Abstract
Conformational changes in the Newcastle disease virus (NDV) fusion (F) protein during activation of fusion and the role of HN protein in these changes were characterized with a polyclonal antibody. This antibody was raised against a peptide with the sequence of the amino-terminal half of the F protein HR1 domain. This antibody immunoprecipitated both F0 and F1 forms of the fusion protein from infected and transfected cell extracts solubilized with detergent, and precipitation was unaffected by expression of the HN protein. In marked contrast, this antibody detected significant conformational differences in the F protein at cell surfaces, differences that depended upon HN protein expression. The antibody minimally detected the F protein, either cleaved or uncleaved, in the absence of HN protein expression. However, when coexpressed with HN protein, an uncleaved mutant F protein bound the anti-HR1 antibody, and this binding depended upon the coexpression of specifically the NDV HN protein. When the cleaved wild-type F protein was coexpressed with HN protein, the F protein bound anti-HR1 antibody poorly although significantly more than F protein expressed alone. Anti-HR1 antibody inhibited the fusion of R18 (octadecyl rhodamine B chloride)-labeled red blood cells to syncytia expressing HN and wild-type F proteins. This inhibition showed that fusion-competent F proteins present on surfaces of syncytia were capable of binding anti-HR1. Furthermore, only antibody which was added prior to red blood cell binding could inhibit fusion. These results suggest that the conformation of uncleaved cell surface F protein is affected by HN protein expression. Furthermore, the cleaved F protein, when coexpressed with HN protein and in a prefusion conformation, can bind anti-HR1 antibody, and the anti-HR1-accessible conformation exists prior to HN protein attachment to receptors on red blood cells.
The entry of Newcastle disease virus (NDV), a prototype paramyxovirus, is directed by two virion glycoproteins, the hemagglutinin-neuraminidase (HN) protein and the fusion (F) protein (12). HN protein, the virus attachment protein, binds to sialic acid-containing receptors, and F protein mediates membrane fusion. In contrast to many viral fusion proteins, paramyxovirus F proteins do not require the acid pH of endosomes to activate fusion activity. As a consequence, infected cells expressing both attachment proteins and F proteins can fuse with adjacent cells to form multinuclear cells, or syncytia, a process that is assumed to mimic virus-cell fusion (12).
The NDV fusion protein is synthesized as a 553-amino-acid precursor, F0 (5, 15). The mature F protein is a homotrimer (24). Fusion activity of the protein requires a proteolytic cleavage of F0 at amino acid 117 to produce disulfide-linked F2 and F1 polypeptides derived from the amino-terminal and carboxyl-terminal domains, respectively (12). The F1 polypeptide has several domains important for fusion activity. The protein has one and perhaps two fusion peptides (20), one located at the amino terminus of the F1 polypeptide (reviewed in reference 12) and the other at a more internal location (20). Upon activation of fusion, fusion peptides are thought to insert into target membranes docking the protein to these membranes (reviewed in references 4, 6, and 21). Paramyxovirus F1 proteins have two heptad repeat (HR) regions, one (HR1) located just carboxyl terminal to the more amino-terminal fusion peptide and the other adjacent to the transmembrane domain (HR2) (reviewed in references 11 and 12). Mutational analyses of both the NDV HR1 and HR2 domains have shown that both domains are important in the fusion activity of the protein (16, 23, 25, 29). These domains have also been implicated in fusion activity by analysis of the structure and function of peptides with sequences of these domains (8, 13, 22, 37-39). Results of these studies, as well as similar studies of human immunodeficiency virus and influenza virus (4), have led to the hypothesis that fusion proteins are synthesized and transported to cell surfaces in a metastable conformation. In this conformation the HR domains are not associated and the fusion peptide is masked (4, 21, 36). Upon fusion activation, the F proteins are thought to undergo a series of conformational changes that result in insertion of fusion peptides into target membranes and the interaction of the HR1 and HR2 domains to form a very stable complex (1, 4, 21). This structure is a six-stranded coiled coil composed of a central core trimer of HR1 domains with three HR2 domains bound to the surfaces of the core trimer (1, 40). The formation of this coiled coil is thought to pull target and attack membranes into close proximity, allowing subsequent fusion events (1, 40).
The attachment protein is required for fusion in most paramyxovirus systems, since most F proteins expressed alone do not mediate membrane fusion (11). It is also clear that HN protein provides more than an attachment function, since mutations in the HN protein stalk domain can retain attachment activity but are defective in fusion promotion (26, 27, 30). The HN protein and the F protein must be from the same virus, with a few exceptions, indicating that virus-specific interactions between HN and F proteins (7) are required for fusion directed by the F protein. It has been proposed that the attachment of HN protein to its receptor serves to activate the F protein (11). The nature of this activation is, however, undefined.
With the goal of defining conformational changes in the F protein during activation of fusion and the role of the HN protein in those changes, we raised a polyclonal antibody against a peptide with the sequence of the amino-terminal half of the HR1 domain. This antibody allowed us to show that the conformation of cell surface NDV F protein is altered by coexpression of the HN protein. Evidence suggesting that HN protein affects the conformation of F protein prior to both F protein cleavage activation and attachment of HN protein to its receptor is presented.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
Cos-7 cells, obtained from the American Type Culture Collection, were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with nonessential amino acids, vitamins, penicillin, and streptomycin, and 10% fetal calf serum. Stocks of NDV strains AV, a virulent strain, and B1, an avirulent strain, (12, 19, 34), were grown in embryonated chicken eggs and purified by standard protocols.
NDV F genes were inserted into pSVL (Pharmacia) as previously described (29). The cleavage mutant F-K115G has been described (14). The Sendai virus HN protein gene, obtained from D. Nayak, and the measles virus hemagglutinin (HA) gene, obtained from M. Oldstone, each were inserted into pSVL.
Anti-NDV antibody was raised in rabbits against UV-inactivated stocks of NDV strain AV by standard protocols. Anti-Ftail was raised against a synthetic peptide with the sequence of the cytoplasmic domain of the F protein as previously described by Wang et al. (35) and prepared by the Peptide Core Facility of the University of Massachusetts Medical School.
