting from structures with 0° or 180° between the axis of the helices and very short distances between hydrophobic residues. However, a variety of other apparently stable structures was formed after a few nanoseconds in other trajectories. Typically these were characterized by low SAS values which in some cases were comparable to the values of the classic coiled coil structures.
Simulations corresponding to two of the most unfavorable initial conformations, aB4 and xB8, were extended for another 100 ns. These trajectories show that increasing the timescale of the simulations by an order of magnitude does not significantly alter the results. A more or less stable structure rapidly forms which slowly evolves to minimize the hydrophobic SAS (data not shown).
Stability of dimers
The SAS and RMSD of the trajectories aB1–aB3 and pB1 suggest that the dimeric coiled coils obtained are very stable. Calculations of the root mean square positional fluctuation show that just the first and last side chains are mobile. The coiled coil structures as represented by Fig. 3, b, c, and e, are extremely rigid in the simulations, especially in the central regions, whereas alternate structures such as the one shown in Fig. 3 f are more mobile. Trajectories aB2 and pB1 were extended by 100 ns to test whether the antiparallel and parallel coiled coils maintained their respective conformations if the timescale was increased by an order of magnitude. No significant differences between the 10 ns trajectories and the 100 ns trajectories were evident.
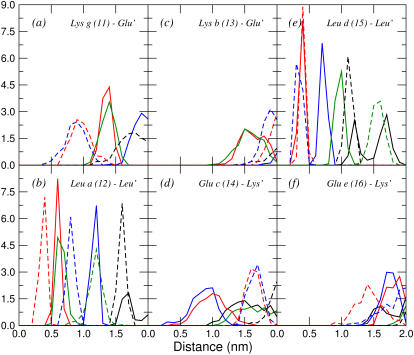
In the idealized schematics of coiled coils presented in Fig. 1, the red and green arrows represent electrostatic interactions between charges of the same and different signs, respectively, and the blue arrows represent hydrophobic interactions. As it is shown in Fig. 1 c, peptide B was designed to optimize the intermolecular interactions in a hypothetical parallel coiled coil. However in three of the four simulations in which a stable coiled coil was obtained, the orientation of the helices was antiparallel. To analyze how the interactions stabilizing the structures obtained from trajectories aB1–aB3 and pB1 compare with the idealized model, the minimal distances between the charged groups of glutamate and lysine, COO− and  respectively, and between leucine-leucine residues of different peptides were calculated. In Figs. 4 and 5, the distributions of some of these distances obtained from the last 5 ns of the trajectories aB2 and pB1 (antiparallel and parallel coiled coils, respectively) are shown. Although only the results from the analysis of aB2 are discussed, the nature of the interactions within the interface was similar in all antiparallel cases. To generate these plots, the six central residues of one of the peptides in each of these trajectories, from lysine 11 at position g to glutamate 16 at position e, were taken as a reference. From Figs. 4 and 5, it is clear that the closest contacts between the hydrophobic residues show relatively narrow distributions compared with the charged residues. Note, the distances in Figs. 4 and 5 correspond to the centers of the interaction sites, which for leucines are the aliphatic carbon atoms given that the nonpolar hydrogen atoms are not treated explicitly.
respectively, and between leucine-leucine residues of different peptides were calculated. In Figs. 4 and 5, the distributions of some of these distances obtained from the last 5 ns of the trajectories aB2 and pB1 (antiparallel and parallel coiled coils, respectively) are shown. Although only the results from the analysis of aB2 are discussed, the nature of the interactions within the interface was similar in all antiparallel cases. To generate these plots, the six central residues of one of the peptides in each of these trajectories, from lysine 11 at position g to glutamate 16 at position e, were taken as a reference. From Figs. 4 and 5, it is clear that the closest contacts between the hydrophobic residues show relatively narrow distributions compared with the charged residues. Note, the distances in Figs. 4 and 5 correspond to the centers of the interaction sites, which for leucines are the aliphatic carbon atoms given that the nonpolar hydrogen atoms are not treated explicitly.
FIGURE 4.
Distributions of minimum distances from the six central residues of the first helix (11–16) to the residues of the second helix with which they are expected to interact, calculated from the last 5 ns of trajectory aB2. The label in each plot specifies the name and position of the residue taken as the reference in the first helix and the residues of the second helix (primed) to which the minimum distances are calculated. Black, blue, red, and green curves represent residues from the first, second, third, and fourth turns, respectively, of the second peptide. Solid lines represent glutamates at position e′, leucines at position d′, or lysines at position g′, and dashed lines represent glutamates at position c′, leucines at position a′, or lysines at position b′, depending on the case. For glutamate and lysine residues, only the charged groups (COO− and 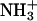 ) were considered. Minimum distances between atoms in the aliphatic chains should be reduced by ∼0.1 nm due to the treatment of the nonpolar hydrogens as united atoms.
