Abstract
This study views each protein structure as a network of noncovalent connections between amino acid side chains. Each amino acid in a protein structure is a node, and the strength of the noncovalent interactions between two amino acids is evaluated for edge determination. The protein structure graphs (PSGs) for 232 proteins have been constructed as a function of the cutoff of the amino acid interaction strength at a few carefully chosen values. Analysis of such PSGs constructed on the basis of edge weights has shown the following: 1), The PSGs exhibit a complex topological network behavior, which is dependent on the interaction cutoff chosen for PSG construction. 2), A transition is observed at a critical interaction cutoff, in all the proteins, as monitored by the size of the largest cluster (giant component) in the graph. Amazingly, this transition occurs within a narrow range of interaction cutoff for all the proteins, irrespective of the size or the fold topology. And 3), the amino acid preferences to be highly connected (hub frequency) have been evaluated as a function of the interaction cutoff. We observe that the aromatic residues along with arginine, histidine, and methionine act as strong hubs at high interaction cutoffs, whereas the hydrophobic leucine and isoleucine residues get added to these hubs at low interaction cutoffs, forming weak hubs. The hubs identified are found to play a role in bringing together different secondary structural elements in the tertiary structure of the proteins. They are also found to contribute to the additional stability of the thermophilic proteins when compared to their mesophilic counterparts and hence could be crucial for the folding and stability of the unique three-dimensional structure of proteins. Based on these results, we also predict a few residues in the thermophilic and mesophilic proteins that can be mutated to alter their thermal stability.
INTRODUCTION
The underlying principles of protein stability and folding, which have not yet been completely understood, have been probed by a variety of analyses on a large number of available protein structures. Theoretical studies of protein structures and experimental protein engineering methods have been used to understand and enhance the stability of proteins (1–7). Further, numerous protein-folding experiments and simulations have been carried out to understand the folding pathway of proteins, and specific residues have been identified in a few proteins that play a role in the folding pathway and the transition state (8–9). This study is focused on understanding the principles of protein structure, stability, and folding by considering the protein structures as networks of noncovalent interactions. We find a novel perspective on how protein structures are formed and stabilized, with the strength of side-chain interactions playing an important role in determining the characteristics of the network.
Protein structure networks have earlier been constructed with varying definitions of nodes and edges (10–16). These investigations have focused on elucidating the network properties such as the shortest path length, clustering coefficient, and other small-world properties. The folding behavior of proteins has also been investigated in some of these studies using the structure of the transition state known in some proteins (10–12). Although this study also considers the protein structures as networks, the method of construction and the analysis of these networks are different from previous studies. Here, the protein structure graphs (PSGs) are constructed by defining the amino acids in the polypeptide chain as the nodes and the noncovalent interactions among them as links. It has been established that such graphs are useful in the identification of clusters of amino acid residues that stabilize the protein structure and protein-protein interfaces (7,17–20). An important feature of such a graph is the definition of edges based on the normalized strength of interaction between the amino acid residues in proteins. Interestingly, we find that the network topology of such PSGs depends on the cutoff of the interaction strength between amino acid residues used in the graph construction.
Apart from analyzing the topological properties of the PSG, two other major findings emerge from the definition of edge-weighted PSG in this work. First, at a critical cutoff of interaction strength, we find a transition as probed by the size of the largest cluster. Interestingly, we find that this critical interaction cutoff, which we have evaluated for more than 200 proteins, falls within a narrow range, emphasizing the fact that this transition is a universal behavior of globular proteins. Second, we are able to identify the amino acid residues, which are highly connected and are crucial for the stability of the protein structure network. In the network terminology, these are the equivalent of “hubs”. In many real-world cases, the networks are known to be less sensitive to random attacks on nodes but much more susceptible to targeted attacks on hubs (21). A similar situation may exist in PSGs, where an inappropriate mutation of the hub residues can destabilize the protein structure. We have also analyzed the role of these hubs in bringing together the different secondary structure elements in the protein tertiary structure. Finally, we have demonstrated that the network parameters are able to account for the additional stability of thermophilic proteins. In a broad sense, this analysis yields novel insights into protein structure and stability by elucidating the role of the amino acid side chains in maintaining the unique topology of protein structures. Thus, we believe that this study will be able to motivate new experiments in protein folding, stability, and design.
MATERIALS AND METHODS
Data set
The data set used in this analysis consists of 232 globular protein structures obtained from the protein data bank (22) and given in the Supplementary Material (Table S1). This is a nonredundant set of proteins with a resolution better than 1.8 Å and sequence identity <20%. The sizes of the proteins considered vary from 50 to 1300 residues. A separate set of 10 pairs of thermophilic and their corresponding mesophilic proteins (given in Table 1) were considered to investigate the thermal stability aspect.
TABLE 1.
