Abstract
Lipid binding to the potassium channel KcsA from Streptomyces lividans has been studied using quenching of the fluorescence of Trp residues by brominated phospholipids. It is shown that binding of phospholipids to nonannular lipid binding sites on KcsA, located one each at the four protein-protein interfaces in the tetrameric structure, is specific for anionic phospholipids, zwitterionic phosphatidylcholine being unable to bind at the sites. The binding constant for phosphatidylglycerol of 3.0 ± 0.7 mol fraction−1 means that in a membrane containing ∼20 mol% phosphatidylglycerol, as in the Escherichia coli inner membrane, the nonannular sites will be ∼37% occupied by phosphatidylglycerol. The binding constant for phosphatidic acid is similar to that for phosphatidylglycerol but binding constants for phosphatidylserine and cardiolipin are about double those for phosphatidylglycerol. Binding to annular sites around the circumference of the KcsA tetramer are different on the extracellular and intracellular faces of the membrane. On the extracellular face of the membrane the binding constants for anionic lipids are similar to those for phosphatidylcholine, the lack of specificity being consistent with the lack of any marked clusters of charged residues on KcsA close to the membrane on the extracellular side. In contrast, binding to annular sites on the intracellular side of the membrane shows a distinct structural specificity, with binding of phosphatidic acid and phosphatidylglycerol being stronger than binding of phosphatidylcholine, whereas binding constants for phosphatidylserine and cardiolipin are similar to that for phosphatidylcholine. It is suggested that this pattern of binding follows from the pattern of charge distribution on KcsA on the intracellular side of the membrane.
INTRODUCTION
The homotetrameric potassium channel KcsA from Streptomyces lividans requires anionic lipid to function; KcsA, when reconstituted into bilayers of phosphatidylcholine or phosphatidylethanolamine, fails to open and only opens when the bilayer also contains an anionic phospholipid such as phosphatidylglycerol, phosphatidylserine, or cardiolipin (1,2). The crystal structure of KcsA, crystallized from solution in decyl maltoside, shows a lipid molecule located at each monomer-monomer interface in the tetrameric structure, modeled as a diacylglycerol because the lipid headgroup was not well resolved (2). This lipid molecule is probably phosphatidylglycerol because KcsA, purified using dodecyl maltoside as detergent, contains ∼0.7 mol of phosphatidylglycerol per monomer (2). Refolding experiments showed that anionic lipid is not required for the formation of the KcsA tetramer (2) and, indeed, KcsA purified using Mega-9 as detergent lacked any significant amount of bound lipid but still forms a stable tetrameric structure (3). It has been suggested that the functional requirement of KcsA for anionic lipid follows from a requirement that the lipid binding sites identified in the crystal structure be occupied by anionic lipid (2).
We have termed lipid binding sites located at protein-protein interfaces or between transmembrane α-helices as nonannular sites, to distinguish them from the boundary or annular lipids that cover the bulk of the hydrophobic surface of a membrane protein (4). The obvious question for KcsA is whether the nonannular sites are empty in the absence of anionic lipid, this leading to the lack of activity, or whether, in the absence of anionic lipid, the nonannular sites are occupied by zwitterionic lipid, the channel not being functional when the nonannular sites are occupied by zwitterionic lipid. It is also important to know how strongly anionic lipid binds to the nonannular sites; the fact that purification from decyl maltoside leads to substoichiometric levels of anionic lipid (2) and that purification from Mega-9 leads to the loss of almost all the bound lipid (3) shows that the affinity is not so high that binding is essentially irreversible. The level of occupancy of the nonannular sites by anionic lipid in the native membrane will depend on the binding constant for anionic lipid and on the concentration of anionic lipid in the outer monolayer of the lipid bilayer where the nonannular binding sites are located.
Here we show how quenching of the fluorescence of Trp residues in KcsA by bromine-containing phospholipids can be used to study the selectivity of binding at the nonannular sites on KcsA. Bromine-containing phospholipids are made by addition of bromine across the double bonds of phospholipids containing two oleyl chains: these lipids behave much like conventional phospholipids with unsaturated fatty acyl chains, because the bulky bromine atoms have effects on lipid packing that are similar to those of a cis double bond (5). The efficiency of quenching of Trp fluorescence by phospholipids containing dibrominated fatty acyl chains depends on the sixth power of the distance between the Trp and the dibromo group with a value for Ro, the distance at which energy transfer is 50% efficient, of 8 Å (6,7). This could either indicate that fluorescence quenching is by a Förster energy transfer mechanism (7) or that quenching is collisional, taking into account the depth distribution of the fluorophore and quencher in the membrane (8). Whichever is the case, the observation that the experimental quenching data fit well to a sixth power dependence on distance suggests that an analysis in these terms can be used to estimate expected efficiencies of quenching by bromine-containing phospholipids. In particular, the short range of the quenching process means that only lipids bound in the immediate vicinity of a Trp residue can affect its fluorescence intensity.
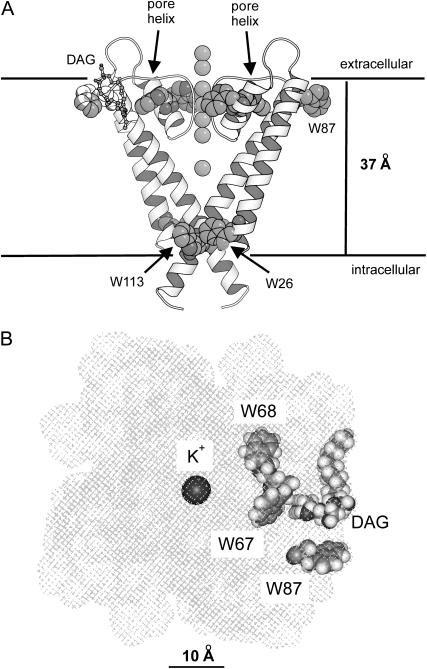
KcsA contains five Trp residues, Trp-26 and Trp-113 exposed to the lipid bilayer on the intracellular side of the membrane, Trp-87 exposed to the lipid bilayer on the extracellular side, and Trp-67 and Trp-68 making up part of the pore structure on the extracellular side (Fig. 1 A). The lipid-exposed Trp residues are not conserved in the potassium channel family, and it is generally found that hydrophobic lipid-exposed residues can be replaced with other hydrophobic residues with no effect on function, as shown, for example, in studies of the Shaker potassium channel (9,10). Replacing the lipid-exposed Trp residues in KcsA with Cys has been shown to have no effect on function (11). In the studies reported here we have replaced Trp-26 and Trp-113 on the intracellular side with Leu, leaving the three Trp residues on the extracellular side. The locations of these three Trp residues with respect to the surface of the KcsA trimer and the nonannular lipid binding site are shown in Fig. 1 B. Distances between the Trp residues and the bromine atoms in the lipid fatty acyl chains can be estimated as follows. Bromination of an oleyl chain gives the corresponding 9,10-dibromostearoyl derivative. The hydrophobic thickness of a bilayer of dioleoylphosphatidylcholine is ∼27 Å (12), so that the bromines in the brominated phospholipids will be ∼7 Å from the glycerol backbone region of the bilayer. Distances along the membrane between the β-carbons of Trp residues 67 and 68 and the closest fatty acyl chains of an annular lipid molecule are ∼12 and 16 Å, respectively, giving Trp-bromine separation distances of ∼14 and 17 Å, respectively. In Förster energy transfer theory, the efficiency of energy transfer E between fluorophore and quencher is related to the distance of separation d by
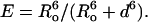 |
(1) |
FIGURE 1.