To raise anti-HR1 antibody, sequences encoding amino acids 132 to173 were prepared by PCR using a primer containing a BamHI site and a primer with an EcoRI site as well as the appropriate F protein gene sequences. The PCR product was cloned into BamHI-EcoRI-cut pGex-2T (Pharmacia), and the ligated product was transformed into BL21 (Stratagene). BL21 cells containing the plasmid were induced with IPTG (isopropyl-β-d-thiogalactopyranoside; 0.1 mM) for 3 h at 37°C. The cells were pelleted and lysed with BugBuster (Novagen) by using protocols recommended by the manufacturer, and the glutathione S-transferase (GST) fusion protein was purified with GST-Bind resin (Novagen) by standard protocols. The purified, concentrated fusion protein was used for production of antisera by Capralogics (Hardwick, Mass.).
Transfections.
Transfections using Lipofectamine (Invitrogen) were done as recommended by the manufacturer. Briefly, Cos-7 cells were plated at 3 × 105 per 35-mm plate. Twenty hours later, the cells were transfected. For each 35-mm plate, mixtures of DNA in 0.1 ml of OptiMEM (BRL/Gibco) and 10 μl of transfection reagent in 0.2 ml of OptiMEM were incubated at room temperature for 40 min, diluted with 0.7 ml of OptiMEM, and added to a plate previously washed twice with 2 ml of OptiMEM. Cells were incubated for 5 h, DNA was removed, and 2 ml of supplemented DMEM was added. Transfection mixtures contained 0.5 μg of each DNA/35-mm plate, but all transfection mixtures contained a total of 1 μg of DNA/35-mm plate. When pSVL-Fwt or pSVL-F-K115G was expressed alone, 0.5 μg of vector was also added to keep DNA concentrations the same in all transfections.
Radiolabeling and immunoprecipitation of protein.
At 48 h posttransfection, cells were radiolabeled for 2 h at 37°C in DMEM lacking methionine but containing 100 μCi of [35S]methionine (Amersham) per ml. For a nonradioactive chase, cells were placed in complete medium. For infected cells, virus was added at a multiplicity of infection of 15. Cells were radioactively labeled at 5 h postinfection as described for transfected cells. At the end of the labeling period, cells were washed in PBS and lysed in RSB buffer (0.01 M Tris-HCl [pH 7.4], 0.01 M NaCl) containing 1% Triton X-100, 0.5% sodium deoxycholate, 2 mg of N-ethyl maleimide per ml, and 0.2 mg of DNase per ml. Immunoprecipitation of NDV proteins was immediately accomplished as previously described (28, 30).
Immunofluorescence of live cells.
Cos-7 cells were plated on 35-mm plates containing glass coverslips and transfected as described above. Forty-eight hours after transfection, cells were incubated at 4°C in primary unlabeled antibody diluted in phosphate-buffered saline (PBS) containing 1% bovine serum albumin (BSA) and 0.02% sodium azide for 1 h. Cells were then washed three times with PBS containing BSA and azide and incubated for 1 h with anti-rabbit immunoglobulin G (IgG) coupled to Alexa-488 (Molecular Probes) diluted in PBS-BSA-azide. Cells were washed in ice-cold PBS-BSA-azide and then fixed in 1% paraformaldehyde. Coverslips were mounted on slides, and cells were photographed immediately with a Nikon Diaphot 300 fluorescence microscope.
Flow cytometry.
Transfected cells were incubated overnight in DMEM without CaCl2. Cells were removed from plates with cell detachment buffer (Sigma Co.), washed in PBS containing 1% BSA and 0.02% azide (fluorescence-activated cell sorting [FACS] buffer) and then incubated with anti-HR1 antibody for 1 h at 4°C (1/200 dilution). After cells had been washed three times with FACS buffer, they were incubated for 1 h at 4°C with goat anti-rabbit IgG coupled to Alexa-488 (Molecular Probes). After incubation, cells were washed, resuspended in PBS containing 2% paraformaldehyde, and subjected to flow cytometry (University of Massachusetts Medical School Flow Cytometry Facility). Cells transfected with vector alone and incubated with both primary and secondary antibody were used as controls.
Fusion assays using R18-labeled RBC.
The protocol used for fusion assays was similar to that previously described (9, 10). Briefly, avian red blood cells (RBC) (Crane Laboratories) were washed in PBS and then incubated with 15 μg of R18 (octadecyl rhodamine B chloride) (Molecular Probes) per ml for 30 min at room temperature in the dark. Three volumes of complete medium (DMEM with 10% fetal calf serum) were added, and incubation was continued for 30 min. The RBC were then washed four times in ice-cold PBS, resuspended in PBS containing CaCl2 (0.01%), and added to transfected cells that had been grown on coverslips and washed in PBS with CaCl2. Transfected cells were incubated with labeled RBC for 30 min on ice. Cells were washed with ice-cold PBS containing CaCl2 and then incubated at 37°C. After incubation, cells were washed in cold PBS containing CaCl2 and immediately visualized and photographed with a Nikon Diaphot 300 fluorescence microscope.
RESULTS
Immunoprecipitation of F protein from cell lysates.
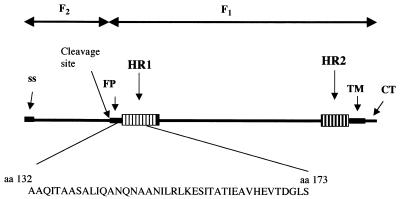
The nucleic acid sequence encoding the amino-terminal region of the NDV F protein HR1 domain (amino acids 132 to 173) (Fig. 1) was cloned in frame at the carboxyl terminus of the gene encoding GST, and the resulting fusion protein was expressed in Escherichia coli. Polyclonal antibodies against the purified fusion protein were raised in rabbits. The HR1 sequences chosen were derived from a region of the NDV F protein that was not resolved in the crystal structure of the F protein (2). However, results of mutagenesis of this region of the F protein showed that it was clearly involved in the fusion activity of the protein (25, 29).
