Abstract
Incorporation of 3-fluorotyrosine and site-specific mutagenesis has been utilized with Fourier transform infrared (FTIR) spectroscopy and x-ray crystallography to elucidate active-site structure and the role of an active-site residue Tyr34 in human manganese superoxide dismutase (MnSOD). Calculated harmonic frequencies at the B3LYP/6-31G** level of theory for L-tyrosine and its 3-fluorine substituted analog are compared to experimental frequencies for vibrational mode assignments. Each of the nine tyrosine residues in each of the four subunits of the homotetramer of human MnSOD was replaced with 3-fluorotyrosine. The crystal structures of the unfluorinated and fluorinated wild-type MnSOD are nearly superimposable with the root mean-square deviation for 198 α-carbon atoms at 0.3 Å. The FTIR data show distinct vibrational modes arising from 3-fluorotyrosine in MnSOD. Comparison of spectra for wild-type and Y34F MnSOD showed that the phenolic hydroxyl of Tyr34 is hydrogen bonded, acting as a proton donor in the active site. Comparison with crystal structures demonstrates that the hydroxyl of Tyr34 is a hydrogen bond donor to an adjacent water molecule; this confirms the participation of Tyr34 in a network of residues and water molecules that extends from the active site to the adjacent subunit.
INTRODUCTION
Superoxide dismutases catalyze the disproportionation of superoxide anion radicals to molecular oxygen and hydrogen peroxide and are classified according to the metal cofactor found in the active site (1,2). Human manganese superoxide dismutase (MnSOD) is a homotetramer of 22 kDa subunits (3) located in mitochondria, where it provides protection against oxidative damage. The crystal structure of human MnSOD shows an apparently hydrogen-bonded network of side chains and water that extends from the manganese-bound solvent molecule at the active site to solvent-exposed residues and to the interface between subunits (4). This network has been suggested to support proton transfer in catalysis (5). In the network, the side chain of Gln143 appears to form hydrogen bonds with the manganese-bound solvent and with the side-chain hydroxyl of Tyr34. The crystal structure shows an ordered water molecule that also appears to form a hydrogen bond with the hydroxyl of Tyr34 (4). The replacement of Tyr34 with Phe (Y34F) has only minor effects on catalysis at low substrate concentrations (6,7); however, near maximal velocity the value of kcat for Y34F is 10-fold lower than for wild-type (7). The ionization of the phenolic side chain of Tyr34 has been identified as the origin of the pH dependence of several spectroscopic and kinetic properties of Mn(III)SOD (7,8).
In this report, Fourier transform infrared (FTIR) spectroscopy and x-ray crystallography have been applied to human MnSOD to better understand the role of Tyr34. Each of the nine tyrosine residues in each subunit of wild-type MnSOD was replaced with 3-fluorotyrosine to help identify vibrational modes associated with tyrosine side chains. To identify vibrational modes that are specific to Tyr34, the FTIR spectrum of wild-type is compared with that of Y34F MnSOD. Our FTIR data show distinct vibrational modes arising from fluorinated tyrosine; based on the frequency of the C-O stretching vibrations in wild-type and Y34F MnSOD, the C-O stretching vibration of the phenolic group of Tyr34 has been identified. The frequency of this absorption is consistent with a phenolic hydroxyl that is hydrogen bonded acting as a proton donor. These data complement the crystal structure and confirm the participation of Tyr34 in the hydrogen-bonded network of residues and water molecules that extends from the active site to the adjacent subunit.
MATERIALS AND METHODS
Reagents
L-tyrosine and 3-fluorotyrosine were purchased from Sigma (St. Louis, MO) and Acros Organics (a subdivision of Fisher Scientific, Pittsburgh, PA).
MnSOD expression, purification, and fluorotyrosine labeling
Escherichia coli cells that express wild-type and Y34F human MnSOD were grown in enriched and minimal media. For minimal media growth, the overnight culture was grown for 17 h at 37°C in 50 mL of minimal media until an OD595 = 1 was reached. The minimal medium (M9) consisted of 0.04 M Na2HPO4, 0.02 M KH2PO4, 8.6 mM NaCl, and 0.02 M NH4Cl. The overnight culture was supplemented with 0.1 mM CaCl2, 1 mM MgSO4, 0.2% glucose, 1 μg/mL of thiamine, 0.2 mg/mL of amino acids (except the aromatic amino acids), 1 mM tryptophan, 1 mM phenylalanine, and ampicillin. The glucose, thiamine, amino acids, tryptophan, and phenylalanine solutions were filter sterilized through a 0.2 μ Corning filter system unit. The CaCl2 and MgSO4 were autoclaved. The overnight growth was then transferred to 11 L of minimal media and supplemented in the same manner as the overnight culture plus the addition of sterile 18 μM MnSO4. The cells were allowed to grow for ∼5 h until an OD595 of 0.3–0.4 was reached. At this point, the cells were induced with 0.3 mM isopropyl-beta-D-thiogalactopyranoside supplemented with 1 mM 3-fluorotyrosine (or unlabeled L-tyrosine as a control) and allowed to grow for an additional 4 h. Due to the low solubility of L-tyrosine and its fluorinated analog in water, these compounds were added as solids to the growing media. The cells were placed at 4°C overnight and harvested the next day by centrifugation. The resulting pellet was frozen at −70°C until lysis was performed. The cells were lysed as previously described (9). The resulting protein solution was filtered, and fast protein liquid chromatography was performed to further separate MnSOD from some cellular proteins still present after the heating and dialysis steps. MnSOD was eluted at ∼40 mM NaCl in the presence of 10 mM Tris-HCl pH 8.2. The purified protein was concentrated and dialysed extensively against the final buffer 20 mM potassium phosphate at pH 7.8. The content of Mn in MnSOD was determined using flame atomic absorption spectroscopy, and the concentration of MnSOD for kinetic study was taken as the manganese concentration.
Fluorotyrosine was incorporated into 67% of all tyrosine sites in human MnSOD, determined by amino acid analysis composition (Protein Chemistry Laboratory, Texas A&M University, College Station, TX), and corroborated by MALDI-TOF mass spectrometry.
