Abstract
Double-stranded RNA (dsRNA) initiates cellular posttranscriptional responses that are collectively called RNA silencing in a number of different organisms, including plants, nematodes, and fruit flies. In plants, RNA silencing has been associated with protection from virus infection. In this study, we demonstrate that dsRNA-mediated interference also can act as a viral defense mechanism in mosquito cells. C6/36 (Aedes albopictus) cells were stably transformed with a plasmid designed to transcribe an inverted-repeat RNA (irRNA) derived from the genome of dengue virus type 2 (DEN-2) capable of forming dsRNA. Clonal cell lines were selected with an antibiotic resistance marker and challenged with DEN-2. The cell lines were classified as either susceptible or resistant to virus replication, based on the percentage of cells expressing DEN-2 envelope (E) antigen 7 days after challenge. Eight out of 18 (44%) cell lines designed to express irRNA were resistant to DEN-2 challenge, with more than 95% of the cells showing no DEN-2 antigen accumulation. One of the DEN-2-resistant cell lines, FB 9.1, was further characterized. DEN-2 genome RNA failed to accumulate in FB 9.1 cells after challenge. Northern blot hybridization detected transcripts containing transgene sequences of both sense and antisense polarity, suggesting that DEN-2-specific dsRNA was present in the cells. In addition, a class of small RNAs 21 to 25 nucleotides in length was detected that specifically hybridized to labeled sense or antisense DEN-2 RNA derived from the target region of the genome. These observations were consistent with RNA silencing as the mechanism of resistance to DEN-2 in transformed mosquito cells.
Dengue (DEN) viruses are mosquito-borne members of the family Flaviviridae, genus Flavivirus, and can cause dengue fever and dengue hemorrhagic fever-dengue shock syndrome. Dengue fever and dengue hemorrhagic fever-dengue shock syndrome represent the most important arthropod-borne viral diseases in the world, affecting 50 to 100 million people every year, with 10 to 20 times as many at risk (13). DEN viruses have a positive-polarity, single-stranded RNA genome approximately 11 kb in length that encodes three structural proteins and seven nonstructural proteins in the order 5′ C-prM-E-NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5 3′. DEN viruses, like other flaviviruses, generate detectable amounts of intracellular double-stranded RNA (dsRNA) as an intermediate of their replication (40).
Aedes aegypti and Aedes albopictus are the principal vectors of DEN virus. DEN virus and other arthropod-borne viruses actively replicate in the mosquito host, but little is known about how these mosquitoes cope with the infection. Like most arthropod-borne viruses, DEN viruses can cause significant damage when they infect vertebrate cells, yet mosquito cells sustain persistent, noncytopathic DEN virus infections; thus, they may have evolved mechanisms to specifically counteract or control RNA virus replication without significantly impairing host function. In infected vertebrate cells, dsRNA triggers a number of antiviral responses by inducing alpha and beta interferons and activating enzymes such as protein kinase R and 2′-5′-oligoadenylate synthetase (11), but these defenses are not known to exist in invertebrates. However, our work has suggested that the dsRNA produced as an intermediate in replication of most arboviruses is a signal that also sets off an invertebrate host defense system (1, 29). Caplen et al. (4) also have recently demonstrated that dsRNA triggers sequence-specific interference with DEN virus type 1 (DEN-1) replication in mosquito cells.
Work with plant viruses has been instrumental in defining the phenomenon of RNA-mediated gene silencing. In the early 1990s, plant virologists demonstrated that a small proportion of transgenic plants engineered to express a part of a plant virus genome were resistant to homologous viruses (23). It was similarly shown elsewhere that expression from a plant virus expression vector of a portion of either a plant endogenous gene or a transgene could very efficiently silence the homologous gene (21). The latter phenomenon was termed virus-induced gene silencing (32). A related phenomenon, in which expression by a virus vector of sequences from a heterologous virus genome induced resistance to the unrelated virus, was termed RNA-mediated cross-protection between viruses (31). Ratcliff et al. (30) provided evidence that viruses were potentially both initiators and targets of gene silencing in plants and concluded that the gene silencing phenomenon likely was a natural defense against viruses. It was subsequently shown that virus-induced gene silencing is one facet of posttranscriptional gene silencing (PTGS), a general phenomenon triggered by dsRNA by which plants detect and defend against infection (14). The dsRNA that triggers gene silencing can be effectively delivered as part of an RNA virus replicative intermediate (32), by transformation with a plasmid that expresses a transcript with an inverted-repeat sequence (39), or after in vitro transcription and annealing (37). Hamilton and Baulcombe (14) showed that 21- to 25-nucleotide (nt) small interfering RNA (siRNA) derived from the dsRNA trigger was a hallmark of PTGS.
