Abstract
Synthetic oligonucleotides with a fluorescent coumarin group replacing a basepair have been used in recent time-resolved Stokes-shift experiments to measure DNA dynamics on the femtosecond to nanosecond timescales. Here, we show that the APE1 endonuclease cleaves such a modified oligonucleotide at the abasic site opposite the coumarin with only a fourfold reduction in rate. In addition, a noncatalytic mutant (D210N) binds tightly to the same oligonucleotide, albeit with an 85-fold reduction in binding constant relative to a native oligonucleotide containing a guanine opposite the abasic site. Thus, the modified oligonucleotide retains substantial biological activity and serves as a useful model of native DNA. In the complex of the coumarin-containing oligonucleotide and the noncatalytic APE1, the dye's absorption spectrum is shifted relative to its spectrum in either water or within the unbound oligonucleotide. Thus the dye occupies a site within the DNA:protein complex. This result is consistent with modeling, which shows that the complex accommodates coumarin at the site of the orphaned base with little distortion of the native structure. Stokes-shift measurements of the complex show surprisingly little change in the dynamics within the 40 ps–40 ns time range.
INTRODUCTION
Polarity is a familiar concept in chemistry that expresses the ability of a solvent to stabilize charge separation (1). Solvent polarity strongly affects rates of reactions involving charge-separated transition states or products (1,2). However, in biochemical reactions, the local environment of the reactions is often not a solvent, but a macromolecule. How do concepts such as polarity translate to such an environment? In solution, it is known that the polarity is generated by subnanosecond motion of solvent molecules. Computer simulations show extensive and complex motion in DNA on those timescales (3–5). However, there has been little experimental data on these motions until recently.
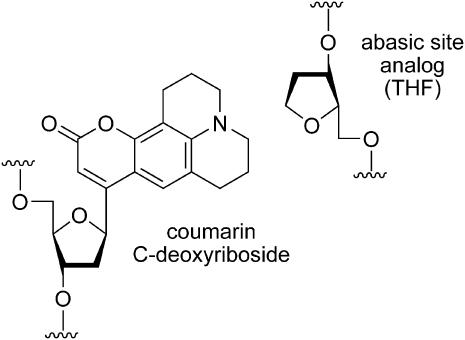
We introduced time-resolved Stokes-shift (TRSS) experiments for the measurement of polarity-related dynamics in DNA (6). This technique relies on synthetic oligonucleotides in which a fluorescent coumarin group replaces a native base (Fig. 1) (7). An abasic-site analog (THF) (8) is placed on the complementary strand opposite the coumarin. [The coumarin is large enough to fill the space between the backbones when the coumarin is opposite an abasic site (6).] The absorption and emission spectra of the coumarin group are sensitive to the polarity of its immediate environment (9,10). The shifts of these spectra in different solvents provide a quantitative measure of solvent polarity (11–13). In a similar fashion, the analysis of time-resolved emission spectra from coumarin-containing oligonucleotides probes the effective polarity in DNA and the mechanisms creating it. The effective polarity of DNA can affect biochemical reactions of DNA if they involve charge separation, just as the polarity of simple solvents affects the rates of nonbiological reactions.
FIGURE 1.
Chemical structure of the coumarin chromophore and the opposing abasic-site analog in the 17-mer oligodeoxynucleotide.
Two important questions arise in using these modified oligonucleotides as models of native biochemistry: How much does the introduction of the nonnative fluorophore affect the properties of the oligonucleotide? Does the formation of a DNA:protein complex, which is necessary for many biochemical reactions, significantly alter the conditions relative to a bare oligonucleotide? To address these questions, this article looks at the interaction of a coumarin-modified oligonucleotide with APE1, an abasic-site endonuclease that is part of the human base-excision repair pathway (14,15). Oligonucleotides containing an abasic site opposite either a coumarin or a normal guanine are compared with regard to both cleavage rate and binding constant. These studies test the degree to which the substitution of a native base by a coumarin alters the functioning of the DNA. They also lead to conditions where a stable complex of the coumarin-containing oligonucleotide and a noncatalytic mutant of APE1 is formed. TRSS measurements of the dynamics in the DNA:protein complex are then compared to similar measurements on uncomplexed DNA. In these measurements, the conditions at the active site of the APE1 are not probed. However, the measurements do address the general question of whether the unusual dynamics seen in bare DNA persist in complexes with proteins.
The TRSS experiment is based on a chromophore, such as coumarin, that has a larger dipole moment in the excited state than in the ground state. Upon excitation of the chromophore, the increased electric field associated with the excited-state dipole extends into the DNA and water surrounding the chromophore. All elements of the local structure that have a charge experience a new force. These elements move into a new configuration at a rate depending on the intrinsic dynamics of the DNA. As the new configuration is formed, the local electric field at the chromophore changes so as to stabilize the excited-state dipole. As the excited-state is stabilized, the fluorescence from the chromophore shifts to lower frequency. The TRSS experiment consists of measuring the frequency of the fluorescence (the Stokes shift) as a function of time after excitation. The size of the Stokes shift directly reflects the progress of the reorganization of the DNA and the nearby water and counterions.
When the TRSS experiment is performed on a chromophore free in solution, the final size of the Stokes shift is a measure of the total ability of the solvent to stabilize a new charge distribution. The total ability of a solvent to stabilize charge is commonly referred to as the “polarity” of the solvent (11–13) and the rate of stabilizing the charge constitutes the “solvation dynamics” (9). DNA is a more complex and structured environment than a simple solvent. Nonetheless, the size and rate of the Stokes shift of coumarin in DNA can be regarded as the effective polarity and solvation dynamics experienced in the interior of DNA. Both factors significantly affect the rates of chemical reactions (1,2). A good example is the recent study of O'Neill and Barton, which showed that freezing the motion in DNA effectively stops long range electron transfer (16).
TRSS experiments in bare DNA have shown that the solvation dynamics in DNA are both strong and qualitatively different than those in a simple solvent. In terms of total polarity, the interior of DNA is similar to ethanol (17). However, it takes a relatively long time for the polarity to respond to changes in the charge distribution. In low viscosity solvents, solvation times typically range from 1 to 100 ps (18). In contrast, solvation in DNA is broadly distributed in time, from <40 fs to >40 ns and follows a power law in time (19,20). Multiple-relaxation times in DNA have also been seen by Zewail's group using 2-aminopurine as the chromophore (21). Lesions to the normal DNA structure can add additional relaxation processes (17).
