Abstract
The interaction of an antimicrobial peptide, MSI-78, with phospholipid bilayers has been investigated using atomic force microscopy, circular dichroism, and nuclear magnetic resonance (NMR). Binding of amphipathic peptide helices with their helical axis parallel to the membrane surface leads to membrane thinning. Atomic force microscopy of supported 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) bilayers in the presence of MSI-78 provides images of the membrane thinning process at a high spatial resolution. This data reveals that the membrane thickness is not reduced uniformly over the entire bilayer area. Instead, peptide binding leads to the formation of distinct domains where the bilayer thickness is reduced by 1.1 ± 0.2 nm. The data is interpreted using a previously published geometric model for the structure of the peptide-lipid domains. In this model, the peptides reside at the hydrophilic-hydrophobic boundary in the lipid headgroup region, which leads to an increased distance between lipid headgroups. This picture is consistent with concentration-dependent 31P and 2H NMR spectra of MSI-78 in mechanically aligned DMPC bilayers. Furthermore, 2H NMR experiments on DMPC-d54 multilamellar vesicles indicate that the acyl chains of DMPC are highly disordered in the presence of the peptide as is to be expected for the proposed structure of the peptide-lipid assembly.
INTRODUCTION
Antimicrobial peptides are important components of the defense mechanism of animals and plants against pathogens (1). The most abundant form of antimicrobial peptide is the linear peptide. Two common properties, cationic and amphipathic structure, are thought to enable them to permeabilize microbial membranes selectively.
Magainins (magainin 2 and PGLa) are well-studied linear antimicrobial peptides that were originally isolated from the skin of the African frog Xenopus laevis (2,3). To increase the antimicrobial activity and selectivity, several synthetic peptides designed based on the naturally occurring magainin 2 and PGLa peptides have been reported (3). One of the most potent peptides, MSI-78 (amino-acid sequence G-I-G-K-F-L-K-K-A-K-K-F-G-K-A-F-V-K-I-L-K-K-NH2, also known as pexiganan), is an analog of magainin 2. This peptide has been the focus of pharmaceutical development largely because of the relative safety of topical therapy and the uncertainty surrounding the long-term toxicology of any new class of drug administered systemically. Previous nuclear magnetic resonance (NMR) studies analyzed the interaction of MSI-78 with various lipid bilayers and revealed the peptide-induced changes in the lipid headgroup conformation (4). The data showed that MSI-78 induces positive curvature strain on lipid bilayers and also predicted toroidal pore formation at higher concentrations (∼10 mol % of peptide), which is consistent with fluorescence studies on magainin 2.
To better understand the mechanism of peptide-lipid interaction, the present study uses atomic force microscopy (AFM), NMR, and circular dichroism (CD) to investigate the interaction of MSI-78 with 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) lipid bilayers. DMPC and POPC lipid bilayers are commonly used for many biophysical studies reported in the literature. The gel-to-liquid-crystalline phase-transition temperatures of these lipids are suitable for CD, NMR, and AFM experiments. Our previous NMR studies also used these lipids (4), the results of which are useful to make comparison with the data reported in this study. DMPC was selected for use in the AFM experiments to allow comparison with the NMR experiments because it forms a fluid-phase supported lipid bilayer at the temperatures employed.
In particular, during AFM experiments, peptide-induced changes in lipid membranes were observed at high lateral resolution (∼10 nm), whereas time-lapse series of images provided information about the dynamics of the process. The resulting data can be interpreted using a simple geometrical model for the peptide-lipid assemblies. The peptide-induced membrane-thinning effect observed by AFM is in excellent agreement with the peptide-induced disorder in the acyl regions of the lipid bilayer measured using 2H NMR experiments.
The mechanism for peptide-induced membrane disruption has been the topic of many previous studies. At low peptide/lipid (P/L) ratios, amphipathic helical segments of these peptides are adsorbed onto lipid bilayers with the helical axis parallel to the membrane surface (5,6). Above a certain critical concentration (P/L*), the peptides disrupt membranes via one of the well-studied mechanisms such as barrel-stave, toroidal pore formation, detergent-type micellization, and induction of nonlamellar phases (1,4–12). Toroidal-pore formation has been well characterized using fluorescence (10) and NMR (4,11) studies. Neutron diffraction experiments have also been valuable in the analysis of these peptide assemblies in lipid membranes (5,8,9,13,14). Interestingly, before the onset of cytolytic activity, peptide-membrane interaction leads to thinning of the membrane in proportion to the peptide concentration (6,15).
At present, the literature offers little information about the spatial distribution of peptides during membrane thinning. Past investigations generally reported the thicknesses averaged over a micrometer or millimeter range. Recently, the group of de Kruijff has used AFM in addition to NMR and differential scanning calorimetry to investigate the structure of different classes of antimicrobial peptides associated with gel-state lipid bilayers (16–18). In the present study, time-lapse scanning-probe imaging provides high resolution visualization of the peptide-membrane interaction. This technique allows the direct measurement of local membrane thickness rather than average properties. The experimental data reveals that the peptide does not induce uniform membrane thinning, but instead leads to the formation of distinct domains in the lipid bilayer.
EXPERIMENTAL PROCEDURES
The MSI-78 peptide sample was originally designed and synthesized by Genaera (Philadelphia, PA). The 15N-Phe16-labeled MSI-78 was synthesized and purified at the University of Michigan using the published procedure (3). For AFM experiments, a stock solution of concentration 1 mg/ml in Tris buffer was diluted with deionized water for a final concentration between 10 and 100 μg/ml (4–40 μM). Lipids, 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), DMPC-d4 and DMPC-d54, and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) were purchased from Avanti Polar Lipids (Alabaster, AL). Chloroform and methanol were procured from Aldrich Chemical (Milwaukee, WI), and naphthalene was from Fisher Scientific (Pittsburgh, PA). All the chemicals were used without further purification.