At the same time that the trigger of PTGS was being elucidated, it was demonstrated that inoculation of Caenorhabditis elegans or Drosophila melanogaster with dsRNA resulted in degradation of cognate mRNA (8, 19, 27, 41); this phenomenon was called RNA interference (RNAi) in invertebrates (34, 41). RNAi, or RNA silencing, is a powerful cellular mechanism that results in the degradation of the introduced dsRNA, as well as homologous mRNA present in the organism, and has been used as a knockdown mechanism to study gene function during development in a number of organisms (24). Recently, Li et al. (22) demonstrated that RNA silencing was induced in Drosophila cells by infection with an animal virus.
The occurrence of RNA silencing in such a wide-ranging group of organisms makes it seem likely that an RNA silencing mechanism exists in A. aegypti and A. albopictus mosquitoes as well and that it might be a mechanism for regulating arbovirus replication in natural hosts. We previously demonstrated virus-induced RNA-mediated resistance to DEN-2 in mosquito cells and mosquitoes by expression of the DEN-2 prM gene in either antisense or sense polarity from a double-subgenomic Sindbis (SIN) virus expression vector (10, 28). We have shown elsewhere that double-subgenomic SIN virus-induced DEN virus interference in mosquitoes has many of the characteristics of PTGS-RNAi (1, 29).
In this study, virus resistance was generated in cultured mosquito cells by transcription of DEN virus-specific RNA from plasmid DNA that was designed to form a hairpin structure in the cells. Cultured mosquito (C6/36) cells were transformed with DNA plasmids designed to express dsRNA derived from the genome of DEN-2. A cell line designated FB 9.1 that was highly resistant to DEN-2 generated siRNA 21 to 25 nt in length, consistent with RNA silencing as the mechanism of resistance. This study demonstrates that dsRNA-mediated interference can serve as a mechanism of virus resistance in mosquito cells. Development of a stably transformed mosquito cell line that exhibits RNA silencing represents a powerful tool for characterizing this endogenous antiviral mechanism and will provide a model system for identifying viral genes that potentially counteract RNA silencing.
MATERIALS AND METHODS
Cells and virus.
C6/36 cells (ATCC no. CRL-1660) were grown in Leibowitz-15 (L-15) medium containing 10% fetal bovine serum (FBS), 100 U of penicillin/ml, and 100 μg of streptomycin/ml at 28°C. Conditioned medium used during the cell selection process consisted of 50% fresh medium and 50% C6/36 cell conditioned medium (medium removed from confluent C6/36 cells and filtered through a 0.2-μm-pore-size filter) containing 10% FBS, 100 U of penicillin/ml, 100 μg of streptomycin/ml, 2 mM l-glutamine, and 300 U of hygromycin B/ml. Hygromycin B-resistant cell lines were grown on L-15 with 10% FBS, 100 U of penicillin/ml, 100 μg of streptomycin/ml, 2 mM l-glutamine, and 300 U of hygromycin B/ml. DEN-2 (Jamaica 1409) was used interchangeably with DEN-2 (New Guinea C) for challenges. After DEN-2 challenge, transformed cells were maintained with L-15 supplemented with 2% FBS, 100 U of penicillin/ml, and 100 μg of streptomycin/ml containing 300 U of hygromycin B/ml.
Plasmid construction.