) were considered. Minimum distances between atoms in the aliphatic chains should be reduced by ∼0.1 nm due to the treatment of the nonpolar hydrogens as united atoms.
FIGURE 5.
The same as Fig. 4 but for trajectory pB1.
In the antiparallel coiled coil, the leucine at position a is juxtaposed with one leucine at position d′ (red dashed line in Fig. 4 b), the distance over the 5 ns being 0.4 ± 0.1 nm (where the error indicates the width of the distribution at one-half height). The distance to the two closest leucines at position a′ (red and green solid lines in Fig. 4 b) is larger, 0.6 ± 0.2 nm, but the distributions are still narrow. The leucine at position d behaves slightly differently since there are three leucines in the second helix, two at position a′ and one at position d′ (red and blue dashed lines and red solid line in Fig. 4 e) at similar distances (0.3 ± 0.2 nm in one case and 0.4 ± 0.1 nm in the other two cases). In contrast, in the parallel coiled coil more regular interactions with the leucines of the second peptide are observed. The distributions of the distances to the three closest leucines at positions a′ and d′ (Fig. 5, b and e) overlap almost exactly, the peak being at 0.4 ± 0.2 for all of them.
In regard to the charged groups, in the antiparallel coiled coil the distance between the lysine at position g and the two closest glutamates of the second peptide at position c′ (blue and red dashed lines in Fig. 4 a) is 0.9 ± 0.4 nm, within the cutoff used in the simulations for the electrostatic interactions. Note, any given lysine interacts with glutamates in two repeats and any given glutamate interacts with lysines in two repeats. This suggests attractive interactions which could contribute to the stability of the dimer. Since both helices have the same sequence of amino acids, the distributions of the distances between residues placed at symmetrical positions, glutamate at c and lysines at g′ (red and blue solid lines in Fig. 4 d), are very similar. The distance between the same lysine at position g and the glutamates at position e′ (red and green solid lines in Fig. 4 a) is also within the cutoff during a significant part of the trajectory (1.4 ± 0.3 nm), indicating a weak interaction. The symmetrical interaction between the glutamate at position e and lysines at position g′ appears to be even weaker, the distance being 1.7 ± 0.3 nm. These residues are almost always separated by more than the cutoff. The distances between all the remaining charged groups for the antiparallel coiled coil are beyond the cutoff over the 5 ns. This means that both lysine at position b (Fig. 4 c) and glutamate at position e (Fig. 4 f) do not have direct attractive electrostatic interactions with any residue of the second helix. The lack of significant attractive interactions involving these two residues in the antiparallel coiled coil was already expected from the model presented in Fig. 1 b. In the parallel coiled coil the distance between the lysine at position g and the glutamates at position e′ (blue and red solid lines in Fig. 5 a) is within the cutoff over almost the 5 ns (0.9 ± 0.4 nm). The distributions of the distances between glutamate at position e and lysines at position g′ (blue and red solid lines in Fig. 5 f) are very similar, showing the symmetrical interaction since both helices have the same sequence of amino acids. As was also expected from the model shown in Fig. 1 c, the residues at positions c and b do not interact significantly with the second helix.
By comparing the distributions of the leucine-leucine distances, it is evident that the arrangement of the hydrophobic residues at the interface between the peptides depends on the orientation of the helices in the dimer. The packing of the leucines in the antiparallel and parallel coiled coils can be appreciated in detail in Fig. 3, b and e, where the side chains of the leucine residues at positions a and d of both peptides are represented with different colors. In the parallel coiled coil all of the leucines are oriented to the core of the dimer, forming a hydrophobic interface with a type of knobs into holes packing. However, in the antiparallel coiled coil formed in the simulations, the leucines at positions a and d′ are oriented perpendicularly to the plane containing the axis of both helices. Only the leucines at positions d and a′ are clearly interacting with each other. It is important to note, however, that the interfaces in both the antiparallel and parallel coiled coils are tightly packed. As discussed in detail later a range of residues, in particular the nonpolar regions of the lysine side chains, form extensive hydrophobic interactions in the antiparallel case.
According to the idealized model presented in Fig. 1 b, interactions between the lysine at position g and the glutamate at position c′ in the antiparallel coiled coil as well as interactions between the glutamate at position c and the lysine at position g′ were not expected. Also no difference in the packing of the hydrophobic core between the parallel and antiparallel orientations was expected. To better understand the role of the lysine residues in the stability of the antiparallel dimer, the minimum distances between the lysine and leucine residues and between the lysine and lysine residues of different peptides along the last 5 ns of the trajectories aB1–aB3 were calculated. In these calculations all of the atoms that comprise the lysine residues were taken into account. In Fig. 6 the distributions of these distances for the lysines 11 and 18 at position g for the trajectory aB2 are plotted. Very narrow distributions for the distances to the leucines were observed (0.4 ± 0.1 nm), suggesting hydrophobic interactions between the side chain of lysines and the hydrophobic core of the dimer (Fig. 6, a and b). The distances from the lysines at position g to the lysines at position g′ have very similar distributions, indicating that the side chains of these residues also participate in the packing at the interface of the dimer (Fig. 6, c and d). Fig. 7 shows the arrangement of the lysine residues at positions g and g′ of both peptides together with the leucines at position d′ of the second peptide. The relative orientation of the lysines in the different helices highlighted in Fig. 7 also suggests hydrophobic interactions between these side chains. Stable lysine-lysine interfaces have been found previously in other designed coiled coils (57) and may also play a role in the unexpected stability of the antiparallel trimer Coil-Ser (33).
