Abstract
Adenovirus serotype 5 (Ad5) has great potential for gene therapy applications. A major limitation, however, is the host immune response against Ad5 infection that often prevents the readministration of Ad5 vectors. In this regard, the most abundant capsid protein, hexon, has been implicated as the major target for neutralizing antibodies. In this study, we sought to escape the host neutralization response against Ad5 via hexon replacement. We constructed a chimeric adenovirus vector, Ad5/H3, by replacing the Ad5 hexon gene with the hexon gene of Ad3. The chimeric viruses were successfully rescued in 293 cells. Compared to that for the control Ad5/H5, the growth rate of Ad5/H3 was significantly slower and the final yield was about 1 log order less. These data indicate that the Ad3 hexon can encapsidate the Ad5 genome, but with less efficiency than the Ad5 hexon. The gene transfer efficacy of Ad5/H3 in HeLa cells was also lower than that of Ad5/H5. Furthermore, we tested the host neutralization responses against the two viruses by using C57BL/6 mice. The neutralizing antibodies against Ad5/H3 and Ad5/H5 generated by the immunized mice did not cross-neutralize each other in the context of in vitro infection of HeLa cells. Preimmunization of C57BL/6 mice with one of the two types of viruses also did not prevent subsequent infection of the other type. These data suggest that replacing the Ad5 hexon with the Ad3 hexon can circumvent the host neutralization response to Ad5. This strategy may therefore be used to achieve the repeated administration of Ad5 in gene therapy applications.
Adenoviruses, especially serotype 5 adenoviruses (Ad5), have attracted tremendous interest as gene therapy vectors on account of their ability to efficiently infect a variety of cells and to be generated to high titers in vitro (2, 19, 32). However, a major limitation is the host humoral immune response against adenovirus that arises from commonly acquired infection with adenoviruses, specifically when infected with the adenovirus-mediated common cold or after the initial administration of adenovirus vectors (6, 16, 27, 29, 37, 38, 41). In this regard, the preexisting antibodies against Ad5 have been shown to be sufficient to neutralize Ad5 infection, thus preventing the subsequent administration of Ad5 vectors (6, 29, 37, 38). This limit thus practically precludes applications requiring repetitive vector-mediated gene delivery.
Of note, the neutralizing antibodies against Ad5 appear to be generated principally against the major capsid proteins (13, 18, 35). The capsid of each adenovirus virion is composed of three major proteins, hexon, fiber, and the penton base, of which hexon is the most abundant. Neutralization of Ad5 by preexisting antibodies could occur in two ways: (i) extracellular neutralization mediated by anti-fiber and anti-penton base antibodies that cause aggregation of virions outside the cells, or (ii) intracellular neutralization mediated by anti-hexon antibody that blocks virion escape from the endosome into the cytoplasm (35). Intracellular neutralization is more effective since anti-hexon neutralization occurs via a single-hit mechanism, that is, one molecule of anti-hexon antibody would be sufficient to block the infection of one virion (35). This is consistent with other studies showing that hexon, not fiber or the penton base, is the major target of host neutralizing antibodies in Ad5 infection (13, 14, 15, 18, 24). On account of this, we hypothesized that hexon modification would be an effective strategy to overcome the host neutralization response.
Hexons of different serotypes are functional homologs and share a certain degree of sequence homology. Hexon monomers are usually composed of more than 900 amino acid residues (4). Electron microscopy and X-ray crystallography of the human Ad2 hexon have revealed that hexon has a dense pedestal base formed by two eight-stranded, antiparallel beta barrels stabilized by an internal loop (L3). Three other loops, L1, L2, and L4, project away from the surface of the virion (1, 23, 25, 30, 31). Analysis of the protein sequences of different hexons has revealed seven discrete hypervariable regions (HVRs) in the L1 and L2 loops, and one or more of these regions may contain the serotype-specific neutralization epitopes (4, 15, 25). Since neutralizing antibodies against adenoviruses have serotype specificity, one strategy to overcome the host neutralization response against Ad5 would be to replace the Ad5 hexon with hexons from different serotypes.
To date, 51 human adenovirus serotypes have been discovered. They are grouped into six subgroups (subgroups A to F) based on their hemagglutinating properties and DNA homologies (3, 7). In principle, when serotypes are more distantly related, they have a lower likelihood of cross-neutralizing. However, their hexons are more likely to fail in the packaging of alternate genomes. This is evidenced by recent studies showing that, although the hexons of about 20 serotypes have been evaluated, only the hexons of Ad1, Ad2, Ad6, and Ad12 could successfully package the Ad5 genome (15, 24, 40). Among them, except Ad12, which belongs to subgroup A, all other serotypes belong to subgroup C, the same subgroup as Ad5. This has led to the assumption that the hexon switch may only work efficiently within a subgroup. However, a recent study using an Ad5 vector that contains a temperature-sensitive (ts) mutation in the hexon protein (Ad5 ts147) showed that coinfection of Ad5 ts147 with other adenovirus serotypes [Ad3 (B), Ad4 (E), or Ad9 (D)] resulted in pseudopackaging of the Ad5 genome, suggesting that the Ad5 genome could be packaged by the capsids of alternate serotypes (21). Nonetheless, the basis of the transcomplementing function was unidentified; thus, it is not known whether all of the capsid proteins or only hexon was supplied by other serotypes. Therefore, compatibility of hexons from Ad3, Ad4, and Ad9, as well as other Ad5 capsid proteins, with the Ad5 genome remains to be investigated in the context of hexon-chimeric viruses.
In this study, we replaced the hexon of Ad5 with that of Ad3 to obtain hexon-chimeric virus Ad5/H3. We rescued the chimeric viruses, characterized them in detail, and examined the host neutralization response against them both in vitro and in vivo. Our data suggested that the Ad3 hexon was capable of packaging the Ad5 genome, but with less efficiency than the Ad5 hexon. The hexon chimera Ad5/H3, along with Ad5 and its native hexon (Ad5/H5), was not cross-neutralized by antibodies against one another as determined by in vitro and in vivo experiments. On these grounds, this strategy may be used to circumvent the host neutralization response against Ad5. Of note, this is the first study that directly demonstrated that hexons of subgroup B adenoviruses could encapsidate the Ad5 genome.
MATERIALS AND METHODS
Antibodies.
Goat polyclonal antibody against Ad2 hexon AB1056 and mouse monoclonal antibodies against Ad2 hexon (MAB8043) and Ad3 hexon (MAB8047) were purchased from Chemicon International, Inc. Alkaline phosphatase (AP)-conjugated donkey anti-goat and goat anti-mouse secondary antibodies were purchased from Jackson ImmunoResearch Laboratories.