Atomic force microscopy (AFM)
Several milligrams of DMPC powder were dissolved in chloroform in a 10-ml round-bottom flask and the solvent was evaporated under vacuum to form a thin lipid film on the glass wall. The lipids were then hydrated for up to 30 min with 20 mM NaCl in a water bath above the main phase-transition temperature of the lipids (24°C for DMPC). To completely suspend the lipid film in the salt solution, the sample was placed in a heated bath sonicator for a few minutes. The milky lipid suspension was then sonicated three times for 5 min using a CZ261 Sonic and Materials Vibra-Cell probe sonicator (Sonics & Materials, Danbury, CT). This procedure resulted in a clear suspension of small unilamellar vesicles (SUVs). Metal particles introduced by the sonicator tip were removed by centrifugation. The SUV solution was diluted with NaCl solution if necessary. Typical concentrations used to prepare bilayer samples for AFM were 0.25–1 mg/ml.
Supported lipid bilayers were formed on pieces of freshly cleaved mica by vesicle fusion. The substrates, 1 cm × 1 cm mica sheets, were attached to metal sample pucks by double-sided tape. After placing a 60–80 μl drop of liposome solution on the mica, the sample was incubated in a closed container for 20–60 min. Samples were then gently rinsed with deionized water and transferred to the AFM for imaging. During AFM scanning, the sample temperature was 28°C, which is above the main phase-transition temperature for DMPC, i.e., the lipid bilayers were in the liquid crystalline state (19).
All AFM images were obtained on a Multimode AFM (Veeco Metrology, Santa Barbara, CA) in tapping mode in aqueous solution at pH 6 using a standard silicon nitride cantilever as described previously (20,21). Typical scan sizes were 1 μm at a resolution of 256 × 256 data points and a scan rate of 2.5–3 Hz. One image was therefore acquired in 1.5–2 min. Bilayer samples were inspected by AFM, and a smooth region with few or no defects was chosen. The AFM tip was then retracted to add 20–30 μl of MSI-78 solution to the sample volume using a syringe needle. The resulting total peptide concentration was between 1 and 10 μM. Imaging continued for up to 1 h to observe the effect of the peptide on the lipid bilayer.
For determining the relative thicknesses of peptide-perturbed and unperturbed bilayer, only depressions wide enough to show the flat part of the P-phase were considered. A zoom box was placed across a step between the higher and the lower level. A histogram of the height distribution within this area then has two clearly distinguishable peaks. The distance between the peak maxima is the average relative height change between the two phases. This procedure was repeated for several areas of five different samples. The resulting average change in thickness was 1.1 ± 0.2 nm.
Circular dichroism (CD)
SUVs were prepared using the following procedure. 10 mM Tris buffer (150 mM NaCl and 0.1 mM EDTA, pH 7.4) was added to a dry film of POPC and subjected to vortex and sonication to obtain a clear dispersion of SUVs. A 40-μM peptide stock solution was prepared in Tris buffer. CD spectra were recorded on an AVIV CD spectropolarimeter (Lakewood, NJ) using samples with peptide/lipid ratios 1:100 and 1:200 over the range from 200 to 250 nm at 25°C. A 5-mm quartz cuvette was used for measurements. Signal from SUVs and the buffer were removed by subtracting the spectra of corresponding control samples without peptide.
Preparation of samples for NMR measurements
Mechanically aligned DMPC bilayer samples for NMR experiments were prepared using the recently published naphthalene procedure (7). Briefly, four milligrams of lipids for 31P and 2H NMR and 100 milligrams of lipids for 15N NMR and an appropriate amount (since the P:L molar ratios are presented in the article, one can calculate the exact quantity of the peptide used in each experiment) of MSI-78 were dissolved in CHCl3/CH3OH (2:1) mixture containing equimolar amounts of naphthalene. The solution was spread on thin glass plates (11 mm × 22 mm × 50 μm, Paul Marienfeld, Bad Mergentheim, Germany) and then dried under N2 gas at room temperature. The lipid films were then dried under vacuum at ∼35°C for ∼8 h to remove naphthalene and any residual organic solvents. The glass plates were placed in a hydration chamber that was maintained at ∼93% relative humidity for 2–3 days at 37°C. Approximately 2–5 μL of deionized water was sprayed onto the surface of the lipid-peptide film on glass plates. The glass plates were stacked, wrapped with parafilm, sealed in plastic bags (Plastic Bagmart, Marietta, GA), and then incubated at 4°C for 6–24 h.
Multilamellar vesicles (MLVs) for 2H NMR experiments were prepared as follows. Approximately 5.0 mg of lipid and the desired amount of peptide were dissolved in CHCl3/CH3OH (2:1) mixture. The solvent was removed under N2 and the lipid-peptide mixture was dried under vacuum overnight. Approximately 55 wt % deuterium-depleted water (Aldrich, Milwaukee, WI) was added and the mixture was subjected to several cycles of freeze-thaw with a gentle vortex to produce MLVs of DMPC.