The structure of KcsA. (A) A side view of the structure with only two of the monomers making up the tetrameric structure being included for clarity. Trp residues and the K+ ions are shown in space-fill format; a bound lipid molecule (DAG) is shown in ball-and-stick format. The horizontal lines show the probable locations of the glycerol backbone regions on the two sides of the lipid bilayer (3). (B) A view of KcsA from the extracellular side of the membrane showing the locations of Trp-67, Trp-68, and Trp-87 around a bound lipid molecule (DAG). The hatches show the surface of KcsA with the K+ ions, the Trp residues, and the bound lipid molecule shown in space-fill format. The coordinates were from PDB 1K4C (31).
The expected levels of quenching of Trp-67 and Trp-68 due to brominated annular lipid will then be ∼3 and 1%, respectively, assuming that quenching is dominated by the nearest brominated fatty acyl chain. In contrast, the distance along the membrane surface between Trp-87 and the nearest annular lipid molecule is ∼3.3 Å, giving a Trp-bromine separation distance of 7.7 Å and an expected level of quenching of ∼56%. With a lipid diameter of ∼9.4 Å the Trp-bromine distance for Trp-87 and the next nearest annular lipid molecule will be 12.1 Å, giving an expected level of quenching of ∼7%. Thus, quenching of the fluorescence of Trp-87 will report on only those one or two lipid binding sites closest to Trp-87. A similar calculation for the effect of a brominated lipid molecule bound to the nonannular site suggests that the level of quenching for Trp-67, for which the Trp-bromine separation distance is ∼7.4 Å, will be ∼61%, with a level of quenching for the more distant Trp-68 and Trp-87 of ∼7%. Thus, quenching of Trp-67 will report on binding at nonannular sites, quenching of Trp-87 will report on binding at annular sites, and Trp-68 will be essentially unquenched by brominated lipid.
EXPERIMENTAL PROCEDURES
Dioleoylphosphatidylcholine (di(C18:1)PC), dioleoylphosphatidylserine (di(C18:1)PS), dioleoylphosphatidic acid (di(C18:1)PA), dioleoylphosphatidylglycerol (di(C18:1)PG), and tetraoleoylcardiolipin (tetra(C18:1)CL) were obtained from Avanti Polar Lipids (Alabaster, AL). Phospholipids were brominated as described by East and Lee (5) to give the corresponding brominated lipids di(9,10-dibromostearoyl)phosphatidylcholine (di(Br2C18:0)PC), di(9,10-dibromostearoyl)phosphatidylserine (di(Br2C18:0)PS), di(9,10-dibromostearoyl)phosphatidic acid (di(Br2C18:0)PA), di(9,10-dibromostearoyl)phosphatidylglycerol (di(Br2C18:0)PG), and tetra(9,10-dibromostearoyl)cardiolipin (tetra(Br2C18:0)CL). Cholate was purified by dissolving cholic acid in the minimum volume of warm methanol, mixing with KOH in methanol, and then precipitation by addition of a large excess of diethyl ether. The precipitate was collected by filtration and then dried under a stream of air.
Mutagenesis and purification of KcsA
A plasmid containing the kcsA gene (13) with a poly-His epitope at the N-terminus was the generous gift of Professor H. Schrempf, University of Osnabruck, Osnabruck, Germany. KcsA was purified according to the method of Williamson et al. (3) with a few modifications. Briefly, cells were washed and resuspended in buffer (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4) and lysed by sonication. The sample was spun at 100,000 × g for 30 min and the membrane pellet was solubilized in the above buffer containing 5 mM dodecyl maltoside (Calbiochem, San Diego, CA) for 1 h at room temperature. Unsolubilized material was removed by centrifugation at 8000 × g for 20 min and the supernatant was loaded onto a 1-ml Ni2+-Sepharose-His-Trap affinity column (Amersham, Buckinghamshire, UK) preequilibrated with buffer containing 20 mM imidazole. After washing the column, the His-tagged KcsA protein was eluted with 300 mM imidazole and stored at −80°C until use. The homogeneity of KcsA was assessed by sodium dodecyl-sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), using the method of Laemmli (14). Concentrations of KcsA were estimated from absorption spectra measured in buffer containing 1% sodium dodecyl sulfate to reduce light scatter, using extinction coefficients of 34,850 M−1 cm−1, 29,160 M−1 cm−1, and 23,470 M−1 cm−1, for wild-type KcsA, W113L, and W26, 113L, respectively, at 280 nm.
Site-directed mutagenesis was performed using the Quick-change protocol from Stratagene (La Jolla, CA). Mutated KcsA were prepared with the Trp residue at position 113 replaced by Leu (W113L) or with the Trp residues at positions 26 and 113 both replaced by Leu (W26,113L). W113L was produced using synthetic oligonucleotide primers 5′-CCGCGCTGGCAACCTTGTTCGTCGGCCGGGAAC-3′ and 5′-CCGACGAACAAGGTTGCCAGCGCGGCGGTCACCAGAC-3′ and W26,113L was pro-duced by mutation of Trp-26 in W113L using the primers 5′-GTGC-GCTTCATTTGAGGGCCGCGGGTGC-3′ and 5′-GGCCCTCAAATGAAGCGCACTGCCGTGGC-3′. After polymerase chain reaction mutagenesis, the native methylated DNA templates were digested with Dpn1 (Promega, Madison, WI) for 2 h at 37°C. The mutations were confirmed by DNA sequencing.