Cells and cell culture.
The human embryonic kidney cell line (293) transformed with Ad5-E1 DNA was purchased from Microbix (Toronto, Ontario, Canada) and cultured in Dulbecco's modified Eagle medium-Ham's F12 medium containing 10% fetal calf serum and 2 mM l-glutamine. Human cervix carcinoma cells (HeLa) were obtained from the American Type Culture Collection and grown in minimum essential Eagle medium containing 10% fetal calf serum and 2 mM l-glutamine. Both 293 and HeLa cells were cultured at 37°C in a 5% CO2 atmosphere.
DNA construction. (i) Construction of the hexon-deleted Ad5 backbone (pAd5/ΔH5).
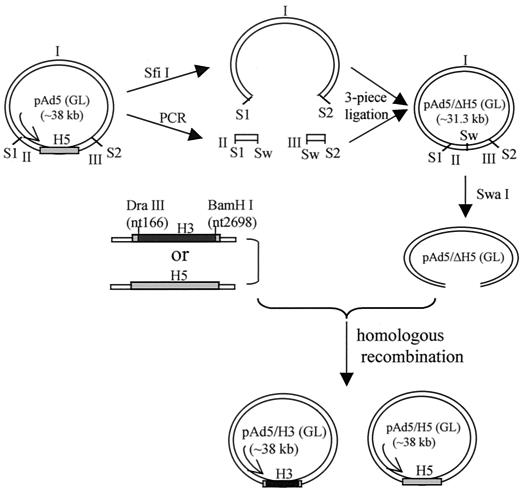
The Ad5 genome containing the cytomegalovirus promoter-driven green fluorescence protein (GFP) and firefly luciferase (luc) genes in the E1 region, pAd5(GL), was used as the template to create pAd5/ΔH5 via a combination of PCR and a three-piece ligation strategy. pAd5(GL) was digested with the SfiI restriction enzyme, and the large fragment (∼31.3 kb) was purified by using a QiaEX II gel extraction kit (Qiagen) (fragment I). By using pAd5(GL) as the template, fragment II (∼2.5 kb), which spans the first SfiI site and the 5′ end of the hexon gene, was amplified by PCR with the primers 5′-GGCGGCCATGCGGGCC-3′ and 5′-GCCATTTAAATGCGGGCGCGCGGCGG-3′ and fragment III (∼1.3 kb), which spans the 3′ end of the hexon gene and the second SfiI site, was amplified by PCR with the primers 5′-GCCATTTAAATCAGCTGCCGCCATGGGC-3′ and 5′-CGCGGCCTGTCCGGCC-3′. The SfiI and SwaI sites (underlined) were introduced in both fragments II and III with the SwaI site proximal to and the SfiI site distal to the original site of the hexon gene. Fragments II and III were then digested with SfiI and SwaI, purified, and ligated with fragment I to generate pAd5/ΔH5, which contains the GFP and Luc reporter genes in the E1 region. Be aware that the two SfiI sites in the Ad5 genome are not identical. The different sticky ends allowed oriented ligation.
(ii) Generation of hexon shuttle vectors.
The shuttle vector for Ad5 hexon replacement, pH5S, was constructed as follows. The 5′ flanking region of the Ad5 hexon (5′ homologous region, ∼1.2 kb) was amplified by PCR with primers 5′-CGGAATTCGCGAAGGAGGCAGGAC-3′ and 5′-GCGGGCGCGCCGCGGCTCAGCAGC-3′, and the 3′ flanking region (3′ homologous region, ∼0.9 kb) was amplified with primers 5′-CTTCTAGAAGAAGCAAGCAACATCAACAAC-3′ and 5′-GCGTTTAAACGCCCGACCTGCTAAACG-3′ by using pAd5(GL) as the template. pH 5S was created by two steps of subcloning: first, the 5′ homologous region was cloned into pNEB193 (New England Biolabs) with EcoRI and AscI, and then the 3′ homologous region was subcloned in with XbaI and PmeI.
To subclone the Ad5 hexon gene (H5) into pH5S, pH5S was digested with EcoRV and KpnI and the large fragment was purified. In the meantime, a fragment containing the H5 gene was obtained as follows. pAd5(GL) was first digested with the SfiI enzyme, and the small fragment (∼6.7 kb) was purified, followed by restriction digestion with EcoRV and KpnI. The resultant large fragment of 4.1 kb was purified and ligated into the digested pH5S vector described above to create H5/pH5S.
To create the shuttle vector containing the Ad3 hexon gene (H3), H3/pH5S, first we obtained H3 by PCR from Ad3 viruses (American Type Culture Collection) with the primers 5′-ACCCACGACGTGACCACCGACCGTAGCCAGCG-3′ and 5′-GGCTCTAGATTATGTTGTGGCGTTGCCGGCC-3′. The H3 PCR product was digested with DraIII and BamHI and ligated into the vector part of DraIII- and BamHI-digested H5/pH5S. The resultant shuttle vector, named H3/pH5S, contained nucleotide (nt) sequences of H3 from nt 166 to nt 2698, and the resultant hexon differed from wild-type full-length H3 at only 7 amino acid residues.
(iii) Homologous recombination to create pAd5/H5 and pAd5/H3.
The corresponding shuttle vectors H3/pH5S and H5/pH5S were digested with EcoRI and PmeI, and the fragment containing the homologous recombination regions and the hexon genes were purified and recombined with SwaI-digested pAd5/ΔH5 in Escherichia coli strain BJ5183. The resultant clones, both of which contained the GFP and Luc genes in the E1 region, were designated pAd5/H3 and pAd5/H5, and the constructs were confirmed by restriction digestions and sequencing.
Virus rescue and preparation.
To rescue modified Ad5 viruses, pAd5/H3 and pAd5/H5 were digested with PacI. Two micrograms of each purified DNA was transfected into 293 cells grown in 60-mm-diameter dishes with Superfect (Qiagen). The transfected cells were coated with agarose after 24 h and continued in culture at 37°C in 5% CO2 until plaques formed. The plaques were then collected and processed for large-scale proliferation.
The viruses were purified with standard CsCl gradient centrifugation. In brief, the viruses collected from infected 293 cells underwent four freeze-thaw cycles. After removal of the cell debris by centrifugation at 3,800 × g for 25 min at 4°C, the supernatant was loaded on the top of CsCl 1.33 to 1.45 discontinuous gradient and centrifuged at 55,000 × g for 3 h at 4°C. The bottom band was collected, loaded on a new CsCl 1.33 to 1.45 gradient, and centrifuged at 100,000 × g overnight at 4°C. The bottom band was then collected, and the viruses were dialyzed with phosphate-buffered saline (PBS) containing 10% glycerol three times. The virus particle titers were determined by spectrophotometry at 260 nm, in which 1 U of optical density at 260 nm equals 1.1 × 1012 viral particles (VPs).