Network parameters of thermophilic and mesophilic proteins
| Thermophile PDBid
|
Mesophile PDBid
|
No. of hubs
|
Total No. of edges
|
Edge/node ratio
|
Largest cluster size
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Serial No. | Protein | Imin | T* | M† | T | M | T | M | T | M | ||
| 1 | TATA box binding protein | 1PCZ | 1VOK | 0 | 59 | 52 | 326 | 325 | 1.75 | 1.69 | 165‡ | 174‡ |
| 2 | 22 | 13 | 220 | 214 | 1.20 | 1.11 | 147 | 101 | ||||
| 4 | 8 | 1 | 139 | 124 | 0.76 | 0.65 | 35 | 32 | ||||
| 2 | Adenylate kinase | 1ZIP | 1AK2 | 0 | 75 | 54 | 376 | 316 | 1.73 | 1.44 | 184 | 170 |
| 2 | 21 | 20 | 238 | 211 | 1.10 | 0.96 | 158 | 121 | ||||
| 4 | 5 | 4 | 134 | 127 | 0.62 | 0.58 | 57 | 29 | ||||
| 3 | Subtilisin | 1THM | 1ST3 | 0 | 95 | 89 | 521 | 481 | 1.87 | 1.78 | 262 | 247 |
| 2 | 26 | 25 | 310 | 300 | 1.12 | 1.10 | 213 | 184 | ||||
| 4 | 2 | 1 | 152 | 150 | 0.56 | 0.54 | 47 | 29 | ||||
| 4 | Carboxy peptidase | 1OBR | 2CTC | 0 | 149 | 135 | 661 | 635 | 2.05 | 2.01 | 298 | 290 |
| 2 | 64 | 61 | 468 | 451 | 1.45 | 1.42 | 278 | 272 | ||||
| 4 | 12 | 8 | 295 | 269 | 0.91 | 0.88 | 214 | 129 | ||||
| 5 | Neutral protease | 1THL | 1NPC | 0 | 116 | 109 | 582 | 560 | 1.84 | 1.77 | 290 | 287 |
| 2 | 41 | 40 | 407 | 384 | 1.29 | 1.21 | 238‡ | 260‡ | ||||
| 4 | 4‡ | 7‡ | 242 | 229 | 0.77 | 0.72 | 101 | 72 | ||||
| 6 | Phosphofructo kinase | 3PFK | 2PFK | 0 | 89 | 87 | 524 | 506 | 1.64‡ | 1.69‡ | 275 | 267 |
| 2 | 20 | 15 | 334 | 332 | 1.05‡ | 1.11‡ | 238 | 232 | ||||
| 4 | 0 | 0 | 165‡ | 180‡ | 0.52‡ | 0.60‡ | 62 | 49 | ||||
| 7 | Lactate dehydrogenase | 1LDN | 1LDM | 0 | 104 | 95 | 541 | 524 | 1.71 | 1.59 | 277 | 267 |
| 2 | 27 | 19 | 353 | 317 | 1.12 | 0.96 | 219 | 209 | ||||
| 4 | 3 | 2 | 198 | 169 | 0.63 | 0.51 | 97 | 95 | ||||
| 8 | Glyceraldehyde-3-phosphate dehydrogenase | 1GD1 | 1GAD | 0 | 108 | 103 | 577 | 553 | 1.74 | 1.69 | 294 | 284 |
| 2 | 38 | 35 | 373 | 352 | 1.13 | 1.08 | 229 | 220 | ||||
| 4 | 9 | 3 | 201 | 188 | 0.61 | 0.57 | 57 | 49 | ||||
| 9 | Phosphoglycerate kinase | 1PHP | 3PGK | 0 | 146 | 80 | 739 | 526 | 1.88 | 1.27 | 363 | 313 |
| 2 | 45 | 24 | 451 | 353 | 1.14 | 0.85 | 282 | 205 | ||||
| 4 | 3‡ | 5‡ | 217‡ | 226‡ | 0.55 | 0.53 | 55 | 52 | ||||
| 10 | Reductase | 1EBD | 1LVL | 0 | 115 | 127 | 725 | 713 | 1.59 | 1.56 | 412 | 367 |
| 2 | 33 | 22 | 461 | 417 | 1.01 | 0.91 | 232 | 181 | ||||
| 4 | 3 | 2 | 236 | 209 | 0.52 | 0.46 | 27 | 22 | ||||
T, thermophile.
M, mesophile.
Cases where the values are higher for the mesophilic protein than the corresponding thermophilic protein.
Construction of the PSG
The PSG is constructed from the three-dimensional atomic coordinates of the protein structures obtained from the protein data bank as follows.
Definition of nodes and edges
Each protein in the data set is represented as a graph consisting of a set of nodes and edges. Each amino acid in the protein structure is represented as a node, and these nodes (amino acids) are connected by edges based on the strength of noncovalent interaction between the side chains of the two amino acid residues. The strength of interaction between two amino acid side chains is evaluated as a percentage given by:
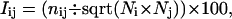 |
(1) |
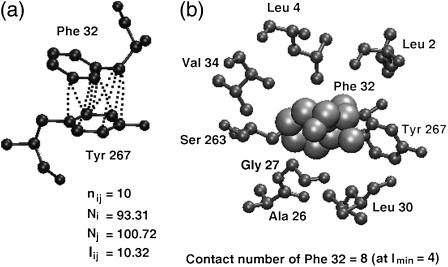
where, nij is the number of distinct atom pairs between the side chains of amino acid residues i and j, which come within a distance of 4.5 Å (23), and Ni and Nj are the normalization factors for residue types i and j and are given in the Supplementary Material (Table S2). An example of a pair of aromatic residues interacting with an Iij value of 10.3% is shown in Fig. 1 a.
FIGURE 1.
Contact number versus interaction strength. An example taken from the protein L-arabinose binding protein (Protein Data Bank (PDB) code: 8abp). (a) Interaction strength: two aromatic residues (shown in ball-and-stick representation) making contact at high interaction strength (Iij = 10.3%). The atom-atom contacts (≤4.5 Å) between the two residues are indicated by dotted lines. (b) Contact number: a phenylalanine residue (shown in van der Waals representation) interacts with eight other residues (shown in ball-and-stick representation) at an interaction cutoff (Imin) of 4%.
The normalization factor was evaluated from a nonredundant set of protein structures for the 20 different amino acids and was taken from the work of Kannan and Vishveshwara (17). This factor takes into account the differences in the sizes of the side chains of the different residue types and their propensity to make the maximum number of contacts with other amino acid residues in protein structures. Since the interaction strength Iij depends on the property of both residues i and j, different combinations of the normalization values, such as (Ni + Nj)/2 and min(Ni,Nj) were explored in Eq. 1. However, they were found to give qualitatively very similar results.
Iij is thus evaluated for all the ij pairs in the protein structure. We then choose a cutoff value, Imin and any ij residue pair with Iij > Imin is connected by an edge in the PSG, which has N nodes, where N is the number of amino acid residues in the protein structure. This cutoff (Imin) is varied from 0% (>0% is denoted as 0%) to 10% (very few nodes interact with a value >10%), and the PSG is constructed for all the proteins in the data set at these varying cutoffs. As the interaction cutoff is increased from 0% to 10%, the number of edges in the PSGs decreases because, at higher cutoff, the number of nodes making the high level of interaction will be less. Thus, we are able to quantify the interactions among the side chains of the residues and thus construct amino acid-based PSGs at varying strengths of interaction using this method. Our definition of amino acid interaction is based purely on the number of distance-based contacts between two amino acid residues. (This could further be refined by other factors such as hydrogen bonds and electrostatic interactions, where the energy of interaction can be directly taken into account). The PSGs of all the proteins in the data set, constructed at different Imin values, have been analyzed using various parameters given below.
Analysis of PSGs
Network properties
The networks are analyzed for the distribution of nodes with k links. For each PSG, the number of nodes n with k edges (links), n(k), is evaluated at various Imin values. The cumulative value (ntot(k)) over all proteins in the data set is taken, and then ntot(k) versus k is plotted at different Imin values. Further, we also evaluate the total number of edges or links in a PSG at a given Imin, referred to as ktotal and the ratio of the total number of edges to the total number of nodes in the PSG at a particular Imin, given by ktotal/N (where N is total number of residues or nodes in the protein structure). Both these parameters (ktotal and ktotal/N) are used in understanding the stability of thermophilic proteins.
Size of the largest cluster
The PSG is represented as an adjacency matrix (A), where
Aij = 1, if i ≠ j and i and j are connected according to the Imin criterion.
