Abstract
A polyepitopic CD8+-T-cell response is thought to be critical for control of hepatitis C virus (HCV) infection. Using transgenic mice, we analyzed the immunogenicity and dominance of most known HLA-A2.1 epitopes presented during infection by using vaccines that carry the potential to enter clinical trials: peptides, DNA, and recombinant adenoviruses. The vaccines capacity to induce specific cytotoxic T lymphocytes and interferon gamma-producing cells revealed that immunogenic epitopes are clustered in specific antigens. For two key antigens, flanking regions were shown to greatly enhance the scope of epitope recognition, whereas a DNA-adenovirus prime-boost vaccination strategy augmented epitope immunogenicity, even that of subdominant ones. The present study reveals a clustered organization of HCV immunogenic HLA.A2.1 epitopes and strategies to modulate their dominance.
The efficacy of the state-of-the art therapy to treat hepatitis C virus (HCV)-infected individuals is greatly dependent on the genotype of the infecting virus. Up to 60 to 80% of genotype 1-infected patients fail to clear the virus after treatment (29). The reasons for this failure are unclear, although mounting evidence suggests that therapeutic, as well as spontaneous viral clearance is associated with vigorous, polyclonal, durable T-helper and cytotoxic-T-lymphocyte (CTL)-mediated responses (8, 11, 12, 17, 21, 22, 38). Surprisingly, however, HCV-specific CTLs are detected in chronically infected individuals, both in the peripheral blood and the liver demonstrating that the virus persists in spite of a specific immune response mounted by the host (8, 20, 35). Higher frequencies of CTLs are found in the liver than in the periphery as recently shown in studies by using soluble tetrameric class I major histocompatibility complex (MHC-I) molecules (15, 17). A recent longitudinal study suggests that viral resolution is associated with a very dynamic immune response that involves the contribution of cells with different effector functions (45). In their study, Thimme et al. observed that during the acute phase of infection CD8+ T cells presenting an activated phenotype (CD38+) but unable to induce the production of gamma interferon (IFN-γ) are detectable. The resolution of infection was observed when CD8+ T cells displayed a nonactivated phenotype (CD38−) but were associated with the production of IFN-γ. In a different study, CD8+ T cells able to induce the production of IFN-γ were indeed hardly detectable in chronically infected patients (16).
A key question in the identification of HCV-immune correlates of resolution remains the definition of the role played by individual viral antigens or epitopes in the nature and vigor of immune responses observed during resolution. CTL epitopes have been identified over the entire HCV polyprotein and in virtually all encoded antigens (36). To date, it has been difficult to link viral clearance with responses mounted against specific class I CD8+-cell-restricted epitopes. A recent study has explored for the first time CTL activities to a large panel of HLA-A2.1 presented epitopes (up to 16) in patients chronically infected and receiving or not antiviral therapy (47). It was observed that, whereas the majority of chronically infected patients had CTL specific for a NS4-derived peptide, individuals receiving and responding to therapy presented mainly or solely a strong activity to an NS3-derived epitope, documenting for the first time a switch in the choice of CTL targets after successful therapy. Thus, to date, not only the role played by specific HLA epitopes in HCV control or clearance, but their relative dominance remains vastly uncharacterized. However, the consensus is that a HCV vaccine should induce vigorous, sustained, and above all broad CD8+-T-cell-mediated responses.
Numerous vaccines, and particularly vector-based candidates, can induce such responses, as demonstrated for a wide variety of antigens in various preclinical and clinical studies (4, 7, 24, 40). Nonetheless, a systematic comparison of the vigor and scope of T-cell mediated responses induced by various vaccines expressing the same epitopes has been reported only in a limited number of studies (see, for example, references 18 and 32), most reports focus on a small number of antigens or epitopes encoded by the target pathogens. This is surprising since such information should be critical for both the choice of the antigenic sequence and the type of vaccine vehicle to be selected before initiation of clinical trials.
In the present study, we have taken advantage of a recently developed transgenic mouse model entirely devoid of murine MHC-I molecules to establish a map of the immunogenicity and, to some extent, the immunodominance, of most known HCV HLA-A2.1 epitopes presented in natural infection in the context of three vaccination strategies based on peptides, naked DNA, or recombinant adenoviruses. The influence of flanking genes and vector combinations on the scope and vigor of epitope recognition after administration of the vaccines was examined.
MATERIALS AND METHODS
Mice.
HDD mice transgenic for HLA-A2.1 (A0201) monochain histocompatibility class I molecule and deficient for both H-2Db and murine β2-microglobulin (β2m) were used. H-2Db and mouse β2m genes of these mice have been disrupted by homologous recombination (31). Six- to eight-week-old mice were used. Mice were housed in appropriate animal care facilities and handled according to international guidelines for experiments with animals.
Recombinant expression vectors.
The Core (amino acids [aa] 1 to 191), E1 (aa 165 to 329), E2 (aa 367 to 673), CoreE1E2 (aa 1 to 746), NS3 (aa 1027 to 1657), NS3-NS4 (aa 1027 to 1972), NS4 (aa 1658 to 1972), NS5A (aa 1973 to 2420), and NS5B (aa 2421 to 3011) genes of the HCV genotype 1b HCV-JA strain (19) were inserted into the commercial pgWiz plasmid (Gene Therapy System, Inc., San Diego, Calif.) to create pgWizCore, pgWizE1, pgWizE2, pgWizCE1E2, pgWizNS3, pgWizNS3NS4, pgWizNS4, pgWizNS5A, and pgWizNS5B. A plasmid, pgWizGFP, expressing the green fluorescent protein (GFP) was also constructed to be used as a control. Cloned fragments were verified by sequencing.
Recombinant adenovirus construction, production, and titration.
All recombinant adenoviral genomes were generated as infectious plasmids by homologous recombination in Escherichia coli as described previously (9). The two plasmids (pTG 13387 and pTG 6624) containing the adenoviral genome were kindly provided by Transgene S.A. In brief, sequences from the HCV-JA strain corresponding to the CE1E2 polyprotein (aa 1 to 746), the Core protein (aa 1 to 191), or the NS3 protein (aa 1027 to 1657) were inserted in the adenoviral shuttle plasmid (pTG 13387) containing a cytomegalovirus-driven expression cassette surrounded by adenoviral sequences (nt 1 to 458 and nt 3511-5788) to allow homologous recombination with the adenoviral sequences of the backbone vector (pTG 6624) (9). The resulting full-length viral genomes contain a deletion in the adenoviral E3 (nucleotides 28592 to 30470), whereas the E1 region (nucleotides 459 to 3327) is replaced by the expression cassette containing, from 5′ to 3′, the cytomegalovirus immediate-early enhancer/promoter, a chimeric human β-globin-immunoglobulin G intron, the HCV sequences, and the bovine growth hormone polyadenylation signal. Recombinant adenoviruses were generated by transfection into the 293 complementation cell line of the corresponding plasmids restricted by PacI digestion. Virus propagation and amplification were performed by successive passages on 293 cells and purification, as well as by titration of infectious units by indirect immunofluorescence of the viral DNA-binding protein, were carried out as described previously (27, 37). Ad(βgal), an adenovirus allowing the production of β-galactosidase (Transgene S.A.), was used as a control.
In vitro studies. (i) Transient transfections.
Murine fibroblasts NIH 3T3 were transfected as previously described with HCV-expressing plasmids (5). At 48 h posttransfection, cells were harvested for analysis of antigenic expression by flow cytometry or immunofluorescence analyses. For immunofluorescence studies, transfected cells were fixed with the methanol-acetone and stained with rabbit or purified murine monoclonal antibodies to Core (antibody 19D9D6; Biomérieux), to E1 (antibody A4) and to E2 (antibody H47; J. L. Dubuisson), to NS3 (antibody 1B6DM) and NS4 (antibody 8DE8E1; A. Cahour), to NS5 (Biodesign), to NS5A (antibody 3G6E1 and 4F3H2; G. Baccala and C. Jolivet [Biomérieux]), and to NS5B (antibody 5B-12B7; D. Moradpour). A goat anti-mouse immunoglobulin monoclonal antibody coupled to fluorescein isothiocyanate-labeled fluorophore was used as a secondary antibody (Dako).
(ii) Adenovirus infections.
Subconfluent monolayers of HepG2 cells were infected with the appropriate recombinant adenoviruses with a multiplicity of infection (MOI) of 50 IU/cell. After the indicated time postinfection (p.i.), cell monolayers were rinsed with Tris-buffered saline (10 mM Tris-HCl, pH 7.5; 150 mM NaCl) and lysed in 300 μl of 2× Laemmli buffer. Cell lysates were passed 10 times through a syringe with a 26-guage needle to break the cellular chromatin, and 4 μl was loaded on a 15% polyacrylamide gel. After separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, proteins were transferred to nitrocellulose membranes (Hybond-ECL; Amersham) by using a Trans-Blot apparatus (Bio-Rad, Ivry-sur-Seine, France) and visualized with monoclonal anti-Core antibodies (J. F. Delagneau), followed by donkey anti-mouse immunoglobulin conjugated to horseradish peroxidase and by enhanced chemiluminescence detection (Amersham) as recommended by the manufacturer.
Synthetic peptides.