Gene transfer assay.
Gene transfer efficacy of Ad5/H3 and control viruses was tested in HeLa cells by measuring luciferase activity. In brief, HeLa cells were plated in 24-well plates with a density of 105 cells per well the day before infection. Cells were infected in 150 μl of culture medium containing 2% fetal bovine serum (FBS) at multiplicities of infection (MOIs) of 10, 100, and 1,000 VPs/cell in triplicate. Two hours later, 350 μl of complete medium was added to each well and the cells were continued in culture for 24 h at 37°C in 5% CO2. After a wash with PBS once, the cells in each well were lysed in 250 μl of reporter lysis buffer (Promega), followed by one freeze-thaw cycle. Five microliters of each sample was used to measure the luciferase activity with a luciferase assay kit (Promega) and a luminometer (Berthold, Gaithersburg, Md.). To test the heat stability of Ad5/H3 and Ad5/H5, viruses with an MOI equivalent to 100 were incubated at 45°C for different time intervals before infecting HeLa cells (10). Luciferase activity in infected cells was analyzed 48 h postinfection as described above.
Growth kinetics.
293 cells were plated in six-well plates with a density of 3 × 105 cells per well the day before infection. The cells were infected with Ad5/H3 or Ad5/H5 at an MOI of 5 (5 VPs/cell) as described above, and one well of infected cells including the medium for each virus was collected daily after infection. The infected cells were subjected to four freeze-thaw cycles, followed by centrifugation at 3,800 × g for 25 min at 4°C. The titers of the supernatants were determined by the plaque assay.
In vitro neutralization.
To obtain neutralizing antibodies for Ad5/H3 and Ad5/H5, the viruses were injected into C57BL/6 mice (Charles River Laboratories) by intravenous (i.v.) injection at a concentration of 1010 VPs/mouse. PBS was injected as the control. Three mice were used for each group. Two weeks later, blood was withdrawn from the hearts of the immunized mice and sera were separated in serum separator tubes (Becton Dickinson). Procedures for these animal experiments complied with all relevant federal guidelines and institutional policies.
To test the neutralization properties of the sera, HeLa cells were plated in 96-well plates with a density of 1.5 × 104 cells per well the day before infection. For each well, Ad5/H3 or Ad5/H5 virus was diluted in 45 μl of 2% FBS medium to achieve an MOI of 10 and incubated with 5 μl of serum at 37°C for 1 h. The viruses were added into each well of HeLa cells, and 2 h later, 50 μl of 10% FBS medium was added. The luciferase activity was measured 24 h after the infection as described above.
In vivo neutralization.
To assess whether Ad5/H3 and Ad5/H5 are cross- neutralized in vivo, each of the C57BL/6 mice was injected with PBS or 109 VPs of Ad5/H3 or Ad5/H5 via i.v. injection. Each group contained nine mice. One month later, each group of mice was subjected to a second i.v. injection of PBS or 1010 VPs of Ad5/H3 or Ad5/H5 per mouse, resulting in three mice for each subgroup. Three days after the second injection, the mice were sacrificed with CO2 inhalation and organs, including the liver, heart, lung, spleen, and kidney, were harvested. The organs were homogenized on dry ice, lysed in passive lysis buffer, and measured for luciferase activity as described above. Procedures for these animal experiments complied with all relevant federal guidelines and institutional policies.
SDS-PAGE and Western blotting.
A total of 1010 VPs of each CsCl-purified virus was dissolved in Laemmli sample buffer with or without boiling and separated on 4 to 15% polyacrylamide via sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels. The gels were either stained with Gelcode blue stain reagent (Pierce) to detect the total viral proteins or transferred to a nitrocellulose membrane (Bio-Rad). For Western blotting, the membranes were incubated with blocking solution containing 5% skim milk and 0.05% Tween 20 in Tris-buffered saline before incubation with primary antibodies against hexon, AB1056, MAB8043, or MAB8047 for 2 h at room temperature. After three washes with Tris-buffered saline and reblocking with blocking solution, the blots were incubated with AP-conjugated secondary antibodies for 1 h at room temperature. After extensive washing, the immunoreactive bands were detected by soaking the blots in 10 ml of AP staining solution (100 mM Tris-HCl [pH 9.5], 100 mM NaCl, 5 mM MgCl2) containing 33 μl of 5-bromo-4-chloro-3-indolylphosphate (BCIP; Sigma) and 16.5 μl of Nitro Blue Tetrazolium (Sigma).
RESULTS
Construction of the hexon chimera.
Previous studies have suggested that replacement of the Ad5 hexon with hexons from different subgroups of adenoviruses rarely resulted in the successful rescue of viable viruses (15, 24, 40). Given that coinfection of the Ad5 hexon ts mutant and Ad3 led to pseudopackaging of the Ad5 genome, it is possible that the Ad3 hexon can encapsidate the Ad5 genome. To investigate this possibility, we created a hexon chimera, Ad5/H3, in which the hexon gene of Ad5 was replaced by that of Ad3. Hexons of adenoviruses share a certain degree of homology. Thus, to avoid recombination between these homologous regions, we first deleted the whole hexon gene of Ad5 from the backbone plasmid pAd5 via three-piece ligation (Fig. 1 and Materials and Methods) and the resultant backbone was named pAd5/ΔH5. The Ad3 hexon (H3) gene was obtained by PCR with purified Ad3 virus as the template, and the DraIII-BamHI fragment of the PCR product was used to replace the corresponding region of the Ad5 hexon (H5) in the shuttle vector H5/pH5S to obtain H3/pH5S (see Materials and Methods). This resulted in the replacement of H5 between amino acid residues 54 and 907 with H3 sequences. Since the N-terminal (residues 1 to 53) and C-terminal (residues 908 to 952) regions are very conserved between H3 and H5 (only 5 and 2 amino acid residues are different in the two regions, respectively), the resultant hexon gene is 99.3% identical to the native H3 gene. The hexon chimera plasmid pAd5/H3 and the control plasmid pAd5/H5 were obtained by homologous recombination between the shuttle vectors and pAd5/ΔH5 in E. coli BJ5183, and viruses were rescued in 293 cells.
FIG. 1.