FTIR spectroscopy
A Midac (Costa Mesa, CA) FTIR spectrometer equipped with a deuterated triglycine sulfate detector was utilized to obtain infrared spectra of fluorinated and unlabeled wild-type and Y34F human MnSOD. FTIR data were acquired on samples prepared in a uniform manner; each contained 30 μL of protein at 0.5 mM in 20 mM potassium phosphate at pH 7.8 that were dehydrated on a ZnSe window by applying a stream of dry nitrogen gas at room temperature. FTIR data were also collected on 1 mM L-tyrosine and 1 mM 3-fluorotyrosine in 20 mM potassium phosphate at pH 7.8. The samples were placed in the sample compartment, which was evacuated through a rotary pump to eliminate water vapor. The spectral parameters were as follows: apodization, Happ-Genzel; resolution, 4 cm−1; number of scans, 215; levels of zero filling, one; and temperature, room temperature. The spectra acquired from protein samples were corrected for an amide I vibration of 0.35 to allow direct comparison. The data shown here represent an average of 860–1075 scans and 4–5 different samples.
Crystallography
Orthorhombic crystals of fluorinated wild-type and Y34F human MnSOD were grown by vapor diffusion from protein solutions at 26 mg/mL, buffered in 20 mM potassium phosphate buffer at pH 7.8 and 22% poly(ethylene glycol) (PEG) 2000 monomethyl ether. X-ray diffraction data collection was conducted at 100 K on flash-frozen single crystals, supplemented with 20% ethylene glycol cryoprotectant in their mother liquor. Data sets were collected at beamline 8.3.1 of the Advanced Light Source, Lawrence Berkeley National Laboratory (Berkeley, CA) on a Q315 ADSC CCD detector. Data sets for the fluorinated wild-type and Y34F MnSOD were collected to 1.85 Å and 1.53 Å, respectively. Data sets were indexed and merged with Denzo and scaled with Scalepack (10). The fluorinated wild-type crystal belongs to space group P21212 with unit cell dimensions of a) 74.0 Å, b) 75.4 Å, and c) 67.8 Å. The data set was 97.8% complete (94.7% for the highest-resolution shell (1.85–1.97 Å); 32,265 unique reflections measured) with an Rsym of 6.7% (30.8% for the highest-resolution shell). The data have an overall I/ÛI of 37.1 with an I/σI in the highest resolution shell of 6.2. The fluorinated mutant Y34F crystal also belongs to space group P21212, and has unit cell dimensions of a) 73.7 Å, b) 75.3 Å, and c) 68.0 Å. The data set was 99.9% complete (94.7% for the highest-resolution shell (1.58–1.53 Å); 57,452 unique reflections measured) with an Rsym of 5% (28.4% for the highest-resolution shell). The data have an overall I/ÛI of 57.9 with an I/σI in the highest resolution shell of 5.3. Phases were obtained for both structures by molecular replacement against the wild-type human MnSOD structure using AMoRe (11). The two structures were refined with several cycles of rigid body and restrained refinement, using Crystallography and NMR Systems, version 1.1 (CNS) (12). Final refinement steps for the higher resolution Y34F-fluorinated MnSOD were completed using SHELX (13). Side-chain orientations were corrected where required, due to potential van der Waals clashes of their hydrogen atoms, using Reduce and Probe (14,15). The structures were fit against calculated 2Fo − Fc, Fo − Fc and composite omit electron density maps in the Xfit module of XtalView (16). The fluorinated wild-type MnSOD structure was refined to an Rwork of 21.2% and an Rfree (17) of 25.2% (with 5% of the reflections flagged for the test set) and the Y34F to a Rwork of 15.3% and an Rfree of 20.3%. Coordinates for the structures are deposited in the Protein Data Bank with accession numbers 1XDC for wild-type fluorinated MnSOD and 1XIL for the fluorinated Y34F mutant MnSOD.
Density functional theory calculations
Geometry optimization and harmonic frequencies were calculated for L-tyrosine and 3-fluorotyrosine and their modified structures with density functional theory (DFT) applying the hybrid Becke3-Lee-Yang-Parr (B3LYP) exchange-correlation function and 6-31G** basis set under the Gaussian 98 suite of programs (18). This level of theory has been demonstrated to be reliable in predicting vibrational frequencies and relative intensities of medium-sized polycyclic aromatic hydrocarbons (19). Zero point-corrected total energies were used to search for the global energy minima of the potential surfaces. A factor of 0.978 was used to scale the harmonic frequencies and zero point energies to account for anharmonicity effects and basis set deficiency. Such a factor was adopted earlier for neutral and cationic fluorene (a medium-sized polycyclic aromatic hydrocarbon) and found to give good agreement between calculated and observed mode frequencies (19).
Modeling of the atomic displacements in the various vibrational modes of the optimized molecular structures was viewed using AniMol software (Innovative Software, Gainesville, FL).
RESULTS
FTIR spectra of tyrosine and 3-fluorotyrosine
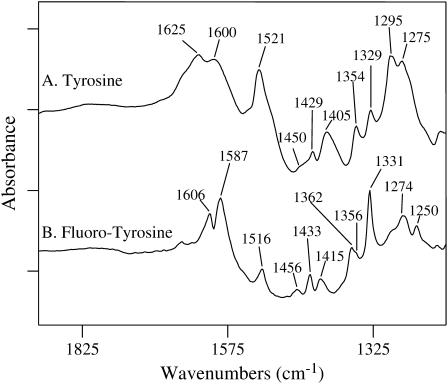
The FTIR absorption spectra of uniformly hydrated tyrosine and 3-fluorotyrosine solutions in phosphate buffer at pH 7.8 were measured at room temperature (Figs. 1 and 2). Only the 1200–1900 cm−1 region is shown due to the broad OH band at 3000 cm−1 and large phosphate contribution below 1200 cm−1. No clear band for the bending vibration of liquid water (at 1650 cm−1) is observed in our experimental spectrum (Fig. 1 A). This indicates that there is no significant nonbonded (i.e., free) water left in our sample. The excellent agreement of the observed IR spectrum with that calculated for hydrogenated tyrosine, as displayed in Fig. 1 B, indicates our samples are nearly fully hydrated. (In our calculations we hydrogenated a carboxyl oxygen to simulate the presence of a water molecule hydrogen bonded to that group.) In addition, the 3300 cm−1 peak, which contains contributions from the OH stretch of water (as well as the NH stretch from the protein) is 0.3 absorbance units in all the spectra. This suggests uniformity of hydration.