The synthetic peptides used were synthesized with a purity of >95% (Neosystem, Strasbourg, France, or Sigma-Genosys, Cambridge, United Kingdom). Peptides were dissolved in dimethyl sulfoxide (20 μl/mg) and subsequently diluted in distilled water to a final concentration of 1 mM. All peptides were derived from the HCV-JA sequence from Core (YLLPRRGPRL [aa 35 to 44] and DLMGYIPLV [aa 132 to 140], respectively, referred to as C35-44 and C132-140), from E1 (IMHTPGCV [aa 220 to 227], TIRRHVDLLV [aa 257 to 266], and SMVGNWAKV [aa 363 to 372], respectively, referred to as E1 220-227, E1 257-266, and E1 363-372), from E2 glycoprotein (SLVSWLSQGPS [aa 401 to 411], RLWHYPCTI [aa 614 to 622], ALSTGLIHL [aa 686 to 694], and FLLLADARV [aa 725 to 734], respectively, referred to as E2 401-411, E2 614-622, E2 686-694, and E2 725-734), from NS3 (CVNGVCWTV [aa 1073 to 1081], LLCPSGHVV [aa 1169 to 1177], KLTGLGLNAV [aa 1406 to 1415], and YLVAYQATV [aa 1585 to 1593], respectively, referred to as NS3 1073-1081, NS3 1169-1177, NS3 1406-1415, and NS3 1585-1593), from NS4B (HMWNFITGI [aa 1769 to 1777], SLMAFTASI [aa 1789 to 1797], LLFNILGGWV [1807 to 1816], and ILAGYGAGV [aa 1851 to 1859], respectively, referred to as NS4B 1769-1777, NS4B 1789-1797, NS4B 1807-1816, and NS4B 1851-1859), from NS5A (SPDADLIEANL [aa 2221 to 2231] and ILDSFDPLR [aa 2252 to 2260], respectively, referred to as NS5A 2221-2231 and NS5A 2252-2260), and from NS5B (RLIVFPDLGV [aa 2578 to 2587], ALYDVVSTL [aa 2594 to 2602], and KLQDCTMLV [aa 2727 to 2735], respectively, referred to as NS5B 2578-2587, NS5B 2594-2602, and NS5B 2727-2735). A T-helper peptide, derived from the HBV nucleocapsid (TPPATRPPNAPIL [aa 128 to 140]) and referred to as the HBV-Core (26), was used for peptide-based immunization. One peptide, referred to as FLP (FLPSDYFPSV), was used in the in vitro binding assay (26).
Peptide binding assay.
Peptide binding to recombinant HLA-A2.1 molecules called SC-A2 was assayed by competition with the radiolabeled peptide FLP as previously described (26). Briefly, SC-A2 molecules (1.5 μg) were incubated 2 h at room temperature with radiolabeled FLP (8 ng) in the presence of various concentrations of competitor peptide. Unbound peptide was eliminated by ultracentrifugation and extensive washing. Radioactivity was measured in a gamma spectrometer (Gammamatic). For each peptide, experiments were performed twice. The percent inhibition was deduced for the highest competitor peptide/FLP ratio tested (typically 30).
Immunizations.
Peptide immunization were carried at the base of the tail with a peptide mixture containing the HCV epitope of interest (60 μM) and the T-helper peptide: HBV-Core (60 μM) emulsified in incomplete Freund adjuvant. Mice received two injections at 2-week intervals. For DNA based-immunization, 100 μg of plasmid was injected in the tibialis anterior muscle of each mouse as previously described (28). For recombinant adenovirus immunization, mice were immunized intramuscularly by one or two injections of 109 IU of recombinant adenovirus in 100 μl of phosphate-buffered saline (PBS). For DNA priming and recombinant adenovirus boosting, mice were immunized intramuscularly with 109 recombinant adenovirus at 2 weeks after one or two injections of 100 μg of DNA (given at 2-week intervals). In DNA or recombinant adenovirus-based vaccination, control animals received either pgWiz GFP plasmid or a wild-type adenovirus expressing the β-galactosidase protein or both (in prime-boost vaccine studies).
The analysis of the cytotoxic activity and of the production of IFN-γ was typically performed 2 weeks after the final immunization.
CTL analysis.
CTL analysis was performed as previously described (5). Briefly, on day 7 peptide-stimulated spleen cells were used as effectors in a CTL assay. Specific cytolytic activity was tested in a standard 51Cr release assay against an EL-4S3−Rob HHD target pulsed or not with a 10 μM concentration of the selected peptide. After a 4-h incubation period, 51Cr release was measured by using a γ-Cobra II counter (Packard, Rungis, France). Spontaneous and maximum release were determined from wells containing either medium alone or lysis buffer (1 N HCl). The percent specific cytotoxicity was calculated by the formula: (release in assay − spontaneous release)/(maximum release − spontaneous release) × 100. Peptide-specific lysis was determined by the difference between the percentage of specific lysis obtained in the presence or in the absence of peptide. For each effector/target (E:T) ratio, the results are expressed as the mean of duplicates.
ELISPOT assay.
IFN-γ-releasing cells were quantified by cytokine-specific enzyme-linked immunospot assay (ELISPOT) as previously described (14). Briefly, splenocytes from individual mice were isolated and cultured in presence of 10 U of recombinant IL-2/ml for 48 h in alpha minimal essential medium with 10 μg of selected peptide or 5 μg of concanavalin A (positive control)/ml or without peptide (negative control) in triplicate wells. Wells were washed three times, respectively, with PBS and PBS-0.05% Tween before a 2-h incubation with biotinylated anti-murine IFN-γ. After being washed, the wells incubated for 1 h with extravidin-PAL conjugate and the enzyme activity was revealed by using the Bio-Rad kit: 100 μl of alkaline phosphatase (AP) color reagent A and 100 μl of AP color reagent B were mixed in 25× AP color development buffer (400 μl added to 9.6 ml of distilled water; Bio-Rad), and the spots were counted (Spot, KS Zweiss; Carl Zeiss Vision GmbH). The background level was measured in wells containing splenocytes in medium only. The number of peptide-specific spots was obtained by subtracting the background from the number of spots appearing after HLA-A2-peptide stimulation. The results are shown as the mean value obtained for triplicate wells.
Sequence alignments.
Analyses were performed by using the IBCP HCV database website facilities (hepatitis.ibcp.fr), which contains all HCV sequences available from the EMBL database. Sequence alignments were carried out with the CLUSTALW program (46). Determination of the percent conservation between the HLA-A2.1 epitopes used in our study and sequences derived from genotype 1b, 1a, and 4 isolates was performed by using the Antheprot program.
RESULTS
Peptide-based studies. (i) HLA-A2.1 binding assay.
We evaluated 22 HLA-A2 epitopes recognized in HCV natural infection (out of a total of 25 described to date) for their capacity to bind to purified recombinant HLA-A2.1 by using a competition-based assay. This assay evaluates the ability of unlabeled peptide epitopes to inhibit the binding of the radiolabeled peptide FLP, a peptide known for its high binding capacity, on recombinant HLA-A2.1. The results shown in Table 1 indicate the percent inhibition obtained with each peptide. Four classes of peptides could be defined. High binders displaying an inhibition percent ranging from 55 to 100 such as peptides C35-44, C132-140, E1 220-227, E2 614-622, E2 686-694, E2 725-734, NS3 1169-1177, NS3 1406-1415, NS3 1585-1593, NS4B 1789-1797, NS4B 1807-1816, NS5B 2578-2587, NS5B2594-2602, and NS5B 2727-2735 (n = 14). Low binders displaying an inhibition percent ranging from 30 to 37 such as peptides E2 401-411, NS4B 1769-1777, NS4B 1851-1859, NS5A2221-2231, and NS5A 2252-2260 (n = 5). Very low binders displaying an inhibition percentage of 16 to 18 such as E1 363-372 and NS3 1073-1081. Finally, some peptides displayed no binding capacities such as the E1 257-266 peptide. Some peptides presented atypical HLA-A2.1-restricted binding motifs (E1 363-372, NS3 1073-1081, NS4B 1769-1777, and NS5A 2221-2231) that could explain their poor in vitro binding capacities. In the case of the NS3 1073-1081 epitope, the presence of cysteine residues potentially creating disulfide bonds might have hampered its binding. Overall, five peptides—E2 614-622, E2 686-694, NS4B 1807-1816, NS5B 2578-2587, and NS5B 2727-2734—were able to compete with FLP binding with an efficacy comprised between 90 and 100%.
TABLE 1.