Diagram depicting the construction of plasmids pAd5/H3 and pAd5/H5. The plasmid pAd5 containing the GFP and luc (GL) reporter genes in the E1 region was used to create pAd5/ΔH5 by three-piece ligation. A SwaI (Sw) site was introduced into the original site of the H5 gene. Fragments containing H3 or H5 were recombined with the SwaI-linearized backbone pAd5/ΔH5 by homologous recombination in E. coli BJ5183, resulting in the formation of pAd5/H3 and pAd5/H5. S1, the first SfiI site; S2, the second SfiI site. Note: the two SfiI sites in the Ad5 genome are not identical. The different sticky ends allowed for oriented ligation.
Confirmation of the hexon chimera.
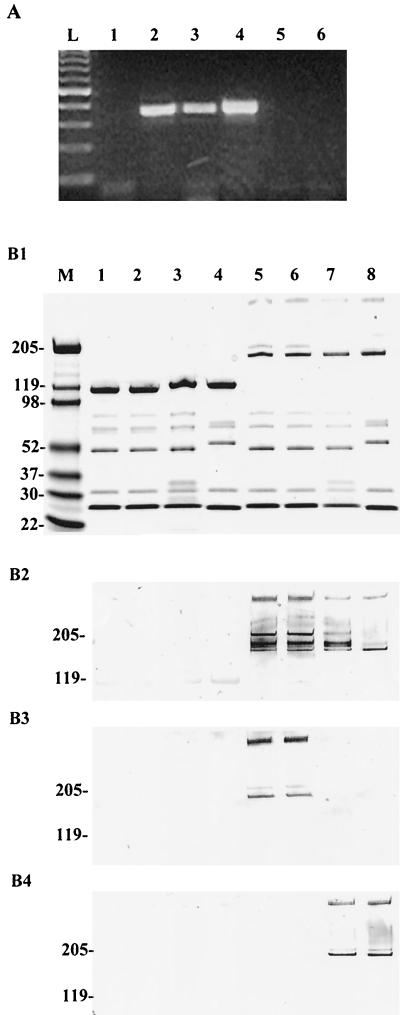
To confirm the chimeric viruses, we first employed a PCR-based assay by using specific PCR primers for H3 and H5. The sense and anti-sense primers are located in the HVRs of hexon, HVR1 and HVR4, respectively. H3-specific PCR generated the right size of PCR products on the pAd5/H3 plasmid and purified Ad5/H3 virus templates but not on the control pAd5 (Fig. 2A). In contrast, H5-specific PCR resulted in specific PCR products only on the template pAd5 (Fig. 2A). This confirmed that H3 sequences were indeed incorporated into the chimeric Ad5/H3 viruses.
FIG. 2.
Confirmation of the hexon chimera Ad5/H3. (A) Serotype-specific PCR analysis confirmed that the chimeric virus indeed contained the H3 gene. The specific PCR primers are located in HVR1 (sense) and HVR4 (anti-sense) of H3 or H5. Plasmids pAd5 and pAd5/H3 and purified Ad5/H3 viruses were used as the template. Lanes 1 to 3, PCR with H3-specific primers resulted in specific PCR products of correct size on templates pAd5/H3 (lane 2) and Ad5/H3 (lane 3) but not on pAd5 (lane 1); lanes 4 to 6, PCR with H5-specific primers produced specific product on template pAd5 (lane 4) but not on pAd5/H3 (lane 5) or Ad5/H3 (lane 6); lane L, 100-bp DNA ladder. (B) SDS-PAGE and Western blotting assays demonstrated that, at the protein level, Ad5/H3 contained the Ad3 hexon. In the assays, 1010 VPs of purified viruses was lysed in Laemmli sample buffer with or without boiling and separated on 4 to 15% polyacrylamide gels. The proteins were either stained with Gelcode blue stain reagent (Pierce) (B1) or transferred to a nitrocellulose membrane and then stained with the anti-hexon antibodies AB1056 (B2), MAB8043 (B3), or MAB8047 (B4). Lane M, molecular mass marker; lanes 1 and 5, Ad5; lanes 2 and 6, Ad5/H5; lanes 3 and 7, Ad5/H3; lanes 4 and 8, Ad3. Lanes 1 to 4 were loaded with boiled samples, while lanes 5 to 8 were loaded with unboiled samples.
We next examined the chimeric viruses at the protein level. Purified viruses (Ad5, Ad5/H5, Ad5/H3, and wild-type Ad3) at 1010 VPs/well were subjected to SDS-PAGE, and the gels were either stained with Gelcode blue stain reagent (Pierce) or transferred to a nitrocellulose membrane for Western blotting. The stained gel showed that the chimeric viruses contained proper viral proteins, compared to those in the original Ad5, the recombined Ad5/H5, and the wild-type Ad3 (Fig. 2B1). To demonstrate that H3 was indeed expressed in the chimeric viruses, we performed Western blot assays with three different anti-hexon antibodies: the polyclonal anti-hexon antibody AB1056, which recognizes most adenovirus hexons; the monoclonal antibody MAB8043, which recognizes only the hexons of Ad1, Ad2, Ad5, and Ad6; and another monoclonal antibody, MAB8047, which recognizes only the Ad3 hexon (Chemicon). As shown in Fig. 2B, AB1056 detected hexons of all of the viruses in the unboiled samples (Fig. 2B2) while the monoclonal antibody MAB8043 detected only the hexons of Ad5 and Ad5/H5 (Fig. 2B3) and MAB8047 detected only the hexons of Ad5/H3 and Ad3 (Fig. 2B4). None of the antibodies recognized hexons in the boiled samples. The detected bands in the unboiled samples corresponded to the molecular masses of the hexon dimer (∼210 kDa) and trimer (∼315 kDa). These data demonstrated that the chimeric Ad5/H3 viruses contained the hexon of Ad3.
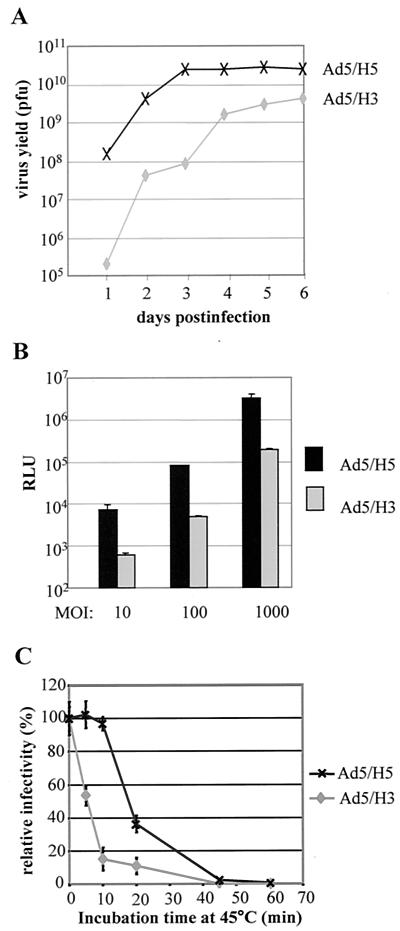
Growth kinetics of Ad5/H3.