FIG. 1.
F protein sequences used to raise antibody. A diagram of the primary sequence of the NDV F protein with important domains is shown. The sequence encoding amino acids 132 to 173 was cloned in frame to the carboxyl terminus of GST. The resulting purified fusion protein was used to raise antibodies. SS, signal sequence; FP, fusion peptide; TM, transmembrane domain; CT, cytoplasmic domain.
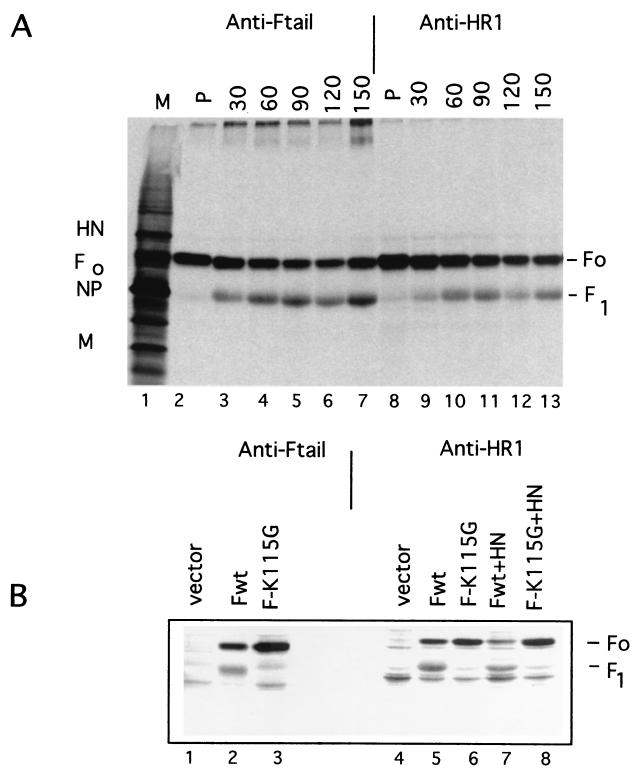
The resulting polyclonal antibodies readily precipitated F proteins from infected cell extracts. Figure 2A shows the precipitation of proteins from infected cell extracts pulse-labeled with [35S]methionine as well as extracts derived from cells that were pulse-labeled and subjected to increasing times of a nonradioactive chase. Precipitation of proteins using a previously characterized antibody specific for the cytoplasmic tail of the F protein is also shown as a positive control. Clearly, the anti-HR1 antibody precipitated the nascent F0 protein as well as the F1-sized polypeptide that appeared during a nonradioactive chase as the protein matured through the cell. The antibody did preferentially precipitate F0.
FIG. 2.
Immunoprecipitation of F protein from cell extracts. (A) Cells infected with NDV strain AV were radioactively labeled for 5 min (P) and then subjected to a nonradioactive chase for the times (in minutes) indicated at the top. Proteins were immunoprecipitated using anti-Ftail or anti-HR1. The proteins precipitated were resolved on 10% polyacrylamide gels in the presence of a reducing agent. Localization of viral proteins in the polyacrylamide gel lane containing infected cell extracts (M, marker proteins) is noted for molecular weight markers. F0, uncleaved F protein; F1, cleaved F protein; NP, nucleocapsid protein; M, membrane protein. (B) Cells were transfected with pSVL (vector), pSVL-Fwt, pSVL-F-K115G, pSVL-Fwt plus pSVL-HN, or pSVL-F-K115G plus pSVL-HN (0.5 μg of each DNA/35-mm plate of Cos cells). After 48 h, cells were radioactively labeled for 2 h and chased in nonradioactive medium for 4 h. Proteins in cells extracts were precipitated with anti-Ftail or anti-HR1.
Similar results were obtained with extracts from transfected cells (Fig. 2B). Cos cells were transfected with a gene encoding the wild-type F protein, a protein with a furin recognition sequence at the cleavage site, and a gene encoding F-K115G, a protein with a mutation in the furin recognition site. Cells transfected with the wild-type protein expressed both F0 and the cleaved form of the fusion protein (F1), while the mutant gene expressed only the F0 protein (14). In addition, both F protein genes were cotransfected with a gene encoding the HN protein. Clearly, anti-HR1 antibody precipitated both F0- and F1-sized polypeptides from extracts expressing the wild-type F protein whether or not the HN protein was expressed. The anti-HR1 antibody also precipitated the F0 protein from extracts expressing the mutant uncleaved F protein, and HN protein coexpression had little effect on the efficiency of precipitation.
Thus, in cell extracts in which membranes are dissolved with detergent, the HR1 domain of the uncleaved F0 protein as well as the cleaved F1 polypeptide was accessible to antibody and coexpression of the HN protein had little effect on the reactivity of this domain to antibody.
Immunofluorescence of intact cells expressing the F protein.
Hypothesized conformational changes in the F protein associated with the onset of fusion occur in a membrane-associated F protein. Therefore, we characterized anti-HR1 antibody binding to F protein expressed on infected or transfected cell surfaces. Figure 3 shows surface immunofluorescence of cells infected with a virulent strain of NDV, which expresses primarily the cleaved F protein (strain AV), and cells infected with an avirulent strain of NDV which expresses only the uncleaved F protein (strain B1). Clearly, cells infected with either virus bound anti-HR1. However, cells infected with the virulent strain of NDV were not as brightly fluorescent as cells infected with the avirulent strain of NDV.
FIG. 3.
Surface immunofluorescence of NDV-infected cells. Cells infected for 6 h with NDV strain AV or strain B1, and uninfected cells were incubated with either anti-HR1 or anti-NDV antibody as described in Materials and Methods. Cells reacted with anti-HR1 were exposed to films for 4 s, while cells reacted with anti-NDV were exposed to film for 2 s. Fields shown are representative of the entire culture, and each field is representative of at least three separate experiments.