FIGURE 1.
(A) Experimental room temperature infrared absorption spectrum of a partially hydrated sample of tyrosine in phosphate buffer (at pH 7.8) and room temperature, and (B) calculated (B3LYP/6-31G**) infrared absorption spectrum of hydrogenated tyrosine at optimized (displayed) geometry. In the calculated spectrum, the harmonic band frequencies were scaled uniformly by a factor of 0.978. Bands of 3 cm−1 bandwidth were constructed using 50% Gaussian and 50% Lorentzian functions. The vertical lines mark the proposed band assignments collected in Table 1.
FIGURE 2.
Infrared absorption spectra of uniformly hydrated samples of (A) tyrosine and (B) 3-fluorotyrosine. FTIR data were collected on 1 mM L-tyrosine and 1 mM 3-fluorotyrosine in 20 mM phosphate buffer at pH 7.8 at 20°C. The tick marks on the y axis represent 0.3 absorbance units.
The lack of any strong bands in the 1700–1800 cm−1 region indicates the absence of significant C = O stretching vibration from the carboxylic group of the tyrosines, consistent with ionization to carboxylate at the pH of 7.8 of the original solution. To mimic this in our calculations, hydrogen atoms were placed in proximity to the carboxyl groups in the tyrosines, as displayed in Fig. 1. The figure also shows a comparison of the calculated and observed FTIR spectra for hydrogenated tyrosine. All bands in the experimental spectrum are now readily assignable, based on agreement of band frequencies and relative intensities. Table 1 gives a similar comparison for hydrogenated 3-fluorotyrosine.
TABLE 1.
Infrared absorption band frequencies (in cm−1) observed and calculated (B3LYP/6-31G**) for tyrosine (Figs. 1 A and 2 A) and 3-fluorotyrosine (Fig. 2 B)
| Tyrosine
|
3-Fluorotyrosine
|
|||
|---|---|---|---|---|
| Mode description* | Observed | Calculated† | Observed | Calculated† |
| HNH bend + CC symmetric ring stretch | 1625 | 1640 + 1637 (0.71) | 1606 | 1643 + 1638 (0.20) |
| CC asymmetric ring stretch | 1600 | 1607 (0.18) | 1587 | 1615 (0.12) |
| CC/CH symmetric ring stretch | 1521 | 1526 (0.98) | 1516 | 1526 (1.00) |
| HCαH bend | 1450sh | 1471 (0.05) | 1456 | 1467 (0.03) |
| CC/CH asymmetric ring stretch | 1429 | 1447 (0.20) | 1433 | 1450 (0.14) |
| CβCγ stretch | 1405 | 1394 (0.60) | 1415 | 1393 (0.29) |
| HCβCγ bend | ∼1390sh | 1381 (0.19) | 1362 | 1378 (0.09) |
| CC asymmetric ring stretch | 1354 | 1345 (0.38) | 1356sh | 1375 (0.12) |
| HCαCβ bend | 1329 | 1340 (0.39) | 1331 | 1338 (0.19) |
| C1CαHα bend | − | 1329 (0.02) | − | 1320 (0.06) |
| C1CαHα bend | − | 1305 (0.05) | − | 1304 (0.24) |
| FC stretch | − | − | 1293sh | 1282 (0.20) |
| CO phenolic stretch | 1275 | 1277 (0.99) | 1250 | 1246 (0.65) |
| HCαH/HCβ Cγ /HOCγ bend | 1295 | 1260 (1.00) | 1274 | 1256 (0.52) |
Major vibrations are noted only.
Frequencies are scaled uniformly by a factor of 0.978. Relative integral intensities are given in parentheses.
A symmetric C-C ring-stretching vibration is assigned to the 1625 cm−1 tyrosine band (and the 1606 cm−1 band in 3-fluorotyrosine) (Figs. 1 and 2). DFT computations place this mode at 1637 cm−1 in tyrosine (and 1638 cm−1 in 3-fluorotyrosine). The asymmetric C-C ring-stretching mode is calculated at 1607 cm−1 for tyrosine (and 1615 cm−1 for fluorotyrosine) and observed at 1600 cm−1 (tyrosine) and 1587 cm−1 (3-fluorotyrosine; Table 1). Note that this region also has a contribution from the carboxylate anion near 1600 cm−1. In the case of 3-fluorotyrosine, a strong coupling of the C-F stretch oscillator to the C-C stretch ring oscillators was identified from viewing the atom motion during vibrations. Also, the electronegative F-atom causes a change in the atomic charge distribution in phenol, which likely affects the interaction of the phenol ring with the surrounding molecular bath (vide infra). These effects account for the change in the observed C-C stretch frequencies in 3-fluorotyrosine. However, in the B3LYP harmonic frequency prediction, the modes coupling and interaction with the surrounding bath are not incorporated into the calculation. This omission certainly affects the calculated band frequencies for the C-C stretch ring modes, causing higher predicted frequencies than observed (Table 1).
Both tyrosines are predicted to have a symmetric ring CC/CH stretching mode at 1526 cm−1 (Table 1). A prominent band is observed at 1521 cm−1 for tyrosine (1516 cm−1 for fluorotyrosine) and is assigned to the symmetric ring CC/CH mode. Although higher in frequency than previously observed (1500 cm−1) (20), this discrepancy could be due to different experimental conditions (see below). A low intensity HCαH bending mode is expected at ∼1471 cm−1 for tyrosine (and ∼1467 cm−1 for fluorotyrosine). A weak shoulder at 1450 cm−1 in tyrosine and more pronounced, though still weak, band in fluorotyrosine at 1456 cm−1 is assigned to this bending mode. These results are in agreement with the literature values for ring modes of the phenolic side chain of tyrosine (21) but are significantly different than ring modes in tyrosinate (20) and phenolate anionic species (22).