Successful rescue of the Ad5/H3 viruses suggested that H3 was able to package the Ad5 genome. However, during the rescuing and purification processes of the viruses, we noticed that (i) it took longer for Ad5/H3-infected 293 cells to reach a complete cytopathic effect (corresponding to the maximal amount of virus production) than cells infected with Ad5/H5, and (ii) a substantial portion of Ad5/H3 viruses was defective—after CsCl centrifugations, the defective viruses (upper band) for Ad5/H3 made up ∼50% of the total particles, while for Ad5/H5, the ratio was only about 5%. These implicated hexon substitutions had an impact on virus packaging. To gain a quantitative understanding of this effect, we compared the growth kinetics of Ad5/H3 with that of Ad5/H5 (Fig. 3A). Under the same condition, while Ad5/H5 was found to reach a complete cytopathic effect at postinfection day 3, Ad5/H3 took about 6 days, suggesting that Ad5/H3 grew slower (Fig. 3A). In addition, the final yield of Ad5/H3 was 1 log order lower than that of Ad5/H5 (Fig. 3A). These data indicate that, although H3 could encapsidate the Ad5 genome, its efficiency was less than that of the native hexon H5.
FIG. 3.
Characterization of Ad5/H3. (A) Growth kinetics of Ad5/H3 and Ad5/H5. 293 cells plated in six-well plates were infected with Ad5/H3 or Ad5/H5 at an MOI of 5, and cells including medium were collected daily after infection. The titers of the viruses were determined by plaque assay. (B) Comparison of the gene transfer efficacy of Ad5/H3 and Ad5/H5. HeLa cells were infected with Ad5/H3 or Ad5/H5 at three different MOIs (10, 100, 1,000) in triplicate, and cell lysates were processed for the luciferase activity assay. The RLU showing the activity of luciferase were detected with a luciferase assay kit (Promega) and a luminometer. (C) Thermostability of Ad5/H3. The viruses at an MOI of 100 were incubated at 45°C for different time intervals before infecting HeLa cells. The relative infectivity was obtained by changing the RLU readings of the heat-treated viruses to the percentage of the readings of untreated Ad5/H3 or Ad5/H5 viruses.
Gene transfer efficacy of Ad5/H3 in HeLa cells was lower than that of Ad5/H5.
We next examined whether Ad5/H3 retained the gene transfer ability of Ad5/H5 by using the reporter gene luciferase. HeLa cells were chosen because they express a high level of the Ad5 receptor coxsackie adenovirus receptor (CAR) and can be effectively infected by Ad5 vectors (26). Ad5/H3 and Ad5/H5 viruses with MOIs equivalent to 10, 100, and 1,000 were used to infect the cells, and the gene transfer efficacy was assessed by detecting the relative light units (RLU) that correspond to luciferase activity by using a luminometer. Ad5/H3 was found to exhibit an approximately 1.2-log order-lower luciferase activity than Ad5/H5 at all MOIs (Fig. 3B), suggesting that the gene transfer ability of Ad5 was reduced by hexon replacement with H3. Similar results were obtained for Ad5 bearing the Ad12 hexon (40). Hexons of adenoviruses are not known to be involved in cell binding and internalization steps during Ad5 infection. How hexon replacement might affect the gene delivery ability of Ad5 is not clear.
Heat stability of Ad5/H3.
Since hexon is the major capsid protein, we tested whether hexon replacement could affect the structural integrity of the virions by comparing the thermostabilities of Ad5/H3 and Ad5/H5. The viruses were incubated at 45°C for different time intervals before infecting HeLa cells, and their relative gene transfer efficiencies were obtained by comparing them to those of unheated viruses. Gene transfer efficiency of Ad5/H5 was not significantly reduced following a 10-min incubation at 45°C, and its activity was retained about 40% even after a 20-min incubation at 45°C (Fig. 3C). In contrast, gene transfer efficiency of Ad5/H3 was reduced by more than 80% following the first 10-min incubation at 45°C (Fig. 3C). This suggested that the chimeric Ad5/H3 viruses were not as stable as Ad5 bearing the native hexon.
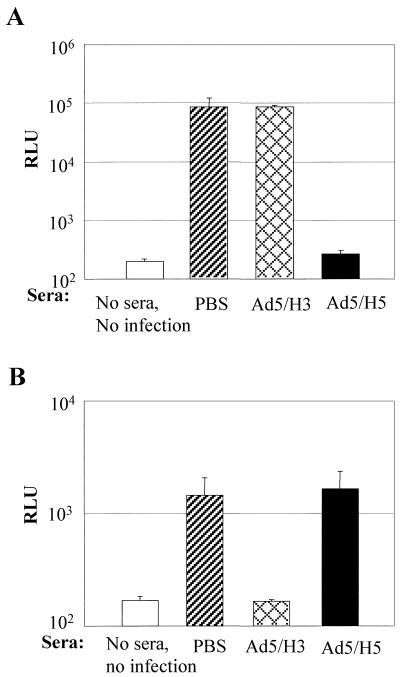
Ad5/H3 and Ad5/H5 were not cross-neutralized in vitro.
Hexons are the major target of the host neutralizing antibodies. To determine whether Ad5/H3 and Ad5/H5 can escape the host neutralization response against one another, we first performed in vitro neutralization assays using sera obtained from C57BL/6 mice that were immunized with PBS or 1010 VPs of Ad5/H3 or Ad5/H5 per mouse. Viruses at an MOI of 10 were preincubated with 5 μl of different sera before infecting HeLa cells, and their gene transfer efficacy was assessed with luciferase activity. We found that sera from Ad5/H3-immunized mice did not inhibit Ad5/H5 infection in HeLa cells, while sera from Ad5/H5-immunized mice almost completely blocked Ad5/H5 infection (Fig. 4A). Similarly, Ad5/H3 infection in HeLa cells was completely inhibited by sera from Ad5/H3-immunized mice, though not by sera from Ad5/H5-immunized mice (Fig. 4B). These data suggested that neutralizing the antibody against one type of the virus did not neutralize the other type. In addition, given that Ad5/H3 and Ad5/H5 differ only in their hexons, these data clearly demonstrated that the neutralizing antibodies against adenoviruses were principally directed against hexons.
FIG. 4.