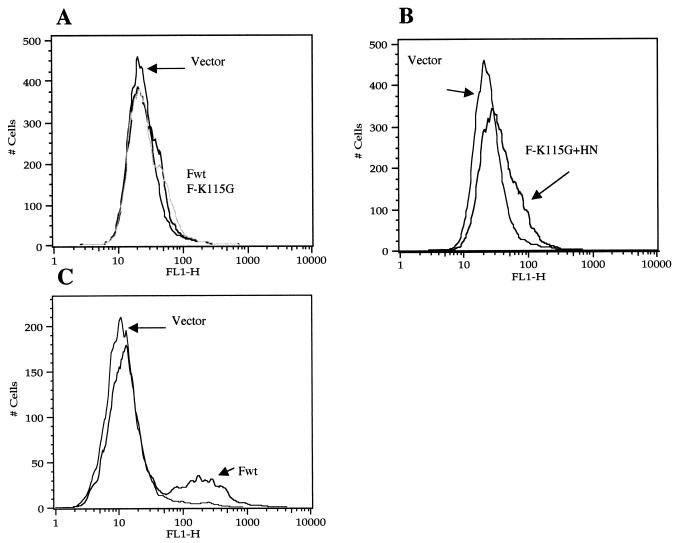
Figure 4 shows immunofluorescence of transfected cells. Cells transfected only with the gene encoding F-K115G are shown in Fig. 4C and D. Cells expressing only the uncleaved F protein bound anti-HR1 antibody very poorly (Fig. 4C), although cells transfected with this gene were clearly expressing F protein, as shown by using anti-NDV antibody (Fig. 4D). Surprisingly, cells expressing both the uncleaved F-K115G protein and the HN protein were strongly positive for anti-HR1 binding (Fig. 4E). This result was verified by FACS analysis (Fig. 5B). Cells expressing the uncleaved F-K115G protein alone were minimally reactive to anti-HR1, while cells expressing both the uncleaved F protein and the HN protein bound antibody. Cells expressing F-K115G were, however, positive by flow cytometry using anti-NDV antibody (data not shown). This result is consistent with the idea that coexpression of HN protein alters the conformation of F protein on cell surfaces.
FIG. 4.
Surface immunofluorescence of transfected cells. Cells transfected with pSVL (vector), pSVL-F-K115G, pSVL-F-K115G plus pSVL-HN, pSVL-Fwt, or pSVL-Fwt plus pSVL-HN (0.5 μg of each DNA/35-mm plate of Cos cells) were incubated with either anti-NDV or anti-HR1 as described in Materials and Methods. Film was exposed for 2 s (B, D, F, H, and J) or 3 s (A, C, E, G, and I). The field shown in panel I was also exposed for 7 s (K). Fields shown are representative of the entire culture, and each is representative of at least four separate experiments.
FIG. 5.
Detection of cell surface F protein by flow cytometry. At 48 h after transfection (0.5 μg of each DNA/35-mm plate), cells were prepared for FACS as described in Materials and Methods. (A and B) Anti-HR1 antibody; (C) anti-NDV antibody. (A) pSVL (vector), pSVL-Fwt, and pSVL-F-K115G; (B) pSVL and pSVL-F-K115G plus pSVL-HN; (C) pSVL (vector) and pSVL-Fwt. FL1-H, fluorescence. The results are representative of four different experiments.
Figures 4G and 5A show that, like uncleaved F protein, cleaved cell surface wild-type F (Fwt) protein expressed alone was unreactive to anti-HR1 antibody. The F protein was, however, accessible to binding anti-NDV antibody, as shown by immunofluorescence (Fig. 4H) and by flow cytometry (Fig. 5C). The protein was also detected by immunofluorescence and flow cytometry using anti-Fu1a, a monoclonal antibody specific for the F protein (18, 27) (data not shown). Coexpression of HN protein with this F protein resulted in syncytia (Fig. 4J). While the syncytia did bind anti-HR1 antibody (Fig. 4I and K), the intensity of fluorescence was significantly lower than that seen with uncleaved F protein coexpressed with HN protein. Flow cytometry of cells expressing HN and F proteins was not attempted because of the fragility of syncytia.
Immunofluorescence of intact cells expressing the F protein in the presence of heterologous attachment proteins.
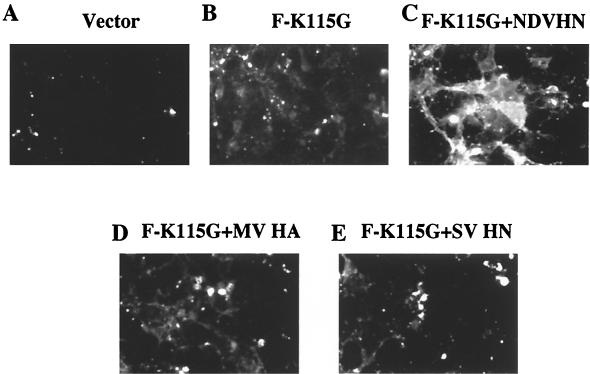
It was possible that the effect of HN protein on accessibility of the F protein to anti-HR1 antibody was unrelated to the fusion promotion activity of the HN protein. For example, the neuraminidase activity of the HN protein could increase the accessibility of the F protein by removal of sialic acid from cell surfaces. We therefore determined if the effect of HN protein expression on F protein conformation was virus specific by coexpressing the NDV F protein with the HA protein of measles virus (Fig. 6D) or the HN protein of Sendai virus (Fig. 6E). Binding of the anti-HR1 antibody to the uncleaved F protein expressed with the heterologous attachment proteins was similar to that observed when F protein was expressed alone (Fig. 6B) and much less than that observed when F protein was expressed with the NDV HN protein (Fig. 6C).
FIG. 6.