Aij = 0, if i ≠ j and i and j are not connected.
Aij = 0, if i = j.
The adjacency matrix is then analyzed using standard graph techniques like the depth first search (DFS) method (24) to identify distinct clusters and the cluster-forming nodes (residues) in the PSG. The largest cluster is then identified, and its size (in terms of the number of amino acid residues) is determined for all the PSGs at different interaction cutoffs. The normalized value of the largest cluster size (with respect to the total number of residues in the protein) is plotted as a function of Imin values for all the proteins in the data set.
Contact number versus interaction strength
It is important to understand the difference between the two parameters, namely, the contact number and the interaction strength, both of which are used in the analysis of the PSGs in this study. The interaction strength is a parameter evaluated between two residues using the number of atom-atom contacts between them as given in either Eq. 1 or 2 (given below). However, the contact number of a residue i is defined as the total number of interactions which it makes with all other residues at a particular cutoff of the interaction strength (Imin). Although the interaction strength is evaluated between a pair of residues i and j and is based on the number of atom-atom contacts between them, the contact number works at a higher level and includes the number of residue-residue contacts made by a residue i at a particular cutoff of the interaction strength. Fig. 1, a and b, elucidates the difference between contact number and interaction strength, where examples of high interaction strengths and high contact number are shown clearly. We obtain the contact number (number of links or edges) of all the residues at varying Imins to analyze the PSGs of all the proteins in the data set. Specifically, we look at the high contact number residues (those which interact with more than four residues in the protein structure), referred to as “hubs” henceforth, at both high and low Imins. As explained earlier, the evaluation of interaction between two residues in a protein structure involves the normalization values of both the residue types. However, for the identification of hubs in a protein structure, it would be accurate to use the normalization value of the hub-forming residue alone. Hence, the interaction equation given in Eq. 1 reduces to the following for hub identification.
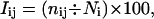 |
(2) |
where, Iij and nij are the same as in Eq. 1 and Ni is the normalization factor of residue type i, whose contact number is being evaluated. (However, we noted that the results did not vary significantly when the Iij definition given in Eq. 1 (sqrt(Ni × Nj)) or the other combinations of normalization values like (Ni + Nj)/2 and min(Ni,Nj) are used.)
Edge distribution profile of amino acids
The contact numbers of each of the 20 amino acid types in all the proteins in the data set (cumulative) were calculated at different Imin values. The number of amino acids of type i with contact numbers varying from 0 (orphans), 1 to 2, 3 to 4, and >4 (hubs) have been obtained using the definition given in Eq. 2 for all the proteins in the data set. The cumulative values have been obtained using all the proteins at desired Imin values for the 20 amino acid types, and the frequency distribution is plotted. This is referred to as the edge distribution profile of amino acids.
The plots presented in this work were obtained using MATLAB (The MathWorks, Natick, MA) and the protein structure figures were generated using VMD (25).
RESULTS
The nature and properties of the PSGs analyzed in this study are found to depend upon the cutoff of the interaction strength between the amino acid residues. The interaction strength is evaluated using a robust method developed earlier in the laboratory, which has provided biologically relevant insights into protein structure, folding, stability, and interactions (7,17–20). The PSGs of 232 proteins are constructed using different cutoffs of the interaction strengths (Imins), varying from a minimum (0%) to 10%. The amino acids interacting at higher Imin values make strong contacts, whereas the ones that interact only at lower Imin values make weak contacts. The network properties of these PSGs and the preferences of amino acid residues to make strong and weak interactions are analyzed. The results of these investigations are presented in the following sections. We discuss the application of the network concepts developed here to understand the thermal stability of thermophilic proteins in the last section.
Distribution of the nodes with k links as a function of the interaction criterion
The plot of the number of nodes (ntot(k)) with k links (cumulative value over all proteins in the data set), as a function of the number of links (k) at various interaction cutoffs is shown in Fig. 2. This plot gives us an idea of the number of orphans (k = 0) and the number of hubs (k > 4) in the PSGs at various interaction cutoffs (Imin). As the interaction cutoff is increased, ntot(k) decreases in general for most of the k values. However, at lower Imin values (0–4%), the number of nodes with less than two links is small, thereby giving rise to a bell-shaped curve. At Imin values ∼4.5–6% a sigmoidal curve is obtained, and at Imin >6% the curves show a steep decay behavior. At Imin = 4.5, the number of orphans in the PSGs exceeds the number of nodes with any k connections with k > 0 and this number keeps increasing when Imin is further increased. Since the nature of the distribution shown in Fig. 2 varies from bell shaped to sigmoidal to decay with increasing Imin, the PSGs certainly show a complex behavior. However, it is a consistent one, seen for a large number of proteins of various sizes and folds. It can be noted that the maximum number of edges made by any node in the PSGs in the complete range of Imin values is 12, and the maximum size of the PSGs is only ∼1500 nodes (this may be higher in the case of multimers). Hence, the PSGs are small networks when compared to most of the real-world networks analyzed (21). The results presented in Fig. 2, represent a cumulative value over all the proteins in the data set. Nevertheless, an examination of the behavior of n(k) versus k for individual proteins qualitatively shows the same behavior of network topology, irrespective of the protein size.
FIGURE 2.
Distribution of number of nodes making k links (cumulative over all proteins in the data set) in the PSGs, which are constructed as described in the Methods section. The frequency distribution of nodes with a particular number of edges at various interaction cutoffs (Imin) in the PSGs is plotted.
Fig. 2 clearly shows a complex behavior of the PSG with the nature of the ntot(k) versus k plot being dependent on Imin values. The nature of these graphs was evaluated by the log-linear and the log-log plots (figures not shown) of ntot(k) versus k at various Imin values. We find that both the log-linear and the log-log plots are nonlinear at almost all Imin values, and hence it is difficult to infer the nature of PSGs from these plots. However, above Imin = 6%, the plots show a power-law tail with the critical exponent γ ranging from 1.2 to 2.3. In essence, the PSGs seem to behave in a complex manner with varied network topologies at different interaction cutoffs.