Several bands in the 1430–1330 cm−1 region arise from additional ring and carboxyl group vibrations as well as from modes containing the Cα, Cβ, and Cγ atoms. They are observed at 1429, 1405, 1390 (shoulder), 1354 and 1329 cm−1 for tyrosine (Figs. 1 A and 2 A), and 1433, 1415, 1362, 1356 (shoulder), and 1331 cm−1 for fluorotyrosine (Fig. 2 B).
The C-O stretching vibration of the phenolic group of tyrosine appears at 1275 cm−1 (Figs. 1 A and 2 A); this is characteristic of protonated phenols with the O-H group involved in hydrogen-bonding interactions and acting as a proton donor (21). In fluorotyrosine, this band is shifted significantly to lower frequencies (to 1250 cm−1). Substitution of an o-hydrogen atom with the more electronegative fluorine atom is expected to redshift the C-O stretch of the phenolic group, an expectation fully consistent with our DFT calculations, which predict a downshift of 31 cm−1 (observed shift: 25 cm−1). Modeling of the atomic displacements for this mode shows that the phenol C-O stretch requires a substantial ring deformation involving F atom movement as well. The large mass difference between fluorine and hydrogen is probably partially responsible for the lowering of the phenol C-O stretching vibrational frequency. The spectrum of fluorotyrosine (Fig. 2 B) shows an additional band at ∼1293 cm−1 (shoulder), which is assigned to a ring mode that includes a large C-F displacement. As expected, this band is not observed in L-tyrosine.
FTIR spectra of unlabeled and fluorinated wild-type human MnSOD
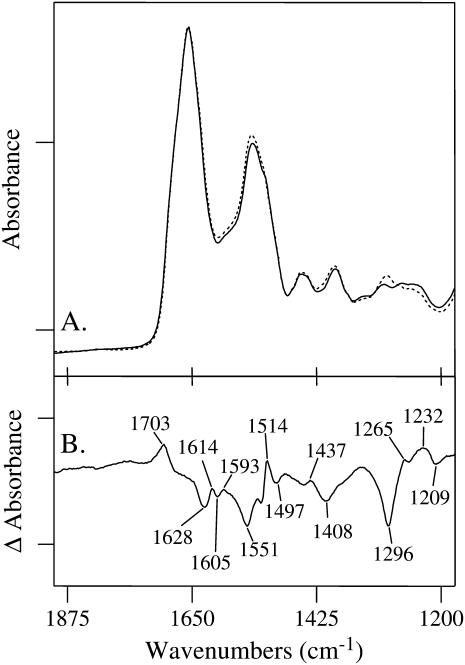
To identify the vibrational modes that arise from the tyrosine moieties, human MnSOD was prepared with each of the nine tyrosine residues in each subunit replaced with 3-fluorotyrosine. Fig. 3 A (solid line) shows the FTIR spectrum of unlabeled wild-type MnSOD in phosphate buffer at pH 7.8; the fluorinated version is shown superimposed as a dotted line. A difference spectrum, given in Fig. 3 B, shows only those vibrational modes that are sensitive to fluorination. Vibrations not altered in frequency or intensity upon fluorination will cancel and not be observed. Any differences in environment of the 3-fluorotyrosine residues will influence their normal modes, and thus affect the IR spectrum.
FIGURE 3.
Infrared absorption spectra of (A, solid line) unlabeled human wild-type MnSOD, and (A, dotted line) wild-type MnSOD in which each tyrosine residue is labeled with fluorine at the 3-position of the phenol ring. The tick marks on the y axis represent 0.2 absorbance units. The difference spectrum (unlabeled-minus-fluorinated) is shown in B. The tick marks on the y axis represent 0.03 absorbance units. FTIR data were acquired at 20°C on 30 μL of protein sample at 0.5 mM in 20 mM phosphate buffer at pH 7.8 and partially dehydrated onto a ZnSe window.
The difference spectrum shows the vibrational modes of unlabeled tyrosine as positive bands and those from fluorinated tyrosine as negative bands. Significant changes are observed in the aromatic ring-stretching regions around 1600 cm−1. The amide I (C = O stretching of the backbone) absorbs at ∼1650 cm−1 and is expected to contribute absorption in this region if there are changes in the secondary structure of the protein upon fluorination. A positive line at 1614 cm−1 can be assigned to the symmetric CC stretching vibration of unlabeled tyrosine. This mode was observed at 1625 cm−1 for the model compound (Table 1). Upon fluorination of the tyrosines in MnSOD, this mode shifts to 1605 cm−1 and is shown as a negative band in Fig. 3 B. The asymmetric CC ring-stretching vibration for unlabeled tyrosine occurs most probably at 1593 cm−1. The band resulting from fluorination is not evident. The negative 1628 and 1551 cm−1 bands are assigned to the amide I and amide II groups, respectively. These modes may indicate small changes in the secondary structure of MnSOD upon fluorination. The exact attribution of the 1703 cm−1 band is uncertain, but it is characteristic of the carbonyl group of the carboxylic acid.
The positive 1514 cm−1 band is assigned to the symmetric ring CC/CH stretching vibration of unlabeled tyrosine (Table 2). The red-shifted band is observed at 1497 cm−1. The HCαH bending and asymmetric counterparts of the ring CC/CH stretching mode are not evident but are predicted to be low intensity modes. Vibrational bands observed at 1437 and 1408 cm−1 could represent vibrational changes involving the Cβ and Cγ atoms. A negative band at 1296 cm−1 is assigned to the C-F stretching modes of fluorotyrosine in wild-type MnSOD. It is observed at ∼1293 cm−1 in the model compound (Fig. 2 B). Two distinct bands are observed for unlabeled MnSOD in this region: at 1265 and 1232 cm−1. The mode at 1265 cm−1 is consistent with the phenol group of tyrosine residues involved as proton donors in hydrogen-bonding interactions (21). Furthermore, the mode at 1232 cm−1 can be assigned to tyrosine residues in which the phenol group is a proton acceptor in hydrogen-bonding interactions (21). These differences in environment give rise to different components of the C-O stretching vibration.