FIGURE 6.
Distributions of minimum distances from the lysine 11 and lysine 18, both at position g, of the first helix to the leucines and lysines of the second helix, calculated from the last 5 ns of trajectory aB2. The label in each plot specifies the name and position of the lysine taken as the reference in the first helix and the residues of the second helix (primed) to which the minimum distances are calculated. Black, blue, red, and green curves represent residues from the first, second, third, and fourth turns, respectively, of the second peptide. Solid lines represent leucines at position d′ or lysines at position g′, and dashed lines represent leucines at position a′, or lysines at position b′, depending on the case. Minimum distances between the atoms in the aliphatic chains should be reduced by ∼0.1 nm due to the treatment of the nonpolar hydrogens as united atoms.
FIGURE 7.
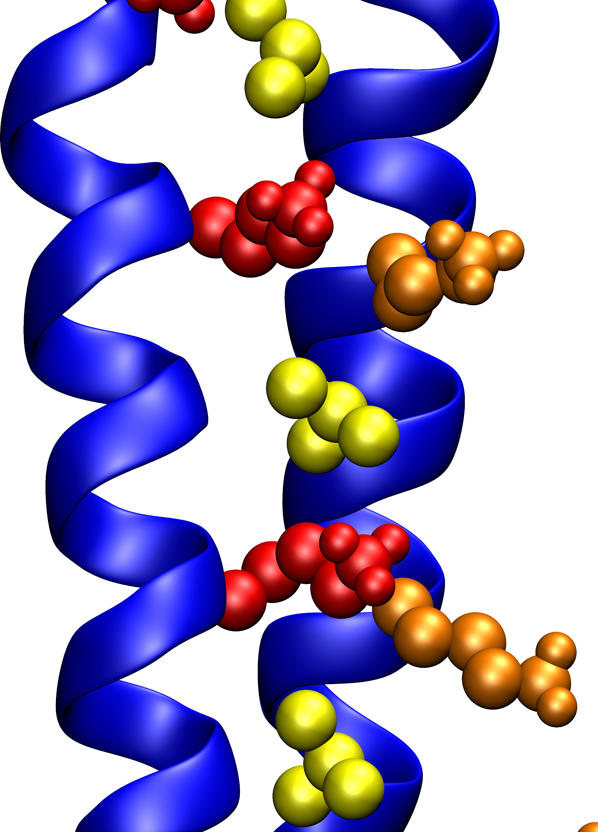
View of the lysine interface in an antiparallel coiled coil (trajectory aB2). Lysines at position g and g′ are highlighted in red and orange, respectively. The side chains of the leucines at position d′ are shown in yellow. The rest of the leucines are not shown for clarity. Figure generated using VMD.
The intramolecular distances between the charged groups of the glutamate and lysine residue within each peptide were also calculated (data not shown). The distributions corresponding to the individual glutamate-lysine pairs have a maximum at ∼0.4 nm. This suggests that the intramolecular electrostatic interactions are more significant than intermolecular ones. The proximity between the glutamate and lysine residues within one peptide might also screen unfavorable charge-charge interactions between the lysines residues which lie in close proximity in the antiparallel orientation.
In light of the findings above, a modified model for the interaction of the residues in the antiparallel B + B coiled coil can be proposed. The electrostatic and the hydrophobic interactions suggested by the MD simulations are illustrated in Fig. 8. These differ significantly from those proposed in Fig. 1 b and on which the design was based. Specifically, we find that in addition to the leucine residues at the interface, lysine-lysine interactions play a central role in stabilizing the structure. Weak hydrophobic interactions between the side chains of glutamate and leucine and/or hydrogen bonds between the serines and glutamates may also contribute to stability but are only of secondary importance.
FIGURE 8.
Proposed model for the interactions in the homodimeric antiparallel coiled coil of peptide B. Attractive and repulsive electrostatic interactions are represented by green and red arrows, respectively. Hydrophobic interactions are represented by blue arrows. Dashed arrows represent the new interactions added to the original model of Fig. 1 b. The italic letters indicate the position of each amino acid in the heptad, and the small capital letters represent the name of the residue.