For these studies to have relevance to biological DNA, the coumarin must occupy a position in the helix typical of a native basepair, and the properties of coumarin-containing DNA must be substantially the same as those of native DNA. Several results suggest that these conditions are fulfilled. The absorption spectrum of the coumarin is red-shifted relative to its position in water, showing that the coumarin does not flip to an extrahelical position (6). Molecular modeling of the coumarin in an oligonucleotide places the coumarin in the interior of DNA with only minor distortions of the overall structure (6). The coumarin is effectively shielded from the addition of a fluorescence quencher to the solution (Supplemental Material), confirming that the coumarin is embedded into the helix.
This study makes a more rigorous test of coumarin-containing DNA as a biologically relevant model system by testing its biological activity with the APE1 endonuclease, also known as HAP1, APEX, and Ref1 (14,15). APE1 is the predominant apurinic/apyrimidinic (AP) endonuclease in humans. It is a key component in base-excision repair, which is the primary defense mechanism against DNA damage such as alkylation, oxidation, or deamination of DNA bases (22,23). The pathway is initiated by DNA glycosylases, which remove the damaged base to create an abasic site in the DNA (24,25). Abasic sites also form by spontaneous or chemically induced hydrolysis of the N-glycosidic bond (14,26,27). APE1 begins the repair of an abasic site by cleaving the phosphodiester bond 5′ to an abasic site, leaving a 3′-OH group and a 5′-deoxyribose-5-phosphate residue (14,15). APE1 also efficiently cleaves the abasic-site analog used in this article (Fig. 1), which lacks the 1′-hydroxyl of a naturally occurring abasic site (28).
This article begins by modeling the DNA:APE1 complex using the known crystal structure. The modeling shows that the coumarin can reside near the position of a native base without significant distortion of the structure.
In the next section, the cleavage rate of coumarin-containing DNA is compared to the rate for native DNA. We will see that native APE1 is still able to specifically cleave at an abasic site when it is opposite a coumarin group instead of a native base.
Site-directed mutagenesis studies have revealed that aspartate 210 is crucial for the catalytic endonuclease activity of APE1. An asparagine substitution (D210N) results in a mutant of APE1 that lacks endonuclease activity, but retains the ability to bind to abasic sites (29,30). In a second set of studies, we measure the binding of this mutant to an abasic site opposite a coumarin group. The binding constant is 85 times lower than to an abasic site opposite a guanine, but the binding remains strong and specific.
Using these results, samples of the complex of the DNA and the mutant APE1 were prepared for spectroscopic studies. The steady-state spectra also show that the coumarin remains embedded in the complex and is not expelled into the surrounding water. The favorable results from all these studies allow us to undertake our final goal of measuring the change in DNA dynamics upon formation of the complex with the APE1 protein.
A number of TRSS experiments have been performed on proteins alone (31–38). Essentially equivalent information can also be obtained from photon-echo based techniques (39,40) or from vibrational-echo methods (41). In proteins, both extrinsic and intrinsic fluorophores have been used, and they have been located both at the protein surface and in the interior of the protein. In general, components of the dynamics slower than normal water are found and are attributed either to protein motions or perturbed dynamics of nearby water.
Only one previous study, by Zewail's group, has looked at the TRSS in a DNA-protein complex (42). Histone 1 was labeled at several amine groups on the histone surface with a fluorescent dye to monitor dynamics at the histone surface. Upon binding to calf thymus DNA, the average TRSS signal from the dye molecules showed a modest slowing. In comparison to that study, these measurements focus on the DNA dynamics, rather than the protein dynamics. The structure being measured is also better defined in this study.
The x-ray structure of the cocrystal of APE1 bound to DNA shows severe disruption of the DNA structure near the abasic site (43). A number of proposals could be made for the effect of protein on the polarity and solvation dynamics experienced within the DNA. Because the relatively low dielectric protein displaces water, the net polarity might be reduced. If mobile counterions are replaced by relatively static protein charges, the solvation dynamics might be slowed. On the other hand, the disruption of the normal helical structure upon binding might open up the interior DNA to create greater exposure to the solvent, or the disruption of basepairing might allow the DNA structure to fluctuate more strongly. Either effect would increase both the size and rate of solvation dynamics.
In reality, our results show little change in the dynamics upon binding to the protein. A small difference in the earliest measured Stokes shift may indicate a change in the dynamics at shorter times. However, from 40 ps to 40 ns, the dynamics with and without the protein are nearly indistinguishable.
MATERIALS AND METHODS
Oligonucleotide preparation
The oligodeoxynucleotide 5′-GCATGCGC(coumarin)CGCGTACG-3′ containing coumarin-102 C-deoxyriboside was synthesized as described earlier (6,7). The complementary strand containing a tetrahydrofuran (THF) abasic-site analog opposite the coumarin was obtained commercially. For the binding and incision experiments, the complementary strand was labeled with 32P at the 5′-end before annealing to the coumarin-modified strand. Identical samples with guanine in place of the coumarin served as controls.
Cleavage and binding studies
The wild-type and D210N mutant of APE1 were purified as described previously (29). For the cleavage studies, wild-type APE1 (0.02 ng) was incubated with the 32P-labeled oligodeoxynucleotide (1 nM in helices) in a total volume of 10 μl at 37°C for various times. The reaction buffer consisted of 50 mM Tris-HCl, 1 mM EDTA, 5% glycerol, 1 mM MgCl2, and 100 ng of bovine serum albumin (BSA) at pH 7.2. After the addition of formamide loading dye, the samples were loaded onto a 20% denaturing polyacrylamide (19:1) gel, and the gel was run at a constant current of 30 mA for ∼1 h. Gels were visualized using a Molecular Imager FX and quantified using Quantity One software (Bio-Rad, Hercules, CA).
Binding was determined by an electromobility shift assay. For these studies, various concentrations of the catalytically inactive D210N mutant of APE1 were incubated with the same oligonucleotides (0.1 nM in helices) as in the cleavage studies in a total volume of 10 μl for 30 min at room temperature in the same reaction buffer. At the end of the incubation, glycerol loading dye was added, the samples were loaded onto a 10% nondenaturing polyacrylamide (37.5:1) gel, and the gel was run at a constant voltage of 150 V for 2 h. Gels were visualized and quantified as above. For the binding competition assays, the conditions were as above, except that unlabeled competitor duplex (guanine:THF or coumarin:THF) were added before the addition of D210N APE1. The gels shown are representative of four independent experiments.