Capacity of HCV-specific HLA-A2.1-restricted peptide epitope to bind to purified recombinant HLA-A2.1 molecules and to induce specific CTL or IFN-γ-producing T cells after direct injection
| Gene | Residue (aa) | Epitope
|
Binding (% inhibition)a | ELISPOT (no. of specific spots/106 cells)b
|
Specific lysis (%) (E/T = 33)c
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sequence | Name | Mc | M1 | M2 | M3 | Mc | M1 | M2 | M3 | |||
| C | 35-44 | YLLPRRGPRL | C35-44 | 58 | 12 | 73 | 160 | 73 | 2 | 5 | 23 | 2 |
| 132-140 | DLMGYIPLV | C132-140 | 71 | 0 | 211 | 206 | 284 | 4 | 44 | 19 | 26 | |
| E1 | 220-227 | IMHTPGCV | E1 220-227 | 55 | 0 | 0 | 55 | 0 | 1 | 2 | 0 | 14 |
| 257-266 | TIRRHVDLLV | E1 257-266 | 0 | ND | ND | ND | ND | ND | ND | ND | ND | |
| 363-372 | SMVGNWAKV | E1 363-372 | 16 | 0 | 172 | 160 | 328 | 0 | 47 | 3 | 66 | |
| E2 | 401-411 | SLVSWLSQGPS | E2 401-411 | 30 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 614-622 | RLWHYPCTI | E2 614-622 | 92 | 0 | 82 | 47 | 200 | 10 | 34 | 26 | 22 | |
| 686-694 | ALSTGLIHL | E2 686-694 | 100 | 0 | 403 | 260 | 112 | 2 | 65 | 10 | 79 | |
| 725-734 | FLLLADARV | E2 725-734 | 66 | 0 | 333 | 215 | 622 | 0 | 83 | 34 | 36 | |
| NS3 | 1073-1081 | CVNGVCWTV | NS3 1073-1081 | 18 | 0 | 5 | 47 | 13 | 2 | 27 | 88 | 0 |
| 1169-1177 | LLCPSGHVV | NS3 1169-1177 | 77 | 2 | 137 | 322 | 160 | 9 | 45 | 7 | 53 | |
| 1406-1415 | KLTGLGLNAV | NS3 1406-1415 | 71 | 0 | 188 | 458 | 230 | 18 | 33 | 15 | 40 | |
| 1585-1593 | YLVAYQATV | NS3 1585-1593 | 76 | 0 | 0 | 173 | 190 | 11 | 7 | 19 | 16 | |
| NS4B | 1769-1777 | HMWNFITGI | NS4B 1769-1777 | 30 | 0 | 162 | 93 | 30 | 0 | 13 | 35 | 0 |
| 1789-1797 | SLMAFTASI | NS4B 1789-1797 | 63 | 71 | 61 | 157 | 177 | 20 | 31 | 15 | 38 | |
| 1807-1816 | LLFNILGGWV | NS4B 1807-1816 | 94 | 0 | 605 | 22 | 658 | 0 | 19 | 12 | 0 | |
| 1851-1859 | ILAGYGAGV | NS4B 1851-1859 | 32 | 0 | 5 | 2 | 0 | 6 | 0 | 0 | 0 | |
| NS5A | 2221-2231 | SPDADLIEANL | NS5A 2221-2231 | 37 | 42 | 102 | 0 | 8 | 4 | 0 | 2 | 2 |
| 2252-2260 | ILDSFDPLR | NS5A 2252-2260 | 34 | 0 | 37 | 0 | 30 | 8 | 0 | 3 | 8 | |
| NS5B | 2578-2587 | RLIVFPDLGV | NS5B 2578-2587 | 100 | 49 | 234 | 116 | 305 | 12 | 41 | 33 | 46 |
| 2594-2602 | ALYDVVSTL | NS5B 2594-2602 | 80 | 39 | 261 | 331 | 326 | 0 | 81 | 44 | 2 | |
| 2727-2735 | KLQDCTMLV | NS5B 2727-2735 | 99 | 25 | 255 | 277 | 297 | 13 | 38 | 52 | 60 | |
Percent inhibition of binding of the FLP-control peptide onto purified HLA.A2.1 molecules for a ratio competitor peptide/control peptide = 30.
ND, not done. Mc, control mouse; M1, M2, and M3, immunized mice.
Percent specific lysis at an E:T ratio of 1:33.
(ii) Peptide-based immunizations.
To evaluate whether the mouse repertoire for the various epitopes was intact and to define a first map of immunogenicity of the epitopes, mice were immunized with the different peptides and induction of specific CTL and IFN-γ production was evaluated. The results are summarized in Table 1. Specific T cells secreting IFN-γ were detected in response to the majority of the peptides, the number of detected spots ranging from 55 to 658 per 106 splenocytes. Some of the highest frequencies of specific IFN-γ producing T cells (up to 658 spots) were detected for peptides E2 686-694, NS4B 1807-1816, NS5B 2578-2587, NS5B 2594-2602, and NS5B 2727-2735 which belong to the “high binder” group described above. The poorest inducers of IFN-γ-producing T cells (<100 spots) were peptides such as E1 220-227, E2 401-411, NS3 1073-1081, NS5A 2221-2231, and NS5A 2252-2260 that are all “low” or “very low” binders to HLA-A2 molecules. The correlation between HLA-A2 binding ability of the peptides, and their capacity to stimulate IFN-γ-producing T cells was nonetheless not systematic. One notable exception was peptide E1 363-372, which displayed a very low binding capacity (16% inhibition) but induced a rather high number of specific spots (160 to 328 per 106 splenocytes).
For most peptides, specific CTL responses were detected and ranged from 23 to 88% lysis for an E:T ratio of 33:1. Peptides which had shown very poor induction of IFN-γ production also induced very limited CTL-mediated lysis (E1 220-227, E2 401-411, NS4B 1851-1859, NS5A 2221-2231, and NS5A 2252-2260). Interestingly, three peptides—NS4B 1807-1816, NS3 1585-1593, and NS4B 1769-1777—displayed one predominant function since they induced IFN-γ-producing T cells in the absence or in the presence of only very weak specific CTL activity. Overall, the correlation between ELISPOT and CTL data was good.
Single genes DNA-based immunization reveals a different use of HLA-A2.1 restricted epitopes than observed after peptide-based immunization.
Seven plasmids expressing various HCV genes (Core to NS5B) of the well-described HCV-JA genotype 1b prototype strain (19) were engineered and used to vaccinate transgenic mice with the goal to define the immunogenicity and potential dominance of the HLA-A2.1 epitopes when expressed endogenously by the host's cells. Only 19 of the 22 epitopes were screened, 3 being absent from the HCV sequences cloned into the plasmids engineered. Comparison of responses following one, two, or three immunizations revealed that most vigorous specific CTLs and highest frequencies of IFN-γ-producing T cells were systematically observed after two immunizations (data not shown). Overall data obtained after two immunizations are summarized in Table 2 and revealed a number of striking observations. The number of epitopes displaying the two effector functions was much smaller than that observed after peptide immunization. Of the 19 epitopes screened, induction of immune responses was observed against only 11 epitopes spanning all of the HCV antigens, with the notable exception of NS5A. These are C132-140, E2 401-411, E2 614-622, NS3 1073-1081, NS3 1169-1177, NS4B 1769-1777, NS4B 1789-1797, NS4B 1807-1816, NS5B 2578-2587, NS5B 2594-2602, and NS5B 2727-2735. The number of epitopes found reactive after Core- and NS3-DNA vaccination was particularly limited. Core-DNA-induced detectable proliferation of IFN-γ producing T cells and a CTL activity, which were specific of the C132-140 epitope only, whereas no reactivity was found against the C35-44 epitope. After NS3-plasmid vaccination, only one of the screened epitopes (NS3 1073-1081) was found highly reactive, able to induce both specific CTL and IFN-γ-producing T cells whereas a second peptide (NS3 1169-1177) induced the proliferation of IFN-γ-secreting cells but had only a very low capacity to induce specific CTL activity (1:3 immunized mice). The two other epitopes (NS3 1406-1415 and NS3 1585-1593) were found to be nonreactive in contrast to observations made after peptide vaccination. High responses, both made in terms of proliferation of the specific IFN-γ-secreting cells (reaching up to 750 spots/106 cells) and CTL activity (reaching up to 70% of specific lysis at an E:T ratio of 1:33) were overall seen for the C132-140, E2 614-622, NS3 1073-1081, NS4B 1807-1816, NS5B 2578-2587, NS5B 2594-2602, and NS5B 2727-2735 epitopes. As observed in the context of peptide-based vaccination, the dominant function displayed by epitopes NS4B 1769-1777 and NS4B 1807-1816 was linked to IFN-γ production. The poor immunogenicity of the NS5A plasmid confirmed that observed with the NS5A peptides themselves (Table 1).
TABLE 2.
Capacity of HCV-DNA vaccines encoding individual antigens to induce specific CTL and IFN-γ-producing T cells
| Plasmid | Epitope | ELISPOT (no. of specific spots/106 cells)
|
Specific lysis (%) (E/T = 33)b
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Mca | M1 | M2 | M3 | Mc | M1 | M2 | M3 | ||
| pgWizCore | C35-44 | 0 | 0 | 0 | 0 | 10 | 20 | 9 | 0 |
| C132-140 | 154 | 650 | 390 | 400 | 5 | 44 | 39 | 70 | |
| pgWizE1 | E1 220-227 | 3 | 66 | 29 | 31 | 4 | 1 | 10 | 5 |
| E1 257-266 | 0 | 76 | 38 | 12 | 23 | 22 | 3 | 5 | |
| pgWizE2 | E2 401-411 | 5 | 25 | 60 | 90 | 12 | 42 | 39 | 37 |
| E2 614-622 | 15 | 65 | 180 | 120 | 10 | 29 | 54 | 55 | |
| pgWizNS3 | NS3 1073-1081 | 10 | 120 | 475 | 120 | 16 | 43 | 48 | 34 |
| NS3 1169-1177 | 10 | 120 | 90 | 145 | 13 | 31 | 22 | 13 | |
| NS3 1406-1415 | 68 | 112 | 102 | 66 | 12 | 17 | 11 | 6 | |
| NS3 1585-1593 | 56 | 90 | 66 | 69 | 9 | 23 | 12 | 23 | |
| pgWizNS4 | NS4B 1769-1777 | 120 | 550 | 330 | 480 | 2 | 1 | 7 | 5 |
| NS4B 1789-1797 | 3 | 35 | 54 | 72 | 9 | 1 | 0 | 0 | |
| NS4B 1807-1816 | 9 | 240 | 360 | 750 | 1 | 40 | 28 | 33 | |
| NS4B 1851-1859 | ND | ND | ND | ND | ND | ND | ND | ND | |
| pgWizNS5A | NS5A 2221-2231 | 105 | 0 | 0 | 0 | 12 | 21 | 19 | 23 |
| NS5A 2252-2260 | 120 | 0 | 0 | 0 | 16 | 22 | 15 | 9 | |
| pgWizNS5B | NS5B 2578-2587 | 0 | 240 | 300 | 195 | 8 | 42 | 23 | 36 |
| NS5B 2594-2602 | 0 | 300 | 380 | 330 | 9 | 41 | 33 | 49 | |
| NS5B 2727-2735 | 15 | 390 | 309 | 198 | 5 | 10 | 17 | 26 | |
Mc, control mouse; M1, M2, and M3, immunized mice. ND, not done.