Fluorescence measurements
Purified KcsA was reconstituted into lipid bilayers by mixing lipid and KcsA in cholate followed by dilution of 50 μl of the detergent-lipid-protein mixture into 3 ml buffer (20 mM HEPES and 1 mM EGTA, at pH 7.2) to decrease the concentration of cholate below its critical micelle concentration, as described in Alvis et al. (15). The final protein concentration was 0.24 μM and the molar ratio of lipid to KcsA was 100:1. Fluorescence was recorded on an SLM 8000C fluorimeter (Urbana, IL) with excitation at 290 nm, at 25°C. Fluorescence emission spectra were corrected for light scatter by subtraction of a blank consisting of lipid alone in buffer. Spectra were corrected for the wavelength dependence of instrumental response using a set of correction factors generated by comparison of an emission spectrum for tryptophan in buffer with the corrected emission spectrum for tryptophan published by Chen (16). In quenching experiments with acrylamide, the inner filter effect was minimized by exciting fluorescence at 295 nm, corrections for the inner filter effect being made by applying the correction factor 10ɛ0.5c, where ɛ is the molar extinction coefficient at 295 nm and c is the molar concentration of acrylamide.
The reported fluorescence intensities represent the average of duplicate measurements from two or three separate reconstitutions.
Analysis of fluorescence quenching results
Quenching of Trp fluorescence in a mixture of a nonbrominated lipid with its brominated analog, where the lipid binds only to the annular sites around KcsA, has been fitted to a lattice model for quenching (3,15,17–19) using the equation
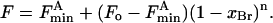 |
(2) |
Here Fo and 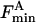 are the fluorescence intensities of KcsA in nonbrominated and brominated lipid, respectively, F is the fluorescence intensity in the phospholipid mixture when the mol fraction of brominated lipid is xBr, and n is the number of annular lipid binding sites on KcsA from which the fluorescence of a Trp residue can be quenched. In a mixture of two classes of lipid A and B where, for example, lipid A is a nonbrominated zwitterionic lipid and lipid B is a brominated anionic lipid, an equilibrium will be established at each of the annular sites:
are the fluorescence intensities of KcsA in nonbrominated and brominated lipid, respectively, F is the fluorescence intensity in the phospholipid mixture when the mol fraction of brominated lipid is xBr, and n is the number of annular lipid binding sites on KcsA from which the fluorescence of a Trp residue can be quenched. In a mixture of two classes of lipid A and B where, for example, lipid A is a nonbrominated zwitterionic lipid and lipid B is a brominated anionic lipid, an equilibrium will be established at each of the annular sites:
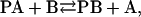 |
where PA and PB are protein bound to lipids A and B, respectively, and the binding constant for B relative to A is given by
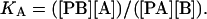 |
(3) |
Fluorescence quenching in the mixture is described by the equation
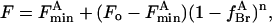 |
(4) |
where 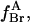 the fraction of annular sites on KcsA occupied by brominated lipid, is given by
the fraction of annular sites on KcsA occupied by brominated lipid, is given by
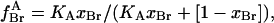 |
(5) |
where xBr is the mol fraction of brominated lipid in the mixture.
If only anionic lipid, lipid B, can bind to the nonannular sites on KcsA then binding to the nonannular sites can be described by a simple binding equation:
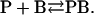 |
The association constant KNA for binding at the nonannular sites is given by
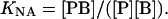 |
(6) |
The fraction of nonannular sites occupied by brominated anionic lipid, 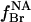 is given by
is given by
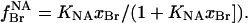 |
(7) |
The fluorescence intensity for a Trp residue close to the nonannular site will be directly related to the probability of occupation of the nonannular site by brominated lipid:
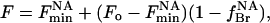 |
(8) |
where 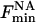 is the fluorescence intensity when the nonannular site is occupied by brominated lipid.
is the fluorescence intensity when the nonannular site is occupied by brominated lipid.
In the special case of the mutant W26,113L that contains three Trp residues with equal fluorescence emission intensities, where one Trp residue is not quenched by brominated lipid, one Trp is quenched by binding of brominated lipid to the annular sites, and one Trp is quenched by binding of brominated lipid to the nonannular sites, the fluorescence intensity will be given by
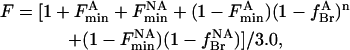 |
(9) |
where the fluorescence intensity has been scaled to 1 for the protein in nonbrominated lipid.
To describe fluorescence quenching for wild-type KcsA from the annular sites it is necessary to consider the possibility that binding constants at the annular sites on the extracellular and intracellular sides of the membrane are different. For wild-type KcsA, Trp-67 and Trp-87 will not be quenched by annular lipid, Trp-87 will be quenched by annular lipid binding on the extracellular side of the membrane, and Trp-26 and Trp-113 will be quenched by annular lipid binding on the intracellular side of the membrane (Fig. 1). The fluorescence intensity in this case will be given by
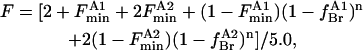 |
(10) |
where the fluorescence intensity has been scaled to 1 for the protein in nonbrominated lipid and 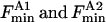 are fluorescence intensities for Trp-87 and for Trp-26 and Trp-113, respectively, in brominated lipid, and
are fluorescence intensities for Trp-87 and for Trp-26 and Trp-113, respectively, in brominated lipid, and 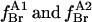 are the fractional occupancies of the annular sites by brominated lipid on the extracellular and intracellular sides of the membrane, respectively, calculated from Eq. 5 with the appropriate value for the relative binding constant KA on the extracellular and intracellular sides of the membrane.
are the fractional occupancies of the annular sites by brominated lipid on the extracellular and intracellular sides of the membrane, respectively, calculated from Eq. 5 with the appropriate value for the relative binding constant KA on the extracellular and intracellular sides of the membrane.
The experimental data were fitted to the above equations using the nonlinear least-squares routine in the SigmaPlot package (SPSS, Chicago, IL).