TABLE 2.
Proposed assignment of infrared absorption band frequencies (in cm−1) observed for wild-type and Y34F human manganese superoxide dismutase
| Wild-type
|
Y34F
|
|||
|---|---|---|---|---|
| Mode description | Unlabeled | Fluorinated | Unlabeled | Fluorinated |
| CC symmetric ring stretch | 1614 | 1605 | — | — |
| CC/CH asymmetric ring stretch | 1593 | — | — | — |
| CC/CH symmetric ring stretch | 1514 | 1497 | 1514 | 1498 |
| HCβCγ bend | 1437 | 1408 | — | — |
| CβCγ stretch | — | — | 1406 | — |
| HCαCβ bend | — | — | 1348 | — |
| FC ring stretch | — | 1296 | — | 1296 |
| C-O phenolic stretch | 1265 | — | 1257 | — |
| C-O phenolic stretch | 1232 | 1209 | 1228 | 1211 |
FTIR spectra of unlabeled and fluorinated Y34F Human MnSOD
FTIR spectroscopy has been applied to the site-specific mutant of human MnSOD in which Tyr34 was replaced with Phe (Y34F). Fig. 4 A shows the FTIR absorption spectra of unlabeled Y34F (solid line) and Y34F in which each of the eight remaining tyrosine residues is labeled with fluorine at the 3-position of the phenol ring (dotted line); the difference spectrum is shown in Fig. 4 B. Again, vibrational modes of unlabeled tyrosine are positive bands, and the modes of fluorinated tyrosine are negative bands. As was observed for wild-type, fluorination of the tyrosine residues in Y34F causes changes in the vibrational modes of tyrosine. The 1600 cm−1 region, which contains contributions from the tyrosine ring-stretching vibrations and from the amide I/II of the protein backbone, shows perturbations upon fluorination. Again, these changes are more likely due to ring vibrations than to changes in the backbone vibrations because of fluorine labeling on the phenol ring. Because of large and overlapping bands in the 1600 cm−1 region, it is difficult to pinpoint particular ring-stretching vibrations that shift upon fluorination.
FIGURE 4.
Infrared absorption spectra of (A, solid line) unlabeled Y34F and (A, dotted line) fluorinated Y34F MnSOD. The difference spectrum (unlabeled-minus-fluorinated) is shown in (B). The tick marks on the y axis represent 0.03 AU. Spectral conditions were as described in Fig. 3.
However, the CC/CH stretching vibrations observed at ∼1500 cm−1 are quite apparent. A positive 1514 cm−1 band is assigned to the symmetric ring CC/CH stretching vibration of unlabeled tyrosine (Table 2). The red-shifted band is observed at 1498 cm−1. The asymmetric counterpart of the ring CC/CH stretching mode is not seen, due to its expected low intensity. A negative mode at 1296 cm−1 is attributed to the C-F stretching modes of 3-fluorotyrosine in Y34F MnSOD. Also, positive bands are observed at 1406 cm−1 and 1348 cm−1 and are tentatively assigned to vibrations involving the Cα, Cβ, and Cγ atoms.
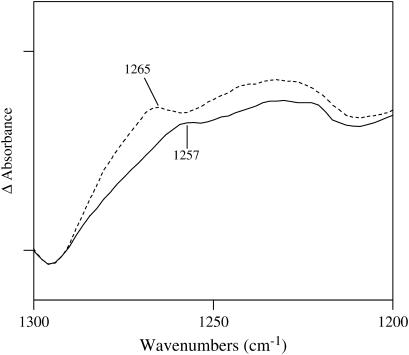
Note that the intensity of the 1265 cm−1 band has decreased significantly and shifted to lower energy by 8 cm−1 in Y34F compared with wild-type (Fig. 4 B). An expanded version of this part of the spectrum is shown in Fig. 5. The difference spectrum shown in Fig. 5 is the average of five spectra obtained on five different samples. Each one of the individual spectra shows the 1265 cm−1 band. The spectra are corrected for protein concentrations by setting the amide I vibration of each absorbance spectrum to 0.35 absorbance units. This is an arbitrary number that was chosen to normalize the spectra that were used to obtain a difference spectrum. This method has been employed extensively for photosystem II (for an example, see Ayala and Barry (23)). By correcting for the amide I vibrational mode, small differences in protein concentration from sample to sample were taken into account. In addition to the correction for protein concentration, baseline correction of the absorbance spectra was performed using the software GRAMS (Galactic Industries, Salem, NH). The absorbance spectra shown in Figs. 3 A and 4 A were corrected using a two-point baseline correction. In this method two baseline points were selected at the ends of the spectral region. A multiple-point baseline correction was not practicable because our absorbance spectra show multiple overlapping bands and it is difficult to select points that belong to the baseline without biasing our data. Difference spectra were constructed using the baseline-corrected absorbance spectra; these difference spectra were indistinguishable from those obtained using no baseline correction.
FIGURE 5.
Comparison of the FTIR difference spectra of wild-type and Y34F MnSOD. The tick marks on the ordinate represent 0.02 absorbance units. Note that the intensity at 1265 cm−1 has decreased significantly in Y34F compared with wild-type. These data show that the C-O stretching vibration of the phenolic group of the tyrosine side chain at residue 34 in wild-type makes an infrared contribution at 1265 cm−1. This frequency is characteristic of phenol groups that act as hydrogen bond donors (21).
One of the components of the C-O vibration at 1228–1232 cm−1 is evident in both Figs. 3 B and 4 B; the other component at 1265 cm−1 is very much smaller in Y34F. These data show that the C-O stretching vibration of the phenolic group of Y34 makes a contribution to the 1265 cm−1 mode. This frequency is characteristic of phenol groups, which hydrogen bond and act as proton donors (21). This band has shifted to 1257 cm−1 and has nearly disappeared in Y34F. A summary of the proposed vibrational mode assignments for wild-type and Y34F are shown in Table 2.