Solid-state NMR
All experiments were performed on a Chemagnetics/Varian Infinity 400 MHz solid-state NMR spectrometer (Chemagnetics, Varian, Palo Alto, CA) operating with resonance frequencies of 400.138, 161.979, 61.424, and 40.55 MHz for 1H, 31P, 2H, and 15N nuclei, respectively. A Chemagnetics temperature-controller unit was used to maintain the sample temperature, and the sample was equilibrated for at least 30 min before starting each experiment. All experiments on oriented samples were performed with the bilayer-normal parallel to the external magnetic field. The 31P spectra of mechanically aligned samples were obtained using a homebuilt double-resonance probe, which has a four-turn square coil (12 mm × 12 mm × 4 mm) constructed using a 2-mm-wide flat-wire with a spacing of 1 mm between turns. 31P and 2H spectra of MLVs were obtained using a Chemagnetics double-resonance probe. A typical 31P 90°-pulse length of 3.1 μs was used in both probes. 31P chemical shift spectra were obtained using a spin-echo sequence (90°-τ-180°-τ-acquire), 30 kHz proton-decoupling radio frequency field, 50 kHz spectral width, and a recycle delay of 5 s. A typical spectrum required the co-addition of 100–500 transients. The 31P chemical shift spectra were referenced relative to 85% H3PO4 on thin glass plates (0 ppm). 2H quadrupole coupling spectra were obtained using a quadrupole-echo sequence (90°-τ-90°-τ-acquire) with a 90°-pulse length of 3.0 μs, a spectral width of 100 kHz, and a recycle delay of 2 s. A typical spectrum required the co-addition of ∼10,000 transients. Data processing was accomplished using the software Spinsight (Chemagnetics/Varian) on a Sun SPARC workstation (Sun Microsystems, Palo Alto, CA). Spectral simulation for the 31P spectra was done using a FORTRAN program. The 2H spectra of DMPC-d54 multilamellar dispersions were processed and de-Paked using MatLab software (The MathWorks, Cambridge, MA) as explained in the literature (22). The 15N chemical shift spectrum was obtained using a ramp cross-polarization (CP) sequence with a 1H π/2 pulse length of 3.5 μs, 35 kHz CP power, and a 1H decoupling of 71 kHz during acquisition. Other experimental parameters include: 2000 scans, 3-s recycle delay, 50 kHz spectral width, and 1.25 ms ramp CP with a 10 kHz ramp on the 1H channel. The 15N chemical shift spectrum was referenced to 15N labeled ammonium sulfate powder at 24.1 ppm relative to liquid ammonia.
RESULTS
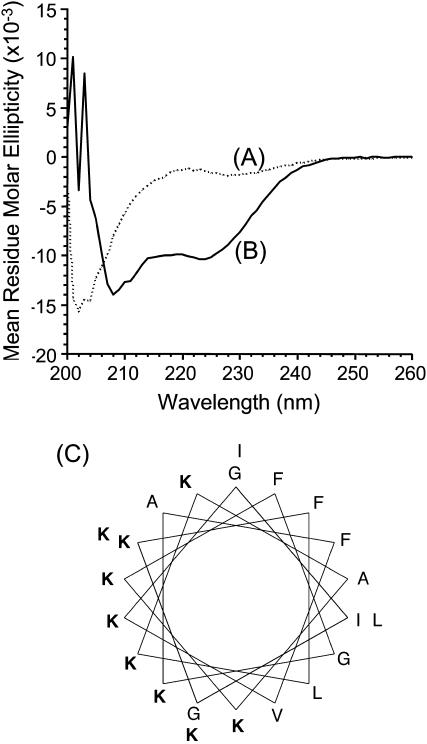
To understand the nature of interaction between MSI-78 and lipid membranes, we examined the secondary structure of MSI-78 in Tris buffer (pH = 7.4) and in the presence of POPC SUVs. The CD spectrum of MSI-78 in aqueous buffer (Fig. 1 A) resembled the one for an unordered structure. However, in the presence of POPC vesicles, the peptide showed two negative minima at ∼207 and 222 nm (Fig. 1 B) suggesting that the peptide folds into an α-helix upon binding to the lipid membrane. The helical wheel projection in Fig. 1 C shows an amphipathic helical structure of the peptide.
FIGURE 1.
Circular dichroism spectra of MSI-78 in (A) Tris-buffer (10 mM Tris-HCl, 150 mM NaCl, 2 mM EDTA, pH = 7.4) and (B) 2 mM POPC in Tris buffer. Peptide concentration was 20 μM and P/L was 1:100. The mean helix contents of vesicle bound MSI-78 as calculated from the mean residual ellipticity value at λ222 nm are ∼45%. (C) Helical wheel projection of MSI-78.
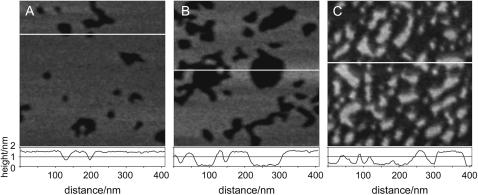
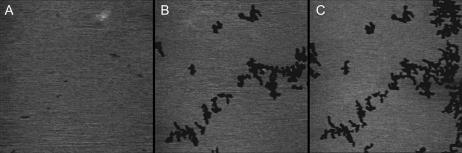
The adsorption of peptide helices to supported DMPC bilayers was observed using AFM. Fig. 2, A–C, shows typical images of different bilayer samples at three different stages of peptide coverage. Initially, the lipid bilayer that contains no peptide was defect-free. After the addition of MSI-78, we did not observe a uniform distribution of peptide binding to the lipid membrane. Instead, within the first 2 min, channel-like depressions began to form in the bilayer, which are seen as dark lines in Figs. 2 and 3. This system of channels grew rapidly in size resulting in a network appearance of this peptide-lipid phase (Fig. 3), which we will refer to as P-phase. Once the P-phase was formed, the shape of the domain boundaries remained fixed for the duration of the entire experiment, up to 1 h.
FIGURE 2.
Representative AFM height images and corresponding line scans of DMPC bilayer at (A) ∼10% coverage, (B) ∼30% coverage, and (C) ∼70% coverage with MSI-78. The amount of peptide coverage increased over time. At the peptide concentrations used, 70% coverage was obtained within ∼45 min. White lines indicate the positions of the line scans.
FIGURE 3.
AFM height images of DMPC bilayer before and after adding MSI-78. Total peptide concentration ∼2 μg/ml = 0.8 μM. Scan size: 1 μm. (A) Pure fluid DMPC bilayers have a smooth surface and are stable without changes for hours, (B) 3 min after adding peptide, and (C) 8 min after adding peptide.