RESULTS
Models for interpretation of fluorescence quenching results
Fluorescence quenching data are fitted using a simple lattice model in which n lipid binding sites are located sufficiently close to a particular Trp residue that occupation of any of the n sites by a brominated lipid molecule will result in fluorescence quenching (3,15,17–19). The value of n is obtained by fitting fluorescence quenching curves in mixtures of a nonbrominated lipid and its brominated analog to Eq. 2. When the fluorescence quenching is <100% efficient this analysis can result in an overestimate of the value of n (20). Yeager and Feigenson (20) showed that in these cases fluorescence quenching is more accurately described by a model in which each quencher contributes additively to the total quenching rate constant for the Trp residue, giving rise to the following equation:
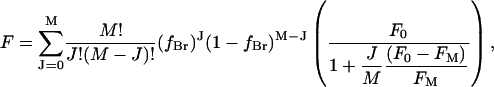 |
(11) |
where M is the number of sites from which the fluorescence of the Trp residue can be quenched, F0 and Fm are the fluorescence intensities in nonbrominated and brominated lipid, respectively, and fBr, the fractional occupancy of the sites by brominated lipid, is given by Eq. 5. In the limit that Fm = 0, Eq. 11 reduces to Eq. 2.
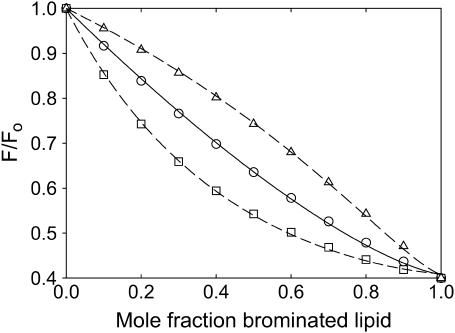
A comparison between Eqs. 2 and 11 is presented in Fig. 2. Fig. 2 shows fluorescence intensities calculated from Eq. 11 for a mixture of a nonbrominated lipid with its brominated analog for values of M and FM of 2 and 0.4, respectively. This calculated curve was then fitted to Eq. 2 giving a value for n of 1.4 (Fig. 2) confirming that Eq. 2 overestimates the number of binding sites from which quenching can be observed, when FM > 0. The important point here, however, is that the use of Eqs. 4 and 11 give the same values for the relative binding constant KA because the values of KA are obtained by comparison of pairs of quenching curves. For example, Fig. 2 shows quenching curves calculated from Eq. 11 again with values of M and FM of 2 and 0.4, respectively, but now for two mixtures of lipids, one where the relative binding constant KA is 2.0 and the other where the relative binding constant KA is 0.5 (Fig. 2). Fitting these two curves to Eq. 4 with the value of n fixed at the value of 1.4 gives relative binding constants KA of 2.0 and 0.5. The fact that Eq. 4 recovers exactly the values of KA used in the simulations justifies the use of the mathematically much more convenient Eq. 4.
FIGURE 2.
Comparison of methods for analysis of quenching data. (○) shows points calculated from Eq. 11 with values for M and FM of 2 and 0.4, respectively, and a value for KA of 1. The solid line shows a fit of the calculated data (○) to Eq. 2 giving a value for n of 1.4. (□, ▵) show points calculated from Eq. 11 with values for M and FM of 2 and 0.4, respectively, with values for KA of 2.0 (□) and 0.5 (▵). The dashed lines show fits of the calculated data (□, ▵) to Eq. 4 with a value for n of 1.4, giving values for KA of 2.0 (□) and 0.5 (▵).
Properties of the Trp mutants of KcsA
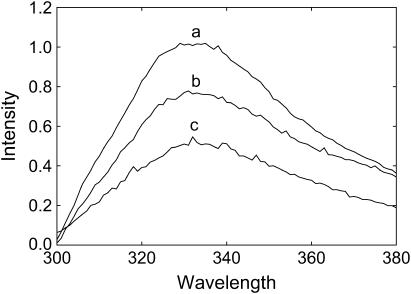
W113L and W26,113L ran on SDS gels as mixtures of monomer and tetramer, as for wild-type KcsA (data not shown). Fluorescence emission maxima for wild-type KcsA and for the mutants W113L and W26,113L are all close to 333 nm (Fig. 3), consistent with a location for all the Trp residues in KcsA close to the glycerol backbone region of the lipid bilayer (3). The decreases in fluorescence intensity on removal of Trp-26 and Trp-113 are similar (Fig. 3), again consistent with similar fluorescence properties for all the Trp residues in KcsA, as previously suggested (3). Quenching of Trp fluorescence by acrylamide fits to identical, linear Stern-Volmer plots for the mutants and wild-type KcsA (data not shown) with values of F/Fo at 0.24 M acrylamide of 0.62, comparable to the value of 0.6 observed by Caputo and London (21) for Trp residues at the ends of a model transmembrane α-helix in a lipid bilayer. Fluorescence emission spectra for W26,113L are identical in di(C18:1)PC, di(C18:1)PA, di(C18:1)PG, di(C18:1)PS, and tetra(C18:1)CL (data not shown), suggesting that binding of anionic lipid to W26,113L results in no large change in conformation.
FIGURE 3.
Fluorescence emission spectra for wild-type and mutant KcsA. Fluorescence emission spectra are shown for wild-type KcsA (a), W113L (b), and W26,113L (c) reconstituted into bilayers of di(C18:1)PC. The concentration of KcsA was 0.24 μM and the molar ratio of lipid/KcsA was 100:1.
Fluorescence quenching by zwitterionic and anionic lipids
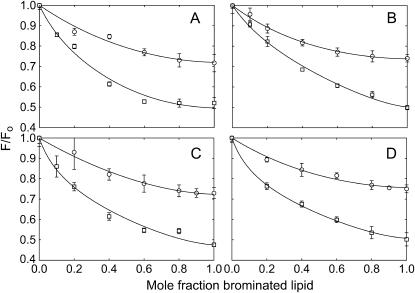
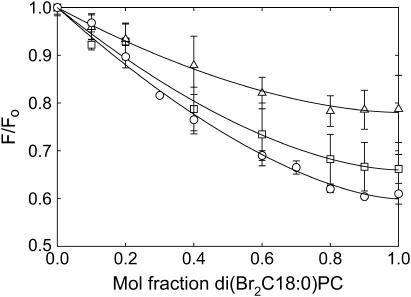
If all phospholipids could bind only to annular sites on W26,113L then the level of fluorescence quenching observed on reconstitution of W26,113L into bilayers of brominated phosphatidylcholine would be the same as in bilayers of brominated anionic lipid, the observed quenching resulting from quenching of the fluorescence of the lipid exposed residue Trp-87. Similarly, if all phospholipids could bind to both the annular and the nonannular sites on W26,113L then reconstitution into bilayers of brominated phosphatidylcholines or brominated anionic lipids would again result in equal fluorescence quenching, corresponding to quenching of the fluorescence of both Trp-67 and Trp-87. If, however, brominated phosphatidylcholine binds only to annular sites but brominated anionic lipids bind to both annular and nonannular sites, then fluorescence quenching of W26,113L in bilayers of brominated anionic lipid would be expected to be about double that seen in bilayers of brominated phosphatidylcholine. As shown in Fig. 4, levels of fluorescence quenching for W26,113L in bilayers of brominated analogs of the anionic phospholipids phosphatidic acid, phosphatidylglycerol, phosphatidylserine, and cardiolipin are about double that in di(Br2C18:0)PC, showing that phosphatidycholines must bind very weakly, if at all, to the nonannular sites on KcsA to which anionic lipids can bind.