Size of the largest cluster as a function of the interaction cutoff
The size of the largest cluster (or the giant component) is often used to understand the nature and properties of graphs (21) and to assess whether there is a phase transition from the percolation point of view (26). Here, we have monitored the variations in the size of the largest cluster with Imin values in all the proteins in the data set. The normalized size of the largest cluster (in terms of the number of nodes) is plotted as a function of Imin for a set of 200 proteins, belonging to various sizes and folds (Fig. 3). It is evident from Fig. 3 that irrespective of the protein size or fold, the size of the largest cluster in each of the proteins undergoes a transition at a particular Imin value. This Imin value at which the size of the largest cluster decreases dramatically (i.e., the midpoint of the transition) is termed Icritical. The plots in Fig. 3 are similar to the phase transition curves described by percolation theory and observed in physical systems (26). Surprisingly, these plots show that Icritical, where this transition occurs, is within a narrow range for proteins of all sizes and folds. The standard deviation of Icritical is 0.9 around a mean of ∼3.9. We find that >85% of the proteins have an Icritical varying between 3.0 and 5.0, which is a significantly narrow range. However, Icritical is a function of the size of the protein and is generally higher for bigger proteins as indicated by the spread of the plots in Fig. 3. Thus, mean Icritical is ∼3.25% in proteins with 100–200 residues, 3.75% in those with 200–300 residues, 4.25% in those with 300–400 residues, and >4.25% in those with 400–1300 residues. When the proteins are segregated into bins of varying sizes, the standard deviation of the Icritical varies from 0.6–0.7, which further confirms the point that Icritical is dependent on protein size to a small extent. The critical Imin values varying from 3.0% to 5.0% are close to the Imin values discussed earlier (4.5%), where the number of orphans in the PSGs exceeds the number of nodes with any k connections with k > 0. In physical terms, a transition from one giant cluster to small disjoint clusters occurs around Imin = Icritical. This transition reveals that there are large numbers of residue pairs in the protein structures, which have an interaction strength value (Iij) around the region of 4%, which is the critical Imin value. Hence, an interaction cutoff (Imin) of 4% or above makes a large number of residues lose a lot of these contacts, thus causing a sudden drop in the size of the largest cluster and leading to the transition seen in Fig. 3. This transition is indicative of the fact that the PSG exists as a completely connected giant cluster at Imin values lower than Icritical (∼4.5%), and these separate into smaller disjoint clusters at higher Imin values.
FIGURE 3.
Plot of the size of the largest cluster normalized by the protein size (N, number of amino acids in the protein) as a function of the Imin values for ∼200 proteins of varying sizes (50–1300).
The edge distribution profile of amino acids in PSG
We have investigated the preferences of different types of amino acids to acquire different numbers of links (contact number). The number of residues of type i, which make k links in all the PSGs in the data set, has been obtained at different Imin values, and a histogram of the normalized values is displayed in Fig. 4, which is referred to as the edge distribution profile of amino acids. The edge distribution profiles are shown for an Imin value less than Icriticial (Imin = 2%) and at around Icritical (Imin = 4%) in Fig. 4, a and b, respectively. The figure shows the number of residues of type i (normalized with respect to the total number of residues of type i in the data set), which make zero edges (orphans), 1–2 edges, 3–4 edges, and >4 edges (hubs). In general, we find that the amino acid preferences versus contact number correlate with the size of the amino acids as seen in the figure. However, the analysis of hub preferences at different Imin values shows an interesting behavior as discussed below.
FIGURE 4.
Edge distribution profile of the 20 different amino acids in PSGs at (a) Imin = 2% and (b) Imin = 4%. The distributions of the number of edges (summed over all 232 proteins and normalized with respect to the total number of amino acids of each type present in the data set) are represented in different shades.
The amino acid preferences in the hubs (>4 edges) show that before the transition (at Imin = 2%), tryptophan, phenylalanine, tyrosine, isoleucine, leucine, and methionine are the highly preferred ones. However, around the transition, i.e., at Imin = 4%, leucine and isoleucine lose a large number of contacts, thus losing their hub status, whereas arginine and histidine gain preference as hubs at Imin = 4%. However, phenylalanine, tyrosine, tryptophan, and methionine retain their hubs status at Imin = 4%. Those hubs that are preferred at higher Imins are called strong hubs, whereas those that are preferred only at lower Imins are referred to as weak hubs. Thus, the charge-delocalized planar side chains of Phe, Tyr, Trp, Arg, and His along with Met are preferred as strong hubs at higher Imins, whereas the hydrophobic side chains of Leu, Ile, and Val, preferred as weak hubs, appear only at lower Imins, in the PSGs. The other residues are not significantly seen as hubs at any Imin, though they are not completely left out. Further, the transition seen in Fig. 3 is mainly due to the loss of a large number of weak interactions contributed mainly by the hydrophobic residues such as leucine, isoleucine, and valine, which largely form the weak hubs. The preference of the charge-delocalized planar side chains (Phe, Tyr, Trp, Arg, His) to form the strong hubs indicates that the planar geometry and the charge delocalization of these residues have facilitated different types of interactions with a large number of other residues. It is noteworthy that the weak hubs involved in the structural transition observed in Fig. 3 are the hydrophobic residues such as leucine, isoleucine, and valine, which mainly contribute to the hydrophobic core of the natively folded protein. Although, in general, the bulkier residues are preferred as hubs, the hub status is dependent on the cutoff of the interaction strength. The dependence of hub status on the size of the amino acid is not completely linear, since bulkier side chains like lysine are overshadowed by relatively smaller ones like leucine and isoleucine at very low Imins. This could be because lysine, being a charged residue, is less buried than the others. Hence, both size and charge distribution play an important role in deciding the amino acid hub preferences. Further, various combinations of the normalization values as mentioned in the Methods section (Ni, sqrt(Ni × Nj), (Ni + Nj)/2, and min(Ni,Nj)) have been used in the evaluation of interaction strength between two residues in the PSGs. We find that the profiles obtained using the various combinations are very similar to the one shown in Fig. 4. Hence, different combinations of the normalization values qualitatively yield the same results, confirming that the hub preferences presented here are genuine and not an artifact of the size effect.
The edge distribution profile (Fig. 4) shows the significant loss of weak interactions when Imin is increased from 0% to 4%, which leads to the transition shown in Fig. 3. A pictorial representation of the hubs and clusters determined in barnase (1RNB) at Imin = 0% and Imin = 6% are shown in the supplementary figure (Fig. S1) to elucidate this aspect. The significance of weak connections in a network has been earlier demonstrated by Granovetter during his quest for understanding social networks (27). Similarly, from the PSGs obtained at lower Imin values, we find that the weak interactions play an important role in maintaining the integrity of the PSGs, whereas the strong interactions are undoubtedly essential for the stability of protein structures.
The role of hubs in integrating secondary structures
We have analyzed the secondary structure preferences of the hubs as well as that of the residues with which the hubs interact. This provides information on the role of hubs in bringing together different secondary structural elements within the protein structure. The secondary structures of the amino acid residues in the protein structures have been obtained using the DSSP program (28). The hubs and the residues with which they interact are classified as belonging to helices (α, 310, π), extended regions, turns (including bends), or unassigned regions (mainly loops). We find that most of the hubs belong to the regular secondary structural regions of helices and sheets though the loops, turns, and the unassigned regions are not excluded at any Imin (data not shown).