The depth of depressions was less than the fully hydrated lipid bilayer thickness which is ∼5 nm (19,23,24) (Fig. 2, A–C). The smallest visible sites of peptide binding were circular dimples with a diameter of ∼25 ± 4 nm as seen in Fig. 2 A. Within 15–30 min after the addition of the peptide to the lipid bilayer, some of the wider depressions could be seen to have a flat bottom (Fig. 2 B). Eventually, after 50–60 min, the P-phase covered most of the substrate (Fig. 2 C) leaving only isolated plateau regions of the original bilayer. The line scans of images in Fig. 2, B and C, reveal that the P-phase had a well-defined thickness that remained constant independent of the total peptide coverage of the bilayer. (Note that the actual depth can only be determined if the width of the depression is >25–40 nm, considering the size of the AFM tip given by the manufacturer.)
The average relative height difference between the two phases was determined to be 1.1 ± 0.2 nm by plotting a histogram of the height distribution within several images of five different samples. For this analysis, only flat plateaulike regions were chosen so as to avoid the problem of the finite tip radius (i.e., small dimples as in Fig. 2 A were excluded). Generally, the thickness of supported lipid bilayers can be determined by measuring the step height from substrate to top of bilayer in bilayer defect regions. By monitoring this depth before and after the addition of MSI-78 it was confirmed that the bilayer thickness outside of the P-phase regions was not affected by the peptide (data not shown).
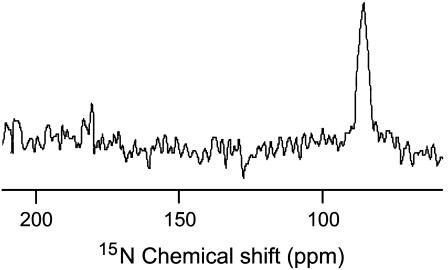
A proton decoupled 15N chemical shift spectrum of mechanically aligned POPC bilayers containing 3 mol % 15N-Phe16 labeled MSI-78 is given in Fig. 4. This spectrum contains a single peak at 86 ppm that corresponds to the perpendicular edge of an unaligned 15N chemical shift anisotropy spectrum. Since the peptide forms a helical structure on lipid bilayers, the 15N data can be interpreted in terms of the orientation of the peptide on the bilayer. Based upon previously reported structural studies on membrane-associated peptides using solid-state NMR (25) this data reveals that the helical MSI-78 is oriented along the bilayer surface, which is perpendicular to the bilayer normal.
FIGURE 4.
Nitrogen-15 chemical shift spectrum of mechanically aligned POPC bilayers containing 3 mol % MSI-78 obtained at 30°C.
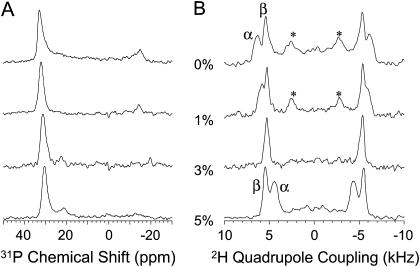
To measure any changes in the conformation of the lipid headgroup due to MSI-78-lipid interactions, 31P and 2H NMR experiments on mechanically aligned DMPC-d4 bilayers were carried out (Fig. 5). A single line ∼30 ppm in the 31P spectra (Fig. 5 A) indicates that the bilayers are aligned and there are no nonlamellar phases due to the presence of MSI-78 in the sample. However, the addition of MSI-78 decreases the chemical shift frequency value of the single peak in a concentration-dependent manner. The presence of the peptide decreases the 2H quadrupole splitting at the Cα site, whereas it increases the 2H quadrupole splitting at the Cβ site (Fig. 5 B). A low intense broad line in 31P spectra (Fig. 5 A) is due to the signal from the unoriented component of the sample which is also marked in Fig. 5 B. The changes in the 31P and 2H spectra are discussed below.
FIGURE 5.
31P (A) and 2H (B) NMR spectra of mechanically aligned DMPC-d4 bilayers containing 0, 1, 3, and 5 mol % MSI-78. Signal from unoriented component of the sample is marked by an asterisk.
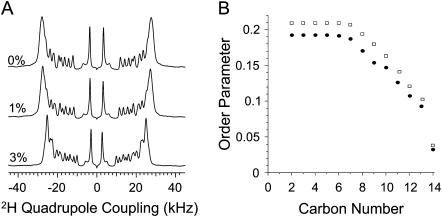
Deuterium NMR experiments were performed on multilamellar vesicles of DMPC-d54 to measure the peptide-induced disorder in the acyl region of the bilayer. Deuterium spectra of unoriented bilayers (data not shown) are de-Paked to obtain aligned spectra of bilayers as explained elsewhere (22). The de-Paked 2H NMR spectra are given in Fig. 6 A. Deuterium quadrupolar couplings were measured from the de-Paked spectra and the order parameters were calculated using published procedures (22). The order parameter for each carbon site of the lipid SN1 acyl chain is given in Fig. 6 B for DMPC with 1 mol % and without MSI-78. Order parameters for the other acyl chain (SN2) of DMPC are not shown as they are similar to that of SN1. The order parameter decreases with the increase of the carbon number as the lower order acyl chain is highly disordered even for the pure lipid bilayer (Fig. 6 B). However, the addition of MSI-78 decreases the order parameter for all carbon sites except the terminal carbons (C14) of the acyl chain that are highly disordered even in the absence of the peptide. This peptide-induced disorder in the hydrophobic region of DMPC bilayers is discussed below.
FIGURE 6.
(A) De-Paked 2H NMR spectra of DMPC MLVs containing 0, 1, and 3 mol % MSI-78. (B) Order parameters (SCD) of the SN1 acyl chain for pure DMPC (rectangles) and bilayers containing 1 mol % MSI-78 (solid circles). Order parameters of SN2 are similar to that of SN1 and for clarity are not shown.