Surface immunofluorescence of cells transfected with heterologous attachment proteins. Cells were transfected with DNA encoding F-K115G or cotransfected with F-K115G and either NDV HN protein, Sendai virus (SV) HN protein, or measles virus (MV) HA protein. After 48 h, cells were incubated with anti-HR1 antibody and then goat anti-rabbit antibody coupled to a fluorescence dye as described in Materials and Methods. (A) Vector; (B) pSVL-F-K115G; (C) pSVL-F-K115G plus pSVL-HN (NDV); (D) pSVL-F-K115G plus pSVL-MV HN; (E) pSVL-F-K115G plus pSVL-SV HA. All panels show 2-s exposures. Fields shown are representative of the entire culture.
We also found that binding of anti-HR1 antibody to F protein that had been either cleaved or uncleaved and expressed alone was not increased by prior treatment of cells with neuraminidase (data not shown).
Inhibition of fusion by anti-HR1 antibody.
The decreased binding of anti-HR1 to cells expressing the cleaved F protein with HN protein may indicate that the antibody will only bind residual uncleaved F protein at cell surfaces. Alternatively, anti-HR1 may bind a cleaved form of the F protein present in smaller amounts, such as F protein in a prefusion conformation. Syncytia expressing both HN protein and the cleaved F protein likely contain a significantly reduced population of F protein molecules that remain in such a prefusion conformation. However, if there are such molecules present and if anti-HR1 can bind to them, then the anti-HR1 antibody may inhibit fusion mediated by these molecules.
To determine whether anti-HR1 antibody could inhibit the fusion activity of the F protein, we characterized the effect of antibody on the fusion of RBC with Cos-7 cells coexpressing the HN and F proteins. Initial steps in fusion can be monitored by the transfer of fluorescence-labeled lipid from RBC membranes to the membranes of cells expressing viral fusion proteins, as shown in Fig. 7. Cos cells transfected with genes encoding the HN protein and the wild-type F protein form large syncytia which will bind RBC (data not shown). Upon incubation at 37°C in the presence of anti-NDV antibody, no fusion was observed (Fig. 7A), while during incubation at 37°C without antisera, transfected cells fused with the RBC and the fluorescence-labeled lipid in RBC spread into the syncytia (Fig. 7B). Over 80% of syncytia were positive for fusion with RBC (Fig. 7E). This result shows that there are F proteins expressed on the surfaces of the syncytia that are capable of directing fusion upon binding of the RBC and are therefore in a prefusion conformation.
FIG. 7.
Inhibition of fusion by anti-HR1 antibody. At 48 h after transfection of Cos cells with pSVL-Fwt and pSVL-HN, monolayers were incubated with R18-labeled RBC on ice in the absence of antibody followed by the addition of anti-NDV and incubation for 1 h at 37°C (A) or on ice followed by incubation at 37°C for 1 h in the absence of antisera (B). (C) Cells coexpressing HN and F proteins that were incubated with anti-HR1 on ice for 30 min, followed by incubation on ice with anti-HR1 and RBC for 30 min and then by incubation at 37°C for 1 h in the presence of anti-HR1. (D) Cells were treated similarly to those in panel C except that a preimmune serum was used. Each field shows a single syncytium. (E) Quantitation of results. Each column represents at least 300 syncytia counted in three separate experiments. Ab, antibody.
To determine if anti-HR1 antibody could inhibit this fusion, the antibody was present prior to, during, and after RBC binding. The antibody did not interfere with RBC binding (data not shown). The antibody clearly inhibited fusion of the RBC with syncytia (Fig. 7C and E). Over 80% of all syncytia were negative for fusion after incubation at 37°C. This inhibition is not due to a nonspecific effect of IgG, since addition of preimmune sera did not inhibit RBC fusion with syncytia (Fig. 7D and E).
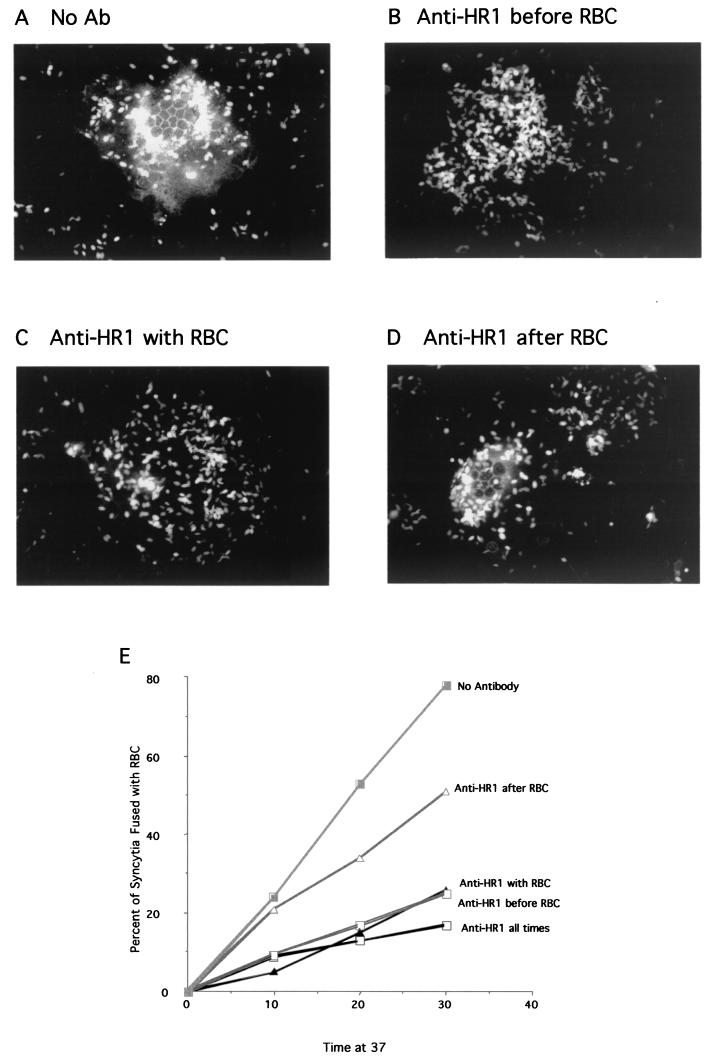
Inhibition of fusion by anti-HR1 antibodies could be due to antibody binding before or after attachment of RBC to cell surfaces. To determine when the binding site was accessible relative to attachment, antibody was added to monolayers of cells cotransfected with HN and Fwt cDNAs only prior to RBC binding, only during RBC binding, or only after RBC binding (Fig. 8). Clearly, if antibody was present only prior to RBC binding, fusion was inhibited (Fig. 8B and E). If antibody was present only during RBC binding, fusion was also inhibited (Fig. 8C and E). However, if antibody was present only after RBC binding, there was less inhibition (Fig. 8D and E). These results were quantitated at three times after incubation at 37°C (Fig. 8E). These results show that antibody can inhibit if bound only prior to the attachment of RBCs to the surfaces of HN and F protein-expressing cells. Furthermore, inhibition is greater than that observed if antibody is added only after RBC.