Percent specific lysis at an E:T ratio of 1:33.
Taken together, these results illustrate the capacity of HCV-DNA vaccines to induce specific and vigorous cell-mediated immune responses targeting HCV epitopes presented in natural infection, as well as the fact that a rather narrow spectrum of epitopes is involved in this recognition.
Optimization of HCV vaccine-induced cellular immune response. (i) Influence of flanking domains on epitope recognition.
Vaccination with some of the plasmids induced immune responses for which the spectrum was unexpectedly narrow in comparison to that obtained after peptide based-vaccination (Table 1). We investigated the hypothesis that an incomplete or biased processing of the antigens might be responsible for the restriction observed in the particular cases of capsid and NS3, two potential key components of an HCV vaccine. More specifically, we looked at the influence of flanking genes such as E1 and E2 (in the case of Core) and NS4 (in the case of NS3) on the efficiency of epitope presentation. To do this, two additional plasmids were constructed, which expressed Core in the context of E1 and E2 (pgWizCoreE1E2) or NS3 in the context of NS4 (pgWizNS3NS4). After transient transfection of the plasmids, protein expression was analyzed by flow cytometry. Both the level of transfection and protein detection were higher for pgWizCore compared with pgWizCoreE1E2 and for pgWizNS3 compared to pgWizNS3-NS4 (up to twofold [data not shown]). These data, as expected, are coherent with the fact that the level of transfection and overall protein expression are typically higher with smaller plasmids.
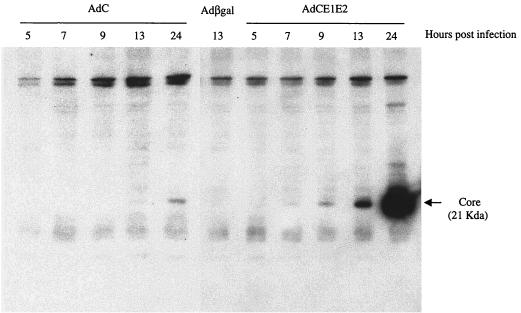
It has recently been shown that coexpression of NS4A influences the intracellular localization of NS3 and is critical for its stability (49). We analyzed here, by Western blot assay, the detection of core when expressed in the presence or absence of E1 and E2 (Fig. 1). Experiments were performed with recombinant adenoviruses and not plasmid DNAs in order to increase transfection efficiency of the HCV sequences. After infection with a recombinant adenovirus expressing Core-E1-E2, a specific band could be observed as early as 7 h p.i., whereas a similar band could only been detected at 13 h P.I. by using a Core-expressing adenovirus. At 24 h p.i., a 25-fold increase in Core detection was observed from Core-E1-E2 adenovirus-infected cells compared with the level observed in cells infected with the Core-expressing virus, suggesting that expression or stability of Core is enhanced in presence of E1 and E2.
FIG. 1.
Western blot analysis of the core protein after infection of HepG2 cells with a recombinant adenovirus expressing Core (AdC) or Core-E1-E2 (AdCE1E2). HepG2 cells were infected with AdC (left panel), AdCE1E2 (right panel), or a control adenovirus expressing β-galactosidase (Adβgal, center) at an MOI of 50 IU/cell and then harvested at the indicated times after infection. Expression of the core protein (21 kDa) was monitored by using the specific monoclonal antibody ACAP27 as described in Materials and Methods.
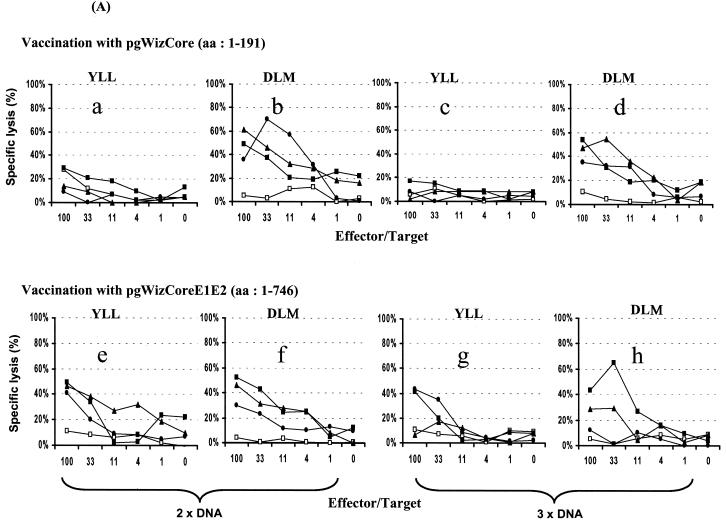
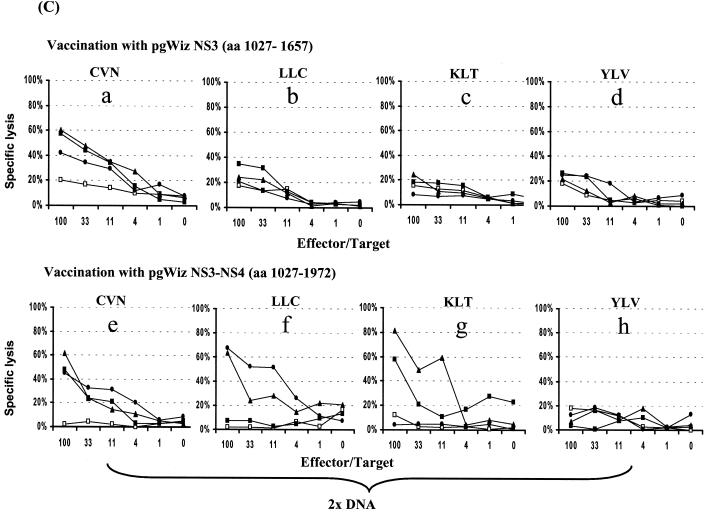
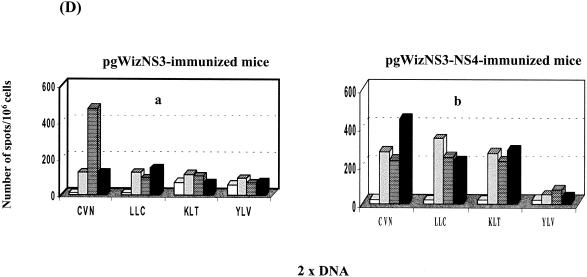
Induction of specific CTL and IFN-γ-producing cells were examined after vaccination with the various plasmids (Fig. 2). In contrast to observation previously reported with pgWizCore (Table 2) and as shown in Fig. 2A (a to d), vaccination with pgWizCoreE1E2 resulted not only in induction of a CTL activity specific of the C132-140 epitope (Fig. 2Af and h) but also in lysis specific to the C35-44 epitope (Fig. 2Ae and g). These data were supported by the detection of specific IFN-γ-producing cells (Fig. 2Bc and d versus a and b). The spectrum of epitopes recognized was also broadened after vaccination of mice with pgWizNS3NS4 in comparison with vaccination performed with pgWizNS3 (Fig. 2C and D). Three of four tested epitopes (NS3 1073-1081, NS3 1169-1177, and NS3 1406-1415) were targets of a specific CTL response (percent specific lysis at E:T of 1:33 ranged from 21 to 52%; Fig. 2Ce, f, and g) and could result in the production of IFN-γ (Fig. 2Da and b). The levels of IFN-γ-producing cells were up to two to three times higher than those seen after vaccination with pgWizNS3 except for one highly reactive mouse in this group (Table 2 and Fig. 2D).
FIG. 2.
Influence of flanking genes on the scope of epitope recognition. HLA-A2.1-transgenic mice were immunized intramuscularly two (2× DNA) or three (3× DNA) times at 2-week intervals with 100 μg of pgWizCore or pgWizCoreE1E2 plasmids (A and B) or pgWizNS3 or pgWizNS3NS4 plasmids (C and D). Spleen cells were collected 2 weeks after the final injection and cultured in the presence of each tested peptides. Bulk CTL analysis and IFN-γ-producing cells were performed after two immunizations (Aa, b, e, and f; Ba to c; C; and D) or three immunizations (Ac, d, g, and h). Each curve or each bar represents the response tested for a single mouse. The Core-specific epitopes tested were YLL and DLM; the NS3-specific epitopes tested were CVN, LLC, KLT, and YLV. Solid or shaded symbols or bars represent vaccinated mice (three per group), whereas open symbols or open bars represent mock-vaccinated mice (one per group).
Specific E1, E2, or NS4 induced immune responses observed when Core-E1-E2 or NS3-NS4 plasmids were used were identical to those seen after vaccination with E1-, E2-, or NS4-expressing plasmids (Table 2 and data not shown).