DISCUSSION
Previous studies have shown that membrane thinning is an intermediate process that is crucial in the overall antimicrobial activity of the peptide (6,15,26). The AFM results presented here lead to new insights about the membrane thinning effect of antimicrobial peptides. Huang et al., have shown that helical peptides initially bind electrostatically with the helical axes parallel to the bilayer surface at low concentrations (6,15,26). Since MSI-78 forms an amphipathic α-helical structure in lipid bilayers as shown by the CD data (Fig. 1 A), it is likely that the hydrophobic part of the helix (see Fig. 1 C) is exposed to the hydrophobic part of the bilayer with the hydrophilic face of the helix interacting with the lipid headgroup in the water phase. This in-plane orientation of peptides in model lipid membranes has been shown for several other antimicrobial peptides using 31P and 15N solid-state NMR experiments and has been confirmed for the case of MSI-78 (Fig. 4) (11,25). This model is supported by the previously reported 31P NMR results that suggested the tilt of lipids in the presence of MSI-78. It is further confirmed by the 31P and 2H NMR experiments on mechanically aligned DMPC-d4 bilayers (Fig. 5) as discussed below. The decrease in the frequency of the 31P peak in Fig. 5 A suggests that the lipid is either tilted and/or the headgroup conformation is altered by the presence of the peptide. The addition of MSI-78 to DMPC-d4 bilayers decreases quadrupole coupling at the Cα site, while it increases at the Cβ site (Fig. 5 B). These changes can be interpreted in terms of modulation of headgroup conformation and/or dynamics. Since any changes in the dynamics of the lipid headgroup (alteration of order or disorder or angular fluctuations) would have the same effect on both the carbon sites, they would cause the quadrupole splittings to change in the same direction for the α- and β-segments. The observed counter-directional changes in the quadrupole splittings cannot therefore be explained by the changes due to dynamics of the headgroup. On the other hand, these results can be interpreted in terms of a conformational change in the phosphocholine headgroup of DMPC. NMR studies have shown that the orientation of the −P-N+ dipole of the phosphocholine headgroup that is almost parallel to the plane of the bilayer surface can be altered in the presence of electric charges. For example, the addition of a cationic amphiphile moves the N+ end of the dipole toward the water phase, whereas an anionic amphiphile has the opposite effect (27). Since MSI-78 is a cationic peptide at neutral pH, it could repel the N+ end of the −P-N+ dipole and move the dipole toward the water phase of the bilayer, thus changing the phosphocholine conformation. This interpretation is in good agreement with the results reported for the effects of cations (27) and a cationic peptide pardaxin (28) on the conformation of the lipid headgroup. Although the interaction of pardaxin with the DMPC headgroup is similar to that of MSI-78, the membrane disruptive mechanism of pardaxin varied with membrane composition (7,28). In addition, the observed NMR results are in excellent agreement with a recent molecular dynamics study that showed hydrogen bonding between positively charged amino acid residues (arginine and lysine) of MSI-78 and the oxygens of the POPC headgroup regions (29). These results are in complete agreement with a model where MSI-78 lies on the bilayer surface.
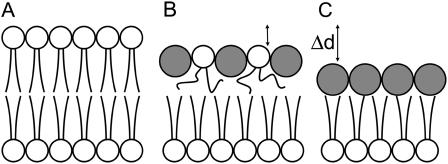
The Huang group has used the following geometric model to describe these peptide-induced changes in the lipid headgroup region and the resulting membrane thinning effect of antimicrobial peptides (6,15,26), (see also Fig. 7, A and B). In the unperturbed bilayer the lipid chains of each molecule occupy a volume v = Ad, where d is the average length of the lipid chains and A the average area available in the plane of the bilayer. Since the peptide helices push apart the lipid headgroups, the cross sectional area to fill by the tails increases by an amount cΔA. Here c is the peptide/lipid ratio in the top monolayer, and ΔA is the cross-sectional area along the axis of the peptide helix (note that we are ignoring the bottom monolayer for the moment; therefore c and P/L are not the same). For a constant density (and therefore constant volume) of the acyl chain region, its thickness is reduced accordingly by an amount Δd. This conservation of volume can be written as Ad = (A+cΔA)(d–Δd). The expected height change Δd is thus
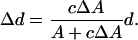 |
(1) |
FIGURE 7.
Schematic representation of membrane thinning effect. (Shaded circles, cross section of peptide helix with its axis perpendicular to this plane; open circles, DMPC headgroups; lipid tails are shown as lines.) (A) Unperturbed lipid bilayer; (B) peptide helices push apart lipid headgroups leading to greater mobility of lipid chains and membrane thinning; and (C) top monolayer has been replaced by peptide layer. The value Δd is the height difference measured by AFM.
AFM allows direct visualization of the peptide adsorption process at high resolution. In this manner, we are able to obtain information at length scales of only several nm rather than measuring average properties. The data reveals that peptide-membrane interaction does not lead to a gradual and uniform thinning of the membrane. Instead, it is observed that the peptide forms distinct domains of a peptide-lipid phase within the lipid bilayer that has a well-defined thickness independent of the total amount of peptide bound to the membrane. As P/L increases, so does the area covered by this P-phase. As a result, the average thickness of the bilayer decreases linearly as a function of area covered by the P-phase. The AFM experiments are therefore consistent with previous data on magainin 2 peptide (15).
To estimate c in the P-phase, we can apply Eq. 1 using d = 1.3 nm for the thickness of a DMPC hydrocarbon monolayer and A = 60 Å2 (30–32). Furthermore, a 22-amino acid peptide helix is expected to be 3.3-nm long (1.5 Å per residue) and 1.1-nm wide (assuming it has similar dimensions to alamethicin (33,34)), which gives ΔA = 360 Å2. The resulting change in membrane thickness is Δd ≤ 1.1 nm, even if c was as high as 1:1. (Given the relative cross-sectional area of peptide and lipid, c is most likely <1:1.) This is in the range of experimentally determined Δd = 1.1 ± 0.2 nm. Another hypothetical structure of the P-phase is shown in Fig. 7 C. In this model the peptide molecules displace all lipid molecules within a region of the bilayer. The resulting change in membrane thickness is 1.1 nm, given a headgroup thickness of 0.9 nm (32,35) and assuming the bottom monolayer is not affected. The above analysis indicates that within the P-phase the molecules arrange as in Fig. 7 B with c close to 1, or the structure resembles that shown in Fig. 7 C. However, the AFM data does not allow us to rule out the case of either Fig. 7 B or Fig. 7 C.