Ad5/H3 and Ad5/H5 were not cross-neutralized in vitro. Ad5/H3 and Ad5/H5 viruses at an MOI of 10 were incubated with 5 μl of sera from PBS-, Ad5/H3-, or Ad5/H5-immunized C57BL/6 mice before infecting HeLa cells plated in 96-well plates. The infectivity was determined by luciferase activity assay. (A) Ad5/H5 infection was blocked by sera from Ad5/H5-immunized mice but not by sera from Ad5/H3-infected mice. (B) In contrast, Ad5/H3 infection was blocked by sera from Ad5/H3-immunized mice but not by sera from Ad5/H5 infected mice. Three mice were used for each group, and all of the infections were done in triplicate.
Ad5/H3 and Ad5/H5 were not cross-neutralized in vivo.
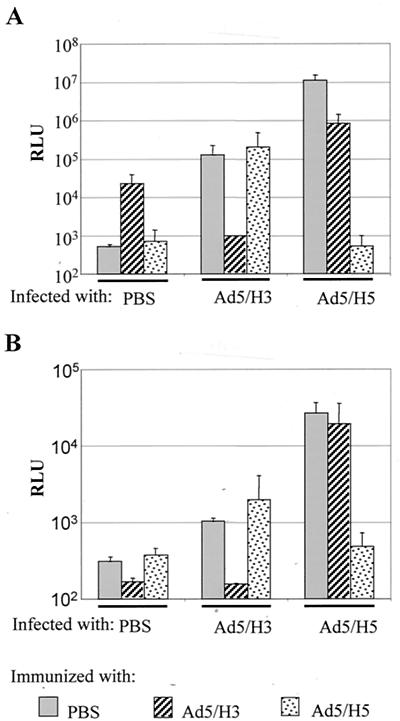
To further investigate whether Ad5/H3 and Ad5/H5 are cross-neutralized, we examined whether the two viruses could escape the host neutralization response against one another in vivo when administrated sequentially. C57BL/6 mice were first injected with PBS or 109 VPs of Ad5/H3 or Ad5/H5 per mouse via the tail vein. This concentration of viruses has been shown to sufficiently prevent reinfection of the Ad5 vector (29). One month later, each group of mice was injected again with PBS or 1010 VPs of Ad5/H3 or Ad5/H5 per mouse via the tail vein. Three days later, the mice were sacrificed and the organs, including the lung, heart, spleen, liver, and kidney, were harvested for luciferase activity measurement. In both the liver (Fig. 5A) and heart (Fig. 5B), preimmunization with Ad5/H5 blocked Ad5/H5 infection but not Ad5/H3 infection. Similarly, preimmunization with Ad5/H3 prevented Ad5/H3 infection but allowed Ad5/H5 infection to a large extent. Other organs exhibited similar patterns (data not shown). These data suggested that Ad5/H3 and Ad5/H5 were not cross-neutralized in vivo and that sequential injections of the two viruses could overcome the host neutralization response.
FIG. 5.
Ad5/H3 and Ad5/H5 were not cross-neutralized in vivo. C57BL/6 mice were preimmunized with PBS or 109 VPs of Ad5/H3 or Ad5/H5 per mouse by i.v. injection. One month later, the mice were injected with either PBS or 1010 VPs of Ad5/H3 or Ad5/H5 per mouse by i.v. injection. Three days later, the mice were sacrificed and their livers and hearts were harvested for the luciferase activity assay. In both the liver (A) and heart (B), Ad5/H5 infection was greatly inhibited by Ad5/H5 preimmunization but was retained to a large degree in Ad5/H3-preimmunized mice. Similarly, Ad5/H3 infection was retained in Ad5/H5-preimmunized mice but was blocked by Ad5/H3 preimmunization. Note: it appears that luciferase expression in mice preimmunized with Ad5/H3 was sustained in liver even after 1 month but not in the heart.
DISCUSSION
In this study, we constructed a novel hexon chimera, Ad5/H3, and characterized its growth kinetics, heat stability, gene transfer efficiency, and neutralization properties. We found that the hexon of Ad3, which belongs to adenovirus subgroup B, was able to encapsidate the Ad5 genome, though with less efficiency than the Ad5 hexon, and that the hexon-chimeric virions were less stable than Ad5 bearing its native hexon. Furthermore, we found that Ad5/H3 and Ad5/H5 escaped the host neutralizing response against one another and thus could be employed sequentially to achieve readministration in gene therapy applications.
Several strategies have been employed to escape the host immunity against Ad5 vectors (22). These include transient suppression of the host immune response by blocking CD4+ T-cell activation (17, 39) or physical masking of the capsid of Ad5 from neutralizing antibodies with polyethylene glycol polymers (5, 20). However, these approaches require additional procedures and their efficacy and safety remain to be tested. Alternatively, since the neutralizing response is serotype specific, different serotypes of adenoviruses can be used sequentially to escape the host immunity. However, this requires the development of genomes of different serotypes and evaluation of the gene transfer efficacy of each adenovirus vector carrying the gene of interest. Switching the hexon, the serotype determinant of adenovirus, on the other hand, would be a much simpler approach to overcoming the host neutralizing antibodies generated in previous administrations.
Many efforts have been made in search of hexon chimeras that can be used to circumvent host immunity against Ad5. However, hexon switch has only succeeded between Ad5 and Ad1, Ad2, Ad6, and Ad12 (15, 24, 40). Among them, except Ad12, which belongs to subgroup A, all others belong to the same subgroup as Ad5, subgroup C. This led to the assumption that hexon switch might only work efficiently within a subgroup. Our study suggests that the hexon of Ad3, a subgroup B member, can also replace the Ad5 hexon, supporting the concept that hexon switch can work between subgroups. In addition, neutralizing antibodies generated by C57BL/6 mice against Ad5/H3 and Ad5/H5 exhibited minimal cross neutralization to each other in vitro and the two viruses successfully escaped the host neutalization responses against each other in vivo, suggesting that the two viruses can be used sequentially to circumvent host neutralization responses.
The inability of many alternate hexons to package the Ad5 genome may be due to the incompatibility of the hexons with other Ad5 capsid proteins that are involved in virion assembly. For example, polypeptides VI, VIII, and IX are associated with the hexon protein and may participate in stabilizing the hexon capsomere lattice (11, 28). Another protein, the 100-kDa protein, is required for hexon trimerization (8, 12). Incompatibility of foreign hexons with these Ad5 components might also explain the fact that the Ad3 hexon encapsidated the Ad5 genome less efficiently than the native Ad5 hexon and that Ad5/H3 was more susceptible to heat inactivation of virions. Although thermostability data are not available for other hexon chimeras with regard to packaging efficiency, similar results were obtained for Ad5 bearing the hexons of Ad1, Ad2, and Ad12 (24, 40). Ad5 with the Ad12 hexon has the lowest efficiency since its yield is about 100-fold lower than that of Ad5, while Ad5 with the Ad1, Ad2, or Ad3 hexon resulted in a yield about 10-fold lower than that for Ad5 (Fig. 3A) (24, 40), indicating that the Ad12 hexon is less compatible with Ad5 components than the hexons of Ad1, Ad2, and Ad3. Considering that HVRs of hexons contain the serotype-specific epitopes and that they do not appear to be involved in binding any other viral proteins, an alternative strategy to construct hexon chimeras could be to switch only the HVRs. This should allow better rescue of hexon chimeras while retaining their ability to circumvent the host neutralization response.