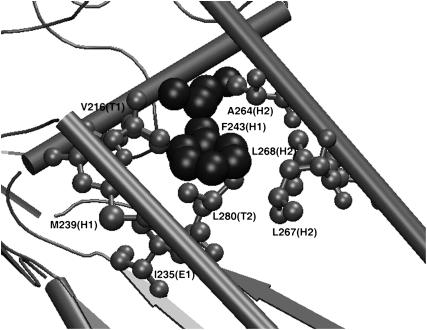
The distribution of the secondary structures of the residues interacting with these hubs at any Imin showed that the hubs interact with residues from both regular and nonregular secondary structural elements. We also find that these structural hub-forming residues form many inter- and intrasecondary structural contacts, thereby integrating different regions of the protein tertiary structure. Fig. 5 shows an example of a hub along with its interacting residues in a protein structure. It can be seen from the figure that the hub-forming phenylalanine residue, which belongs to a helix, interacts with residues belonging to different secondary structures, including a strand, another helix, and some loop regions. Hence, there is a clear indication of the stitching together of different secondary structures through the side-chain interactions of the hubs. Therefore, these hubs play a significant role in intersecondary structural interactions in the folded tertiary structure of the protein.
FIGURE 5.
Example of a hub along with the residues interacting with it in a protein structure. A fragment of phosphoglycerate kinase (16pk) is shown here with the hub-forming residue phenylalanine (Phe-243) and the residues with which it interacts at Imin = 4%. The secondary structure adopted by the protein backbone is shown in gray cartoon representation. The phenylalanine residue (Phe-243), which forms the hub, belongs to a helix and is shown in black van der Waals representation. The other residues with which this hub interacts are shown in gray ball-and-stick representation. The residue names and numbers along with the secondary structural elements to which they belong are indicated within parentheses in the figure.
Correlation of protein stability with network parameters
Proteins in thermophilic organisms are found to be stable at higher temperatures compared to their mesophilic counterparts. Various theoretical and experimental studies carried out earlier by different groups have implicated different factors like hydrogen bonds, salt bridges, aromatic interactions, hydrophobic interactions, etc. for the additional stability of thermophilic proteins (1–7). In this study, we have considered a set of 10 protein structures with counterparts in both a mesophilic organism (stable at moderate temperatures) and a thermophilic organism (stable at higher temperatures) so as to understand whether the concepts of the protein structure networks discussed above provide insights into protein stability. We had earlier carried out a similar analysis on a set of thermophilic and mesophilic proteins using a similar graph representation. However, that analysis was restricted to identifying aromatic residue clusters in these proteins, and we found that the numbers of aromatic clusters are higher in the thermophilic protein than the mesophilic protein (7). The 10 proteins chosen for this analysis are a subset of the proteins studied earlier (7) and have been chosen so as to include the ones that gave varied results in that investigation. The aim of this study is to verify whether the network concepts discussed above are able to distinguish the thermophiles and mesophiles and thus elucidate the factors responsible for the additional stability of the thermophilic proteins. In this study, we have obtained the number of hubs, total number of edges or links (ktotal), the edge/node ratio (ktotal/N, where N is the number of residues in the protein structure), and the size of the largest cluster for the 10 pairs of thermophilic and mesophilic proteins. The results of this analysis are summarized in Table 1, which gives all four parameters for the 10 pairs of proteins considered in this study at three different Imins, 0%, 2%, and 4%. The values of the parameters in all the protein sets are very similar since they all have sizes in the range of 200–450 amino acid residues. However, it is relevant to compare the values between the thermophilic and the corresponding mesophilic protein. In general, we find that all four parameters are significantly higher for the thermophilic protein than the corresponding mesophilic one. However, the values are less discriminatory at Imin = 4%, probably because of the drastic reduction of these parameters at higher Imins.
There are a few exceptions where the mesophilic protein performs better than the thermophilic one as indicated in Table 1. For example, in neutral protease, the number of hubs at 4% and the size of the largest cluster at 2% show a discrepancy, with the mesophilic protein having a higher value than the thermophilic protein. However, in this case, the total number of edges and the edge/node ratio show a better profile for the thermophilic protein than the mesophilic protein at all Imins. Further, the size of the largest cluster at 4% is significantly higher in the thermophilic protein than the mesophilic protein, thus compensating for the other losses by making many stronger interactions. Similarly, in the case of phopshoglycerate kinase, the number of hubs and the total number of edges in mesophilic protein are higher than that in the thermophilic one at Imin = 4%, though the edge/node ratio and the size of the largest cluster are not. However, in this case, all four parameters at 0% and 2% show a much higher percentage in the thermophilic protein than the mesophilic one. This may indicate that the lack of strong interactions at a higher Imin in the thermophile is made up significantly of a very large number of weak interactions at lower Imin. Phosphofructo kinase and TATA box-binding protein also exhibit some deviations from the trend in some parameters; however, the thermophilic counterparts of these proteins score better with some other parameters. In all the other proteins shown in Table 1, the trend observed in the number of hubs, total number of edges, the edge/node ratio, and the size of the largest cluster are quite straightforward, with the numbers being higher for the thermophilic counterpart than the mesophilic protein at all Imins. Thus, in general, there is very good correlation between the network parameters evaluated here and the additional stability of thermophilic proteins, with reasonably valid explanation for the few cases of exception. This analysis clearly shows that the network representation of protein structures presented in this work and the hubs identified are extremely useful in understanding protein stability.
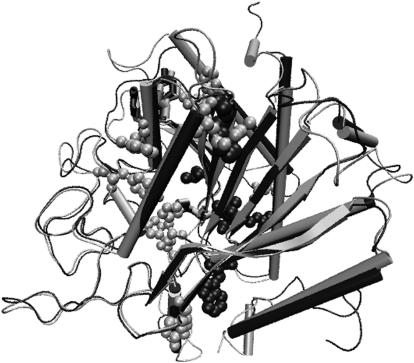
A cartoon representation of the differences in the hubs (at Imin = 4%) of the thermophilic and the mesophilic carboxy peptidase is depicted in Fig. 6, which clearly shows more hubs in the thermophile than the mesophile. It should be noted that the common hubs in the thermophilic and mesophilic proteins are limited and the additional ones in the two proteins are not present in structurally identical positions. Further, the figure also shows that the backbone topologies of both the thermophilic and mesophilic proteins are very similar and hence it is the interactions involving the side chains that impart additional stability to the thermophilic proteins, which is what has been considered in the PSG representation presented in this work. Hubs, which are conserved in sequence, are likely to be more important from the biological perspective, and hence, this aspect is analyzed in the following subsection.