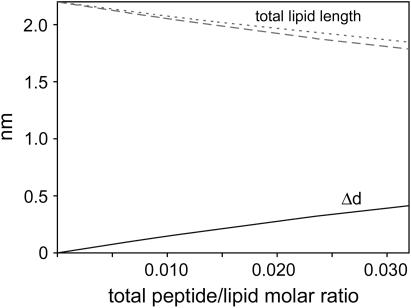
This information can be used to compare the AFM data to the literature (15). In the following paragraphs it will be assumed that all peptides are located in the top monolayer. The total number of lipids obviously includes both the lipids between peptide helices in the top monolayer as well as the lipids in the bottom monolayer. In each case one lipid is assumed to occupy an area of 60 Å2, i.e., one-sixth that of the peptide helix. The relationship between c and P/L is then c = (2 P/L)/(1–6 P/L). With this, Eq. 1 gives Δd as a function of P/L, see the solid line in Fig. 8. The corresponding lipid layer thickness including the lipid headgroup is shown as a dashed line. This is the same analysis as in Fig. 8 of Ludtke et al. (15) and is in good agreement with that data. For this analysis, it makes no difference if the peptide is distributed uniformly over the entire area or concentrated in certain domains as the AFM images show. For comparison, the dotted line in Fig. 8 is the theoretical average monolayer thickness assuming the P-phase structure has the form of Fig. 7 C with a Δd of 1.1 nm. For example, if P/L = 0.056 then the ratio of P-phase to non-P-phase would be 50%, giving an average monolayer thickness of 1.65 nm and an average Δd of 0.55 nm. Fig. 8 shows that both our AFM data and Fig. 8 of Ludtke et al. (15) are in agreement with regard to average membrane thickness.
FIGURE 8.
Height change Δd (solid line) according to Eq. 1 and corresponding DMPC monolayer thickness (dashed line). Monolayer thickness calculated by averaging the thickness of a DMPC monolayer with distinct domains of pure peptide as in Fig. 7 C (dotted line).
For each of the AFM images shown in Figs. 2 and 3, the fraction of the area covered by the P-phase can be determined using the bearing analysis of the NanoScope software (Veeco Metrology). Using this and either model in Fig. 7 B or Fig. 7 C, we can then estimate P/L in each image. Values for P/L in Fig. 2, A and B, and Fig. 3, B and C, range from 0.006 to 0.028. This is probably below the threshold concentration P/L* for peptide insertion (see Introduction). For magainin in DMPC/DMPG bilayers, this threshold was determined to be close to P/L = 0.033 (36), but the exact ratio depends both on peptide and lipid composition. Given the coverage in Fig. 2 C, P/L = 0.06 for the model in Fig. 7 B and P/L = 0.09 for the model in Fig. 7 C. Thus the sample in Fig. 2 C is probably close to or above the insertion limit. We do caution the reader that these estimates based on P/L* are very uncertain, since they are derived from values obtained for magainin in a mixed DMPC/DMPG bilayer, whereas we are a studying MSI-78 interacting with a pure DMPC bilayer. However, we include the estimates because they may help to provide comparison points with previously published studies.
Using the known amount of peptide added to the sample and the approximate area covered by the bilayer, we can put an upper limit on P/L in the AFM sample volume; this number is ∼0.5. Note, however, that the entire amount of peptide will not find its way to the lipid bilayer. A certain fraction will be lost to adsorption to other surfaces such as the syringe, tip holder, and cantilever. As shown above, even if most of the peptide eventually diffuses toward the bilayer, at least in the early stages of the experiment P/L will be below the insertion limit. We are therefore confident that the changes seen in Fig. 3, B and C, and Fig. 2, A and B, are formed by peptides oriented parallel to the membrane plane as expected for concentrations below P/L*.
If the peptide causes membrane thinning as shown in Fig. 7 B, the acyl chains of the lipids should be highly disordered in the presence of the peptide. To confirm this effect, 2H NMR experiments were performed on DMPC-d54 lipid bilayers, and order parameters were measured (Fig. 6). As seen from Fig. 6 B, the acyl chains of DMPC are highly disordered in the presence of the peptide. Since the helical peptide interacts with the headgroup region, as seen from previous 31P solid-state NMR results from POPC bilayers (4), as well as from the 31P and 2H data given in Fig. 5, the acyl chains have more free-area to explore as shown in Fig. 7 B and, therefore, they are more disordered in the presence of the peptide. These results are in excellent agreement with a recent molecular dynamics simulation study on POPC bilayers containing helical MSI-78 peptides (29). This effect has also been reported for another amphipathic α-helical antimicrobial peptide, LL37 (22). Interestingly, studies have shown that the peptide-induced disorder is similar to the increase in the temperature of the sample that also causes membrane thinning (22). Therefore, the NMR data is in agreement with the model in Fig. 7 B and rules out the possibility given in Fig. 7 C, since it would not decrease the order parameter of the acyl chain, but instead would increase the order parameter at least for the lower end of the acyl chain. The NMR data supporting model in Fig. 7 B provides the key support for our assumption that the limit on c is ∼1. This can be understood as follows. In the derivation of Eq. 1, the hydrocarbon region of the lipid bilayer is treated as a continuous fluid that fills the space between the lipid headgroup/peptide region and the bottom lipid layer. In reality, however, the hydrocarbon region consists of two lipid tails per headgroup of finite length and width. The model therefore breaks down once Δd approaches 1.3 nm. At this point the structure will look more like that shown in Fig. 7 C. To apply Eq. 1 to our data, the lipid tails have to be able to fill the space underneath the area covered by lipid headgroups and the peptide helices. Without trying to set a rigorous limit on the value of c, the relative dimensions of lipid tails and peptide helix point to it being close to 1 (two hydrocarbon chains of 1.3-nm length and one α-helix of 3.3 nm × 1.1 nm). With more than one peptide per lipid there will be “empty spaces” below the peptide helices, resulting in a structure (Fig. 7 C) that is inconsistent with our NMR data.