FIGURE 4.
Quenching of the fluorescence of W26,113L in mixtures with anionic lipid. W26,113L was reconstituted into mixtures of di(C18:1)PC and brominated anionic lipid (□) or into mixtures of di(Br2C18:0)PC with nonbrominated anionic lipid (○). Fluorescence intensities are expressed as F/Fo where Fo is the fluorescence intensity in the nonbrominated lipid. The lines show fits to Eqs. 4 and 9 giving values for the binding constants listed in Table 2. The anionic lipids were: (A) di(C18:1)PA; (B) di(C18:1)PG; (C) di(C18:1)PS; (D) tetra(C18:1)CL. For experiments with cardiolipin the mol fraction of cardiolipin was calculated on a chain basis to account for the fact that cardiolipin contains four fatty acyl chains and phosphatidylcholine two.
If Trp-87 in W26,113L is the only one of the three Trp residues to be quenched by di(Br2C18:0)PC then the observed value for F/Fo of 0.78 for W26,113L in di(Br2C18:0)PC (Fig. 5; Table 1) corresponds to an F/Fo value of 0.36 for Trp-87, in good agreement with the value of ∼0.44 estimated above from the expected distance of separation between Trp-87 and the nearest annular lipid molecule. Similarly, if only Trp-26 and Trp-87 in W113L are quenched by di(Br2C18:0)PC then the observed value for F/Fo of 0.66 for W113L corresponds to an average F/Fo value of 0.32 for Trp-26 and Trp-87, and if only Trp-26, Trp-87, and Trp-113 in wild-type KcsA are quenched by di(Br2C18:0)PC then the observed value of F/Fo of 0.61 for wild-type KcsA corresponds to an average F/Fo for the three quenched Trp residues of 0.35. The similarity of the values for F/Fo calculated in this way for the three lipid-exposed Trp residues is consistent with the suggestion that Trp-26, Trp-87, and Trp-113 are the only three Trp residues quenched significantly by di(Br2C18:0)PC. The quenching observed in brominated anionic lipid is ∼25% greater than that observed in di(Br2C18:0)PC (Fig. 4), corresponding to a value for F/Fo for Trp-67 of ∼0.27, compared to the value of ∼0.39 estimated above from the expected distance of separation between Trp-67 and the nearest fatty acyl chain of a bound nonannular lipid molecule.
FIGURE 5.
Quenching of the fluorescence of mutant and wild-type KcsA by di(Br2C18:0)PC. KcsA (○), W113L (□), and W26,113L (▵) were reconstituted into mixtures of di(C18:1)PC and di(Br2C18:0)PC. Fluorescence intensities are expressed as F/Fo where Fo is the fluorescence intensity in di(C18:1)PC. The lines show fits to Eq. 2 giving the parameters listed in Table 1.
TABLE 1.
Fluorescence quenching of KcsA by di(Br2C18:0)PC
| Mutant | F/Fo | n |
|---|---|---|
| Wild type | 0.61 ± 0.01 | 1.66 ± 0.12 |
| W113L | 0.66 ± 0.01 | 1.68 ± 0.20 |
| W26,113L | 0.78 ± 0.01 | 1.73 ± 0.12 |
Determination of annular and nonannular binding constants
Plots of fluorescence quenching as a function of mol fraction of brominated lipid in the membrane (Fig. 4) can be used to determine binding constants at the nonannular sites and at the annular sites on the extracellular side of the membrane using Eqs. 4 and 9. Because phosphatidylcholines bind weakly to nonannular sites, analysis of the quenching curves for mixtures of di(Br2C18:0)PC with an anionic lipid gives the relative binding constant of the anionic lipid at the annular site if n in Eq. 2, that approximates to the number of annular sites from which the fluorescence of the Trp residue can be quenched, is known. As described above, the appropriate value of n in Eq. 2 can be determined by fitting quenching curves for W26,113L in mixtures of di(Br2C18:0)PC and di(C18:1)PC to Eq. 2. Fig. 5 compares fluorescence quenching for wild-type KcsA and the mutants W113L and W26,113L in mixtures of di(Br2C18:0)PC and di(C18:1)PC. The fluorescence quenching curves fit well to Eq. 2 with the same value of n for W113L, W26,113L, and wild-type KcsA (Table 1).
Data for quenching of the fluorescence of W26,113L in mixtures of di(Br2C18:0)PC and anionic lipid (Fig. 4) were analyzed in terms of Eq. 4 with a value for n of 1.69, the average value from Table 1, giving the relative binding constants for the anionic lipids at the annular sites on the extracellular side of KcsA listed in Table 2; the fact that all values were close to 1 shows that there is little selectivity in binding to the annular sites on the extracellular side of the membrane.
TABLE 2.