FIGURE 6.
Hubs in carboxy peptidase from Thermoactinomyces vulgaris (1OBR, thermophile) and Bos taurus (2CTC, mesophile). The superposed backbone structures (using ALIGN (32)) for the thermophilic (gray) and the mesophilic (black) proteins are shown in cartoon representation. The hubs at Imin = 4% are shown in the figure. The hubs common to both the proteins (H196me–H204th, Y206me–Y214th, and Y259me–Y266th) are shown in gray/black bond representation. The hubs seen only in the thermophilic protein (R42, H68, H69, Y149, R189, F233, W264, F272, N301) are shown in gray ball-and-stick representation, and those seen only in the mesophilic protein (W73, F189, L201, L271, R272) are shown in black van der Waals representation. Some of the hubs are not seen due to the orientation.
Hub conservation in thermophiles and mesophiles
Multiple sequence alignments of each of the 10 families of thermophilic-mesophilic proteins mentioned above have been obtained from HOMSTRAD ((29), proteins with both known and unknown structures are considered), and the sequence conservation of the hubs identified at Imin = 4% within the thermophilic and mesophilic proteins in these families have been examined. It is important to mention that the numbers of mesophilic sequences are much higher than the numbers of thermophilic sequences in each family, and in some cases there is only one thermophilic protein sequence in the alignment. The average sequence identities in these alignments vary from 30% to 60% in all the families, as given by HOMSTRAD.
On mapping the strong hubs obtained at Imin = 4% (82 in total) onto the multiple sequence alignments of the thermophiles and mesophiles in each of the 10 families, we find that these hubs fall into four distinct categories according to their conservation. These include the common hubs, the exclusive hubs, the nonexclusive hubs, and the nonconserved hubs. The definitions and features of these four types of hubs are described below, and the relevant results are summarized in Table 2.
The common hubs are those residues which are hubs in both the thermophile and mesophile and are also conserved in both. These are significant for the tertiary structure of protein, irrespective of whether it is a thermophilic or a mesophilic one. We find eight such common hubs in the whole data set, distributed among 4 of the 10 families (Table 2).
The exclusive hubs are those residues which form hubs exclusively in the thermophiles or mesophiles and are conserved only within the thermophiles or mesophiles. Hence, these are specific for the thermophiles or mesophiles in the family. Further, the exclusive hubs in the thermophiles are likely to play a very significant role imparting additional stability to the thermophilic proteins since they form hubs and are conserved within the thermophiles only. There are 16 exclusive hubs in the thermophilic and 10 in the mesophilic proteins (Table 2), which is ∼30% of the total hubs obtained at Imin = 4%. The only family without any exclusive hub is the neutral protease, whereas all others have at least one exclusive hub, which is specific to the thermophile or the mesophile. The common and exclusive hubs together are referred to as conserved hubs. We find that the aromatic and charged residues are preferred in these conserved hubs in both thermophilic and mesophilic proteins (Table 2).
The nonexclusive hubs are those residues which form hubs either in the thermophile or mesophile but are conserved in both the thermophiles and mesophile. There are 24 nonexclusive hubs in the thermophilic proteins and 13 in the mesophilic proteins.
The nonconserved hubs are those residues which are not conserved even within the thermophiles or mesophiles, though they form hubs in either of them. This category is insignificant with only one example in thermophiles and two in mesophiles. The multiple sequence alignment of the carboxypeptidases marked with the different types of hubs is shown as an example in the Supplementary Material (Fig. S2).
TABLE 2.
| Protein (No. of hubs in thermophile, No. of hubs in mesophile at Imin = 4%)
|
Common hubs‡ in thermophile and mesophile
|
Exclusive hubs‡
|
|
|---|---|---|---|
| Thermophile | Mesophile | ||
| TATA box bindingprotein (1,8) | – | F94, F107,L124, E177 | F185 |
| Adenylate kinase (4,5) | – | F81, Y191 | H146, D166 |
| Subtilisin (1,2) | – | I78 | D121 |
| Carboxy peptidase (8,12) | H204(H196), Y214(Y206), Y266(Y259)§ | H68, Y149, R189, F233, N301 | W73, L271, R272 |
| Neutral protease (4,7) | Y83(Y84) | –¶ | –¶ |
| Phosphofructo kinase (0,0)‖ | – | – | – |
| Lactate dehydrogenase (2,3) | – | Y145 | N164 |
| Glyceraldehyde-3-phosphate dehydrogenase (3,9) | F16(F16), H108(H108), N236(N236) | R195 | – |
| Phosphoglycerate kinase (3,5) | – | H153 | F194 |
| Reductase (2,3) | K399(390R) | F364 | L214 |
Note that the numbers of mesophilic sequences are much higher than the thermophilic sequences in each family.
All hubs are strong hubs identified at Imin = 4%.
The definition of common hubs and exclusive hubs are given in the text.
Residue name and number of the common hubs in the mesophile is given within parentheses.
No exclusive hubs in neutral protease at Imin = 4%.
No hubs are found in phosphofructo kinase at Imin = 4%.
The small number of common hubs and the large number of nonexclusive hubs found in the 10 sets of thermophilic and mesophilic proteins considered in this analysis indicate that although the overall sequence identities are high and the tertiary structures at the backbone level are almost identical (Fig. 6) among the thermophiles and mesophiles of a particular family, the specific orientations and the mutual packing of side chains within the thermophilic and mesophilic protein structures are different. This leads to the differences in the hubs identified in the thermophiles and the mesophiles. The nonconserved hubs in both thermophilic and mesophilic proteins are very small in number (three in total), indicating that the hubs identified using this method (with Imin = 4%) in general are biologically significant and may be important for the formation and stabilization of the protein tertiary structure. Finally, the exclusive hubs are the most significant ones, which impart the specific characteristics to the thermophilic and the mesophilic proteins, and those present in the thermophiles are bound to be important for the additional thermal stability of thermophilic proteins. Although, the nature of the residues forming the exclusive hubs is similar between the thermophiles and the mesophiles, their positions in the sequence and structures make them important for the protein. Hence, such exclusive hubs (Table 2) can be valuable mutation targets for altering the thermal stability of the protein, which can be tested experimentally.