Taken together, these results suggest that in the case of the core and NS3 an incomplete or altered peptide processing might take place when these antigens are expressed out of a more natural context.
Influence of a DNA priming and recombinant adenovirus-boosting vaccine regimen on the vigor of epitope recognition.
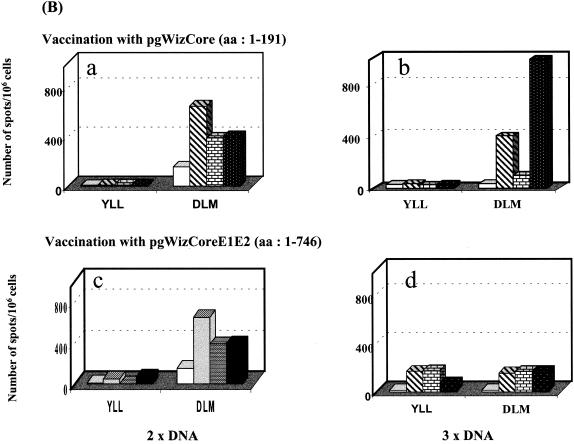
While selection of sequences to be expressed by a vaccine vector appears critical for an optimal presentation of epitopes, we observed that the choice of the vector itself seems to play a less important role. When we compared immunization with recombinant adenoviruses expressing core or NS3 with vaccination with the corresponding DNA vaccines, no differences could be observed in terms of the scope of epitopes recognized (data not shown). On the other hand, a clear increase in the vigor of specific cellular immune responses could be observed when combined DNA and adenovirus vaccination was implemented. This is demonstrated in Fig. 3, which shows results obtained by using splenocytes from mice immunized either one, two, or three times with pgWizCE1E2 DNA or once with a recombinant adenovirus (expressing the same CE1E2 sequence) preceded by zero, one, or two DNA primings. Six epitopes were screened, two within core (C35-44 and C132-140) and four within E2 (E2 401-411, E2 614-622, E2 686-694, and E2 725-734). Specific IFN-γ-producing T cells were observed for all six epitopes after two or three DNA vaccinations (Fig. 3b and c). A detectable but weak response was observed after a single vaccination with adenovirus (Fig. 3d) and was not boosted by a second injection of the virus (data not shown). A single priming with the DNA vaccine, followed by an adenovirus booster, had a significant effect on the frequencies of IFN-γ-producing T cells induced against all epitopes (Fig. 3e). The number of spots/106 cells doubled to tripled on average for all six epitopes tested, including the otherwise subdominant E2 401-411 and E2 725-734 epitopes. These responses were not enhanced if two DNA primings were performed (Fig. 3f). Increase in the vigor of CTL activity specific for all six epitopes was also observed in agreement with the ELISPOT data (data not shown).
FIG. 3.
Influence of a DNA-priming and recombinant adenovirus-boosting regimen on the vigor of the induced immune response. HLA-A2.1-transgenic mice were immunized intramuscularly one, two, or three (1× DNA, 2× DNA, and 3× DNA) times at 2-week intervals with 100 μg of gWizCoreE1E2 or with pgWizGFP control plasmid (a, b, and c). Mice were also immunized with a single injection of 109 IU of recombinant adenovirus CoreE1E2 after no DNA priming (d), one DNA priming (e), or two DNA priming (f) with 100 μg of the pgWizCE1E2. The results are shown as the number of spots per 106 cells specific for the two core epitopes (YLL and DLM) or the four E2 epitopes (SLV, RLW, ALS, and FLL). Mock-vaccinated mice are represented by open bars, whereas the three immunized mice per group are represented by shaded or solid bars.
Taken together, these results indicate the capacity of a DNA prime/recombinant adenovirus boost regimen to potentiate the specific cellular response directed against multiple HLA-A.2.1-restricted epitopes. The dominance of epitopes tends to “even out” in a prime-boost vaccination context compared with single-vectored vaccine regimens.
Immunogenicity and immunodominance of HCV HLA-A2.1-restricted epitopes recognized during natural infection.
Table 3 summarizes, for each epitope, the level of immune responses that can be obtained with the different vaccine formulations tested in our study. Four types of epitopes were defined based on whether they induced strong specific CTL and high frequencies of IFN-γ-producing T cells (type 1) or weak specific CTL and low frequencies of IFN-γ-producing T cells (type 2) or whether they were unable to induce either specific CTL or proliferation of IFN-γ-producing T cells (type 3) or induced predominantly IFN-γ-producing T cells with only a weak or undetectable CTL activity (type 4). Although the immunogenicity of a given epitope was altered depending on the vaccine strategy selected (e.g., C35-44 or E1 363-372), most epitopes displayed the same immunogenicity independent of the immunization strategy implemented (e.g., see E2 614-622, E2 686-694, E2 725-734, NS5A 2221-2231, and NS5A 2252-2260). Most surprisingly, epitopes belonging to one type were found clustered within specific antigens and not randomly scattered. The three antigens containing most highly immunogenic epitopes are E2, NS3, and NS5B, followed by NS4B and Core. Overall, E1 and NS5A appear as very weak immunogens. Unexpectedly, NS4B had a particular profile since it contains the two epitopes identified by all vaccine strategies as being able to induce mainly specific IFN-γ-producing T cells in the absence of or with only a limited specific lytic activity.
TABLE 3.
Comparative classification of HCV HLA-A2.1 epitopes according to their immunogenicity in different vaccination contexts
| Gene | Epitope | Vaccination strategyc
|
|||
|---|---|---|---|---|---|
| Peptide | DNAa | Adenovirusb | DNA-priming/adenovirus-boosting | ||
| C | C35-44 | Type 3 | Type 2 | Type 2 | Type 1 |
| C132-140 | Type 1 | Type 1 | Type 1 | Type 1 | |
| E1 | E1 220-227 | Type 3 | Type 3 | Type 3 | Type 3 |
| E1 257-266 | ND | Type 3 | Type 3 | Type 3 | |
| E1 363-372 | Type 1 | Type 2 | Type 2 | Type 2 | |
| E2 | E2 401-411 | Type 3 | Type 2 | Type 2 | Type 2 |
| E2 614-622 | Type 1 | Type 1 | Type 1 | Type 1 | |
| E2 686-694 | Type 1 | Type 1 | Type 1 | Type 1 | |
| E2 725-734 | Type 1 | Type 1 | Type 1 | Type 1 | |
| NS3 | NS3 1073-1081 | Type 2 | Type 1 | Type 1 | ND |
| NS3 1169-1177 | Type 1 | Type 1 | Type 4 | ND | |
| NS3 1406-1415 | Type 1 | Type 1 | Type 4 | ND | |
| NS3 1585-1593 | Type 4 | Type 3 | Type 3 | ND | |
| NS4B | NS4B 1769-1777 | Type 4 | Type 4 | ND | ND |
| NS4B 1789-1797 | Type 1 | Type 3 | ND | ND | |
| NS4B 1807-1816 | Type 4 | Type 4 | ND | ND | |
| NS4B 1851-1859 | Type 3 | ND | ND | ND | |
| NS5A | NS5A 2221-2231 | Type 3 | Type 3 | ND | ND |
| NS5A 2252-2260 | Type 3 | Type 3 | ND | ND | |
| NS5B | NS5B 2578-2587 | Type 1 | Type 1 | ND | ND |
| NS5B 2594-2602 | Type 1 | Type 1 | ND | ND | |
| NS5B 2727-2735 | Type 1 | Type 1 | ND | ND | |
Combined data from immunizations performed with plasmids expressing single (Table 2) or multiple (Core-E1-E2 or NS3-NS4; Fig. 2) genes. Optimal immunogenicity that could be observed for each epitope with either a single or combined gene vaccination is indicated.
Data obtained after one immunization with recombination adenoviruses expressing Core, Core-E1-E2, or NS3 (data not shown; Fig. 3).
Type 1, able to induce both strong specific CTL (typically >30% lysis at on E:T ratio of 1:33) and high frequencies of IFN-γ-secreting cells (typically >100 for 106 cells, up to 650); type 2, able to induce very weak specific CTL (typically <30% at an E:T ratio of 1:33) and low frequencies of IFN-γ-secreting cells (typically <100 for 106 cells, down to 0); type 3, unable to induce specific CTL or IFN-γ-secreting cells; type 4, able to predominantly induce IFN-γ-secreting cells.
Evaluation of epitope dominance was deduced either from data obtained after single gene-based vaccination (Table 2 and Fig. 2) or, when possible, when vaccination with larger sequences was performed (Fig. 2 and 3). After all observations were taken into account, epitopes such as E1 220-227, E1 257-266, NS5A 2221-2231, and NS5A 2252-2260 could be considered as subdominant (since they belong to the type 3 whatever the vaccination strategy implemented). C132-140, E2 614-622, E2 686-694, E2 725-734, NS5B 2578-2587, NS5B 2594-2602, and NS5B 2727-2735 would be dominant (since they belong to type 1 whatever the vaccination strategy implemented). Although these data provide a first and original insight into HCV HLA-A2.1 epitope dominance, additional studies are still required to establish a definite and exhaustive map of dominance at the level of the full-length polyprotein.
Epitope sequence conservation.