Lipid binding constants for W26,113L determined from fluorescence quenching plots
| Lipid | Annular sites on extracellular side Relative binding constant | Nonannular sites Binding constant (mol fraction−1) | Annular sites on intracellular side Relative binding constant |
|---|---|---|---|
| Phosphatidic acid | 0.80 ± 0.17 | 4.6 ± 2.1 | 2.93 ± 0.77 |
| Phosphatidylglycerol | 0.68 ± 0.07 | 3.0 ± 0.7 | 1.88 ± 0.10 |
| Phosphatidylserine | 0.94 ± 0.11 | 7.1 ± 2.7 | 0.70 ± 0.30 |
| Cardiolipin | 0.77 ± 0.10 | 7.3 ± 1.2 | 0.70 ± 0.16 |
Annular lipid binding constants relative to di(C18:1)PC on the extracellular side were determined by fitting quenching data for W26,113L to Eq. 4 using a value for n of 1.69. Nonannular lipid binding constants were determined by fitting quenching data for W26,113L to Eq. 9 using the values for the relative binding constant at the annular sites given in the table with a value for n of 1.69 for all lipids except phosphatidic acid for which n = 2.49 (15). Annular lipid binding constants relative to di(C18:1)PC on the intracellular side were determined by fitting quenching data for wild-type KcsA to Eq. 10. The mol fraction of cardiolipin was calculated on a chain basis, to account for the fact that cardiolipin contains four fatty acyl chains whereas phosphatidylcholine contains only two.
It was shown in Alvis et al. (15) that n values for anionic phospholipids were the same as for phosphatidylcholines except for cardiolipin where the value of n was half that for the other phospholipids due to the four-chain nature of cardiolipin, and for phosphatidic acid for which the value of n was higher (n = 2.49) possibly related to the small size of the phosphatidic acid headgroup. Data for mixtures of di(C18:1)PC and brominated anionic lipid (Fig. 4) were then fitted to Eq. 9 using these values for n together with the determined values for the annular binding constant on the extracellular side of KcsA, giving the values for the nonannular binding constant listed in Table 2.
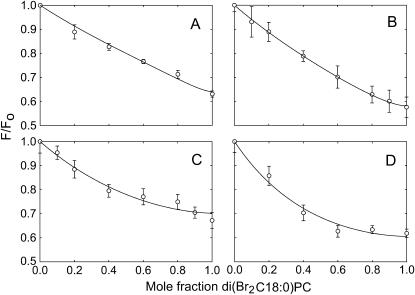
Because quenching by di(Br2C18:0)PC follows just from binding to annular sites, the binding constants for binding at the annular sites on the intracellular side of KcsA can be obtained from the previously published quenching data for wild-type KcsA (15) using the binding constants for binding at the annular sites on the extracellular side of KcsA determined above. Quenching data for wild-type KcsA in mixtures of anionic lipid and di(Br2C18:0)PC were therefore analyzed in terms of Eq. 10 (Fig. 6) to give the annular binding constants on the intracellular side given in Table 2.
FIGURE 6.
Quenching of the fluorescence of wild-type KcsA in mixtures of anionic lipid with di(Br2C18:0)PC. KcsA was reconstituted into mixtures of di(Br2C18:0)PC and anionic lipid. Fluorescence intensities are expressed as F/Fo where Fo is the fluorescence intensity in the nonbrominated lipid. The lines show fits to Eq. 10 with the annular binding constant on the extracellular side of KcsA fixed at the value given in Table 2, giving values for the annular binding constants on the intracellular side listed in Table 2. The anionic lipids were: (A) di(C18:1)PA; (B) di(C18:1)PG; (C) di(C18:1)PS; (D) tetra(C18:1)CL. For experiments with cardiolipin the mol fraction of cardiolipin was calculated on a chain basis to account for the fact that cardiolipin contains four fatty acyl chains and phosphatidylcholine two.
DISCUSSION
An advantage of the fluorescence quenching technique for studying lipid binding to membrane proteins is that the short range of the quenching process means that it is possible to study binding events in the immediate vicinity of a particular Trp residue in the protein. To study binding of lipids at the nonannular sites on KcsA identified in the crystal structure (2), we mutated the two Trp residues on the intracellular side of KcsA, leaving the lipid exposed Trp residue (Trp-87) and the two Trp residues that are part of the pore structure (Trp-67 and Trp-68) on the extracellular side (Fig. 1), thus minimizing the possibility of significant perturbation of the protein structure. The locations of the Trp residues are such that Trp-87 will be quenched mainly by brominated lipids binding to the annular sites on the extracellular side of the protein whereas Trp-67 will be quenched mainly by brominated lipids binding to the nonannular sites (Fig. 1).
Binding to annular sites
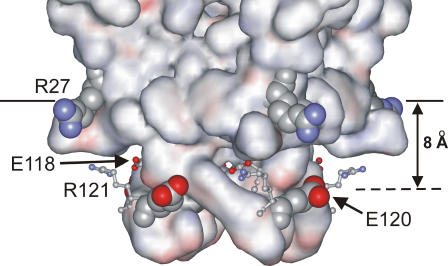
Studies of fluorescence quenching of W26,113L in mixtures of anionic lipids and di(Br2C18:0)PC show that anionic lipids bind to annular sites on the extracellular side of KcsA with an affinity very similar to that of phosphatidylcholine (Table 1). Using these values for the annular binding constants on the extracellular side of KcsA, data for fluorescence quenching of wild-type KcsA by di(Br2C18:0)PC can be analyzed to obtain annular binding constants on the intracellular side of KcsA (Table 1; Fig. 6). In contrast to binding on the extracellular side of KcsA, the strength of binding on the intracellular side of KcsA is structurally specific (Table 2); whereas binding constants for phosphatidylserine and cardiolipin are close to those for phosphatidylcholine, binding of phosphatidic acid and phosphatidylglycerol is approximately three- and twofold stronger, respectively, than phosphatidylcholine. The lack of specificity in binding at annular sites on the extracellular side of the membrane is consistent with the lack of any marked cluster of charged residues on KcsA close to the membrane surface on the extracellular side of the membrane that might interact with the phospholipid headgroups. This contrasts with a distinct pattern for the charged residues on the intracellular side of the membrane (Fig. 7). The location of the glycerol backbone region of the lipid bilayer on the intracellular side of the membrane is marked by the location of the Trp residues on this side of the protein (3). Arg-27 can then be seen to snorkel up to the glycerol backbone region to give a girdle of four positively charged residues (Fig. 7). Located ∼8 Å from this girdle of positive charge is a girdle of charged residues made up of Glu-118, Glu-120, and Arg-121, Glu-118 and Arg-121 forming a charged pair (Fig. 7). Thus, it is possible that favorable binding of phosphatidic acid and phosphatidylglycerol follows from favorable interaction between Arg-27 and the lipid phosphate group, the only charged group present in these lipids. An unfavorable interaction between Glu-120 and the negatively charged carboxyl group in phosphatidylserine or the additional negatively charged phosphate group present in cardiolipin could then explain the less favorable binding of these more complex anionic lipids.