Steady-state and time-resolved spectroscopy
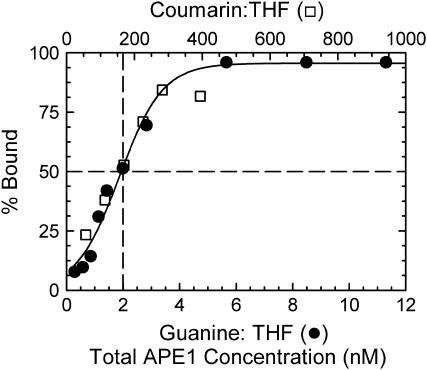
Samples for optical spectroscopy required both higher DNA concentrations and larger volumes than the samples for gel studies. A stock solution of double stranded DNA was prepared by annealing the two single strands in 100 mM, pH 7.2 sodium phosphate buffer at a DNA concentration of 14 μM (in helices). The concentration of DNA was calculated from the absorbance of the solution at 260 nm. A first sample of the DNA:APE1 complex was prepared by adding D210N-APE1 stock solution (28 μM APE1 in 50 mM HEPES, 150 mM KCl, 10% glycerol, pH 7.4) and reaction buffer to a dried aliquot of DNA stock solution. The same reaction buffer was used as in the binding studies, but without the BSA. The final concentrations of D210N-APE1 and DNA oligonucleotide were 7 μM and 5 μM (in helices), respectively. Binding studies indicate that the DNA will be fully bound at these concentrations, i.e., 2 μM free protein (see Fig. 5 below).
FIGURE 5.
Data on the binding of oligonucleotides by D210N APE1 (see also Fig. 4). The data on coumarin:THF (open squares, top scale) have been shifted 85-fold relative to guanine:THF (solid circles, bottom scale). Dashed lines mark the 50% binding concentrations. The solid curve is a guide to the eye.
The measurements were repeated on a second sample. This sample was made in the same way, except that the final concentration of D210N-APE1 was 10 μM. Thus, the free protein concentration was 2½ times higher than in the first sample. Spectra and TRSS measurements on the two samples were within experimental error of each other, confirming that the DNA is fully bound in both samples. The reported results are averages of the two samples.
A corresponding sample of uncomplexed DNA was prepared in the same way as the complex, except that a protein-free aliquot of the protein buffer was substituted for the APE1 aliquot.
A control sample was prepared with the protein and the DNA buffer, but no DNA. This sample showed no detectable fluorescence under the conditions of either the steady-state or time-resolved measurements.
Steady-state emission and excitation spectra were collected using magic-angle polarization. Emission spectra were collected by exciting the samples at 390 nm. Excitation spectra were collected by detecting at 500 nm. Excitation and collection bandpasses were 4 nm in all cases. The sample cells were silanized to reduce scattered light from DNA and protein stuck to the cell walls. The raw emission spectra were corrected for instrumental sensitivity by using a correction factor obtained from a quinine-sulfate standard (44). All spectra were converted to susceptibility versus frequency before analysis (17).
Steady-state spectra in a rigid glassy matrix were measured by adding glycerol to the samples to make a 1:3 solution of buffer and glycerol. The room-temperature absorption spectrum shifted by <2 nm upon addition of the glycerol. The samples were frozen in a dry-ice/acetone bath.
Time-resolved emission spectra were measured by time-correlated single-photon counting as described previously (17). In brief, ∼100 fs pulses from a mode-locked Ti:sapphire laser were frequency doubled in a 1-mm BBO crystal to 390 nm. The 80-MHz repetition rate of the laser was reduced to 6 MHz by an acoustooptic pulse selector. Fluorescence was collected at right angles to the excitation beam and passed through a Glan-laser polarizer at magic-angle (54.7°) to the excitation polarization. The fluorescence passed through a 2-nm bandpass single monochromator before detection on a microchannel plate and timing by single-photon counting electronics. The typical instrument response of the system was 45 ps (full width at half-maximum). Transients were collected at 13 different wavelengths ranging from 425 to 620 nm for each sample. Using the transients in conjunction with the steady-state emission spectrum, time-resolved emission spectra were reconstructed by standard methods (45). The frequency points were interpolated, and the spectral positions were characterized as the first moments as described before (17).
With a sample volume of 30 μL, sample degradation could be observed as changes in the absorption spectrum after prolonged exposure to 3 mW of excitation power. The total data collection time was limited to ∼3 h to avoid significant degradation. Nonetheless, a total of 107 counts were collected at each wavelength, except the two most extreme wavelengths, where 106 counts were collected.
Modeling
The coordinates for the DNA:APE1 cocrystal structures were obtained from the Brookhaven database (1DE8 and 1DE9) (43). Structure visualization and modeling was carried out on an SGI Indigo2 Impact workstation with an IRIX 6.5 operating system and Turbo-Frodo (version 5.5) software (Silicon Graphics, Mountain View, CA). Energy minimizations were carried out using SYBYL 7.0 (Tripos, St. Louis, MO).
RESULTS
Modeling of coumarin within the DNA:APE1 complex
In general, if a native DNA base is replaced by a group with a similar size and shape, the modified DNA retains much of the structure and even function of native DNA (46,47). Like a native base, coumarin is a flat aromatic molecule, but it is larger than a single base. Modeling of coumarin-modified DNA shows that it replaces an entire basepair, effectively filling the otherwise empty opposing abasic site (6). The DNA structure is not changed significantly by the modification.
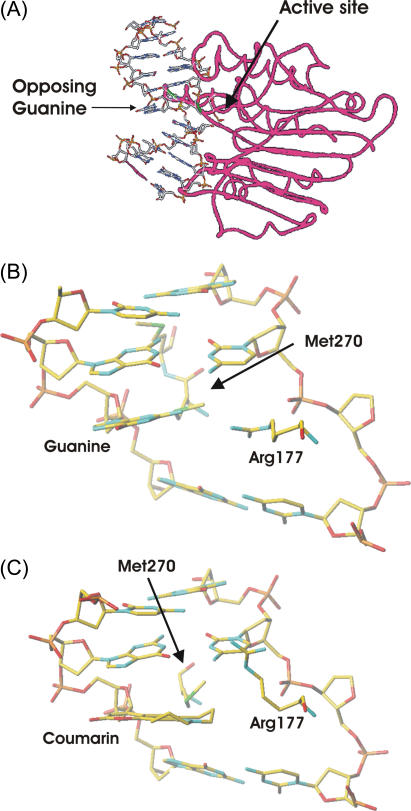
In the DNA:APE1 complex, there is a concern that the large coumarin group might not be compatible with the native structure of the complex. If so, the binding might be weak, the coumarin might be expelled from the normal position of the orphan base, or the complex might be otherwise distorted. As a first step toward addressing these concerns, we constructed models based on the cocrystal structure of APE1 bound to DNA containing a native abasic-site and an orphaned guanine (43). This structure shows that the DNA is kinked sharply near the abasic site as the abasic sugar is twisted into the catalytic pocket (Fig. 2 A). In Fig. 2 A, the orphaned guanine remains stacked against the cytosine immediately above (5′) and is near the surface of the protein. However, the kink in the DNA leaves the orphaned guanine exposed to solvent on its major-groove face and below.