We performed alignments of all 22 epitope sequences used in our study with existing genotype 1b, 1a, and 4 sequences present in the database since these genotypes are currently the most resistant to antiviral therapy and may represent priority targets for a therapeutic vaccine. Percentages of conservation for each epitope are shown in Table 4. These ranged from 0 to 100%. Remarkably, some of the highest percent of conservation were found, not only in the expected highly conserved core antigen but also in E2 (80 to 98%), NS4 (93 to 97%) and NS5B (87 to 99%). For most peptides, major diverging amino acids were not anchor amino acid residues typically found at position 2 and 8 (data not shown). As indicated in Table 3, these conserved epitope sequences are also among the most immunogenic ones. Among these, C132-140, E2 686-694, E2 725-734, NS3 1585-1593, and NS4B 1807-1816 displayed a high degree of conservation not only when aligned with genotype 1b sequences but also with 1a and 4 sequences.
TABLE 4.
Amino acid sequence comparisons of HCV HLA-A2 epitopes sequences with counterparts derived from genotype 1b, 1a, and 4 isolates
| Protein | Epitope | % Conservation genotypea
|
||
|---|---|---|---|---|
| 1b | 1a | 4 | ||
| C | C35-44 | 95 (195/206) | 100 (56/56) | 100 (11/11) |
| C132-140 | 96 (197/206) | 97 (58/60) | 78 (8/11) | |
| E1 | E1 220-227 | 78 (178/229) | 0 (0/397) | 0 (0/15) |
| E1 257-266 | 83 (191/229) | 0 (0/397) | 0 (0/15) | |
| E1 363-372 | 84 (189/224) | 91 (365/401) | 0 (0/10) | |
| E2 | E2 401-411 | 0 (0/143) | 0 (0/391) | 0 (0/11) |
| E2 614-622 | 80 (115/143) | 88 (15/17) | 0 (0/1) | |
| E2 686-694 | 98 (138/141) | 94 (15/16) | 100 (1/1) | |
| E2 725-734 | 90 (127/141) | 94 (15/16) | 100 (1/1) | |
| NS3 | NS3 1073-1081 | 62 (84/136) | 0 (0/14) | 0 (0/1) |
| NS3 1169-1177 | 28 (38/136) | 0 (0/14) | 0 (0/1) | |
| NS3 1406-1415 | 15 (20/135) | 0 (0/14) | 0 (0/1) | |
| NS3 1585-1593 | 96 (130/135) | 100 (14/14) | 100 (1/1) | |
| NS4 | NS4B 1769-1777 | 0 (0/30) | 0 (0/14) | 0 (0/2) |
| NS4B 1789-1797 | 93 (28/30) | 100 (14/14) | 0 (0/2) | |
| NS4B 1807-1816 | 97 (29/30) | 100 (14/14) | 100 (1/1) | |
| NS4B 1851-1859 | 93 (28/30) | 100 (14/14) | 0 (0/1) | |
| NS5A | NS5A 2221-2231 | 83 (115/139) | 0 (0/14) | 0 (0/1) |
| NS5A 2252-2260 | 40 (56/139) | 0 (0/14) | 0 (0/1) | |
| NS5B | NS5B 2578-2587 | 96 (136/142) | 100 (14/14) | 0 (0/1) |
| NS5B 2594-2602 | 99 (141/142) | 0 (0/14) | 0 (0/1) | |
| NS5B 2727-2735 | 87 (123/142) | 0 (0/14) | 0 (0/1) | |
That is the number of isolates presenting a homology of 100% with the epitope sequences used in our study (derived from the HCV-JA isolate)/the total number of isolate sequences aligned.
DISCUSSION
This study provides the first exhaustive analysis of the immunogenicity and, although to a lesser extent, the immunodominance of most known HLA-A2.1 epitopes described in HCV infection in the context of vaccines that have the potential to reach clinical evaluation. Four major conclusions can be drawn from the study. First, only a limited number of CD8+ epitopes are the target of a strong cytotoxic activity and can induce high frequencies of IFN-γ-producing T cells and, for the majority, independently of the vaccine regimen implemented (Table 3, type 1 epitopes). Remarkably, these epitopes are not randomly distributed but cluster essentially within three antigens: E2, NS3, and NS5B. Immune responses specific of these sequences have been described in chronically infected patients (2, 8, 33, 35, 47), as well as in cases of resolved hepatitis (16, 44, 45, 47). No case of viral resolution has been associated with the development of a response directed specifically at any of the E2 or NS5B epitopes. On the contrary, two of the most recent studies that have followed HCV-induced T-cell immune responses either from the onset of infection (45) or after administration of therapy (47), have identified NS3 as the main target of resolution-associated induced responses. In both studies, resolution of infection correlated with a detectable NS3-specific CD8+-cell-mediated immune response, particularly in association with IFN-γ production (45). Interestingly, the CTL activity observed appears to target different immunodominant epitopes: the NS3 1073-1081 epitope in Vertuani et al. (47) and the NS3 1406-1415 epitope in Thimme et al. (45). The capacity of a vaccine to induce a vigorous anti-NS3 CD8+-cell-mediated response could thus be a critical feature. As for E1 and NS5A, none of the vaccine strategies that we evaluated were capable to reveal highly imunogenic epitopes (except for E1 363-372). Apparently a low binding efficiency to HLA-A2 may explain why these epitopes are so poorly immunogenic. Interestingly, some of the most conserved sequences were found among E2, NS4B and NS5B highly immunogenic epitopes (Table 4). As recently suggested, the establishment of HCV chronicity may be linked to the early appearance of escape mutations in MHC-I-restricted epitopes due to the host immune pressure (13). Nonvariable epitopes may be so by escaping this pressure because they are, in the natural infection, subdominant. We show here that a number of these nonvariable, highly conserved epitopes can be highly immunogenic and dominant in different vaccine contexts. Their inclusion in a therapeutic vaccine formulation may thus be attractive.
Another major observation of our study is that the selection of the antigenic sequence to be expressed by a vaccine vector is of critical importance with respect to the scope of epitope recognition, whereas the choice of the vector itself appears to be much less important in this regard. Our data suggest that for two of HCV key vaccine candidates, Core and NS3 (Core because it is the most conserved viral antigens and NS3 for the reasons outlined above), coexpression of flanking genes is essential to efficiently present otherwise subdominant epitopes. Three studies, performed in HLA-A2.1-transgenic mice, have so far shown that vaccines expressing the NS3 protein alone (based on DNA, recombinant Semliki Forest virus particles, or recombinant Salmonella enterica serovar Typhimurium or recombinant adenoviruses [6, 48]) indeed induce both CTL and IFN-γ-producing cells specific of a single dominant epitope (NS3 1073-1081). Epitope flanking sequences have been described to have the capacity to modulate proteasome processing and thus epitope presentation (30, 41). We show here that flanking gene sequences or domains, although located distantly, can have an influence on the immunogenicity of certain epitopes. For NS3, it has been documented that coexpression with NS4A plays a major role in the stability of the antigen (49). As for Core, our in vitro results demonstrate that coexpression of E1 and/or E2 increase the amount of Core detected. This may be explained by an increase of Core stability, although this hypothesis remains to be confirmed to distinguish between a difference in expression levels, RNA stability, or protein stability. When expressed in the context of the full-length polyprotein or of various polyprotein precursors, Core has been shown to display an endoplasmic reticulum (ER)-bound localization (similar to what has been reported for NS3 when coexpressed with NS4A), whereas it translocates into the nucleus when expressed alone (25). An increased stability of the proteins located in the ER may explain why epitope processing and consequently presentation is more efficient, as observed in our transgenic mice when larger plasmids were used. It is well known that efficient presentation of class I-restricted viral peptides and therefore induction of optimized immune responses depends on multiple factors. These include peptide affinity for class I molecules and stability of the MHC-peptide complex, as well as upstream events controlling peptide production in the cytosol and their transport to the ER (10, 23). Antiviral immune responses often focus CTL responses to a narrow spectrum of immunodominant epitopes (51) Our data further support recent literature describing that although DNA vaccines have long been described as potent inducers of cell-mediated immune responses; unless such responses are carefully scrutinized in terms not only of their vigor but also of the breadth of epitope recognition, these vaccines may not be able to induce the kind of immunity needed to achieve protection against or resolution of infection (39, 50).
As shown, and in agreement with recent data obtained in the human immunodeficiency virus or Ebola virus models (42, 43), another stategy for potentiating vaccine-induced T-cell responses is to combine priming by DNA vaccination with a booster injection of a recombinant adenovirus. We demonstrated here that when both types of vectors express antigenic sequences designed to allow a maximun number of epitopes to be presented (such as Core-E1-E2), a potent immune response can be observed that is simultaneously directed at multiple epitopes, including epitopes that are otherwise subdominant in the context of single-vector vaccinations (Fig. 3).
Finally, our study reveals the unique nature of two HCV-encoded epitopes (NS4B 1769-1777 and NS4B 1807-1816) in that these epitopes, in all of the vaccine contexts tested, displayed a predominant and particularly potent capacity to induce the proliferation of IFN-γ-producing cells. Interestingly, these two epitopes are clustered in a single antigen (NS4B). These observations have been corroborated in at least one study performed with patients' cells (33). The design of a vaccine capable to induce the secretion of IFN-γ or other cytokines in association with only a limited or no T-cell cytolytic activity may be particularly attractive in the context of a therapeutic intervention in which exacerbation of inflamation and liver cell lysis should be avoided (3). In other viral models, IFN-γ has been shown to influence viral load levels by inhibiting directly viral replication in infected cells (34).