Crystal structures
Crystal structures were determined for human wild-type MnSOD and Y34F MnSOD at 1.85 and 1.53 Å, respectively, in which the tyrosine residues were replaced with 3-fluorotyrosine. The electron density maps show the presence of 3-fluorotyrosine side chains in the two structures, with each of the nine 3-fluorotyrosine side chains in a single orientation. The fluorine groups were mostly refined at 70% occupancy. This is consistent with the percentage incorporation of fluorine tyrosine and resulted in reasonable B values for the fluorine atoms and the surrounding side chain atoms. We had expected to see a distribution of rotamers for 3-fluorotyrosine side chains, which would be clearly evident in these relatively high resolutions structures. However, it was immediately apparent from the initial electron density maps after molecular replacement that this was not the case. In the refined structures, each of the side chains of the nine 3-fluorotyrosine residues was observed clearly to adopt a preferred orientation, most likely reflecting local environments. The B values for these fluorine atoms were refined to reasonable B values, which were very similar to or the same as the surrounding atoms of the side chain.
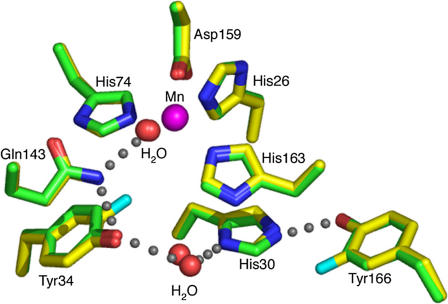
Incorporation of 3-fluorotyrosine does not significantly alter the structures of the fluorinated wild-type and Y34F MnSOD, as they are structurally superimposable with their nonfluorinated counterparts; the average root mean-square deviation for all 198 α-carbon atoms between unfluorinated and fluorinated wild-type MnSOD is 0.3 Å. An apparent hydrogen-bonding scheme extending from the metal-bound solvent is maintained in the structure of the wild-type MnSOD containing 3-fluorotyrosines (Fig. 6). Analysis of potential van der Waals contacts of the hydrogen atoms (14,15) of Gln143, which is part of the hydrogen-bonding scheme, indicates that the amide group is oriented toward the hydroxyl of Tyr34; a 180° flip of the Gln143 side chain (about χ3) would cause steric clashes with neighboring atoms, including Asn73, Trp123, and the protein backbone. This suggests the directionality of the hydrogen bond chain emanating from the metal-bound solvent. The amide of the carboxamide side chain of Gln143 acts as a hydrogen bond donor to the phenolic hydroxyl of fluorinated Tyr34, which in turn appears to be in a hydrogen bond with the water molecule between the side chains of Tyr34 and His30 (Fig. 6; Table 3). This is the same observation as made for the wild-type human MnSOD (4). However, this hydrogen-bonding network is altered in the Y34F mutant structure. The Y34F mutation results in another water molecule being present in the active site, in a position approximately equivalent to the hydroxyl group of the wild-type Tyr34 (Fig. 7; Table 3). Thus, the His30 ND1-water-water-Gln143 NE2 hydrogen bonds may partially restore the hydrogen-bonding relay in Y34F.
FIGURE 6.
Crystal structure of the active site of human MnSOD, in which tyrosine residues have been replaced with 3-fluorotyrosine (yellow), superimposed upon wild-type human MnSOD (green). The apparent hydrogen-bond scheme between the manganese-bound solvent molecule, the side chains of Gln143, Tyr34, a water molecule, and the His30 and Tyr166 side chains is conserved in both structures and is depicted by gray spheres.
TABLE 3.
Bond distances and angles of the active-site hydrogen bond network in fluorinated wild-type and fluorinated Y34F human MnSOD
| Fluorinated wild-type
|
Fluorinated Y34F
|
|||
|---|---|---|---|---|
| Bond distances, Å | Subunit A | Subunit B | Subunit A | Subunit B |
| Mn-bound OH-Gln143 NE2 | 2.8 | 2.8 | 2.7 | 2.7 |
| Gln143 NE2-Tyr34 OH | 2.9 | 3.0 | — | — |
| Tyr34 OH-H2O | 2.9 | 2.8 | — | — |
| Gln143 NE2-H2O | — | — | 3.1 | 3.0 |
| H20-H2O | — | — | 2.7 | 2.7 |
| H20-His30 ND1 | 2.7 | 2.5 | 2.8 | 2.8 |
| His30 NE2-Tyr166 OH | 2.8 | 2.7 | 2.8 | 2.7 |
| Bond angles, degrees | ||||
| Mn-bound OH-Gln143 NE2-Y34OH | 116.0 | 112.2 | — | — |
| Gln143-Y34OH-H2O | 120.7 | 122.1 | — | — |
| Ty34OH-H2O-His30 ND1 | 147.8 | 144.7 | — | — |
| Mn-bound OH-Gln143 NE2-H2O | — | — | 95.9 | 91.1 |
| Gln143 NE2-H2O-H2O | — | — | 140.8 | 137.9 |
| H2O-H2O-His30 ND1 | — | — | 122.0 | 115.3 |
| His30 NE2-Tyr166 OH-Ty166 CZ | 123.8 | 115.8 | 120.5 | 122.7 |
FIGURE 7.
Crystal structure of the active site of Y34F mutant of human MnSOD that has tyrosines replaced by 3-fluorotyrosine (salmon), superimposed upon wild-type human MnSOD (green). The active-site hydrogen-bonding scheme, present in the wild-type enzyme, is altered in the Y34F mutation. Gray spheres depict the Y34F active-site hydrogen bonds, where an extra water molecule is observed in the active site, which is likely to partially restore the hydrogen bond relay.