FIGURE 2.
(A) Large-scale view of the DNA:APE1 complex, showing a strong kink in the DNA (43). The protein is represented by red tubes and the DNA by sticks. The positions of the orphaned guanine opposing the abasic site and active site are marked by arrows. (B) Closeup of the local structure near the native orphaned base (guanine). (C) Closeup of the local structure after replacement of the guanine by coumarin and after relaxation of the structure. Only one of the three rotamers of Arg-177 is shown.
We modeled the modified complex by replacing the guanine in the native structure (Fig. 2 B) with coumarin and allowing rotation about the glycosidic bond. The coumarin fits comfortably in place of the native guanine within the DNA. Only two amino acids from APE1 are in proximity to the coumarin. A methionine (Met-270) comes no closer than 3.89 Å from the coumarin, too far to create a steric clash. An arginine (Arg-177) comes within 1.7 Å of the coumarin group, creating the only steric problem in this structure. Interestingly, this arginine may also have important steric interactions in the native structure. Mutating Arg-177 to alanine improves the catalytic efficiency of APE1, and it may play a role in moderating the catalytic activity of APE1 (43).
Arginine is a relatively flexible amino acid, so we considered the feasibility of Arg-177 moving to accommodate the coumarin. Energy minimizations were performed allowing Arg-177 and the DNA to move, but holding the rest of the protein fixed. Little change occurred in the DNA structure. However, three different rotamers of Arg-177 of approximately equal energy were found. All three are easily accommodated with no further rearrangement of the structure (Fig. 2 C).
Thus the modeling suggests that the large coumarin group is accommodated in the DNA:APE1 complex with only minor changes in the native structure. In particular, it suggests that the coumarin will adopt a position in the complex similar to the position of the native orphaned guanine. To test this conclusion experimentally, the sections below look at the change in both the cleavage rate and binding constant due to the coumarin modification.
Native APE1 cleaves coumarin-containing DNA
Fig. 3 shows the results from incision experiments with wild-type APE1 acting on oligonucleotides with either guanine opposite the abasic site (guanine:THF, Fig. 3 A) or coumarin opposite the abasic site (coumarin:THF, Fig. 3 B). The incision rate for guanine:THF (0.465 min−1) was four times greater than the incision rate for coumarin:THF (0.115 min−1). In either case, APE1 exhaustively cleaved the DNA over time. The coumarin modification only mildly reduces the ability of APE1 to incise the opposing THF. This result is consistent with the model structure of the DNA:APE1 complex, in which the coumarin is well separated from the active site (Fig. 2 A).
FIGURE 3.
Data for incision of oligonucleotides by APE1: (A) with guanine opposite the abasic site; (B) with coumarin opposite the abasic site. The dashed curves are fits to first-order kinetics. Each point is the average of four independent experiments.
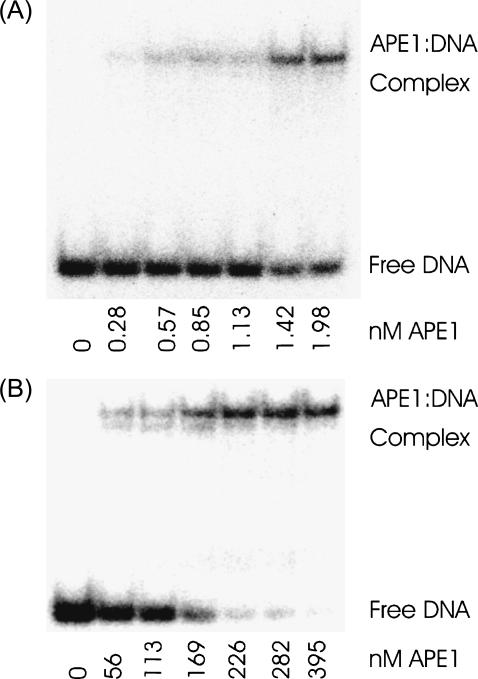
Noncatalytic APE1 mutant binds specifically to modified DNA
Fig. 4 shows representative gels comparing the binding of the D210N-APE1 mutant to DNA containing either guanine:THF (A) or coumarin:THF (B). Quantitative results from the gels are shown in Fig. 5. The 50% binding concentrations are 2 nM for guanine:THF and 170 nM for coumarin:THF, an 85-fold reduction in binding affinity. Nonetheless, APE1 still binds strongly to the coumarin-containing DNA with submicromolar affinity.
FIGURE 4.
Representative gels showing binding of oligonucleotides by D210N APE1 at various concentrations: (A) with guanine opposite the abasic-site analog; (B) with coumarin opposite the abasic-site analog. See also Fig. 5.
The reduction in binding affinity to the coumarin-containing DNA represents an 11.5 kJ/mol (2.7 kcal/mol) reduction in binding free energy. The reduction in binding free energy is due to a combination of two factors: i), decreased stability of the DNA:APE1 complex containing coumarin relative to the complex containing guanine; and ii), any increased stability of the uncomplexed coumarin:THF oligonucleotide relative to the guanine:THF oligonucleotide.
The relative stabilities of the uncomplexed oligonucleotides can be estimated from the change in melting temperature of each abasic oligonucleotide relative to the corresponding guanine:cytosine oligonucleotide. Coumarin:THF shows a melting-point reduction of 13°C (6); guanine:THF shows a melting-point reduction of 11–19°C depending on sequence context (48,49). The destabilization of coumarin:THF relative to guanine:cytosine is primarily due to the introduction of the abasic site. The switch from guanine:THF to coumarin:THF causes only a small change in stability. Most of the measured change in binding free energy is due to a reduction in the stability of the coumarin-containing complex relative to the guanine-containing complex.
The larger change in substrate-binding constant than in overall rate initially seems surprising, but the same effect has been seen for other mutants of APE1 (50). APE1 tightly binds its incised DNA product (51). Under conditions where the enzyme is undergoing multiple turnovers, the overall rate may be dominated by the product-release rate and thus be independent of the substrate-binding constant.