Whether an HCV vaccine will be based on synthetic peptides or on some other form of immunogen, it is obvious that an in-depth knowledge of epitopes composing such a vaccine will be a prerequisite for a most rational vaccine design. In contrast to the human immunodeficiency virus situation (1), it is not yet clear whether the dominant epitopes targeted by the responses to HCV in the acute setting are different from those recognized during chronic disease. It is now critical to identify epitopes recognized by those early responses and to evaluate them in combination with already-identified ones in the setting of various vaccine strategies.
.
Acknowledgments
We are grateful to C. Bain, C. Brinster, C. Wychowsky, M. L. Michel, and J. Dubuisson for constructive discussion during the course of this work. We thank J. P. Lavergne for help with sequence alignments.
This study was supported by a grant from the ARC and the European Community (QLRT-PL 1999-00356).
REFERENCES
- 1.Altfeld, M., E. S. Rosenberg, R. Shankarappa, J. S. Mukherjee, F. M. Hecht, R. L. Eldridge, M. M. Addo, S. H. Poon, M. N. Phillips, G. K. Robbins, P. E. Sax, S. Boswell, J. O. Kahn, C. Brander, P. J. Goulder, J. A. Levy, J. I. Mullins, and B. D. Walker. 2001. Cellular immune responses and viral diversity in individuals treated during acute and early HIV-1 infection. J. Exp. Med. 193:169-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Battegay, M., J. Fikes, A. M. Di Bisceglie, P. A. Wentworth, A. Sette, F. Celis, W. M. Ching, A. Grakoui, C. M. Rice, and K. Kurokohchi. 1995. Patients with chronic hepatitis C have circulating cytotoxic T cells which recognize hepatitis C virus-encoded peptides binding to HLA-A2.1 molecules. J. Virol. 69:2462-2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertoletti, A., and M. K. Maini. 2000. Protection or damage: a dual role for the virus-specific cytotoxic T lymphocyte response in hepatitis B and C infection? Curr. Opin. Immunol. 12:403-408. [DOI] [PubMed] [Google Scholar]
- 4.Boyer, J. D., B. Wang, K. E. Ugen, M. Agadjanyan, A. Javadian, P. Frost, K. Dang, R. A. Carrano, R. Ciccarelli, L. Coney, W. V. Williams, and D. B. Weiner. 1996. In vivo protective anti-HIV immune responses in non-human primates through DNA immunization. J. Med. Primatol. 25:242-250. [DOI] [PubMed] [Google Scholar]
- 5.Brinster, C., S. Muguet, Y. C. Lone, D. Boucreux, N. Renard, A. Fournillier, F. Lemonnier, and G. Inchauspe. 2001. Different hepatitis C virus nonstructural protein 3 (NS3)-DNA-expressing vaccines induce in HLA-A2.1 transgenic mice stable cytotoxic T lymphocytes that target one major epitope. Hepatology 34:1206-1217. [DOI] [PubMed] [Google Scholar]
- 6.Brinster, C., M. Chen, D. Boucreux, G. Paranhos-Baccala, P. Liljestrom, F. Lemmonier, and G. Inchauspe. 2002. Hepatitis C virus non-structural protein 3-specific cellular immune responses following single or combined immunization with DNA or recombinant Semliki Forest virus particles. J. Gen. Virol. 83:369-381. [DOI] [PubMed] [Google Scholar]
- 7.Calarota, S., G. Bratt, S. Nordlund, J. Hinkula, A. C. Leandersson, E. Sandstrom, and B. Wahren. 1998. Cellular cytotoxic response induced by DNA vaccination in HIV-1-infected patients. Lancet 2:1320-1325. [DOI] [PubMed] [Google Scholar]
- 8.Cerny, A., J. G. McHutchison, C. Pasquinelli, M. E. Brown, M. A. Brothers, B. Grabscheid, P. Fowler, M. Houghton, and F. V. Chisari. 1995. Cytotoxic T lymphocyte response to hepatitis C virus-derived peptides containing the HLA A2.1 binding motif. J. Clin. Investig. 95:521-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chartier, C., E. Degryse, M. Gantzer, A. Dieterle, A. Pavirani, and M. Mehtali. 1996. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J. Virol. 70:805-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, W., S. Khilko, J. Fecondo, D. H. Margulies, and J. McCluskey. 1994. Determinant selection of major histocompatibility complex class I-restricted antigenic peptides is explained by class I-peptide affinity and is strongly influenced by nondominant anchor residues. J. Exp. Med. 180:1471-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper, S., A. L. Erickson, E. J. Adams, J. Kansopon, A. J. Weiner, D. Y. Chien, M. Houghton, P. Parham, and C. M. Walker. 1999. Analysis of a successful immune response against hepatitis C virus. Immunity 10:439-449. [DOI] [PubMed] [Google Scholar]
- 12.Diepolder, H. M., J. T. Gerlach, R. Zachoval, R. M. Hoffmann, M. C. Jung, E. A. Wierenga, S. Scholz, T. Santantonio, M. Houghton, S. Southwood, A. Sette, and G. R. Pape. 1997. Immunodominant CD4+ T-cell epitope within nonstructural protein 3 in acute hepatitis C virus infection. J. Virol. 71:6011-6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erickson, A. L., Y. Kimura, S. Igarashi, J. Eichelberger, M. Houghton, J. Sidney, D. McKinney, A. Sette, A. L. Hughes, and C. M. Walker. 2001. The outcome of hepatitis C virus infection is predicted by escape mutations in epitopes targeted by cytotoxic T lymphocytes. Immunity 15:883-895. [DOI] [PubMed] [Google Scholar]
- 14.Fournillier, A., C. Wychowski, D. Boucreux, T. F. Baumert, J. C. Meunier, D. Jacobs, S. Muguet, E. Depla, and G. Inchauspe. 2001. Induction of hepatitis C virus E1 envelope protein-specific immune response can be enhanced by mutation of N-glycosylation sites. J. Virol. 75:2088-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grabowska, A. M., F. Lechner, P. Klenerman, P. J. Tighe, S. Ryder, J. K. Ball, B. J. Thomson, W. L. Irving, and R. A. Robins. 2001. Direct ex vivo comparison of the breadth and specificity of the T cells in the liver and peripheral blood of patients with chronic HCV infection. Eur. J. Immunol. 31:2388-2394. [DOI] [PubMed] [Google Scholar]
- 16.Gruener, N. H., F. Lechner, M. C. Jung, H. Diepolder, T. Gerlach, G. Lauer, B. Walker, J. Sullivan, R. Phillips, G. R. Pape, and P. Klenerman. 2001. Sustained dysfunction of antiviral CD8+ T lymphocytes after infection with hepatitis C virus. J. Virol. 75:5550-5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He, X.-S., B. Rehermann, F. X. Lopez-Labrador, J. Boisvert, R. Cheung, J. Mumm, H. Wedemeyer, M. Berenguer, T. L. T. L., T. L. Wright, M. M. Davis, and H. B. Greenberg. 1999. Quantitative analysis of hepatitis C virus-specific CD8+ T cells in peripheral blood and liver using peptide-MHC tetramers. Proc. Natl. Acad. Sci. USA 96:5692-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hel, Z., Tsai, W. P., A. Thornton, J. Nacsa, L. Giuliani, E. Tryniszewska, M. Poudyal, J. Tartaglia, G. N. Pavlakis, and G. Franchini. 2001. Potentiation of simian immunodeficiency virus (SIV)-specific CD4+ and CD8+ T-cell responses by a DNA-SIV and NYVAC-SIV prime/boost regimen. J. Immunol. 167:7180-7191. [DOI] [PubMed] [Google Scholar]
- 19.Kato, N., M. Hijikata, Y. Ootsuyama, M. Nakagawa, S. Ohkoshi, T. Sugimura, and K. Shimotohno. 1990. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc. Natl. Acad. Sci. USA 87:9524-9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koziel, M. J., D. Dudley, N. Afdhal, A. Grakoui, C. M. Rice, Q. L. Choo, M. Houghton, and D. B. Walker. 1995. HLA class I-restricted cytotoxic T lymphocytes specific for hepatitis C virus: identification of multiple epitopes and characterization of patterns of cytokine release. J. Clin. Investig. 96:2311-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lechner, F., D. Wong, P. Dunbar, R. Chapman, R. Chung, P. Dohrenwend, G. Robbins, R. Philips, P. Klenerman, and B. Walker. 2000. Analysis of a successful immune response in persons infected with hepatitis C virus. Exp. Med. 191:1499-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lechner, F., N. H. Gruener, S. Urbani, J. Uggeri, T. Santantonio, A. R. Kammer, A. Cerny, R. Philips, C. Ferrari, G. R. Pape, and P. Klenerman. 2000. CD8+ T lymphocyte responses are induced during acute hepatitis C virus infection but are not sustained. Eur. J. Immunol. 30:2479-2487. [DOI] [PubMed] [Google Scholar]
- 23.Lehner, P. J., and P. Cresswell. 1996. Processing and delivery of peptides presented by MHC class I molecules. Curr. Opin. Immunol. 8:59-67. [DOI] [PubMed] [Google Scholar]