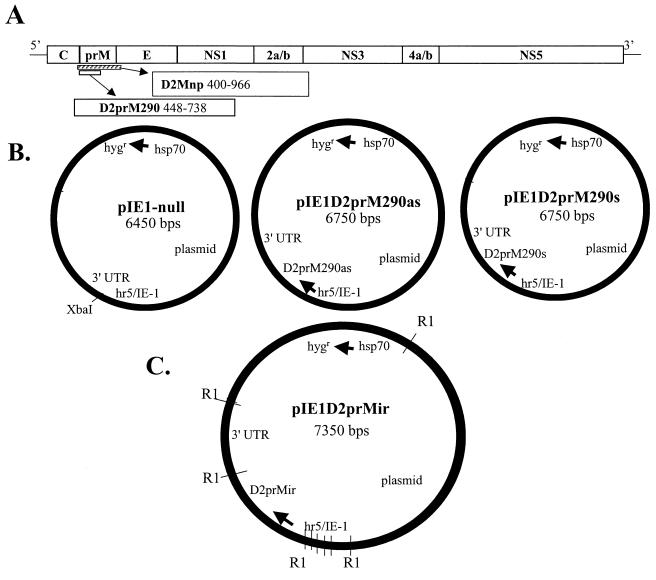
pIE1-null was generated by modifying the pIE1-3 plasmid (Novagen, Madison, Wis.) (17) to contain the hygromycin resistance gene under the control of the Drosophila heat shock protein 70 (hsp70) promoter before the insertion of the inverted-repeat DEN virus sequences. The hsp70 promoter constitutively expresses hygromycin in C6/36 cells (5). DEN-2 sequences (Fig. 1A) were amplified by reverse transcription-PCR and subcloned into modified pBluescript (Stratagene, La Jolla, Calif.) before insertion into the XbaI site of pIE1-null (1), which yielded plasmids pIE1D2prM290as and pIE1D2prM290s (Fig. 1B). pIE1D2prMir was constructed by inserting the Mnp sequence (np, no protein) in sense orientation into the XbaI site of pIE1D2prM290as (Fig. 1B). Mnp was isolated from pBMnp as an NheI/XbaI fragment, as described by Gaines et al. (10). The orientation of all inserted DEN-2 sequences was confirmed by sequencing. The nucleotide sequence of the D2prMir insert and the structure of the expected mRNA are shown in Fig. 2.
FIG. 1.
Maps of pIE1-null plasmid and derivatives used to confer DEN-2 resistance in transformed C6/36 mosquito cells. (A) Map of the DEN-2 genome showing the locations of the prM sequences used in this study. Nucleotide position numbers for the Mnp and prM290 sequences are given. (B and C) The pIE1-null plasmid was used as a backbone for insertion of DEN-2 prM sequences, and the three D2prM-containing constructs are shown. DEN-2 prM sequences were cloned into the XbaI site in sense, antisense, or inverted-repeat orientations. Arrows indicate the hsp70 promoter of the hygromycin resistance gene and the IE-1 promoter driving the prM inserts. The XbaI site downstream from the baculovirus hr5/IE-1 enhancer-promoter, which was used for inserts, and the EcoRI (R1) restriction sites are shown. UTR, untranslated region.
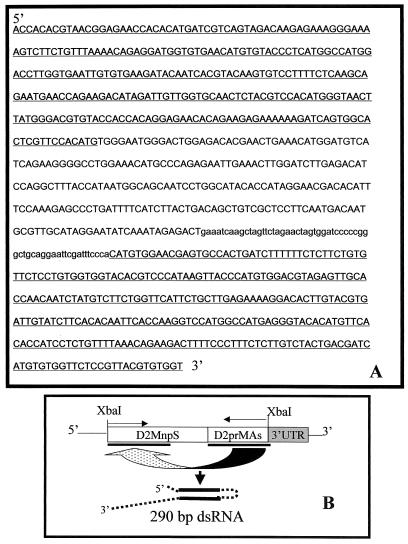
FIG. 2.
Nucleotide sequence of the D2Mnp-D2prM290as insert in plasmid pIE1D2prMir. (A) Sequence of nt 400 to 966 of DEN-2 (New Guinea C) genome RNA (uppercase), followed by a linker region (lowercase), followed by the complementary sequence to DEN-2 RNA nt 448 to 738. Self-complementary regions are underlined. (B) Diagram of the insert transcript, showing formation of a 290-bp double-stranded region. Complementary regions are underlined, and arrows indicate sequence polarity. The 3′ untranslated region (UTR) is a baculovirus genome sequence from the pIE1-3 cloning vector.