We performed competition experiments with either direct or nonspecific competitors that lacked the 32P label (Fig. 6). The direct competitor (unlabeled guanine:THF) reduced the binding of APE1 to either guanine:THF or coumarin:THF by >3-fold (Lane 3). The nonspecific competitor was the same double-stranded oligonucleotide as guanine:THF, except that cytosine replaced the THF, eliminating the abasic site. Within experimental error, the nonspecific competitor did not reduce APE1 binding to coumarin:THF (18% Lane 2 versus 22% Lane 4), even in the presence of a 100-fold excess of the competitor. Thus, the APE1 specifically recognizes and binds to the abasic-site analog in both guanine:THF and coumarin:THF. We conclude that the binding mode to the coumarin-modified DNA is essentially the same as the native binding mode, albeit with a reduced affinity.
FIGURE 6.
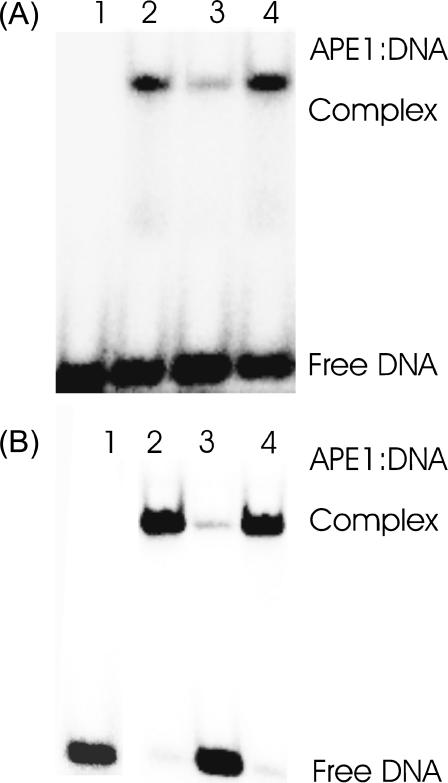
Competitive binding assay of D210N-APE1 to DNA containing either (A) coumarin:THF or (B) guanine:THF. Lane 1 shows the labeled DNA in the absence of the protein. Lane 2 shows the protein binding in the absence of competitor (18% A, 98% B). Lane 3 shows binding in the presence of 100-fold excess of unlabeled, specific oligodeoxynucleotide duplex that has THF opposite coumarin (5%, A) or guanine (3%, B). Lane 4 shows the binding in the presence of 100-fold excess of unlabeled, nonspecific competitor DNA that has guanine opposite cytosine (22%, A) (97%, B).
Steady-state spectra of the DNA:APE1 complex
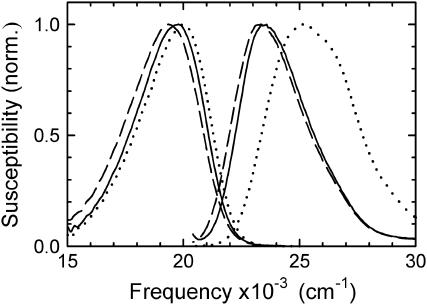
The right hand side of Fig. 7 shows absorption spectra of coumarin in several environments as measured by fluorescence excitation. The dotted curve shows the spectrum of coumarin 102 in pure water. The spectrum of coumarin in uncomplexed DNA is shown as the solid curve; there is a shift of ∼2000 cm−1 to lower frequency. The fact that the coumarin spectrum in DNA is strongly shifted relative to the spectrum in water is one piece of evidence that the coumarin is buried in the helix and not flipped out of the helix into an aqueous environment. Solvent-accessibility measurements also support this conclusion (Supplemental Material).
FIGURE 7.
Fluorescence excitation (right) and emission (left) spectra of coumarin 102 free in water (dotted line), the coumarin-containing oligonucleotide (solid line), and the complex of D210N-APE1 with the coumarin-containing oligonucleotide (dashed line). The vertical susceptibility scale facilitates quantitative comparison of absorption and emission spectra (17).
Our interest is the spectrum of coumarin in the DNA:APE1 complex, which is shown as the dashed curve in Fig. 7. The spectrum is very similar to the one in bare DNA (solid curve), although there is a small additional shift to lower frequency of 225 cm−1. This small shift can be attributed to a change in the preexisting electric field in the DNA:APE1 complex. Given the large structural changes near the coumarin upon forming the complex, some change in the local electric field is not surprising. However, given the large number of charged groups that move when forming the complex, it is difficult to assign the shift to any single group.
The main significance of the spectrum for this article is in relation to the concern that upon forming the complex, the coumarin might be expelled from the complex and swing out into the surrounding water. However, the excitation spectrum in the complex is typical of a DNA-like environment and not of an aqueous environment. In addition, there is no evidence of a shoulder on the high frequency side of the spectrum of the complex. Such a shoulder would be expected if there were a minority population of extrahelical coumarin. These results are consistent with the modeling, which indicates that there is no impediment to retaining the coumarin near the position of the native base.
Fig. 7 also shows the steady-state emission spectra of coumarin in water, in DNA and in the DNA:APE1 complex. These spectra are needed for the analysis of the transient data. However, a qualitative preview of the time-resolved results can be obtained from these spectra. The shift between the excitation and emission spectra (the Stokes shift) reflects the increased polarization of the local environment of the coumarin due to the increased dipole moment in the excited state of the coumarin.
The steady-state Stokes shift in DNA is significantly smaller than in water. This result is consistent with the conclusion of more extensive time-resolved studies, which conclude that the effective polarity in DNA is comparable to the polarity of ethanol (17).
On the other hand, the steady-state Stokes shift in DNA is nearly the same as in the DNA:APE1 complex, suggesting that the effective polarities are similar in both environments. The steady-state results are not conclusive in themselves, because they do not capture stabilization occurring after the fluorescence lifetime. However, the more detailed time-resolved results presented in the next section will continue to show a great deal of similarity between the complexed and uncomplexed DNA.
Time-resolved Stokes-shift measurements on the DNA:APE1 complex
Fig. 8 shows the results of TRSS measurements in two formats. In Fig. 8 A, the time-resolved Stokes shifts in complexed and uncomplexed DNA have been shifted vertically in frequency to maximize their overlap. Over the time window of the experiment, the Stokes shifts are nearly identical. The small difference in the curves at early time is definitely within the limits of reproducibility. The slightly larger difference at long times is near the error limits and may or may not be real. In either case, the primary result is that the dynamics in complexed and uncomplexed DNA are very similar in this time range.
FIGURE 8.
Time-resolved Stokes-shift measurements of the uncomplexed DNA (open circles) and the DNA:APE1 complex (squares). (A) Relative Stokes shifts. The data have been shifted on the vertical axis to highlight differences within the measurement time window. (B) Absolute Stokes shifts relative to the emission in the glass.