Another factor which could favor an antiparallel orientation of any coiled coil is the dipole moment of the helices which arises from the peptide backbone (58,59). For example, this has been proposed to explain the stability of the antiparallel trimer Coil-Ser (33). However, the stability of the parallel trimer Coil-VaLd (34,35), a mutant of Coil-Ser in which the leucines at position a have been changed by valine, suggests that the packing of the hydrophobic residues at the interface of the dimer can be sufficient to determine the orientation of the helices. It should also be noted that although the helix dipole favors an antiparallel orientation in vacuum or in a nonpolarizable implicit solvent, in a highly polarizable medium such as water, the helix dipole will be in part compensated by an induced dipole in the environment. In addition, in simulations performed under periodic boundary conditions it is possible that this effect could be enhanced and parallel orientations stabilized artificially due to the interaction between neighboring periodic images.
Peptide E was designed such that it could not form complementary electrostatic interactions with itself in either a parallel or antiparallel orientation but would form complementary electrostatic interactions with peptide B in an antiparallel orientation (see Fig. 1 d). In the simulations performed with homodimers of peptide E, it was not possible to obtain a stable coiled coil. In all the trajectories, regardless of the initial orientation of the helices both monomers start to unfold within picoseconds. These simulations strongly suggest that in this case the effect of the intramolecular electrostatic forces dominate. It should be noted in this regard that although the peptide E carries a net charge of −4 e no counterions were included in the simulation. The reasons for this are listed in the Methods section. Certainly, counterions might moderate the electrostatic interactions. However, at the concentrations at which these peptides are studied experimentally (10 mM NaCl), the volume of the simulation box would contain on average only between 1 and 2 Na+ ions. In addition, these ions would not necessarily be directly associated with either of the peptides.
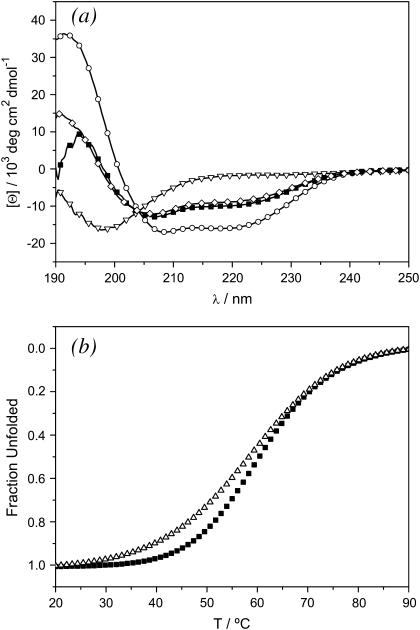
Experimental investigations by CD spectroscopy clearly demonstrate that at pH 7.4 peptide E does not form a coiled coil (triangles, Fig. 9 a). Although peptide E does not form a coiled coil in a 250 μM solution at pH 7.4, the CD curve of peptide B shows two minima at 208 nm and 222 nm, respectively, which are characteristic of an α-helical coiled coil fold (circles, Fig. 9 a). The mixture of peptide E and peptide B in a ratio of 1:1 suggests some formation of a heterodimeric α-helical coiled coil (solid squares, Fig. 9 a). The CD spectrum of the mixture is not simply the sum of the spectra of the B and E peptides in isolation. The diamonds in Fig. 9 a correspond to the sum of the experimental curves for the isolated B and E peptides corrected for concentration. Although the curves are similar, they deviate sharply at low wavelengths. The measured and the calculated spectra also differ in the ratio of the local minima at 208 nm and 222 nm ([θ]222/[θ]208 = 0.72 for addition of single spectra, [θ]222/[θ]208 = 0.76 for the measured spectra of the B + E mixture), indicating an increase in helical content. In addition, thermal unfolding profiles of the B peptide and the BE mixture were recorded (Fig. 9 b). Different curves and different melting temperatures of TM = 58.3°C (BE mixture) and TM = 60.3°C (B peptide) show that the BB homodimer is not the sole α-helical structure in the BE mixture. Rather peptide B as either a monomer or higher aggregate is able to act as a template for peptide E and possibly induce a transformation from random coil to a helical structure.
FIGURE 9.
(a) CD spectra measured in 10 mM Tris-HCl buffer, pH 7.4; 250 μM peptide B (circles), 250 μM peptide E (triangles); a mixture of 125 μM peptide B and 125 μM peptide E (squares). Diamonds correspond to the sum of the curves for peptide B (circles) and peptide E (triangles) curves corrected to a concentration of 125 μM for comparison to the measured curve of the mixture. All spectra were recorded at 298 K. (b) Thermal unfolding profiles of 125 μM peptide B (solid squares; melting temperature 60.3°C) and a mixture of 125 μM peptide B and 125 μM peptide E (triangles; melting temperature 58.3°C). Melting curves were recorded by observing the CD signal at 222 nm at pH 7.4 in 2 M guanidinium chloride. The thermal melting profiles were fitted by using a 5 parameter sigmoidal curve.