FIG.8.
Anti-HR1 antibody bound prior to RBC addition inhibits fusion. Shown is fusion of R18-labeled RBC to cells expressing HN and F proteins in the presence of anti-HR1 antibody (Ab) added at different times relative to the RBC. At 48 h after transfection of Cos cells with pSVL-Fwt and pSVL-HN, monolayers were incubated with R18-labeled RBC with or without antibody followed by incubation at 37°C. (A) Cells without added antibody. (B) Cells incubated with anti-HR1 antibody on ice for 30 min only prior to addition of RBC. These cells were washed three times to remove unbound antibody and then were incubated with R18-labeled RBC on ice for 30 min followed by incubation at 37°C. (C) Cells incubated with RBC and antibody together. Both unbound antibody and RBC were removed in three washes, and cells were incubated at 37°C. (D) Cells incubated with RBC on ice. Unbound RBC were removed, anti-HR1 antibody was added, and then the cells were shifted to 37°C. (E) Cells transfected with HN and F protein cDNAs were treated as for panels A to D, and the percentage of syncytia fused to RBC was determined after 10, 20, or 30 min at 37°C. Each point is the result for over 100 syncytia. The data are the combined results of three separate experiments.
DISCUSSION
It has been hypothesized that, like other fusion proteins, the paramyxovirus F protein is folded into a metastable conformation, and, upon activation of fusion, the F protein undergoes a cascade of conformational changes resulting in the formation of a very stable, six-stranded coiled coil composed of HR1 and HR2 domains (2). To explore potential conformational changes in the F protein during onset of fusion and the role of the HN protein in that process, we characterized the binding and fusion inhibition of antibody specific for the HR1 domain of the NDV F protein. This antibody immunoprecipitated the F0 and F1 forms of the protein from cell extracts solubilized with detergent, and precipitation of F protein was unaffected by expression of the HN protein. Antibody against a comparable domain of the simian virus 5 (SV5) F protein was raised and would precipitate only the uncleaved form of the protein (3). This different result may be attributed to differences in the conformation of the NDV F protein and the SV5 F protein in cell lysates. The anti-HR1 antibody raised against NDV sequences did preferentially precipitate the uncleaved F protein. Furthermore, we observed that precipitation of F1 was preferentially lost as cell extracts “aged,” suggesting conformational changes in the cleaved F protein after cell lysis masked the HR1 domain in the cleaved form of the protein. For these reasons, immunoprecipitation was performed immediately upon cell lysis.
In contrast to results obtained with cell extracts, anti-HR1 antibody minimally detected F protein expressed on cell surfaces, either cleaved or uncleaved, in the absence of HN protein expression. This result suggested that without HN protein expression, the membrane-associated F protein was folded so that the HR1 domain was inaccessible to antibody binding. Perhaps in the absence of HN protein, the F protein folds into a more stable conformation in which the HR1 domain is masked rather than the metastable form that can initiate fusion. Such an interpretation is consistent with the observation that wild-type NDV F protein cannot initiate fusion, even at low levels, in the absence of HN protein expression (17, 27).
In striking contrast, the uncleaved F protein coexpressed with HN protein bound the anti-HR1 antibody, and this binding depended specifically upon the coexpression of NDV HN protein. Binding was not observed in the presence of the measles virus HA protein or the Sendai virus HN protein. This result suggests that F protein expressed with HN protein is folded so that the HR1 domain is accessible to antibody and is, therefore, in a conformation different from that of F protein expressed alone. Since fusion cannot occur prior to F protein cleavage, this result also suggests that the conformational differences in the F protein due to coexpression of HN protein are manifested prior to cleavage activation of fusion. Indeed, it has been reported that HN protein and uncleaved F protein coimmunoprecipitate, a result that suggests an interaction between HN protein and uncleaved F protein (31).
The wild-type, cleaved F protein, coexpressed with HN protein, bound anti-HR1 antibody poorly, although significantly more than cleaved F protein expressed alone. The low level of binding may be due to residual uncleaved F protein on cell surfaces. Another possibility (not mutually exclusive with the first) was that only one of several forms of the cleaved F protein was accessible to anti-HR1 binding. Cleaved F protein in the presence of HN protein may exist in at least two forms on cell surfaces, one in a prefusion form and the other in a postfusion form. It is proposed that in the postfusion conformation, the HR1 domain forms the interior trimer of the six-stranded HR1-HR2 complex (1, 3); thus, it is reasonable to suppose that the HR1 domain is inaccessible to antibody in this conformation. Indeed, Dutch et al. have presented evidence suggesting that the SV5 HR1 domain is inaccessible to a peptide antibody in the six-stranded coiled-coil complex (3). If much of the cleaved F protein exists in this postfusion conformation, there would be only low-level anti-HR1 binding, as was observed. To explore the possibility that the prefusion form of the protein could bind antibody, we determined if anti-HR1 antibody could block fusion. Indeed, RBC fused with syncytia expressing the HN and wild-type F proteins; thus, a population of F protein on syncytium surfaces is capable of directing fusion. Importantly, anti-HR1 antibody inhibited this fusion. This result is consistent with the idea that the fusion-competent, cleaved F protein on the syncytium surfaces was accessible to anti-HR1 antibody in the presence of HN protein.