FIGURE 7.
The structure of KcsA on the intracellular side of the membrane. The surface of the KcsA tetramer is shown colored by electrostatic potential. The location of the glycerol backbone region of the bilayer is shown by the solid line. Arg-27, shown in space-fill format, is seen snorkeling up to the surface. Above the plane defined by the Arg-27 are the charged residues Glu-118, Glu-120, and Arg-121. Glu-118 and Arg-121, shown in ball-and-stick format, form an ion pair on each subunit. The four Glu-120 residues, shown in space-fill format, form a plane ∼8 Å above the plane formed by the Arg-27 residues; favorable interactions between the headgroups of anionic lipids and the Arg-27 residues could be balanced by unfavorable interactions of the headgroups with the Glu-120 residues.
Favorable interaction between Arg-27 and phosphatidic acid or phosphatidylglycerol on the intracellular side of the membrane could give rise to a hot spot for lipid binding in the vicinity of Arg-27, with binding constants for phosphatidic acid and phosphatidylglycerol at sites close to Arg-27 greater than the average value for all the sites on the intracellular side of the membrane given in Table 2. The existence of a hot spot for binding anionic lipid has been detected previously on the mechanosensitive channel of large conductance MscL, corresponding to a cluster of positively charged residues (22).
Binding to nonannular sites
Binding of lipid to the nonannular sites on KcsA shows marked selectivity for anionic lipid. The observation that fluorescence quenching in di(Br2C18:0)PC is approximately half that in brominated anionic lipid (Fig. 4) suggests that only anionic lipid can bind to the nonannular sites and quench the fluorescence of Trp-67. Selectivity of the nonannular sites for anionic lipid probably follows from the presence of two Arg residues, Arg-64 and Arg-89, close to the site, one coming from each of the monomers at the interface (23).
Binding of anionic lipids at the nonannular sites varies by only a factor of 2 with anionic headgroup structure (Table 2) and is of only moderate affinity, with a standard free-energy change for binding ΔGo of ∼0.9–1.2 kcal mol−1. The energy of interaction U between two ions is given by
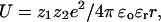 |
(12) |
where z1 and z2 are the charges on the two ions, ɛr is the relative permittivity (dielectric constant) of the medium, and r is the distance between the two ions. Assuming a dielectric constant of 78.5 (water), an energy of interaction of 1 kcal mol−1 corresponds to a distance of separation of two monovalent ions of 4.2 Å. Thus, close contact between the charged groups on the phospholipid headgroup and charged groups on KcsA at the nonannular site is sufficient to explain the observed binding. Binding of this type, with the lipid headgroup not being tightly constrained within a binding pocket, would be consistent with the observation that the anionic lipid headgroup is not resolved in the crystal structure of KcsA (2).
Fluorescence emission spectra for wild-type KcsA (15) and W26,113L are very similar in bilayers of phosphatidylcholine and anionic phospholipids suggesting that binding of anionic lipid to the nonannular sites on KcsA results in no major conformational changes on KcsA, despite the presence of anionic lipid being essential for channel opening.
It is not known whether all four nonannular sites in the tetrameric KcsA structure have to be occupied by anionic lipid for the channel to open. The Escherichia coli cell membrane in which KcsA is expressed contains ∼20 mol % anionic lipid, mostly phosphatidylglycerol (24). In mammalian cells the distribution of lipid species between the two faces of the plasma membrane is highly asymmetric, but it is not known if this is also the case in E. coli (25). If the phosphatidylglycerol content of the outer leaflet of the E. coli membrane were 20 mol %, then, with the nonannular binding constant given in Table 2, the nonannular sites would be ∼37% occupied by phosphatidylglycerol, and the probability that at least one of the four nonannular sites was occupied by phosphatidylglycerol would be ∼85%. If the phosphatidylglycerol were to be concentrated in the outer leaflet of the membrane, then the fraction of nonannular sites occupied by phosphatidylglycerol would rise to ∼55% and the probability that at least one of the four nonannular sites was occupied by phosphatidylglycerol would be ∼96%. The lipid composition of the cell membrane of the Gram positive S. lividans appears not to have been determined, but other species of Streptomyces are rich in cardiolipin (26,27). For a cardiolipin content of 20 mol% in the outer leaflet of the membrane, the nonannular binding sites would be ∼75% occupied by cardiolipin.
High-resolution structures are available for an inwardly rectifying K+ channel (28) and for two voltage gated K+ channels (29,30). None of these structures shows lipid molecules bound at protein-protein interfaces in the tetrameric structures. In the bacterial voltage gated K+ channel structure, Arg-64 and Arg-89 in KcsA are replaced by Asp-185 and Lys-210, forming a salt bridge (23) and in the inwardly rectifying K+ channel there are no charged residues close to the region corresponding to the nonannular binding site on KcsA. Further, the deep cleft at each monomer-monomer interface in KcsA corresponding to the nonannular binding site is less uniform and open in the other K+ channel structures, suggesting that binding of anionic lipid at the monomer-monomer interfaces may not be a general phenomenon for all K+ channels.
Acknowledgments
We thank Professor H. Schrempf, University of Osnabruck, Osnabruck, Germany, for the generous gift of the KcsA construct.
We thank the Wellcome Trust for financial support, and the Biotechnology and Biological Sciences Research Council for a studentship to S.J.A.