In Fig. 8 B, the absolute Stokes shift has been calculated as the difference between the first moment of the emission spectrum at a given time and the first moment of the steady-state emission spectrum in a frozen glass. The spectrum in the glass is used as an approximation to the position of the emission in the absence of any diffusive, i.e., nonvibrational, motion in the system. The position of the glass emission is affected by shifts in the absorption spectrum, changes in the vibronic envelope of the spectra and by vibrational dynamics of the DNA or the glassy solvent. Thus, these effects are removed from the absolute Stokes shift.
In the absence of additional complications, the absolute Stokes shift should go to zero near 50 fs, where diffusive dynamics begin. Measurements with high time resolution on uncomplexed DNA confirm this prediction (20). The fact that there is a substantial Stokes shift at 40 ps suggests that there has been substantial shifting within the 50 fs–40 ps range.
The earliest Stokes shift measured for the DNA:APE1 complex is slightly larger than for uncomplexed DNA (Fig. 8 A). These results suggest that there may be a difference in the dynamics of the two samples at earlier times. Higher time resolution measurements are needed to confirm this inference.
The results above rely on characterizing the time-dependent emission spectrum by a single parameter, the mean frequency. This assumption is only valid if the shape of the emission spectrum is constant as it shifts. An example of a violation of this assumption is the case of heterogeneous dynamics. This case would occur if different helices adopted different conformations with different relaxation rates and if those conformations persisted for a substantial fraction of the experimental time window. Evidence for this type of heterogeneity has been reported in electron-transfer experiments in DNA (52).
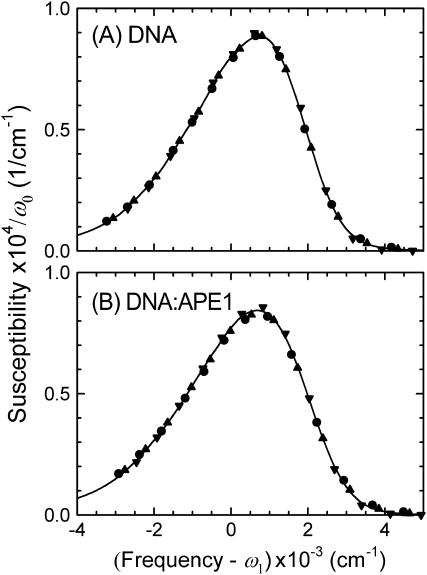
Fig. 9 tests the assumption of unchanging shape as the Stokes shift occurs. The emission spectra at different times have been shifted horizontally by their mean frequencies. They have been scaled vertically to constant area under the spectrum, to compensate for the decay of the emission intensity with time. The figure shows that the points comprising each of the time-resolved spectra all fall on a common spectral shape. There is no evidence of dynamical heterogeneity, and the mean frequency is sufficient to characterize the spectral dynamics.
FIGURE 9.
Time-evolution of the shape of the emission spectrum. Spectra from different times have been shifted in frequency by their first moment ω1 and the susceptibilities scaled by the spectral area ω0 (17). Points: 40 ps (•), 400 ps (▴), and 40 ns (▾). (A) Uncomplexed DNA. (B) DNA:APE1 complex. In each case, the points fit a single, common spectral shape (solid curve). The lack of significant spectral shape change indicates that all molecules relax at the same rate, i.e., the relaxation is homogeneous.
DISCUSSION AND CONCLUSIONS
This article better defines the ability of TRSS experiments on coumarin-containing, bare oligonucleotides to comment on the conditions in native DNA and in more complex DNA:protein assemblies. Despite the replacement of a native base by an unnatural fluorophore, APE1 is able to bind strongly and selectively to the coumarin-containing oligonucleotide and to cleave it. Both the binding constant and cleavage rate are decreased by a moderate amount, but the fundamental biochemical function of the system is retained. This result supports the idea that the dynamics in coumarin-containing oligonucleotides may differ quantitatively from those in native DNA, but the qualitative phenomena and mechanisms acting in native DNA are unchanged.
In previous work, we have seen that solvation dynamics in bare DNA have a large magnitude and persist for an unusually long time. These features make DNA an unusual environment for chemical reactions. This study has shown that both the large magnitude and slow relaxation can persist in DNA:protein complexes. Our results show small changes in the polarity of the coumarin's environment that are reflected in shifts of the steady-state spectrum and an overall difference in the absolute Stokes shift. However, the total range of the Stokes shift and the rate of its relaxation are almost unchanged in the DNA:APE1 complex.
Of course, DNA forms a wide variety of protein complexes with very different structures, so the results with the specific DNA:APE1 complex cannot be assumed to be universal for all DNA-interacting proteins. However, these results do show that it is possible for the large polarity and slow solvation dynamics seen in bare DNA to persist in DNA:protein complexes, where they can influence the stability of transition states and the rates of reactions.
Given the large structural changes in the complex, the small change in dynamics might seem puzzling. However, it should be kept in mind that the TRSS experiment only measures the dynamics of local electric fields. Movement of uncharged structural elements is not seen directly in this experiment. In addition, electrostatic interactions are relatively long range, so the net electric field averages the contribution from many individual structural elements in the neighborhood of the probe. Although the model structure of the complex shows strong distortion of the backbone and flanking bases, it also shows that the probe remains near the surface of the protein, with unperturbed solvent forming much of its local environment. Recent molecular dynamics simulations suggest that counterion motion may dominates the long time dynamics in DNA (53). Because a large fraction of the counterion atmosphere remains in this complex, this idea would be consistent with a small change in the TRSS dynamics in the complex.
Another interesting result of this study is that APE1 is able to recognize and act on an abasic site, even with an unnaturally large opposing group. Thus, the “empty space” left in a helix by the excision of a base is not an essential element for recognition of the abasic site.
In conclusion, the unusual properties of DNA dynamics can persist in more complex assemblies with proteins. An open question is whether other complexes that shield the DNA more completely from the solvent produce greater changes in the dynamics. Fortunately, we have shown that TRSS experiments are a viable means to address this question.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Supplementary Material
Acknowledgments
We thank Leslie Lovelace and Dr. Salvatore Profeta for help with the modeling, and Joseph Stember for the fluorescence quenching data.
This work was supported by the National Institutes of Health through grants GM-61292 and ES-00333. N.A.P. was supported by the Research Experience for Undergraduates Program in Nanoscience at the University of South Carolina, funded by the National Science Foundation (CHE-0139143).
References
- 1.Reichardt, C. 1988. Solvents and Solvent Effects in Organic Chemistry. VCH Publishers, Weinheim, FRG.