Two models have been proposed for the role of HN protein in fusion (Fig. 9), models that differ in the role of attachment in inducing conformational changes in F protein. Model 1 proposes that HN and F proteins interact only after HN protein receptor binding and this interaction initiates F protein conformational changes required for fusion (11, 17). In this model, conformational differences in F protein in the presence and absence of HN protein should be manifested only after HN protein attachment. An alternative model is that HN and F proteins form a metastable complex prior to HN protein attachment (31-33), a complex that prevents the HR1-HR2 interactions that would occur in F proteins expressed without HN protein. HN protein attachment with a concomitant conformational change releases the F protein and thus stimulates the cascade of F protein conformational changes required for fusion, changes that result in the formation of the six-stranded coiled coil composed of HR1 and HR2 domains. In this model, conformational differences in F protein in the presence and absence of HN protein should be manifested prior to HN protein attachment.
FIG. 9.
Proposed relationship of F protein conformational changes to HN protein attachment. (Model 1) HN and F proteins interact only after attachment of HN protein to a receptor. Interaction stimulates a conformational change in F protein that unmasks the HR1 domain and activates fusion. (Model 2) HN and F proteins form a complex that prevents the association of HR1 and HR2 domains, leaving the HR1 domain accessible to antibody binding. Attachment of the HN protein to a receptor releases the F protein, allowing the HR1 and HR2 domains to complex, masking the HR1 domain from antibody binding.
Since anti-HR1 antibody detected a conformational difference between F proteins expressed with and without HN protein, we asked when this conformational difference was manifested, before or after HN protein attachment. We asked when the antibody could inhibit fusion relative to the binding of RBC. Clearly, inhibition occurred if antibody was bound prior to RBC addition. This result is consistent with the idea that the HR1 domain is accessible prior to attachment. Furthermore the inhibition was greater than that observed when antibody was added after the RBC, particularly during the earlier times of incubation at 37°C. That antibody added only prior to RBC binding could inhibit fusion is more consistent with model 2. However, we cannot exclude the possibility that antibody bound to postactivation forms of the F protein indirectly interferes with conformational shifts in the prefusion form of the protein.
These combined results are most consistent with model 2 (Fig. 9). F protein, either cleaved or uncleaved and coexpressed with HN protein, is folded into a conformation in which the HR1 domain is accessible to antibody binding. Upon attachment of HN protein to its receptor, the F protein initiates conformational changes necessary for the close approach of the two membranes, conformational changes that mask the HR1 domain. In contrast, F protein expressed alone folds into a conformation in which the HR1 domain is inaccessible to anti-HR1 antibody.
Acknowledgments
This work was supported by a grant from the National Institutes of Health, AI 30572.
We thank D. Nayak and M. Oldstone for the cDNAs encoding the SV and MV attachment proteins.
REFERENCES
- 1.Baker, K. A., R. E. Dutch, R. A. Lamb, and T. S. Jardetzky. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3:309-319. [DOI] [PubMed] [Google Scholar]
- 2.Chen, L., J. J. Gorman, J. McKimm-Breschkin, L. J. Lawrence, P. A. Tulloch, B. J. Smith, P. M. Colman, and M. C. Lawrence. 2001. The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure 9:255-266. [DOI] [PubMed] [Google Scholar]
- 3.Dutch, R. E., R. N. Hagglund, M. A. Nagel, R. G. Paterson, and R. A. Lamb. 2001. Paramyxovirus fusion (F) protein: a conformational change on cleavage activation. Virology 281:138-150. [DOI] [PubMed] [Google Scholar]
- 4.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 5.Gorman, J. J., A. Nestorowicz, S. J. Mitchell, G. L. Corino, and P. W. Selleck. 1988. Characterization of the sites of proteolytic activation of Newcastle disease virus membrane glycoprotein precursors. J. Biol. Chem. 263:12522-12531. [PubMed] [Google Scholar]
- 6.Hernandez, L. D., L. R. Hoffman, T. G. Wolfsberg, and J. M. White. 1996. Virus-cell and cell-cell fusion. Annu. Rev. Cell Biol. 12:627-661. [DOI] [PubMed] [Google Scholar]
- 7.Hu, X., R. Ray, and R. W. Compans. 1992. Functional interactions between the fusion protein and hemagglutinin-neuraminidase of human parainfluenza viruses. J. Virol. 66:1528-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joshi, S. B., R. E. Dutch, and R. A. Lamb. 1998. A core trimer of the paramyxovirus fusion protein: parallels to influenza virus hemagglutinin and HIV-1 gp41. Virology 248:20-34. [DOI] [PubMed] [Google Scholar]
- 9.Kemble, G. W., T. Danieli, and J. W. White. 1994. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell 76:383-391. [DOI] [PubMed] [Google Scholar]
- 10.Kemble, G. W., Y. I. Henis, and J. M. White. 1993. GPI- and transmembrane-anchored influenza hemagglutinin differ in structure and receptor binding activity. J. Cell Biol. 122:1253-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamb, R. A. 1993. Paramyxovirus fusion: a hypothesis of changes. Virology 197:1-11. [DOI] [PubMed] [Google Scholar]
- 12.Lamb, R. A., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 1305-1340. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 13.Lambert, D. M., S. Barney, A. L. Lambert, K. Guthrie, R. Medinas, D. E. Davis, T. Bucy, J. Erickson, G. Merutka, and S. R. Petteway. 1996. Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion. Proc. Natl. Acad. Sci. USA 93:2186-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, Z., T. Sergel, E. Razvi, and T. Morrison. 1998. Effect of cleavage mutants on syncytium formation directed by the wild-type fusion protein of Newcastle disease virus. J. Virol. 72:3789-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGinnes, L. W., and T. G. Morrison. 1986. Nucleotide sequence of the gene encoding the Newcastle disease virus fusion protein and comparisons of paramyxovirus fusion protein sequences. Virus Res. 5:343-356. [DOI] [PubMed] [Google Scholar]