DISCUSSIONS
Properties of PSGs and comparison with other real-world networks
The PSGs show a complex network topology as mentioned earlier. Recently, the nature and properties of many different kinds of real networks including social, economic, computer, and biological networks as well as the world wide web have been analyzed in detail (21,30). It has been observed that many of the real-world networks fall into one of the three classes (30), namely, a), scale-free, b), broad-scale, and c), single-scale. We find that the PSGs constructed using our definition exhibit a complex behavior with combinations of Gaussian-like, sigmoidal, and exponential/power-law decay for different interaction cutoffs. One of the differences between the PSGs and the other networks lies in the covalent connectivity between the adjacent amino acids in the protein structure, which already restricts the nature of the network in the PSGs. The global tertiary fold adopted by the protein chain is, therefore, constrained by the primary covalent linkages between the adjacent amino acid residues. They are further restricted due to the inherent property of polymer chains to adopt secondary structures such as helices and sheets (31). The constraints imposed by the primary and secondary structures lead to a limited number of folded topologies in the case of tertiary protein structures. Within this restricted framework, the side-chain interactions give rise to more specificity, resulting in a unique three-dimensional structure for the protein sequences selected by nature. Furthermore, there is an inherent steric constraint in biomolecules, which restricts the number of atoms within a given interaction distance. Such a constraint does not seem to exist in other real-world networks. Due to this constraint, the maximum number of links found in an amino acid node in the residue-based PSGs is ∼12, which is very low when compared to the other real-world networks, where there are no restraints with respect to the number of connections acquired by a single node.
The PSGs also differ from many other complex networks in regard to the network growth. Most of the real-world networks are known to grow with time, i.e., the number of nodes in the network generally increases with time (21). In case of the PSGs, the sizes of the proteins selected by nature range from ∼50 to 1500 amino acids. This range is fairly constant and has been stabilized during the course of evolution. Though the size of proteins range from ∼50 to 1500 amino acid residues, the bigger proteins form multiple structural modules (called domains) of similar size of ∼150–200 amino acids. As a result, the larger proteins are made up of modules of individual domains. Thus, the protein domain networks have attained their size limits, and therefore the network growth aspect in the PSG is no longer a relevant factor. Apart from the analysis of the network topology of PSGs, this study has also provided insights into the role of amino acid hubs as sources of robustness and stability in protein structure as discussed in the following section.
PSGs and stability of thermophilic proteins
Various theoretical (from analysis of protein sequences and structures) and experimental (using protein engineering methods) studies have attributed the thermal stability of thermophilic proteins to different factors like higher salt bridges, hydrogen bonds, hydrophobic interactions, aromatic interactions, and better internal packing (1–7). One of the conclusions from all these studies has been that the additional stability of different thermophilic proteins is not a consequence of a single factor. Instead it is a combined effect of various subtle interactions characteristic of each protein. Hence, we thought it appropriate to combine all these factors under a single umbrella and then study the thermophilic proteins from a broader perspective. This we achieve using a network representation of protein structures presented in this work, which considers all kinds of interactions in the protein structure without any discrimination and also takes into account the global topology of the protein structure. Although the strengths of individual interactions are not considered, a crude estimate of the interaction strength is incorporated on the basis of the number of atom-atom contacts between the interacting side chains. We then evaluate different well-known network parameters like the size of the largest cluster, total number of hubs, edge/node ratio, and the total number of edges in a set of 10 thermophilic proteins and their mesophilic counterparts. The analysis of these network parameters showed that in general, the thermophilic proteins have a higher magnitude of these network parameters than the mesophilic proteins. Even in cases where the mesophilic proteins performed better than the thermophilic proteins, we find that the losses in the thermophilic proteins are compensated in various ways, as discussed in the Results section. Though the analysis of the thermophilic proteins from an overall network perspective has given a better picture of the factors involved in their stability and though we find that the network parameters correlate well with the stability of these proteins, we also find that there is no single parameter that can be used as a measure to predict their stability. Some thermophilic proteins make more weak interactions, whereas some make more numbers of stronger interactions. Some of these proteins spread these interactions across the protein structure, giving rise to large interconnected clusters with many weak hubs, whereas some others concentrate their interactions in a particular location of the structure, thereby giving rise to smaller and stronger clusters with more numbers of stronger hubs. It only seems to emphasize the fact that each protein has its own way of achieving the additional stability, and hence a combination of all the network parameters presented here gives a better knowledge of the factors responsible for the stability of these proteins. Hence, the network representation of protein structures and the analysis of the network parameters have significantly improved the understanding of the principles involved in stabilizing the folded three-dimensional structure of proteins.
Hubs in protein structures
From the network perspective, it is known that the role of hubs in a network is to provide robustness to the network against random attacks (21). Moreover, protein structures are made up of a significant number of strongly and weakly interacting amino acid hubs, which integrate different regions of the polypeptide chain, thereby stabilizing the tertiary structure of the protein. These hubs possibly provide robustness to the protein structures against random mutations. Hence, in protein structures, mutation of a single residue chosen randomly may not affect the protein structure or stability unless it is a very crucial hub. Therefore, it is important to carry out mutations of multiple residues (specifically the hub-forming amino acids) simultaneously to significantly destabilize the amino acid networks involved in stabilizing the protein structures. Our study offers a rational method for choosing these important residues in the protein structure by identifying the hubs. Further, this study also shows how the hubs aid in stabilizing the thermophilic proteins in comparison to their mesophilic counterparts.
CONCLUSIONS
The protein structure graphs (PSGs) are constructed as a function of cutoff of noncovalent interaction strength (Imin) between the amino acid nodes in the protein structure. Analyses of such graphs show a complex network topology dependent on the Imin used. A remarkable similarity is seen in proteins of various folds and sizes, where a transition is observed in the size of the largest cluster versus Imin plot. This transition occurs within a very narrow range of Imin for all the proteins and is mediated by the loss of a large number of weak interactions contributed by hydrophobic residues. Further, the identification and characterization of the highly connected nodes (called hubs) as a function of Imin show that charge-delocalized planar residues like phenylalanine, tyrosine, tryptophan, histidine, and arginine along with methionine are preferred as strong hubs, whereas the hydrophobic residues like leucine, isoleucine, and valine are preferred as weak hubs in the PSGs. The study also highlights the role of amino acid hubs in integrating different secondary structural elements in the tertiary structure of the protein, thus stabilizing the protein structure. Hence, the identification of structural hubs provides a rationale for designing mutants so as to understand the factors influencing the formation and stabilizing the protein structures. Further, the network properties analyzed in this study account for the additional thermal stability of the thermophilic proteins compared to their mesophilic counterparts. Moreover, the hub analysis in the thermophilic and mesophilic proteins predicts a set of residues in these proteins that can be mutated to alter their thermal stability and awaits experimental verification. Hence, this study, which involves viewing protein structures as a network of noncovalent connections between amino acid side chains, has provided a new direction in understanding protein structure, stability, and folding.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Supplementary Material
Acknowledgments
We thank Rakesh K. Pandey for the DFS program and Smitha Vishveshwara for useful discussions.