- 2.Hynes, J. T. 1985. Chemical reaction dynamics in solution. Annu. Rev. Phys. Chem. 36:573–597. [Google Scholar]
- 3.Beveridge, D. L., and K. J. McConnell. 2000. Nucleic acids: theory and computer simulation, Y2K. Curr. Opin. Struct. Biol. 10:182–196. [DOI] [PubMed] [Google Scholar]
- 4.Cheatham, T. E., and P. A. Kollman. 2000. Molecular dynamics simulation of nucleic acids. Annu. Rev. Phys. Chem. 51:435–471. [DOI] [PubMed] [Google Scholar]
- 5.Norberg, J., and L. Nilsson. 2003. Advances in biomolecular simulations: methodology and recent applications. Q. Rev. Biophys. 36:257–306. [DOI] [PubMed] [Google Scholar]
- 6.Brauns, E. B., M. L. Madaras, R. S. Coleman, C. J. Murphy, and M. A. Berg. 1999. Measurement of local DNA reorganization on the picosecond and nanosecond time scales. J. Am. Chem. Soc. 121:11644–11649. [Google Scholar]
- 7.Coleman, R. S., and M. L. Madaras. 1998. Synthesis of a novel coumarin C-riboside as a photophysical probe of oligonucleotide dynamics. J. Org. Chem. 63:5700–5703. [Google Scholar]
- 8.Takeshita, M., C. N. Chang, F. Johnson, S. Will, and A. P. Grollman. 1987. Oligodeoxynucleotides containing synthetic abasic sites. Model substrates for DNA polymerases and apurinic/apyrimidinic endonucleases. J. Biol. Chem. 262:10171–10179. [PubMed] [Google Scholar]
- 9.Maroncelli, M. 1993. The dynamics of solvation in polar liquids. J. Mol. Liq. 57:1–37. [Google Scholar]
- 10.Moog, R. S., W. W. Davis, S. G. Ostrowski, and G. L. Wilson. 1999. Solvent effects on electronic transitions in several coumarins. Chem. Phys. Lett. 299:265–271. [Google Scholar]
- 11.Buncel, E., and S. Rajagopal. 1990. Solvatochromism and solvent polarity scales. Acc. Chem. Res. 23:226–231. [Google Scholar]
- 12.Reichardt, C. 1994. Solvatochromic dyes as solvent polarity indicators. Chem. Rev. 94:2319–2358. [Google Scholar]
- 13.Katritzky, A. R., D. C. Fara, H. Yang, K. Tämm, R. Tamm, and M. Karelson. 2004. Quantitative measures of solvent polarity. Chem. Rev. 104:175–198. [DOI] [PubMed] [Google Scholar]
- 14.Demple, B., and L. Harrison. 1994. Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem. 63:915–948. [DOI] [PubMed] [Google Scholar]
- 15.Wilson, D. M., and D. Barsky. 2001. The major human abasic endonuclease: formation, consequences and repair of abasic lesions in DNA. Mutat. Res.-DNA Repair. 485:283–307. [DOI] [PubMed] [Google Scholar]
- 16.O'Neill, M. A., and J. K. Barton. 2004. DNA-mediated charge transport requires conformational motion of the DNA bases: elimination of charge transport in rigid glasses at 77 K. J. Am. Chem. Soc. 126:13234–13235. [DOI] [PubMed] [Google Scholar]
- 17.Somoza, M. I., D. Andreatta, R. S. Coleman, C. J. Murphy, and M. A. Berg. 2004. Effect of lesions on the dynamics of DNA on the picosecond and nanosecond time scales using a polarity sensitive probe. Nucleic Acids Res. 32:2495–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castner, E. W., M. Maroncelli, and G. R. Fleming. 1987. Subpicosecond resolution studies of solvation dynamics in polar aprotic and alcohol solvents. J. Chem. Phys. 86:1090–1097. [Google Scholar]
- 19.Brauns, E. B., M. L. Madaras, R. S. Coleman, C. J. Murphy, and M. A. Berg. 2002. Complex local dynamics in DNA on the picosecond and nanosecond time scales. Phys. Rev. Lett. 88:158101. [DOI] [PubMed] [Google Scholar]
- 20.Andreatta, D., J. L. Pérez Lustres, S. A. Kovalenko, N. P. Ernsting, C. J. Murphy, R. S. Coleman, and M. A. Berg. 2005. Power-law solvation dynamics in DNA over six decades in time. J. Am. Chem. Soc. 127:7270–7271. [DOI] [PubMed] [Google Scholar]
- 21.Pal, S. K., L. Zhao, T. Xia, and A. H. Zewail. 2003. Site- and sequence-selective ultrafast hydration of DNA. Proc. Natl. Acad. Sci. USA. 100:13746–13751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindahl, T., and R. D. Wood. 1999. Quality control by DNA repair. Science. 286:1897–1905. [DOI] [PubMed] [Google Scholar]
- 23.Hoeijmakers, J. H. 2001. Genome maintenance mechanisms for preventing cancer. Nature. 411:366–374. [DOI] [PubMed] [Google Scholar]
- 24.McCullough, A. K., M. L. Dodson, and R. S. Lloyd. 1999. Initiation of base excision repair: glycosylase mechanisms and structures. Annu. Rev. Biochem. 68:255–285. [DOI] [PubMed] [Google Scholar]
- 25.Scharer, O. D., and J. Jiricny. 2001. Recent progress in the biology, chemistry and structural biology of DNA glycosylases. Bioessays. 23:270–281. [DOI] [PubMed] [Google Scholar]
- 26.Lindahl, T. 1993. Instability and decay of the primary structure of DNA. Nature. 362:709–715. [DOI] [PubMed] [Google Scholar]
- 27.Loeb, L. A., and B. D. Preston. 1986. Mutagenesis by apurinic/apyrimidinic sites. Annu. Rev. Genet. 20:201–230. [DOI] [PubMed] [Google Scholar]
- 28.Wilson 3rd, D. M., M. Takeshita, A. P. Grollman, and B. Demple. 1995. Incision activity of human apurinic endonuclease (APE) at abasic site analogs in DNA. J. Biol. Chem. 270:16002–16007. [DOI] [PubMed] [Google Scholar]
- 29.Erzberger, J. P., and D. M. Wilson 3rd. 1999. The role of Mg2+ and specific amino acid residues in the catalytic reaction of the major human abasic endonuclease: new insights from EDTA-resistant incision of acyclic abasic site analogs and site-directed mutagenesis. J. Mol. Biol. 290:447–457. [DOI] [PubMed] [Google Scholar]