- 16.McGinnes, L. W., T. Sergel, H. Chen, L. Hamo, S. Schwertz, and T. G. Morrison. 2001. Mutational analysis of the membrane proximal heptad repeat of Newcastle disease virus fusion protein. Virology 289:343-352. [DOI] [PubMed] [Google Scholar]
- 17.Morrison, T. G., C. McQuain, and L. McGinnes. 1991. Complementation between avirulent Newcastle disease virus and a fusion protein expressed from a retrovirus vector: requirements for membrane fusion. J. Virol. 65:813-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morrison, T. G., M. E. Peeples, and L. W. McGinnes. 1987. Conformational change in a viral glycoprotein during maturation due to disulfide bond disruption. Proc. Natl. Acad. Sci. USA 84:1020-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagai, Y., and H. D. Klenk. 1977. Activation of precursors to both glycoproteins of Newcastle disease virus by proteolytic cleavage. Virology 77:125-134. [DOI] [PubMed] [Google Scholar]
- 20.Peisajovich, S. G., O. Samuel, and Y. Shai. 2000. Paramyxovirus F1 protein has two fusion peptides: implications for the mechanism of membrane fusion. J. Mol. Biol. 296:1353-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peisajovich, S. G., and Y. Shai. 2002. New insights into the mechanism of virus-induced membrane fusion. Trends Biochem. Sci. 27:183-190. [DOI] [PubMed] [Google Scholar]
- 22.Rapaport, D., M. Ovadia, and Y. Shai. 1995. A synthetic peptide corresponding to a conserved heptad repeat domain is a potent inhibitor of Sendai virus-cell fusion: an emerging similarity with functional domains of other viruses. EMBO J. 14:5524-5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reitter, J., T. Sergel, and T. Morrison. 1995. Mutational analysis of the leucine zipper motif in the Newcastle disease virus fusion protein. J. Virol. 69:5995-6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russell, R., R. G. Paterson, and R. A. Lamb. 1994. Studies with crosslinking reagents on the oligomeric form of the paramyxovirus fusion protein. Virology 199:160-168. [DOI] [PubMed] [Google Scholar]
- 25.Sergel, T., L. McGinnes, and T. Morrison. 2001. Mutations in the fusion peptide and adjacent heptad repeat inhibit folding or activity of the Newcastle disease virus fusion protein. J. Virol. 75:7934-7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sergel, T., L. W. McGinnes, and T. G. Morrison. 1993. The fusion promotion activity of the NDV HN protein does not correlate with neuraminidase activity. Virology 196:831-834. [DOI] [PubMed] [Google Scholar]
- 27.Sergel, T., L. W. McGinnes, M. E. Peeples, and T. G. Morrison. 1993. The attachment function of the Newcastle disease virus hemagglutinin-neuraminidase protein can be separated from fusion promotion by mutation. Virology 193:717-726. [DOI] [PubMed] [Google Scholar]
- 28.Sergel, T., and T. Morrison. 1995. Mutations in the cytoplasmic domain of the fusion glycoprotein of Newcastle disease virus depress syncytia formation. Virology 210:264-272. [DOI] [PubMed] [Google Scholar]
- 29.Sergel-Germano, T., C. McQuain, and T. Morrison. 1994. Mutations in the fusion peptide and heptad repeat regions of the Newcastle disease virus fusion protein block fusion. J. Virol. 68:7654-7658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stone-Hulslander, J., and T. Morrison. 1999. Mutational analysis of heptad repeats in the membrane-proximal region of Newcastle disease virus HN protein. J. Virol. 73:3630-3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stone-Hulslander, J., and T. G. Morrison. 1997. Detection of an interaction between the HN and F proteins in Newcastle disease virus-infected cells. J. Virol. 71:6287-6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanaka, Y., B. R. Heminway, and M. S. Galinski. 1996. Down-regulation of paramyxovirus hemagglutinin-neuraminidase glycoprotein surface expression by a mutant fusion protein containing a retention signal for the rough endoplasmic reticulum. J. Virol. 70:5005-5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tong, S., and R. Compans. 1999. Alternative mechanisms of interaction between homotypic and heterotypic parainfluenza virus HN and F proteins. J. Gen. Virol. 80:107-115. [DOI] [PubMed] [Google Scholar]
- 34.Toyoda, T., T. Sakaguchi, H. Hirota, B. Gotoh, K. Kuma, T. Miyata, and Y. Nagai. 1989. Newcastle disease virus evolution. II. Lack of gene recombination in generating virulent and avirulent strains. Virology 169:273-282. [DOI] [PubMed] [Google Scholar]
- 35.Wang, C., G. Raghu, T. Morrison, and M. E. Peeples. 1992. Intracellular processing of the paramyxovirus F protein: critical role of the predicted amphipathic alpha helix adjacent to the fusion domain. J. Virol. 66:4161-4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wiessenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp-41. Nature (London) 387:426-430. [DOI] [PubMed] [Google Scholar]
- 37.Yao, Q., and R. W. Compans. 1996. Peptides corresponding to the heptad repeat sequence of human parainfluenza virus fusion protein are potent inhibitors of virus infection. Virology 223:103-112. [DOI] [PubMed] [Google Scholar]
- 38.Young, J. K., R. P. Hicks, G. E. Wright, and T. G. Morrison. 1997. Analysis of a peptide inhibitor of paramyxovirus (NDV) fusion using biological assays, NMR, and molecular modeling. Virology 238:291-304. [DOI] [PubMed] [Google Scholar]
- 39.Young, J. K., D. Li, M. C. Abramowitz, and T. G. Morrison. 1999. Interaction of peptides with sequences from the Newcastle disease virus fusion protein heptad repeat regions. J. Virol. 73:5945-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao, X., M. Singh, V. N. Malashkevich, and P. S. Kim. 2000. Structural characterization of the human respiratory syncytial virus fusion protein core. Proc. Natl. Acad. Sci. USA 97:14172-14177. [DOI] [PMC free article] [PubMed] [Google Scholar]