It should be noted that the peptide concentrations (1 or 3 mol %) used in NMR samples are not sufficient to perturb all of the lipids in the sample. Therefore, NMR samples should contain peptide-free (or unperturbed) and peptide-rich (or perturbed) domains, but the lipids must be under fast exchange during the NMR timescale to result in an average 2H quadrupole coupling as observed by a single Pake doublet for each CD2 group in the 2H spectra (or a single 31P peak in a mechanically aligned sample as shown in Fig. 5 A). This is unlike the AFM data that provides the evidence for the presence of two different domains in the supported bilayer sample. We also would like to point out that whereas the P/L used in NMR is higher than that used for AFM experiments, it is unlikely that such an increase in the concentration of the peptide would change the lipid-peptide interactions from the model in Fig. 7 C to that in Fig. 7 B. Therefore, it is most likely the peptide is causing membrane-thinning, which could be a crucial step in its antimicrobial activity.
CONCLUSIONS
The process of membrane thinning due to antimicrobial peptide binding has been observed for the first time using AFM. The data shows that the interaction of MSI-78 with supported DMPC bilayers on mica leads to the formation of domains of decreased bilayer thickness. As more and more peptide binds to the membrane, the area covered by those domains grows, whereas their thickness remains constant. NMR data is used to confirm that α-helical peptide segments interact with the lipid headgroup region and disorder the hydrophobic core of the lipid bilayer. This results in a greater distance between headgroups and increased mobility of the acyl chains. The experimental data is consistent with a model for the membrane-thinning effect of antimicrobial peptides oriented parallel to the membrane.
Acknowledgments
We thank Dr. Lee Maloy (Genaera Pharmaceuticals) for providing us with unlabeled MSI-78 peptide.
This project has been funded with federal funds from the National Cancer Institute, National Institutes of Health, under contract No. NOI-CO-97111 (M.M.B.H. and B.G.O.), and by National Institutes of Health grant No. AI054515 to A.R.
References
- 1.Zasloff, M. 2002. Antimicrobial peptides of multicellular organisms. Nature. 415:389–395. [DOI] [PubMed] [Google Scholar]
- 2.Zasloff, M. 1987. Magainins, a class of antimicrobial peptides from Xenopus skin—isolation, characterization of 2 active forms, and partial cDNA sequence of a precursor. Proc. Natl. Acad. Sci. USA. 84:5449–5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maloy, W. L., and U. P. Kari. 1995. Structure-activity studies on magainins and other host-defense peptides. Biopolymers. 37:105–122. [DOI] [PubMed] [Google Scholar]
- 4.Hallock, K. J., D.-K. Lee, and A. Ramamoorthy. 2003. MSI-78, an analogue of the magainin antimicrobial peptides, disrupts lipid bilayer structure via positive curvature strain. Biophys. J. 84:3052–3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ludtke, S. J., K. He, W. T. Heller, T. A. Harroun, L. Yang, and H. W. Huang. 1996. Membrane pores induced by magainin. Biochemistry. 35:13723–13728. [DOI] [PubMed] [Google Scholar]
- 6.Wu, Y., K. He, S. J. Ludtke, and H. W. Huang. 1995. X-ray diffraction study of lipid bilayer membranes interacting with amphiphilic helical peptides: diphytanoyl phosphatidylcholine with alamethicin at low concentrations. Biophys. J. 68:2361–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hallock, K. J., D. K. Lee, J. Omnaas, H. I. Mosberg, and A. Ramamoorthy. 2002. Membrane composition determines pardaxin's mechanism of lipid bilayer disruption. Biophys. J. 83:1004–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He, K., S. J. Ludtke, H. W. Huang, and D. L. Worcester. 1995. Antimicrobial peptide pores in membranes detected by neutron in-plane scattering. Biochemistry. 34:15614–15618. [DOI] [PubMed] [Google Scholar]
- 9.He, K., S. J. Ludtke, D. L. Worcester, and H. W. Huang. 1996. Neutron scattering in the plane of membranes: structure of alamethicin pores. Biophys. J. 70:2659–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuzaki, K. 1998. Magainins as paradigm for the mode of action of pore-forming polypeptides. Biochim. Biophys. Acta Rev. Biomembr. 1376:391–400. [DOI] [PubMed] [Google Scholar]
- 11.Henzler Wildman, K. A., D.-K. Lee, and A. Ramamoorthy. 2003. Mechanism of lipid bilayer disruption by the human antimicrobial peptide, LL-37. Biochemistry. 42:6545–6558. [DOI] [PubMed] [Google Scholar]
- 12.Shai, Y. 2002. Mode of action of membrane active antimicrobial peptides. Biopolymers. 66:236–248. [DOI] [PubMed] [Google Scholar]
- 13.Yang, L., T. A. Harroun, W. T. Heller, T. M. Weiss, and H. W. Huang. 1998. Neutron off-plane scattering of aligned membranes. I. Method of measurement. Biophys. J. 75:641–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang, L., T. M. Weiss, T. A. Harroun, W. T. Heller, and H. W. Huang. 1999. Supramolecular structures of peptide assemblies in membranes by neutron off-plane scattering: method of analysis. Biophys. J. 77:2648–2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ludtke, S., K. He, and H. W. Huang. 1995. Membrane thinning caused by magainin 2. Biochemistry. 34:16764–16769. [DOI] [PubMed] [Google Scholar]