Establishment of clonal cell lines.
Lipofectin (Gibco BRL, Rockville, Md.) was used to transfect plasmids pIE1-null, pIE1D2prM290s, pIE1D2prM290as, and pIE1D2prMir into C6/36 mosquito cells. Following selection for transformants by growth in hygromycin-containing medium, colonies were picked by using extra-long gel-loading micropipette tips and inoculated into 25-cm2 flasks containing 5 ml of conditioned medium. Upon colony formation, this process was repeated, with all clonal cell lines established after at least two rounds of purification.
DEN-2 challenge of clonal cell lines.
Cell lines were challenged with DEN-2 between 4 and 10 passages after establishment. Cells were challenged with DEN-2 (Jamaica 1409) at a multiplicity of infection (MOI) of 0.01. At 7 days postchallenge cells were fixed in 3:1 acetone-phosphate-buffered saline for 5 min at −20°C and stored in phosphate-buffered saline. Immunofluorescence assays (IFA) were performed with anti-DEN virus envelope monoclonal antibody 4G2 (16), a biotinylated sheep anti-mouse antibody, and a streptavidin-fluorescein conjugate as previously described (12). Three microscopic fields of view from each coverslip were photographed with an Olympus DP10 digital camera. All digital files were modified in Adobe Photoshop 5.0 by adjusting the brightness by +20 and the contrast by +75. Cells containing DEN-2 antigen were counted and expressed as a ratio to the total number of cells counted. All experiments were performed with duplicate coverslips, and the percent DEN-2-infected cells was expressed as the average of all trials. Cell lines were considered stably resistant to DEN-2 if several independent challenge experiments showed that less than 5% of cells contained DEN-2 antigen at 7 days postchallenge. Challenges with the heterologous flavivirus West Nile virus were performed in the same manner.
Southern blot hybridization analysis.
High-molecular-weight DNA was isolated from clonal cell lines with a DNeasy minikit (Qiagen Inc., Chatsworth, Calif.). Genomic DNA was digested with the appropriate restriction enzyme, loaded onto an 0.7% agarose-1× Tris-acetate EDTA gel, and electrophoresed at 100 V for 2 to 3 h. DNA was transferred to a positively charged nylon membrane (Brightstar; Ambion, Austin, Tex.) by standard Southern blotting protocols (33). Probes were labeled by randomly primed incorporation of [α-32P]dCTP (3,000 Ci/mmol) with the Megaprime DNA labeling system (Amersham-Pharmacia Biotech, Piscataway, N.J.) and with the use of linearized pIE1D2prMir as template. The specific activity of probes was 4.0 × 108 dpm/μg. Probes were heat denatured and added to hybridization buffer (7.0% sodium dodecyl sulfate [SDS], 0.47 M Na2HPO4, 1.7 mM H3PO4). Hybridization proceeded overnight at 65°C, followed by two washes in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS at 20 min each and two washes in 0.2× SSC-0.1% SDS for 30 min each. Blots were exposed to Kodak Biomax MR film at −70°C with an intensifying screen for 1 to 7 days.
RNA analysis.
For standard Northern blot analysis, total cellular RNA was extracted from clonal cell lines with the Qiashredder and RNeasy minikits (Qiagen Inc.). Fifty micrograms of total RNA was electrophoresed in a 1.25% agarose-1× MOPS (morpholinepropanesulfonic acid)-0.66 M formaldehyde gel and blotted to a positively charged nylon membrane. RNA probes corresponding to the D2prM290 sense and antisense strands were transcribed in vitro by using T3 or T7 RNA polymerase with the MAXIscript kit (Ambion) including 20 mCi of [α-32P]UTP/ml with an activity of 800 Ci/mmol. The specific activity of RNA probes was determined to be 1.4 × 109 cpm/μg. Blots were hybridized with 2.0 × 108 cpm of RNA probe/ml in UltraHyb (Ambion) hybridization buffer at 68°C for 12 to 16 h and washed twice with 2× SSC-0.1% SDS for 20 min each, followed by two washes in 0.2× SSC-0.1% SDS for 30 min each. Blots were exposed to Kodak Biomax MR film at −70°C with an intensifying screen for 1 to 7 days.