- 30.Rothwell, D. G., B. Hang, M. A. Gorman, P. S. Freemont, B. Singer, and I. D. Hickson. 2000. Substitution of Asp-210 in HAP1 (APE/Ref-1) eliminates endonuclease activity but stabilizes substrate binding. Nucleic Acids Res. 28:2207–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Changenet-Barret, P., C. T. Choma, E. F. Gooding, W. F. DeGrado, and R. M. Hochstrasser. 2000. Ultrafast dielectric response of proteins from dynamics Stokes shifting of coumarin in calmodulin. J. Phys. Chem. B. 104:9322–9329. [Google Scholar]
- 32.Toptygin, D., R. S. Savtchenko, N. D. Meadow, and L. Brand. 2001. Homogeneous spectrally- and time-resolved fluorescence emission from single-tryptophan mutants of IIAGlc protein. J. Phys. Chem. B. 105:2043–2055. [Google Scholar]
- 33.Mandal, D., S. Sen, D. Sukul, K. Bhattacharrya, A. K. Mandal, R. Banerjee, and S. Roy. 2002. Solvation dynamics of a probe covalently bound to a protein and in an AOT microemulsion: 4-(n-bromoacetylamino)-phthalimide. J. Phys. Chem. B. 106:10741–10747. [Google Scholar]
- 34.Cohen, B. E., T. B. McAnaney, E. S. Park, Y. N. Jan, S. G. Boxer, and L. Y. Jan. 2002. Probing protein electrostatics with a synthetic fluorescent amino acid. Science. 296:1700–1703. [DOI] [PubMed] [Google Scholar]
- 35.Pal, S. K., J. Peon, and A. H. Zewail. 2002. Biological water at the protein surface: dynamical solvation probed directly with femtosecond resolution. Proc. Natl. Acad. Sci. USA. 99:1763–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lampa-Pastirk, S., and W. F. Beck. 2004. Polar solvation dynamics in Zn(II)-substituted cytochrome c: diffusive sampling of the energy landscape in the hydrophobic core and solvent-contact layer. J. Phys. Chem. B. 108:16288–16294. [Google Scholar]
- 37.Buzady, A., J. Savolainen, J. Erostyak, P. Myllyperkio, B. Somogyi, and J. Korppi-Tommola. 2003. Femtosecond transient absorption study of the dynamics of acrylodan in solution and attached to human serum albumin. J. Phys. Chem. B. 107:1208–1214. [Google Scholar]
- 38.Petushkov, V. N., I. H. M. van Stokkum, B. Gobets, F. van Mourik, J. Lee, R. van Grondelle, and A. Visser. 2003. Ultrafast fluorescence relaxation spectroscopy of 6,7-dimethyl-(8-ribityl)-lumazine and riboflavin, free and bound to antenna proteins from bioluminescent bacteria. J. Phys. Chem. B. 107:10934–10939. [Google Scholar]
- 39.Jordanides, X. J., M. J. Lang, X. Y. Song, and G. R. Fleming. 1999. Solvation dynamics in protein environments studied by photon echo spectroscopy. J. Phys. Chem. B. 103:7995–8005. [Google Scholar]
- 40.Jimenez, R., G. Salazar, K. K. Baldridge, and F. E. Romesberg. 2003. Flexibility and molecular recognition in the immune system. Proc. Natl. Acad. Sci. USA. 100:92–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fayer, M. D. 2001. Fast protein dynamics probed with infrared vibrational echo experiments. Annu. Rev. Phys. Chem. 52:315–356. [DOI] [PubMed] [Google Scholar]
- 42.Zhong, D., S. K. Pal, and A. H. Zewail. 2001. Femtosecond studies of protein-DNA binding and dynamics: Histone I. ChemPhysChem. 2:219–227. [DOI] [PubMed] [Google Scholar]
- 43.Mol, C. D., T. Izumi, S. Mitra, and J. A. Tainer. 2000. DNA-bound structures and mutants reveal abasic DNA binding by APE1 DNA repair and coordination. Nature. 403:451–456. [DOI] [PubMed] [Google Scholar]
- 44.Velapoldi, R. A., and K. D. Mielenz. 1980. Standard Reference Materials: A Fluorescence Standard Reference Material: Quinine Sulfate Dihydrate. Nat. Bur. Stand. (U.S.), Spec. Publ. 260-64. U.S. Government Printing Office, Washington, DC.
- 45.Maroncelli, M., and G. R. Fleming. 1987. Picosecond solvation dynamics of coumarin 153: The importance of molecular aspects of solvation. J. Chem. Phys. 86:6221–6239. [Google Scholar]
- 46.Kool, E. T. 2002. Replacing the nucleobases in DNA with designer molecules. Acc. Chem. Res. 35:936–943. [DOI] [PubMed] [Google Scholar]
- 47.Kool, E. T. 2001. Hydrogen bonding, base stacking, and steric effects in DNA replication. Annu. Rev. Biophys. Biomol. Struct. 30:1–22. [DOI] [PubMed] [Google Scholar]
- 48.Gelfand, C. A., G. E. Plum, A. P. Grollman, F. Johnson, and K. J. Breslauer. 1998. Thermodynamic consequences of an abasic lesion in duplex DNA are strongly dependent on base sequence. Biochemistry. 37:7321–7327. [DOI] [PubMed] [Google Scholar]
- 49.Pompizi, I., A. Häberli, and C. J. Leumann. 2000. Oligodeoxynucleotides containing conformational constrained abasic sites: a UV and fluorescence spectroscopic investigation of duplex stability and structure. Nucleic Acids Res. 28:2702–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nguyen, L. H., D. Barsky, J. P. Erzberger, and D. M. Wilson 3rd. 2000. Mapping the protein-DNA interface and the metal-binding site of the major human apurinic/apyrimidinic endonuclease. J. Mol. Biol. 298:447–459. [DOI] [PubMed] [Google Scholar]
- 51.Masuda, Y., R. A. O. Bennett, and B. Demple. 1998. Dynamics of the interaction of human apurinic endonuclease (Ape1) with its substrate and product. J. Biol. Chem. 273:30352–30359. [DOI] [PubMed] [Google Scholar]
- 52.Fiebig, T., C. Wan, and A. H. Zewail. 2002. Femtosecond charge transfer dynamics of a modified DNA base: 2-aminopurine in complexes with nucleotides. ChemPhysChem. 3:781–788. [DOI] [PubMed] [Google Scholar]
- 53.Ponomarev, S. Y., K. M. Thayer, and D. L. Beveridge. 2004. Ion motions in molecular dynamics simulations on DNA. Proc. Natl. Acad. Sci. USA. 101:14771–14775. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.