DISCUSSION
Each of the nine tyrosine residues of wild-type human MnSOD was replaced with 3-fluorotyrosine. Interestingly, this had rather little effect on the structure of the enzyme; the fluorinated and unfluorinated structures are closely superimposable. Significantly, the apparent hydrogen-bonded network, extending from the manganese-bound solvent, appears unchanged in the fluorinated wild-type MnSOD (Fig. 6). The crystal structure of unfluorinated wild-type human Mn(III)SOD with the highest resolution (Protein Data Bank accession code 1LUV (24)) shows that in subunit A of the asymmetric subunit dimer the hydroxyl oxygen of Tyr34 is located 3.0 Å from the side-chain carboxamide (NE2) of Gln143 and 2.8 Å from the oxygen of a water molecule, both suggesting hydrogen bonding. However, the corresponding distances in subunit B are 2.9 Å for the carboxamide (Gln143) to hydroxyl (Tyr34) and 3.3 Å for the hydroxyl to water. The fluorinated wild-type human Mn(III)SOD structure shows that in subunit A the hydrogen bond distancing between hydroxyl oxygen of Tyr34 and the side-chain carboxamide of Gln143 is 2.9 Å and also 2.9 Å to the oxygen of the water molecule. Distances in subunit B are 3.0 Å for the Gln143 carboxamide to Tyr34 hydroxyl and 2.8 Å for Tyr34 hydroxyl to water. Thus, these crystal structures suggest that the hydroxyl of Tyr34 is a hydrogen bond acceptor from NE2 of Gln143; however, the nature of the interaction of the hydroxyl of Tyr34 with the adjacent water molecule is ambiguous.
In this work, FTIR spectroscopy is used to study MnSOD; thorough studies of MnSOD using resonance Raman spectroscopy are reported (8). To the best of our knowledge, this is the first FTIR study of fluorinated tyrosine in a protein with the express goal of assessing the structural characteristics of a particular tyrosine residue. The addition of the electronegative fluorine atom at the 3-position of the phenol ring of tyrosine caused the ring-stretching vibrations to be lower in frequency, indicating weaker bonds. Also, the phenolic C-O stretching vibration of 3-fluorotyrosine was found at lower frequency than in tyrosine, suggesting a weaker C-O bond in 3-fluorotyrosine than in tyrosine. This was confirmed in both tyrosine as a model compound and DFT calculations (Figs. 1 and 2; Table 1) and in the protein data (Figs. 3 and 4; Table 2).
The FTIR spectra revealed additional differences between fluorinated and unfluorinated MnSOD (Figs. 3 and 4). Although the crystal structures of the unfluorinated and fluorinated wild-type MnSOD were closely superimposable (Fig. 6), the FTIR data showed changes in the amide I and II vibrational bands, suggesting some changes in backbone conformation which are apparently not within the detection limits of the crystallography. Among other notable differences was the C-O stretch of the phenol group of tyrosine, the intensity and frequency of which is sensitive to the status of the hydoxyl as a proton donor or acceptor (21). The difference FTIR spectra of Figs. 3 and 4 for fluorinated and unfluorinated MnSOD identify this tyrosine C-O stretch, and the comparison of these spectra for wild-type and Y34F MnSOD identify this vibrational mode in the active-site residue Tyr34.
Although the phenolic side chain of Tyr34 can participate as both a hydrogen bond acceptor and donor, the FTIR analysis of human Mn(III)SOD in this study shows a prominent absorption at 1265 cm−1 (Fig. 5), indicating that the C-O stretch in the side chain of Tyr34 is dominated by the OH group acting as a hydrogen bond donor. This then confirms the protonation state of Tyr34 in Mn(III)SOD and the presence of a hydrogen bond between the hydroxyl of Tyr34 and the adjacent water molecule. Moreover, the data complement the crystal structure, which suggests this hydrogen bond, although somewhat ambiguously. These hydrogen bond interactions of Tyr34 have implications for the spectroscopic and catalytic properties of the enzyme. They affect the visible absorption spectrum of MnSOD at 480 nm, with Tyr34 being the source of the pKa near 9.5 (7,8,25). The ionization of Tyr34 also explains the pH dependence of catalysis observed in wild-type and mutants in which kcat/Km shows a pKa near 9.5 (8). Tyrosine34, most likely through hydrogen-bonded interactions with Gln143 and a water molecule, provides an active-site framework that supports proton transfer in catalysis, although Tyr34 may not be the source of the protons that eventually form the H2O2 product (5).
Acknowledgments
We thank Professor I. Fridovich for helpful suggestions on growing bacteria in minimal media and Max Iurcovich for excellent technical assistance.
This work was supported by grants from the National Institutes of Health: GM54903 to D.N.S. and AI054945 to J.A.T.