- 16.Rinia, H. A., R. A. Kik, R. A. Demel, M. M. E. Snel, J. A. Killian, J. P. J. M. van der Eerden, and B. de Kruijff. 2000. Visualization of highly ordered striated domains induced by transmembrane peptides in supported phosphatidylcholine bilayers. Biochemistry. 39:5852–5858. [DOI] [PubMed] [Google Scholar]
- 17.Rinia, H. A., J.-W. P. Boots, D. T. S. Rijkers, R. A. Kik, M. M. E. Snel, R. A. Demel, J. A. Killian, J. P. J. M. van der Eerden, and B. de Kruijff. 2002. Domain formation in phosphatidylcholine bilayers containing transmembrane peptides: specific effects of flanking residues. Biochemistry. 41:2814–2824. [DOI] [PubMed] [Google Scholar]
- 18.van Kan, E. J. M., D. N. Ganchev, M. M. E. Snel, V. Chupin, A. van der Bent, and B. de Kruijff. 2003. The peptide antibiotic clavanin A interacts strongly and specifically with lipid bilayers. Biochemistry. 42:11366–11372. [DOI] [PubMed] [Google Scholar]
- 19.Tokumasu, F., A. J. Jin, and J. A. Dvorak. 2002. Lipid membrane phase behaviour elucidated in real time by controlled environment atomic force microscopy. J. Electron Microsc. (Tokyo). 51:1–9. [DOI] [PubMed] [Google Scholar]
- 20.Hong, S., A. U. Bielinska, A. Mecke, B. Keszler, J. L. Beals, X. Shi, L. Balogh, B. G. Orr, J. R. Baker Jr., and M. M. Banaszak Holl. 2004. Interaction of poly(amidoamine) dendrimers with supported lipid bilayers and cells: hole formation and the relation to transport. Bioconjug. Chem. 15:774–782. [DOI] [PubMed] [Google Scholar]
- 21.Mecke, A., S. Uppuluri, T. M. Sassanella, D. K. Lee, A. Ramamoorthy, J. R. Baker Jr., B. G. Orr, and M. M. Banaszak Holl. 2004. Direct observation of lipid bilayer disruption by poly(amidoamine) dendrimers. Chem. Phys. Lipids. 132:3–14. [DOI] [PubMed] [Google Scholar]
- 22.Henzler Wildman, K. A., G. V. Martinez, M. F. Brown, and A. Ramamoorthy. 2004. Perturbation of the hydrophobic core of lipid bilayers by the human antimicrobial peptide LL-37. Biochemistry. 43:8459–8469. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, S. J., T. M. Bayerl, D. C. McDermott, G. W. Adam, A. R. Rennie, R. K. Thomas, and E. Sackmann. 1991. Structure of an adsorbed dimyristoylphosphatidylcholine bilayer measured with specular reflection of neutrons. Biophys. J. 59:289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie, A. F., R. Yamada, A. A. Gewirth, and S. Granick. 2002. Materials-science of the gel to fluid phase transition in a supported phospholipid bilayer. Phys. Rev. Lett. 89:246103. [DOI] [PubMed] [Google Scholar]
- 25.Lee, D. K., J. A. Santos, and A. Ramamoorthy. 1999. Application of one-dimensional dipolar shift solid-state NMR spectroscopy to study the backbone conformation of membrane-associated peptides in phospholipid bilayers. J. Phys. Chem. B. 103:8383–8390. [Google Scholar]
- 26.He, K., S. J. Ludtke, W. T. Heller, and H. W. Huang. 1996. Mechanism of alamethicin insertion into lipid bilayers. Biophys. J. 71:2669–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scherer, P. G., and J. Seelig. 1989. Electric charge effects on phospholipid headgroups. Phosphatidylcholine in mixtures with cationic and anionic amphiphiles. Biochemistry. 28:7720–7728. [DOI] [PubMed] [Google Scholar]
- 28.Porcelli, F., B. Buck, D.-K. Lee, K. J. Hallock, A. Ramamoorthy, and G. Veglia. 2004. Structure and orientation of pardaxin determined by NMR experiments in model membranes. J. Biol. Chem. 279:45815–45823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kandasamy, S. K., and R. G. Larson. 2004. Binding and insertion of α-helical anti-microbial peptides in POPC bilayers studied by molecular dynamics simulations. Chem. Phys. Lipids. 132:113–132. [DOI] [PubMed] [Google Scholar]
- 30.Koenig, B. W., H. H. Strey, and K. Gawrisch. 1997. Membrane lateral compressibility determined by NMR and x-ray diffraction: effect of acyl chain polyunsaturation. Biophys. J. 73:1954–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petrache, H. I., S. Tristram-Nagle, and J. F. Nagle. 1998. Fluid phase structure of EPC and DMPC bilayers. Chem. Phys. Lipids. 95:83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petrache, H. I., S. W. Dodd, and M. F. Brown. 2000. Area per lipid and acyl length distributions in fluid phosphatidylcholines determined by 2H NMR spectroscopy. Biophys. J. 79:3172–3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fox, R. O., and F. M. Richards. 1982. A voltage-gated ion channel model inferred from the crystal-structure of alamethicin at 1.5 Å resolution. Nature. 300:325–330. [DOI] [PubMed] [Google Scholar]
- 34.Yang, L., T. A. Harroun, T. M. Weiss, L. Ding, and H. W. Huang. 2001. Barrel-stave model or toroidal model? A case study on melittin pores. Biophys. J. 81:1475–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson, S. J., T. M. Bayerl, W. Weihan, H. Noack, J. Penfold, R. K. Thomas, D. Kanellas, A. R. Rennie, and E. Sackmann. 1991. Coupling of spectrin and polylysine to phospholipid monolayers studied by specular reflection of neutrons. Biophys. J. 60:1017–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ludtke, S. J., K. He, Y. Wu, and H. W. Huang. 1994. Cooperative membrane insertion of magainin correlated with its cytolytic activity. Biochim. Biophys. Acta Biomembr. 1190:181–184. [DOI] [PubMed] [Google Scholar]