It is noteworthy that Ad5/H3 exhibited a gene transfer efficacy lower than that for Ad5/H5. Similar results were obtained with Ad5 containing the Ad12 hexon, but not with Ad5 containing the Ad1, Ad2, and Ad6 hexons (40). This implies that hexons from different subgroups—which are thus more distantly related—interfered with Ad5 infectivity. Hexons are not known to be involved in the cell-binding or internalization steps of adenoviruses; thus, it is not clear whether hexons interfere with these pre-cell entry steps directly. Nonetheless, these hexons could indirectly interfere with penton base-mediated internalization by their interaction with the penton base. Another possibility is that the hexon may interfere with virus disassembly at the post-cell entry steps. In the disassembly process, viral components including polypeptides VIII, VI, and IX dissociate in an orderly manner from the hexon capsid and DNA is finally freed into the nucleus (28). Interactions of these components with the hexon capsid may be disturbed by hexon replacement, which in turn could affect the efficacy of the dissociation process. Reduction in infectivity caused by hexon chimerism is a drawback for gene therapy application. However, this may be overcome by replacing only the HVRs of hexon, and/or by combining the hexon switch strategy with infectivity enhancement technology that can be achieved via fiber modifications (2, 9, 33, 34, 36).
Acknowledgments
This study was supported by National Institutes of Health and National Cancer Institute grants R01 HL67962, R01 CA86881-01, and P50 CA89019, as well as a grant from the Susan G. Komen Breast Cancer Foundation, BCTR01-00481.
REFERENCES
- 1.Athappilly, F. K., R. Murali, J. J. Rux, Z. Cai, and R. M. Burnett. 1994. The refined crystal structure of hexon, the major coat protein of adenovirus type 2, at 2.9 Å resolution. J. Mol. Biol. 242:430-455. [DOI] [PubMed] [Google Scholar]
- 2.Barnett, B. G., C. J. Crews, and J. T. Douglas. 2002. Targeted adenoviral vectors. Biochim. Biophys. Acta 1575:1-14. [DOI] [PubMed] [Google Scholar]
- 3.Benko, M., B. Harrach, and W. C. Russell. 1999. Family adenoviridae, p. 227-238. In M. H. Van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Seventh Report of the International Committee on Taxonomy of Viruses. Academic Press, Inc., New York, N.Y.
- 4.Crawford-Miksza, L., and D. P. Schnurr. 1996. Analysis of 15 adenovirus hexon proteins reveals the location and structure of seven hypervariable regions containing serotype-specific residues. J. Virol. 70:1836-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Croyle, M. A., N. Chirmule, Y. Zhang, and J. M. Wilson. 2001. “Stealth” adenoviruses blunt cell-mediated and humoral immune responses against the virus and allow for significant gene expression upon readministration in the lung. J. Virol. 75:4792-4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dai, Y., E. M. Schwarz, D. Gu, W. W. Zhang, N. Sarvetnick, and I. M. Verma. 1995. Cellular and humoral immune responses to adenoviral vectors containing factor IX gene: tolerization of factor IX and vector antigens allows for long-term expression. Proc. Natl. Acad. Sci. USA 92:1401-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Jong, J. C., A. G. Wermenbol, M. W. Verweij-Uijterwaal, K. W. Slaterus, P. Wertheim-Van Dillen, G. J. Van Doornum, S. H. Khoo, and J. C. Hierholzer. 1999. Adenoviruses from human immunodeficiency virus-infected individuals, including two strains that represent new candidate serotypes Ad50 and Ad51 of species B1 and D, respectively. J. Clin. Microbiol. 37:3940-3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D'Halluin, J. C. 1995. Virus assembly. Curr. Top. Microbiol. Immunol. 199:47-66. [PubMed] [Google Scholar]
- 9.Dmitriev, I., V. Krasnykh, C. R. Miller, M. Wang, E. Kashentseva, G. Mikheeva, N. Belousova, and D. T. Curiel. 1998. An adenovirus vector with genetically modified fibers demonstrates expanded tropism via utilization of a coxsackievirus and adenovirus receptor-independent cell entry mechanism. J. Virol. 72:9706-9713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dmitriev, I. P., E. A. Kashentseva, and D. T. Curiel. 2002. Engineering of adenovirus vectors containing heterologous peptide sequences in the C terminus of capsid protein IX. J. Virol. 76:6893-6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Everitt, E., B. Sundquist, U. Pettersson, and L. Philipson. 1973. Structural proteins of adenoviruses. X. Isolation and topography of low molecular weight antigens from the virion of adenovirus type 2. Virology 52:130-147. [DOI] [PubMed] [Google Scholar]
- 12.Frost, E., and J. Williams. 1978. Mapping temperature-sensitive and host-range mutations of adenovirus type 5 by marker rescue. Virology 91:39-50. [DOI] [PubMed] [Google Scholar]
- 13.Gahery-Segard, H., F. Farace, D. Godfrin, J. Gaston, R. Lengagne, T. Tursz, P. Boulanger, and J. G. Guillet. 1998. Immune response to recombinant capsid proteins of adenovirus in humans: antifiber and anti-penton base antibodies have a synergistic effect on neutralizing activity. J. Virol. 72:2388-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gall, J., A. Kass-Eisler, L. Leinwand, and E. Falck-Pedersen. 1996. Adenovirus type 5 and 7 capsid chimera: fiber replacement alters receptor tropism without affecting primary immune neutralization epitopes. J. Virol. 70:2116-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gall, J. G., R. G. Crystal, and E. Falck-Pedersen. 1998. Construction and characterization of hexon-chimeric adenoviruses: specification of adenovirus serotype. J. Virol. 72:10260-10264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harvey, B. G., N. R. Hackett, T. El-Sawy, T. K. Rosengart, E. A. Hirschowitz, M. D. Lieberman, M. L. Lesser, and R. G. Crystal. 1999. Variability of human systemic humoral immune responses to adenovirus gene transfer vectors administered to different organs. J. Virol. 73:6729-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jooss, K., L. A. Turka, and J. M. Wilson. 1998. Blunting of immune responses to adenoviral vectors in mouse liver and lung with CTLA4Ig. Gene Ther. 5:309-319. [DOI] [PubMed] [Google Scholar]