To test this and the basic design hypothesis, six simulations of heterodimeric B + E coiled coils were performed. The starting configurations varied slightly between the simulations as can be appreciated from Table 1 and Fig. 10. Two of the antiparallel heterodimeric B + E coiled coils maintained their initial structure during 10 ns, demonstrating that although a helical peptide E is not stable in isolation the heterodimeric B + E coiled coil is stable in the simulations and might account for the experimental results. In Fig. 10 the initial and final structures of the trajectories corresponding to an antiparallel orientation of the helices are shown. In all cases where the starting orientation was parallel, no coiled coils were obtained. After 10 ns the structure of peptide E was effectively random. These results suggest that antiparallel B + E heterodimers are more stable than parallel dimers. It should be noted, however, that in general a wide variety of structures could give rise to the CD signal observed experimentally. These include structures such as that shown in Fig. 3 f, which have significant α-helical content as well as the possibility of trimers or higher order oligomers.
FIGURE 10.
Initial and last structures (after 10 ns) of MD for the three simulations of antiparallel heterodimers consisting of one peptide B (black) and one peptide E (gray); trajectories aBE1 (a), aBE2 (b), and aBE3 (c). Figures generated using VMD.
Oligomerization state
Trimers
To investigate the possible stability of trimers, conformations consisting of bundles of three B peptides in a parallel and an antiparallel orientation were constructed and simulated for 10 ns. The trajectories starting from antiparallel and parallel arrangements were labeled 3aB1–3aB3 and 3pB1–3pB3, respectively. Three simulations were performed starting from a parallel orientation. In all cases the trimer disassociated, leaving a coiled coil consisting of no more than two helices. In contrast, in two of the three antiparallel cases the trimeric coiled coil remained stable. Although it was not possible to obtain a stable parallel trimer, α-helical parallel dimers similar to that seen in pB1 (Fig. 3 e) were observed in 3pB1 and 3pB3. Nevertheless, the results still strongly suggest a preference for an antiparallel arrangement. Fig. 11, a and b, shows two views of the trimer formed in trajectory 3aB3. The side chains of the leucines of each helix are shown in different colors to highlight the packing of the hydrophobic core.
FIGURE 11.
Two views of conformation of the trimer of α-helices at the end of trajectory 3aB3 with the side chain of the leucines highlighted with a different color for each peptide (a and b). Configuration at 10 ns from trajectories 4aB1 (c), 4aB4 (d), and 4pB4 (e). In the antiparallel structures, the helices with different orientation are in dark blue. Figures generated using VMD.
Tetramers
Simulations were also performed with bundles of four helices. The trajectories starting from antiparallel and parallel arrangements were labeled 4aB1–4aB4 and 4pB1–4pB4, respectively. In Fig. 11, c–e, the last structures corresponding to trajectories 4aB1, 4aB4, and 4pB4 are shown. For each helix the side chains of the leucines are highlighted in different colors. Only in trajectory 4aB4 did an antiparallel tetrameric coiled coil form and remain stable. No parallel tetrameric coiled coils were observed. Some dimeric coiled coils were obtained starting from the parallel tetramer, as for instance the ones shown in Fig. 11 e, and one antiparallel trimer interacting loosely with the final monomer was obtained from 4aB1. The antiparallel tetramer obtained from trajectory 4aB4 was very stable during the 10 ns the RMSD remaining almost constant over the whole trajectory (data not shown). This suggests that coiled coils consisting of four α-helices in an antiparallel orientation may exist in equilibrium with other structures, including antiparallel trimers and parallel or antiparallel dimers. The interaction between the dipole moments of the helices appears to prevent the formation of parallel coiled coils consisting of more than two helices for these peptides. As noted earlier, there was a danger that interactions between the total dipole of the systems due to the imposition of periodic boundary conditions could have artificially favored parallel orientations. Although this clearly proved not to be the case, as this effect was potentially larger in the case of the tetrameric systems, larger box dimensions were used in this case as a precautionary measure.
CONCLUSIONS
This study had three aims: i), to simulate the spontaneous formation of coiled coils, ii), to characterize the arrangement of the residues in stable dimeric parallel and antiparallel coiled coils, and iii), to determine if it is possible to discriminate between different stoichiometries/orientations.
The classic coiled coil configuration could only be obtained using 10 ns simulations if the initial structure was very close to the expected final configuration. Alternative starting configurations led to a wide variety of alternative dimer metastable conformations. These nevertheless contained a high degree of α-helix. The work suggests either that the coiled coils form by a two-state process forming an initial complex which only slowly rearranges to form the classic coiled coil or that a high helical content as inferred from CD spectra does not necessarily imply the formation of a classic coiled coil structure.
Although peptide B had been designed to form a parallel coiled coil in the simulations, an antiparallel configuration was clearly favored. This was associated with the formation of an asymmetric lysine interface. Factors which may stabilize the antiparallel dimer include the favorable interactions between the helix dipoles and the effect of the packing of the lysines and leucines at the hydrophobic interface.