We acknowledge the Computational Genomics Initiative at the Indian Institute of Science, funded by the Department of Biotechnology, India, for support. K.V.B. thanks the Council of Scientific and Industrial Research, India, for the award of a fellowship.
References
- 1.Jaenicke, R., and G. Bohm. 1998. The stability of proteins in extreme environments. Curr. Opin. Struct. Biol. 8:738–748. [DOI] [PubMed] [Google Scholar]
- 2.Ladenstein, R., and G. Antranikian. 1998. Proteins from hyperthermophiles: stability and enzymatic catalysis close to the boiling point of water. Adv. Biochem. Eng. Biotechnol. 61:37–82. [DOI] [PubMed] [Google Scholar]
- 3.Nicholson, H., W. J. Becktel, and B. J. Matthews. 1988. Enhanced protein thermostability from designed mutations that interact with α-helix dipoles. Nature. 336:651–656. [DOI] [PubMed] [Google Scholar]
- 4.Serrano, L., A. G. Day, and A. R. Fersht. 1993. Step-wise mutation of barnase to binase. A procedure for engineering increased stability of proteins and an experimental analysis of the evolution of protein stability. J. Mol. Biol. 233:305–312. [DOI] [PubMed] [Google Scholar]
- 5.Querol, E., J. A. Perez-Pons, and A. Mozo-Villarias. 1996. Analysis of protein conformational characteristics related to thermostability. Protein Eng. 9:265–271. [DOI] [PubMed] [Google Scholar]
- 6.Szilagyi, A., and P. Zavodszky. 2000. Structural differences between mesophilic, moderately thermophilic and extremely thermophilic protein sub-units: results of a comprehensive survey. Structure. 8:493–504. [DOI] [PubMed] [Google Scholar]
- 7.Kannan, N., and S. Vishveshwara. 2000. Aromatic clusters: a determinant of thermal stability of thermophilic proteins. Protein Eng. 13:753–761. [DOI] [PubMed] [Google Scholar]
- 8.Onuchic, J. N., and P. G. Wolynes. 2004. Theory of protein folding. Curr. Opin. Struct. Biol. 14:70–75. [DOI] [PubMed] [Google Scholar]
- 9.Fersht, A. R., and V. Daggett. 2002. Protein folding and unfolding at atomic resolution. Cell. 108:1–20. [DOI] [PubMed] [Google Scholar]
- 10.Vendruscolo, M., N. V. Dokholyan, E. Paci, and M. Karplus. 2002. Small-world view of the amino acids that play a key role in protein folding. Phys. Rev. E. 65:061910. [DOI] [PubMed] [Google Scholar]
- 11.Vendruscolo, M., E. Paci, C. M. Dobson, and M. Karplus. 2001. Three key residues form a critical contact network in a protein folding transition state. Nature. 409:641–645. [DOI] [PubMed] [Google Scholar]
- 12.Dokholyan, N. V., L. Li, F. Ding, and E. I. Shakhnovich. 2002. Topological determinants of protein folding. Proc. Natl. Acad. Sci. USA. 99:8637–8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amitai, G., A. Shemesh, E. Sitbon, M. Shklar, D. Netanely, I. Venger, and S. Pietrokovski. 2004. Network analysis of protein structures identifies functional residues. J. Mol. Biol. 344:1135–1146. [DOI] [PubMed] [Google Scholar]
- 14.Atilgan, A. R., P. Akan, and C. Baysal. 2004. Small-world communication of residues and significance for protein dynamics. Biophys. J. 86:85–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greene, L. H., and V. A. Higman. 2003. Uncovering network systems within protein structures. J. Mol. Biol. 334:781–791. [DOI] [PubMed] [Google Scholar]
- 16.Bagler, G., and S. Sinha. 2005. Network properties of protein structures. Physica A. 346:27–33. [Google Scholar]
- 17.Kannan, N., and S. Vishveshwara. 1999. Identification of side-chain clusters in protein structures by a graph spectral method. J. Mol. Biol. 292:441–464. [DOI] [PubMed] [Google Scholar]
- 18.Kannan, N., P. Chander, P. Ghosh, S. Vishveshwara, and D. Chatterji. 2001. Stabilizing interactions in the dimer interface of alpha-subunit in Escherichia coli RNA polymerase: a graph spectral and point mutation study. Protein Sci. 10:46–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brinda, K. V., N. Kannan, and S. Vishveshwara. 2002. Analysis of homodimeric protein interfaces by graph-spectral methods. Protein Eng. 4:265–277. [DOI] [PubMed] [Google Scholar]
- 20.Vishveshwara, S., Brinda K. V., and N. Kannan. 2002. Protein structure: insights from graph theory. J. Theor. Comput. Chem. 1:187–211. [Google Scholar]
- 21.Barabasi, A. L. 2002. Linked: The New Science of Networks. Persues Publishing, Cambridge, MA.
- 22.Berman, H. M., J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, and P. E. Bourne. 2000. The protein data bank. Nucleic Acids Res. 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henringa, J., and P. Argos. 1991. Side-chain clusters in protein structures and their role in protein folding. J. Mol. Biol. 220:151–171. [DOI] [PubMed] [Google Scholar]
- 24.West, D. B. 2000. Introduction to Graph Theory. Prentice-Hall of India Private Limited, New Delhi, India.
- 25.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:27–28, 33–38. [DOI] [PubMed] [Google Scholar]
- 26.Stauffer, D. 1985. Introduction to Percolation Theory. Taylor and Francis, London.
- 27.Granovetter, M. S. 1973. The strength of weak ties. AJS. 78:1360–1380. [Google Scholar]
- 28.Kabsch, W., and C. Sander. 1983. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 22:2577–2637. [DOI] [PubMed] [Google Scholar]
- 29.Mizuguchi, K., C. M. Deane, T. L. Blundell, and J. P. Overington. 1998. HOMSTRAD: a database of protein structure alignments for homologous families. Protein Sci. 7:2469–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amaral, L. A. N., A. Scala, M. Barthélémy, and H. E. Stanley. 2000. Classes of small-world networks. Proc. Natl. Acad. Sci. USA. 97:11149–11152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoang, T. X., A. Trovato, S. Flavio, J. R. Banavar, and A. Maritan. 2004. Geometry and symmetry presculpt the free-energy landscape of proteins. Proc. Natl. Acad. Sci. USA. 101:7960–7964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen, G. H. 1997. ALIGN: a program to superimpose protein coordinates, accounting for insertions and deletions. J. Appl. Crystallogr. 30:1160–1161. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.