References
- 1.Holm, R. H., P. Kennepohl, and E. I. Solomon. 1996. Structural and functional aspects of metal sites in biology. Chem. Rev. 96:2239–2314. [DOI] [PubMed] [Google Scholar]
- 2.Beyer, W., J. Imlay, and I. Fridovich. 1991. Superoxide dismutases. Prog. Nucleic Acid Res. Mol. Biol. 40:221–253. [DOI] [PubMed] [Google Scholar]
- 3.Fridovich, I. 1989. Superoxide dismutases: an adaptation to a paramagnetic gas. J. Biol. Chem. 264:7761–7764. [PubMed] [Google Scholar]
- 4.Borgstahl, G. E. O., H. E. Parge, M. J. Hickey, W. F. Beyer, R. A. Hallewell, and J. A. Tainer. 1992. The structure of human mitochondrial manganes superoxide dismutase reveals a novel tetrameric interface of two 4-helix bundles. Cell. 71:107–118. [DOI] [PubMed] [Google Scholar]
- 5.Silverman, D. N., and H. S. Nick. 2002. The catalytic pathway of manganese superoxide dismutase by direct observation of superoxide. Methods Enzymol. 349:61–74. [DOI] [PubMed] [Google Scholar]
- 6.Whittaker, M. M., and J. W. Whittaker. 1997. Mutagenesis of a proton linkage pathway in Escherichia coli manganese superoxide dismutase. Biochemistry. 36:8923–8931. [DOI] [PubMed] [Google Scholar]
- 7.Guan, Y., M. J. Hickey, G. E. O. Borgstahl, R. A. Hallewell, J. A. Lepock, D. O'Connor, Y. Hsieh, H. S. Nick, D. N. Silverman, and J. A. Tainer. 1998. The crystal structure of Y34F mutant human mitochondrial manganese superoxide dismutase and the functional role of tyrosine 34. Biochemistry. 37:4722–4730. [DOI] [PubMed] [Google Scholar]
- 8.Maliekal, J., A. Karapetian, C. Vance, E. Yikilmaz, Q. Wu, T. Jackson, T. C. Brunold, T. G. Spiro, and A.-F. Miller. 2002. Comparison and contrasts between the active site pKs of MnSOD and those of FeSOD. J. Am. Chem. Soc. 124:15064–15075. [DOI] [PubMed] [Google Scholar]
- 9.Beck, Y., D. Bartfield, Z. Yavin, A. Levanon, M. Gorecki, and J. R. Hartman. 1988. Efficient production of active human manganese superoxide dismutase in Escherichia coli. Biotechnology. 6:930–935. [Google Scholar]
- 10.Otwinowski, Z., and W. Minor. 1997. Processing of x-ray diffraction data collected in oscillation mode. In Methods in Enzymology, Vol. 276. C. W. Carter Jr. and R. M. Sweet, editors. 307–326. Academic Press, New York. [DOI] [PubMed]
- 11.Navaza, J. 2001. Implementation of molecular replacement in AMoRe. Acta Crystallogr. D. 57:1367–1372. [DOI] [PubMed] [Google Scholar]
- 12.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D54:905–921. [DOI] [PubMed] [Google Scholar]
- 13.Sheldrick, G. M., and T. R. Schneider. 1997. SHELXL: high-resolution refinement. Methods Enzymol. 277:319–343. [PubMed] [Google Scholar]
- 14.Word, J. M., S. C. Lovell, J. S. Richardson, and D. C. Richardson. 1999. a. Asparagine and glutamine: using hydrogen atom contacts in the choice of side-chain amide orientation. J. Mol. Biol. 285:1735–1747. [DOI] [PubMed] [Google Scholar]
- 15.Word, J. M., S. C. Lovell, T. H. LaBean, H. C. Taylor, M. E. Zalis, B. K. Presley, J. S. Richardson, and D. C. Richardson. 1999. b. Visualizing and quantifying molecular goodness-of-fit: small-probe contact dots with explicit hydrogen atoms. J. Mol. Biol. 285:1711–1733. [DOI] [PubMed] [Google Scholar]
- 16.McRee, D. E. 1999. XtalView/Xfit: a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125:156–165. [DOI] [PubMed] [Google Scholar]
- 17.Adams, P. D., N. S. Pannu, R. J. Read, and A. T. Brunger. 1997. Cross-validated maximum likelihood enhances crystallographic simulated annealing refinement. Proc. Natl. Acad. Sci. USA. 94:5018–5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frisch, M. J., G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, V. G. Zakrzewski, J. A. Montgomery Jr., R. E. Stratmann, J. C. Burant, S. Dapprich, J. M. Millam, A. D. Daniels, K. N. Kudin, M. C. Strain, O. Farkas, J. Tomasi, V. Barone, M. Cossi, R. Cammi, B. Mennucci, C. Pomelli, C. Adamo, S. Clifford, J. Ochterski, G. A. Petersson, P. Y. Ayala, Q. Cui, K. Morokuma, D. K. Malick, A. D. Rabuck, K. Raghavachari, J. B. Foresman, J. Cioslowski, J. V. Ortiz, B. B. Stefanov, G. Liu, A. Liashenko, P. Piskorz, I. Komaromi, R. Gomperts, R. L. Martin, D. J. Fox, T. Keith, M. A. Al-Laham, C. Y. Peng, A. Nanayakkara, C. Gonzalez, M. Challacombe, P. M. W. Gill, B. Johnson, W. Chen, M. W. Wong, J. L. Andres, C. Gonzalez, M. Head-Gordon, E. S. Replogle, and J. A. Pople. 1998. Gaussian 98, Revision A.3. Gaussian Inc., Pittsburgh, PA.
- 19.Szczepanski, J., J. Banisaukas, M. Vala, S. Hirata, R. J. Bartlett, and M. Head-Gordon. 2002. Vibrational and electronic spectroscopy of the fluorene cation. J. Phys. Chem. A. 106:63–73. [Google Scholar]
- 20.Ayala, I., K. Range, D. York, and B. A. Barry. 2002. Spectroscopic properties of tyrosyl radicals in dipeptides. J. Am. Chem. Soc. 124:5496–5505. [DOI] [PubMed] [Google Scholar]
- 21.Takeuchi, H., N. Watanabe, Y. Satoh, and I. Harada. 1989. Effects of hydrogen-bonding on the tyrosine Raman bands in the 1300–1150 cm−1 region. J.Raman Spectrosc. 20:233–237. [Google Scholar]
- 22.Mukherjee, A., M. L. McGlashen, and T. G. Spiro. 1995. Ultraviolet resonance Raman spectroscopy and general valence force field analysis of phenolate and phenoxyl radical. J. Phys. Chem. 99:4912–4917. [Google Scholar]
- 23.Ayala, I., and B. A. Barry. 2002. Histidine 190–D1 and glutamate 189–D1 provide structural stabilization in photosystem II. Biochemistry. 41:11456–11465. [DOI] [PubMed] [Google Scholar]
- 24.Hearn, A. S., M. E. Stroupe, D. E. Cabelli, C. A. Ramilo, J. P. Luba, J. A. Tainer, H. S. Nick, and D. N. Silverman. 2003. Catalytic and structural effects of amino acid substitution at His 30 in human manganese superoxide dismutase. Biochemistry. 42:2781–2789. [DOI] [PubMed] [Google Scholar]
- 25.Hsu, J.-L., Y. Hsieh, C. Tu, D. O'Connor, H. S. Nick, and D. N. Silverman. 1996. Catalytic properties of human manganese superoxide dismutase. J. Biol. Chem. 271:17687–17691. [DOI] [PubMed] [Google Scholar]