It did prove possible to distinguish between different stoichiometries and/or orientations. In agreement with experiment, it was found that B + B or B + E did form stable coiled coils, whereas E + E did not. In the case of peptide B, the simulations suggest that parallel and antiparallel dimers as well as antiparallel trimers and tetramers are potentially stable and may even be in equilibrium in solution. Parallel trimers and tetramers were not stable.
In summary, despite the apparent strong tendency of the B peptide to form a coiled coil in solution it was not possible to simulate the spontaneous formation of a coiled coil on a nanosecond timescale unless the starting structure was very close to the target structure. Nevertheless, it was possible to distinguish between alternative orientations and stoichiometries using relatively short simulation times. This suggests MD simulations could have an important role to play in elucidating the properties of this widespread structural motif.
Acknowledgments
We thank Prof. Stephan Berger from the University of Leipzig for stimulating discussions.
Financial support from the VolkswagenStiftung (project No. I/77 986) and European Community Training and Mobility of Researchers (project No. HRPN-CT-2002-00241) is gratefully acknowledged.
Ángel Piñeiro's present address is Laboratorio de Termofísica, Facultad de Química, Universidad Nacional Autónoma de México, México D. F. 04510.
Alessandra Villa's present address is J. W. Goethe University, Institute for Physical and Theoretical Chemistry, Marie Curie Str. 11, 60439 Frankfurt am Main, Germany.
References
- 1.Gruber, M., and A. N. Lupas. 2003. Historical review: another 50th anniversary—new periodicities in coiled coils. Trends Biochem. Sci. 28:679–685. [DOI] [PubMed] [Google Scholar]
- 2.Lupas, A. 1996. Coiled coils: new structures and new functions. Trends Biochem. Sci. 21:375–382. [PubMed] [Google Scholar]
- 3.Mason, J. M., and K. M. Arndt. 2004. Coiled coil domains: stability, specificity, and biological implications. ChemBioChem. 5:170–176. [DOI] [PubMed] [Google Scholar]
- 4.Lupas, A. 1997. Predicting coiled coils regions in proteins. Curr. Opin. Struct. Biol. 7:388–393. [DOI] [PubMed] [Google Scholar]
- 5.Lupas, A., M. Van Dyke, and J. Stock. 1991. Predicting coiled coils from protein sequences. Science. 252:1162–1164. [DOI] [PubMed] [Google Scholar]
- 6.Berger, B., D. B. Wilson, E. Wolf, T. Tonchev, M. Milla, and P. S. Kim. 1995. Predicting coiled coils by use of pairwise residue correlations. Proc. Natl. Acad. Sci. USA. 92:8259–8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirst, J. D., M. Vieth, J. Skolnich, and C. L. Brooks 3rd. 1996. Predicting leucine zippers structures from sequence. Protein Eng. 9:657–662. [DOI] [PubMed] [Google Scholar]
- 8.Woolfson, D. N., and T. Alber. 1995. Predicting oligomerization states of coiled coils. Protein Sci. 4:1596–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolf, E., P. S. Kim, and B. Berger. 1997. Multicoil: a program for predicting two- and three-stranded coiled coils. Protein Sci. 6:1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walshaw, J., and D. J. Woolfson. 2001. SOCKET: a program for identifying and analysing coiled coil motifs within protein structures. J. Mol. Biol. 307:1427–1450. [DOI] [PubMed] [Google Scholar]
- 11.Walshaw, J., and D. J. Woolfson. 2003. Extended knobs-into-holes packing in classical and complex coiled coil assemblies. J. Struct. Biol. 144:349–361. [DOI] [PubMed] [Google Scholar]
- 12.Walshaw, J., and D. J. Woolfson. 2001. Open-and-shut cases in coiled coil assembly: α-sheets and α-cylinders. Protein Sci. 10:668–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landschulz, W. H., P. F. Johnson, and S. L. McKnight. 1988. The leucine zipper: a hypothetical structure common to a new class of DNA-binding proteins. Science. 240:1759–1764. [DOI] [PubMed] [Google Scholar]
- 14.Harbury, P. A. B. 1998. Springs and zippers: coiled coils in SNARE-mediated membrane fusion. Structure. 6:1487–1491. [DOI] [PubMed] [Google Scholar]
- 15.Weis, W. I., and R. H. Scheller. 1998. SNARE the rod, coil the complex. Nature. 395:328–329. [DOI] [PubMed] [Google Scholar]
- 16.Yu, Y., O. D. Monera, R. S. Hodges, and P. L. Privalov. 1996. Investigation of electrostatic interactions in two-stranded coiled coils through residue shuffling. Biophys. Chem. 59:299–314. [DOI] [PubMed] [Google Scholar]