- 18.Molinier-Frenkel, V., R. Lengagne, F. Gaden, S. S. Hong, J. Choppin, H. Gahery-Segard, P. Boulanger, and J. G. Guillet. 2002. Adenovirus hexon protein is a potent adjuvant for activation of a cellular immune response. J. Virol. 76:127-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nemerow, G. R. 2000. Adenoviral vectors—new insights. Trends Microbiol. 8:391-394. [DOI] [PubMed] [Google Scholar]
- 20.O'Riordan, C. R., A. Lachapelle, C. Delgado, V. Parkes, S. C. Wadsworth, A. E. Smith, and G. E. Francis. 1999. PEGylation of adenovirus with retention of infectivity and protection from neutralizing antibody in vitro and in vivo. Hum. Gene Ther. 10:1349-1358. [DOI] [PubMed] [Google Scholar]
- 21.Ostapchuk, P., and P. Hearing. 2001. Pseudopackaging of adenovirus type 5 genomes into capsids containing the hexon proteins of adenovirus serotypes B, D, or E. J. Virol. 75:45-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ritter, T., M. Lehmann, and H. Volk. 2002. Improvements in gene therapy: averting the immune response to adenoviral vectors. BioDrugs 16:3-10. [DOI] [PubMed] [Google Scholar]
- 23.Roberts, M. M., J. L. White, M. G. Grutter, and R. M. Burnett. 1986. Three-dimensional structure of the adenovirus major coat protein hexon. Science 232:1148-1151. [DOI] [PubMed] [Google Scholar]
- 24.Roy, S., P. S. Shirley, A. McClelland, and M. Kaleko. 1998. Circumvention of immunity to the adenovirus major coat protein hexon. J. Virol. 72:6875-6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rux, J. J., and R. M. Burnett. 2000. Type-specific epitope locations revealed by X-ray crystallographic study of adenovirus type 5 hexon. Mol. Ther. 1:18-30. [DOI] [PubMed] [Google Scholar]
- 26.Seki, T., I. Dmitriev, K. Suzuki, E. Kashentseva, K. Takayama, M. Rots, T. Uil, H. Wu, M. Wang, and D. T. Curiel. 2002. Fiber shaft extension in combination with HI loop ligands augments infectivity for CAR-negative tumor targets but does not enhance hepatotropism in vivo. Gene Ther. 9:1101-1108. [DOI] [PubMed] [Google Scholar]
- 27.Sharpe, S., A. Fooks, J. Lee, K. Hayes, C. Clegg, and M. Cranage. 2002. Single oral immunization with replication deficient recombinant adenovirus elicits long-lived transgene-specific cellular and humoral immune responses. Virology 293:210-216. [DOI] [PubMed] [Google Scholar]
- 28.Shenk, T. 1996. Adenoviridae: the viruses and their replication, p. 2111-2148. In B. N. Fields, D. M. Knipe, P. M. Howley, et al. (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 29.Smith, T. A., B. D. White, J. M. Gardner, M. Kaleko, and A. McClelland. 1996. Transient immunosuppression permits successful repetitive intravenous administration of an adenovirus vector. Gene Ther. 3:496-502. [PubMed] [Google Scholar]
- 30.Stewart, P. L., and R. M. Burnett. 1995. Adenovirus structure by X-ray crystallography and electron microscopy. Curr. Top. Microbiol. Immunol. 199:25-38. [DOI] [PubMed] [Google Scholar]
- 31.van Oostrum, J., P. R. Smith, M. Mohraz, and R. M. Burnett. 1987. The structure of the adenovirus capsid. III. Hexon packing determined from electron micrographs of capsid fragments. J. Mol. Biol. 198:73-89. [DOI] [PubMed] [Google Scholar]
- 32.Vorburger, S. A., and K. K. Hunt. 2002. Adenoviral gene therapy. Oncologist 7:46-59. [DOI] [PubMed] [Google Scholar]
- 33.Wickham, T. J., P. W. Roelvink, D. E. Brough, and I. Kovesdi. 1996. Adenovirus targeted to heparan-containing receptors increases its gene delivery efficiency to multiple cell types. Nat. Biotechnol. 14:1570-1573. [DOI] [PubMed] [Google Scholar]
- 34.Wickham, T. J., E. Tzeng, L. L. Shears II, P. W. Roelvink, Y. Li, G. M. Lee, D. E. Brough, A. Lizonova, and I. Kovesdi. 1997. Increased in vitro and in vivo gene transfer by adenovirus vectors containing chimeric fiber proteins. J. Virol. 71:8221-8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wohlfart, C. 1988. Neutralization of adenoviruses: kinetics, stoichiometry, and mechanisms. J. Virol. 62:2321-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu, H., T. Seki, I. Dmitriev, T. Uil, E. Kashentseva, T. Han, and D. T. Curiel. 2002. Double modification of adenovirus fiber with RGD and polylysine motifs improves coxsackievirus-adenovirus receptor-independent gene transfer efficiency. Hum. Gene Ther. 13:1647-1653. [DOI] [PubMed] [Google Scholar]
- 37.Yang, Y., S. E. Haecker, Q. Su, and J. M. Wilson. 1996. Immunology of gene therapy with adenoviral vectors in mouse skeletal muscle. Hum. Mol. Genet. 5:1703-1712. [DOI] [PubMed] [Google Scholar]
- 38.Yang, Y., Q. Li, H. C. Ertl, and J. M. Wilson. 1995. Cellular and humoral immune responses to viral antigens create barriers to lung-directed gene therapy with recombinant adenoviruses. J. Virol. 69:2004-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang, Y., Q. Su, I. S. Grewal, R. Schilz, R. A. Flavell, and J. M. Wilson. 1996. Transient subversion of CD40 ligand function diminishes immune responses to adenovirus vectors in mouse liver and lung tissues. J. Virol. 70:6370-6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Youil, R., T. J. Toner, Q. Su, M. Chen, A. Tang, A. J. Bett, and D. Casimiro. 2002. Hexon gene switch strategy for the generation of chimeric recombinant adenovirus. Hum. Gene Ther. 13:311-320. [DOI] [PubMed] [Google Scholar]
- 41.Zaiss, A. K., Q. Liu, G. P. Bowen, N. C. Wong, J. S. Bartlett, and D. A. Muruve. 2002. Differential activation of innate immune responses by adenovirus and adeno-associated virus vectors. J. Virol. 76:4580-4590. [DOI] [PMC free article] [PubMed] [Google Scholar]