For slot blot analysis, total cellular RNA was extracted from cells at each time point postinfection by the acid guanidinium thiocyanate-phenol-chloroform method described by Chomczynski and Sacchi (6). Five micrograms of total RNA was applied to each well of a slot blot apparatus (Schleicher & Schuell, Keene, N.H.) containing a nylon membrane as described previously (1). Labeled antisense RNA probe corresponding to the NS2B-NS3 region of the DEN-2 genome was used as described above. Blots were exposed to phosphor storage screens for 1.5 h, and the storage screens were scanned on a STORM phosphorimager (Molecular Dynamics, Inc.; Amersham Pharmacia Biotech Ltd.). Quantitation and analysis of phosphorimager data were performed with ImageQuant software (Molecular Dynamics, Inc.) and Microsoft Excel (Redmond, Wash.).
To detect siRNAs, total RNA was extracted with acid guanidinium thiocyanate-phenol-chloroform and ethanol precipitated. The redissolved precipitate was heated to 65°C and then placed on ice, and high-molecular-weight RNA and DNA were precipitated by addition of polyethylene glycol (molecular weight, 8,000) to a final concentration of 5% and NaCl to a final concentration of 0.5 M (14). Low-molecular-weight RNA was precipitated from the supernatant by addition of 3 volumes of ethanol. The low-molecular-weight RNA was further purified by binding to a DEAE-Sepharose CL-6b column or used directly after formamide denaturation for electrophoresis in a 16% polyacrylamide-7 M urea gel. Separated RNA was blotted and cross-linked to a positively charged nylon membrane. Hybridization was performed in Ultra-Hyb hybridization buffer with labeled strand-specific DEN-2 prM RNA probes (15). Hybridization occurred at 40°C overnight followed by washes at 50°C in 2× SSC-0.2% SDS. Hybridization signal was detected after 5 days of exposure to a phosphor storage screen with a STORM phosphorimager.
RESULTS
Identification of DEN-2-resistant cell lines.
After selection for hygromycin resistance and cell colony purification, four types of transformed cell lines were obtained: 10 IE1-null (null [H]) cell lines containing no DEN-2 sequence, 10 IE1D2prM290S (sense [S]) cell lines designed to transcribe a 290-nt segment of the DEN-2 prM gene in sense orientation, 18 IE1D2prM290As (antisense [As]) cell lines designed to transcribe a 290-nt segment of the DEN-2 prM gene in antisense orientation, and 18 IE1D2prMir (fold-back [FB]) cell lines designed to express dsRNA derived from the DEN-2 prM gene (Fig. 1 and 2) (Materials and Methods).
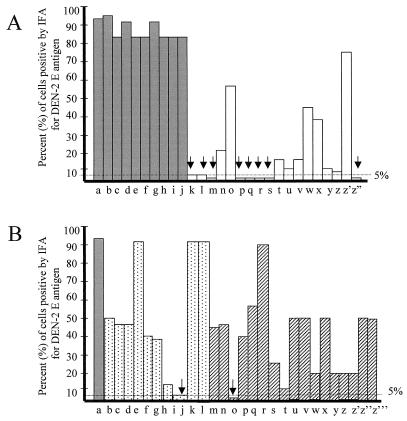
Resistance to DEN-2, defined as 5% or fewer DEN-2 E-positive cells by IFA per trial, was observed in one sense line (S 9.3), one antisense line (As 4.2), and eight inverted-repeat RNA (irRNA) lines (Fig. 3). No resistance to DEN-2 was observed for any of the null cell lines (Fig. 3A). Only 14 and 7% of cell lines transformed with a plasmid designed to express sense or antisense RNA, respectively, derived from DEN-2 were found to be resistant to DEN-2. In contrast, 44% of cell lines transformed with a plasmid designed to express dsRNA derived from DEN-2 were resistant to DEN-2 challenge.
FIG. 3.