- 17.Zitzewitz, J. A., O. Bilsel, J. Luo, B. E. Jones, and C. R. Matthews. 1995. Probing the folding mechanism of a leucine zipper peptide by stopped-flow circular dichroism spectroscopy. Biochemistry. 34:12812–12819. [DOI] [PubMed] [Google Scholar]
- 18.Wendt, H., L. Leder, H. Härmä, I. Jelesarov, A. Baici, and H. R. Bosshard. 1997. Very rapid, ionic strength-dependent association and folding of a heterodimeric leucine zipper. Biochemistry. 36:204–213. [DOI] [PubMed] [Google Scholar]
- 19.Zhu, H., S. A. Celinski, J. M. Scholtz, and J. C. Hu. 2001. An engineered leucine zipper a position mutant with an unusual three-state unfolding pathway. Protein Sci. 10:24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dragan, A. I., and P. L. Privalov. 2002. Unfolding of a leucine zipper is not a simple two-state transition. J. Mol. Biol. 321:891–908. [DOI] [PubMed] [Google Scholar]
- 21.Doerr, A. J., M. A. Case, I. Pelczer, and G. L. McLendon. 2004. Design of a functional protein for molecular recognition: specificity of ligand binding in a metal-assembled protein cavity probed by 19F NMR. J. Am. Chem. Soc. 126:4192–4198. [DOI] [PubMed] [Google Scholar]
- 22.Moll, J. R., S. B. Ruvinov, I. Pastan, and C. Vinson. 2001. Designed heterodimerizing leucine zippers with a range of pIs and stabilities up to 10−15 M. Protein Sci. 10:649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Junius, F. K., S. I. O'Donoghue, M. Nilges, A. S. Weiss, and G. F. King. 1996. High resolution NMR solution structure of the leucine zipper domain of the c-Jun homodimer. J. Biol. Chem. 271:13663–13667. [DOI] [PubMed] [Google Scholar]
- 24.Rasmussen, R., D. Benvegnu, E. K. O'Shea, P. S. Kim, and T. Alber. 1991. X-ray scattering indicates that the leucine zipper is a coiled coil. Proc. Natl. Acad. Sci. USA. 88:561–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki, K., H. Hiroaki, D. Kohda, and T. Tanaka. 1998. An isoleucine zipper peptide forms a native-like triple stranded coiled coil in solution. Protein Eng. 11:1051–1055. [DOI] [PubMed] [Google Scholar]
- 26.Wendt, H., C. Berger, A. Baici, R. M. Thomas, and H. R. Bosshard. 1995. Kinetics of folding of leucine zipper domains. Biochemistry. 34:4097–4107. [DOI] [PubMed] [Google Scholar]
- 27.Potekhin, S. A., T. N. Melnik, V. Popov, N. F. Lanina, A. A. Vazina, P. Rigler, A. S. Verdini, G. Corradin, and A. V. Kajava. 2001. De novo design of fibrils made of short α-helical coiled coil peptides. Chem. Biol. 8:1025–1032. [DOI] [PubMed] [Google Scholar]
- 28.Vu, C., J. Robblee, K. M. Werner, and R. Fairman. 2001. Effects of charged amino acids at b and c heptad positions on specificity and stability of four-chain coiled coils. Protein Sci. 10:631–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Howard, K. P., J. D. Lear, and W. F. DeGrado. 2002. Sequence determinants of the energetics of folding of a transmembrane four-helix-bundle protein. Proc. Natl. Acad. Sci. USA. 99:8568–8572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jelesarov, L., and H. R. Bosshard. 1996. Thermodynamic characterization of the coupled folding and association of heterodimeric coiled coils (leucine zippers). J. Mol. Biol. 263:344–358. [DOI] [PubMed] [Google Scholar]
- 31.Mohanty, D., A. Kolinski, and J. Skolnick. 1999. De novo simulations of the folding thermodynamics of the GCN4 leucine zipper. Biophys. J. 77:54–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gorfe, A. A., P. Ferrara, A. Caflisch, D. N. Marti, and H. R. Bosshard. 2002. Calculation of protein ionization equilibria with conformational sampling: pKa of a model leucine zipper, GCN4 and barnase. Proteins. 46:41–60. [DOI] [PubMed] [Google Scholar]
- 33.Lovejoy, B., S. Choe, D. Cascio, D. K. McRorie, W. F. DeGrado, and D. Eisenberg. 1993. Crystal structure of a synthetic triple-stranded α-helical bundle. Science. 259:1288–1293. [DOI] [PubMed] [Google Scholar]
- 34.Betz, S., R. Fairman, K. O'Neill, J. Lear, and W. DeGrado. 1995. Design of two-stranded and three-stranded coiled coil peptides. Philos. Trans. R. Soc. Lond. B Biol. Sci. 348:81–88. [DOI] [PubMed] [Google Scholar]
- 35.Ogihara, N. L., M. S. Weiss, W. F. DeGrado, and D. Eisenberg. 1997. The crystal structure of the designed trimeric coiled coil coil-VaLd: implications for engineering crystals and supramolecular assemblies. Protein Sci. 6:80–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oakley, M. G., and J. J. Hollenbech. 2001. The design of antiparallel coiled coils. Curr. Opin. Struct. Biol. 11:450–457. [DOI] [PubMed] [Google Scholar]