References
- 1.Heginbotham, L., L. Kolmakova-Partensky, and C. Miller. 1998. Functional reconstitution of a prokaryotic K+ channel. J. Gen. Physiol. 111:741–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valiyaveetil, F. I., Y. Zhou, and R. Mackinnon. 2002. Lipids in the structure, folding and function of the KcsA K+ channel. Biochemistry. 41:10771–10777. [DOI] [PubMed] [Google Scholar]
- 3.Williamson, I. M., S. J. Alvis, J. M. East, and A. G. Lee. 2002. Interactions of phospholipids with the potassium channel KcsA. Biophys. J. 83:2026–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simmonds, A. C., J. M. East, O. T. Jones, E. K. Rooney, J. McWhirter, and A. G. Lee. 1982. Annular and non-annular binding sites on the (Ca2+ + Mg2+)-ATPase. Biochim. Biophys. Acta. 693:398–406. [DOI] [PubMed] [Google Scholar]
- 5.East, J. M., and A. G. Lee. 1982. Lipid selectivity of the calcium and magnesium ion dependent adenosinetriphosphatase, studied with fluorescence quenching by a brominated phospholipid. Biochemistry. 21:4144–4151. [DOI] [PubMed] [Google Scholar]
- 6.Bolen, E. J., and P. W. Holloway. 1990. Quenching of tryptophan fluorescence by brominated phospholipid. Biochemistry. 29:9638–9643. [DOI] [PubMed] [Google Scholar]
- 7.Powl, A. M., J. M. East, and A. G. Lee. 2003. Lipid-protein interactions studied by introduction of a tryptophan residue: the mechanosensitive channel MscL. Biochemistry. 42:14306–14317. [DOI] [PubMed] [Google Scholar]
- 8.Ladokhin, A. S. 1999. Analysis of protein and peptide penetration into membranes by depth-dependent fluorescence quenching: theoretical considerations. Biophys. J. 76:946–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong, K. H., and C. Miller. 2000. The lipid-protein interface of a Shaker K+ channel. J. Gen. Physiol. 115:51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monks, S. A., D. J. Needleman, and C. Miller. 1999. Helical structure and packing orientation of the S2 segment in the Shaker K+ channel. J. Gen. Physiol. 113:415–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perozo, E., D. M. Cortes, and L. G. Cuello. 1998. Three-dimensional architecture and gating mechanism of a K+ channel studied by EPR spectroscopy. Nat. Struct. Biol. 5:459–469. [DOI] [PubMed] [Google Scholar]
- 12.Lewis, B. A., and D. M. Engelman. 1983. Lipid bilayer thickness varies linearly with acyl chain length in fluid phosphatidylcholine vesicles. J. Mol. Biol. 166:211–217. [DOI] [PubMed] [Google Scholar]
- 13.Schrempf, H., O. Schmidt, R. Kummerlen, S. Hinnah, D. Muller, M. Betzler, T. Steinkamp, and R. Wagner. 1995. A prokaryotic potassium ion channel with two predicted transmembrane segments from Streptomyces lividans. EMBO J. 14:5170–5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227:680–685. [DOI] [PubMed] [Google Scholar]
- 15.Alvis, S. J., I. M. Williamson, J. M. East, and A. G. Lee. 2003. Interactions of anionic phospholipids and phosphatidylethanolamines with the potassium channel KcsA. Biophys. J. 85:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, R. F. 1967. Fluorescence quantum yields of tryptophan and tyrosine. Anal. Lett. 1:35–42. [Google Scholar]
- 17.London, E., and G. W. Feigenson. 1981. Fluorescence quenching in model membranes. 2. Determination of local lipid environment of the calcium adenosinetriphosphatase from sarcoplasmic reticulum. Biochemistry. 20:1939–1948. [DOI] [PubMed] [Google Scholar]
- 18.Caffrey, M., and G. W. Feigenson. 1981. Fluorescence quenching in model membranes. 3. Relationship between calcium adenosinetriphosphatase enzyme activity and the affinity of the protein for phosphatidylcholines with different acyl chain characteristics. Biochemistry. 20:1949–1961. [DOI] [PubMed] [Google Scholar]
- 19.O'Keeffe, A. H., J. M. East, and A. G. Lee. 2000. Selectivity in lipid binding to the bacterial outer membrane protein OmpF. Biophys. J. 79:2066–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeager, M. D., and G. W. Feigenson. 1990. Fluorescence quenching in model membranes: phospholipid acyl chain distributions around small fluorophores. Biochemistry. 29:4380–4392. [DOI] [PubMed] [Google Scholar]
- 21.Caputo, G. A., and E. London. 2003. Using a novel dual fluorescence quenching assay for measurement of tryptophan depth within lipid bilayers to determine hydrophobic α-helix locations within membranes. Biochemistry. 42:3265–3274. [DOI] [PubMed] [Google Scholar]
- 22.Powl, A. M., J. M. East, and A. G. Lee. 2005. Heterogeneity in the binding of lipid molecules to the surface of a membrane protein: hot-spots for anionic lipids on the mechanosensitive channel of large conductance MscL and effects on conformation. Biochemistry. 44:5873–5883. [DOI] [PubMed] [Google Scholar]
- 23.Lee, A. G. 2004. How lipids affect the activities of integral membrane proteins. Biochim. Biophys. Acta. 1666:62–87. [DOI] [PubMed] [Google Scholar]
- 24.Harwood, J. L., and N. J. Russell. (1984) Lipids in Plants and Microbes. George Allen & Unwin, London, UK.
- 25.Huijbregts, R. P. H., A. I. P. M. de Kroon, and B. de Kruijff. 2000. Topology and transport of membrane lipids in bacteria. Biochim. Biophys. Acta. 1469:43–61. [DOI] [PubMed] [Google Scholar]
- 26.Schauner, C., A. Dary, A. Lebrihi, P. Leblond, B. Decaris, and P. Germain. 1999. Modulation of lipid metabolism and spiramycin biosynthesis in Streptomyces ambofaciens unstable mutants. Appl. Microbiol. Biotechnol. 65:2730–2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoischen, C., K. Gura, C. Luge, and J. Gumpert. 1997. Lipid and fatty acid composition of cytoplasmic membranes from Streptomyces hygroscopicus and its stable protoplast-type L form. J. Bacteriol. 179:3430–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuo, A., J. M. Gulbis, J. F. Antcliff, T. Rahman, E. D. Lowe, J. Zimmer, J. Cuthbertson, F. M. Ashcroft, T. Ezaki, and D. A. Doyle. 2003. Crystal structure of the potassium channel KirBac1.1 in the closed state. Science. 300:1922–1926. [DOI] [PubMed] [Google Scholar]
- 29.Jiang, Y., A. Lee, J. Chen, V. Ruta, M. Cadene, B. T. Chait, and R. MacKinnon. 2003. X-ray structure of a voltage-dependent K+ channel. Nature. 423:33–41. [DOI] [PubMed] [Google Scholar]
- 30.Long, S. B., E. B. Campbell, and R. MacKinnon. 2005. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 309:897–903. [DOI] [PubMed] [Google Scholar]
- 31.Zhou, Y., J. H. Morals-Cabral, A. Kaufman, and R. Mackinnon. 2001. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 Å resolution. Nature. 414:43–48. [DOI] [PubMed] [Google Scholar]