- 24.Leitner, W. W., M. C. Seguin W. R. Ballou, J. P. Seitz, A. M. Schultz, M. J. Sheehy, and J. A. Lyon. 1997. Immune responses induced by intramuscular or gene gun injection of protective deoxyribonucleic acid vaccines that express the circumsporozoite protein from Plasmodium berghei malaria parasites. J. Immunol. 15:6112-6119. [PubMed] [Google Scholar]
- 25.Liu, Q., C. Tackney, R. A. Bhat, A. M. Prince, and P. Zhang. 1997. Regulated processing of hepatitis C virus core protein is linked to subcellular localization. J. Virol. 71:657-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lone, Y.-C., I. Motta, E. Mottez, Y. Guilloux, A. Lim, F. Demay, J.-P. Levraud, P. Kourilsky, and J.-P. Abastado. 1998. In vitro induction of specific cytotoxic T lymphocytes using recombinant single-chain MHC class I/peptide complexes. J. Immunother. 21:283-294. [DOI] [PubMed] [Google Scholar]
- 27.Lusky, M., M. Christ, K. Rittner, A. Dieterle, D. Dreyer, B. Mourot, H. Schultz, F. Stoeckel, A. Pavirani, and M. Mehtali. 1998. In vitro and in vivo biology of recombinant adenovirus vectors with E1, E1/E2A, or E1/E4 deleted. J. Virol. 72:2022-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Major, M. E., L. Vitvitski, J. Dubuisson, A. Fournillier, G. De Martynoff, C. Trepo, and G. Inchauspe. 1997. Immunization with plasmid DNA encoding hepatitis C virus envelope E2 antigenic domains induces antibodies whose immune reactivity is linked to the injection mode. J. Virol. 71:7101-7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manns, M. P., J. G. McHutchison, S. C. Gordon, V. K. Rustgi, M. Shiffman, R. Reindollar, Z. D. Goodman, K. Koury, M. Ling, and J. K. Albrecht. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958-965. [DOI] [PubMed] [Google Scholar]
- 30.Niedermann, G., E. Geier, M. Lucchiari-Hartz, N. Hitziger, A. Ramsperger, and K. Eichmann. 1999. The specificity of proteasomes: impact on MHC class I processing and presentation of antigens. Immunol. Rev. 172:24-29. [DOI] [PubMed] [Google Scholar]
- 31.Pascolo, S., N. Bervas, J. M. Ure, A. G. Smith, F. A. Lemonnier, and B. Perarnau. 1997. HLA-A2.1-restricted education and cytolytic activity of CD8+ T lymphocytes from β2 microglobulin (β2m) HLA-A2.1 monochain transgenic H-2Db β2m double knockout mice. J. Exp. Med. 185:2043-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peet, N. M., J. A. McKeating, B. Ramos T. Klonisch, J. B. De Souza, P. J. Delves, and T. Lund. 1997. Comparison of nucleic acid and protein immunization for induction of antibodies specific for HIV-1 gp120. Clin. Exp. Immunol. 109:226-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prezzi, C., M. A. Casciaro, V. Francavilla, E. Schiaffella, L. Finocchi, L. V. Chircu, G. Bruno, A. Sette, S. Abrignani, and V. Barnaba. 2001. Virus-specific CD8+ T cells with type 1 or type 2 cytokine profile are related to different disease activity in chronic hepatitis C virus infection. Eur. J. Immunol. 31:894-906. [DOI] [PubMed] [Google Scholar]
- 34.Ramsay, A. J., J. Ruby, and I. A. Ramshaw. 1993. A case for cytokines as effector molecules in the resolution of virus infection. Immunol. Today 14:155-157. [DOI] [PubMed] [Google Scholar]
- 35.Rehermann, B., and F. V. Chisari. 2000. Cell mediated immune response to the hepatitis C virus. Curr. Top. Microbiol. Immunol. 242:299-325. [DOI] [PubMed] [Google Scholar]
- 36.Rehermann, B., K. M. Chang, J. G. McHutchison, R. Kokka, M. Houghton, and F. V. Chisari. 1996. Quantitative analysis of the peripheral blood cytotoxic T lymphocyte response in patients with chronic hepatitis C virus infection. J. Clin. Investig. 98:1432-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reich, N. C., P. Sarnow, E. Duprey, and A. J. Levine. 1983. Monoclonal antibodies which recognize native and denatured forms of the adenovirus DNA-binding protein. Virology 30:480-484. [DOI] [PubMed] [Google Scholar]
- 38.Rosen, H. R., J. G. McHutchison, A. J. Conrad, J. J. Lentz, G. Marousek, S. L. Rose, A. Zaman, K. Taylor, and S. Chou. 2002. Tumor necrosis factor genetic polymorphisms and response to antiviral therapy in patients with chronic hepatitis. Am. J. Gastroenterol. 97:714-720. [DOI] [PubMed] [Google Scholar]
- 39.Santra, S., J. E. Schmitz, M. J. Kuroda, M. A. Lifton, C. E. Nickerson, C. I. Lord, R. Pal, G. Franchini, and N. L. Letvin. 2002. Recombinant canarypox vaccine-elicited CTL specific for dominant and subdominant simian immunodeficiency virus epitopes in rhesus monkeys. J. Immunol. 168:1847-1853. [DOI] [PubMed] [Google Scholar]
- 40.Seth, A., I. Ourmanov, J. E. Schmitz, M. J. Kuroda, M. A. Lifton, C. E. Nickerson, L. Wyatt, M. Carroll, B. Moss, D. Venzon, N. L. Letvin, and V. M. Hirsch. 2000. Immunization with a modified vaccinia virus expressing simian immunodeficiency virus (SIV) Gag-Pol primes for an anamnestic Gag-specific cytotoxic T-lymphocyte response and is associated with reduction of viremia after SIV challenge. J. Virol. 74:2502-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimbara, N., K. Ogawa, Y. Hidaka, H. Nakajima, N. Yamasaki, S. Niwa, N. Tanahashi, and K. Tanaka. 1998. Contribution of proline residue for efficient production of MHC class I ligands by proteasomes. J. Biol. Chem. 273:23062-23071. [DOI] [PubMed] [Google Scholar]
- 42.Shiver, J. W., T. M. Fu, L. Chen, D. R. Casimiro, M. E. Davies, R. K. Evans, Z. Q. Zhang, A. J. Simon, W. L. Trigona, S. A. Dubey, L. Huang, V. A. Harris, R. S. Long, X. Liang, L. Handt, W. A. Schleif, L. Zhu, D. C. Freed, N. V. Persaud, L. Guan, K. S. Punt, A. Tang, M. Chen, K. A. Wilson, K. B. Collins, G. J. Heidecker, V. R. Fernandez, H. C. Perry, J. G. Joyce, K. M. Grimm, J. C. Cook, P. M. Keller, D. S. Kresock, H. H., Mach, R. D. Troutman, L. A. Isopi, D. M. Williams, Z. Xu, K. E. Bohannon, D. B. Volkin, D. C. Montefiori, A. Miura, G. R. Krivulka, M. A. Lifton, M. J. Kuroda, J. E. Schmitz, N. L. Letvin, M. J. Caulfield, A. J. Bett, R. Youil, D. C. Kaslow, and E. A. Emini. 2002. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature 17:331-335. [DOI] [PubMed] [Google Scholar]
- 43.Sullivan, N. J., A. Sanchez, P. E. Rollin, Z. Y. Yang, and G. J. Nabel. 2000. Development of a preventive vaccine for Ebola virus infection in primates. Nature 30:605-609. [DOI] [PubMed] [Google Scholar]
- 44.Takaki, A., M. Wiese, G. Maertens, E. Depla, U. Eifert, A. Liebetrau, J. L. Miller, M. P. Manns, and B. Rehermann. 2000. Cellular immune responses persist and humoral responses decrease two decades after recovery from a single-source outbreak of hepatitis C. Nat. Med. 6:578-582. [DOI] [PubMed] [Google Scholar]
- 45.Thimme, R., D. Oldach, K. M. Chang, C. Steiger, S. C. Ray, and F. V. Chisari. 2001. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J. Exp. Med. 19:1395-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vertuani, S., M. Bazzaro, G. Gualandi, F. Micheletti, M. Marastoni, C. Fortini, A. Canella, M. Marino, R. Tomatis, S. Traniello, and R. Gavioli. 2002. Effect of interferon-alpha therapy on epitope-specific cytotoxic T lymphocyte responses in hepatitis C virus-infected individuals. Eur. J. Immunol. 32:144-154. [DOI] [PubMed] [Google Scholar]
- 48.Wedemeyer, H., S. Gagneten, A. Davis, R. Bartenschlager, S. Feinstone, and B. Rehermann. 2001. Oral immunization with HCV-NS3-transformed Salmonella: induction of HCV-specific CTL in a transgenic mouse model. Gastroenterology 121:1158-1166. [DOI] [PubMed] [Google Scholar]
- 49.Wolk, B., D. Sansonno, H. G. Krausslich, F. Dammacco, C. M. Rice, H. E. Blum, and D. Moradpour. 2000. Subcellular localization, stability, and trans-cleavage competence of the hepatitis C virus NS3-NS4A complex expressed in tetracycline-regulated cell lines. J. Virol. 74:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yasutomi, Y., S. Koenig, R. M. Woods, J. Madsen, N. M. Wassef, C. R. Alving, H. J. Klein, T. E. Nolan, L. J. Boots, J. A. Kessler, et al. 1995. A vaccine-elicited, single viral epitope-specific cytotoxic T lymphocyte response does not protect against intravenous, cell-free simian immunodeficiency virus challenge. J. Virol. 69:2228-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yewdell, J. W., and J. R. Bennink. 1999. Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu. Rev. Immunol. 17:51-88. [DOI] [PubMed] [Google Scholar]