- 37.Sajot, N., and M. Genest. 2000. Structure prediction of the dimeric neu/ErbB-2 transmembrane domain from multi-nanosecond molecular dynamics simulations. Eur. Biophys. J. 28:648–662. [DOI] [PubMed] [Google Scholar]
- 38.Briki, F., J. Doucet, and C. Etchebest. 2002. A procedure for refining a coiled coil protein structure using x-ray fiber diffraction and modeling. Biophys. J. 83:1774–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caballero-Herrera, A., and L. Nilsson. 2003. Molecular dynamics simulations of the E1/E2 transmembrane domain of the Semliki Forest virus. Biophys. J. 85:3646–3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang, P. K., W.-S. Tzou, and M.-J. Hwang. 1999. Restraint-driven formation of α-helical coiled coils in molecular dynamics simulations. Biopolymers. 50:667–677. [DOI] [PubMed] [Google Scholar]
- 41.Magaña, S., A. M. Rubio, and A. Rey. 2002. Influence of the hydrophobic face width on the degree of association of coiled coil proteins. J. Chem. Phys. 117:10321–10328. [Google Scholar]
- 42.Tieleman, D. P., H. J. C. Berendsen, and M. S. P. Sansom. 1999. An alamethicin channel in a lipid bilayer: molecular dynamics simulations. Biophys. J. 76:1757–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Law, R. J., D. P. Tieleman, and M. S. P. Sansom. 2003. Pores formed by the nicotinic receptor M2δ peptide: a molecular dynamics simulation study. Biophys. J. 84:14–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berendsen, H. J. C., D. van der Spoel, and R. van Drunen. 1995. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Comm. 95:43–56. [Google Scholar]
- 45.Lindahl, E., B. Hess, and D. van der Spoel. 2001. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J. Mol. Model. (Online). 7:306–317. [Google Scholar]
- 46.http://www.gromacs.org
- 47.van Gunsteren, W. F., S. R. Billeter, A. A. Eising, P. H. Hünenberger, P. Krüger, A. E. Mark, W. R. P. Scott, and I. G. Tironi. 1996. Biomolecular Simulation: GROMOS96 Manual and User Guide. Hochschulverlag AG an der ETH Zürich, Zurich, Switzerland.
- 48.Schuler, L. D., and W. F. van Gunsteren. 2000. On the choice of dihedral angle potential energy functions for n-alkanes. Mol. Simulat. 25:301–319. [Google Scholar]
- 49.Berendsen, H. J. C., J. P. M. Postma, W. F. van Gunsteren, and J. Hermans. 1981. Interaction models for water in relation to protein hydration. In Intermolecular Forces. B. Pullman, editor. Reidel D. Publishing Company, Dordrecht, The Netherlands. 331–342.
- 50.Berendsen, H. J. C., J. P. M. Postma, W. F. van Gunsteren, A. DiNola, and J. R. Haak. 1984. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81:3684–3690. [Google Scholar]
- 51.Miyamoto, S., and P. A. Kollman. 1992. SETTLE: an analytical version of the SHAKE and RATTLE algorithms for rigid water models. J. Comput. Chem. 13:952–962. [Google Scholar]
- 52.Hess, B., H. Bekker, H. J. C. Berendsen, and J. G. E. M. Fraaije. 1997. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 18:1463–1472. [Google Scholar]
- 53.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33–38. [DOI] [PubMed] [Google Scholar]
- 54.Vriend, G. 1990. WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 8:52–56. [DOI] [PubMed] [Google Scholar]
- 55.Sayle, R. A., and E. J. Milner-White. 1995. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 20:374–376. [DOI] [PubMed] [Google Scholar]
- 56.Eisenhaber, F., P. Lijnzaad, P. Argos, C. Sander, and M. Scharf. 1995. The double cubic lattice method: efficient approaches to numerical integration of surface area and volume and to dot surface contouring of molecular assemblies. J. Comput. Chem. 16:273–284. [Google Scholar]
- 57.Schnarr, N. A., and A. J. Kennan. 2003. Specific control of peptide assembly with combined hydrophilic and hydrophobic interfaces. J. Am. Chem. Soc. 125:667–671. [DOI] [PubMed] [Google Scholar]
- 58.Hol, W. G. J., P. T. van Duijnen, and H. J. C. Berendsen. 1978. The α-helix dipole and the properties of proteins. Nature. 273:443–446. [DOI] [PubMed] [Google Scholar]
- 59.Hol, W. G. J. 1985. The role of the α-helix dipole in protein function and structure. Prog. Biophys. Mol. Biol. 45:149–195. [DOI] [PubMed] [Google Scholar]