Resistance of clonal transformed C6/36 cell lines to DEN-2 challenge. Clonal cell lines were selected by resistance to hygromycin and challenged with DEN-2 infection at an MOI of 0.01. The height of bars shows the percentage of cells containing DEN-2 E antigen by IFA 7 days postchallenge. (A) Bars a to j (shaded), cell lines transformed with pIE1-null (H cell lines); bars k to z" (open), cells transformed with pIE1D2prMir (FB cell lines). (B) Bar a, H cell line; bars b to l (stippled), cell lines transformed with pIE1D2prM290s (S cell lines); bars m to z‴ (striped), cells transformed with pIE1D2prMas (As cell lines). Cell lines with <5% E-antigen-containing cells (arrows) were considered DEN-2 resistant.
Assays for DEN-2 resistance.
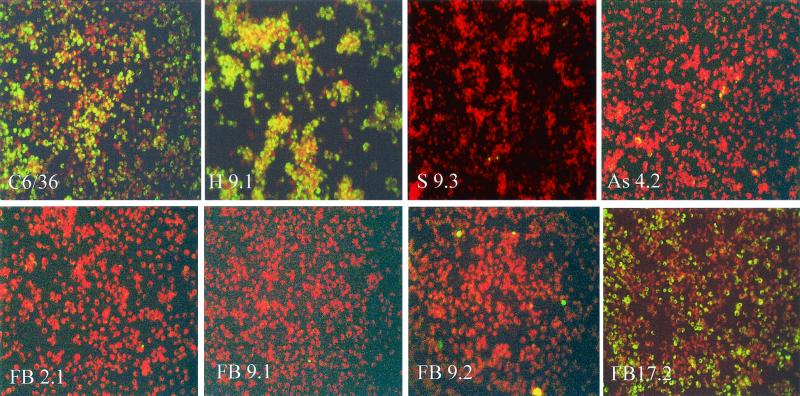
Several of the resistant cell lines had 1% or fewer cells containing DEN-2 E antigen by IFA. Results of IFA performed 7 days after DEN-2 challenge on untransformed C6/36 cells; null cell line H 9.1; sense cell line S 9.3; antisense cell line As 4.2; and irRNA cell lines FB 2.1, FB 9.1, FB 9.2, and FB 17.2 are shown in Fig. 4. Although untransformed C6/36, H 9.1, and FB 17.2 cells all contained a high proportion of DEN-2-positive cells, cell lines S 9.3, As 4.2, FB 2.1, FB 9.1, and FB 9.2 were ≥99% resistant.
FIG. 4.
IFA. Untransformed C6/36 cells; null cell line H 9.1; sense cell line S 9.3; antisense cell line As 4.2; and fold-back cell lines FB 2.1, FB 9.1, FB 9.2, and FB 17.2 were stained for DEN-2 E antigen 7 days after challenge with DEN-2. Cells were counterstained with Evans blue so that uninfected cells appear red, while infected cells appear yellow-green.
In addition to IFA, resistance to DEN-2 was measured by comparing the amounts of infectious virus in the cell culture medium of untransformed and transformed cells after DEN-2 challenge. Cell culture medium was collected from C6/36, H 9.1, S 9.3, As 4.2, FB 2.1, and FB 9.1 cells at 7 days post-DEN-2 (Jamaica 1409) challenge and titrated in Vero cells by plaque assay (25). C6/36 cells and cell line H 9.1 had DEN-2 titers of 1.7 × 105 and 4.3 × 105 PFU/ml, respectively. Cell lines S 9.3 (5.1 × 103 PFU/ml), As 4.2 (2.3 × 102 PFU/ml), FB 2.1 (2.1 × 103 PFU/ml), and FB 9.1 (1.2 × 103 PFU/ml) all produced 100- to 1,000-fold-less infectious DEN-2 than did the H 9.1 line. These titration data corresponded well with the sparse number of cells fluorescing with E antigen in S 9.3, As 4.2, FB 2.1, and FB 9.1 cell lines 7 days after DEN-2 challenge (Fig. 4). Resistance continued to be observed in these cell lines after 40 to 50 passages, indicating stability of the resistant phenotype (data not shown). We tested the specificity of resistance by infecting FB 9.1 cells with the NY99 strain of West Nile virus (Flaviviridae), and more than 60% of the cells displayed West Nile virus E antigen by IFA at 7 days postinfection, suggesting that the silencing response was sequence specific (data not shown).
Physical state of pIE1D2prMir in cell line FB 9.1.
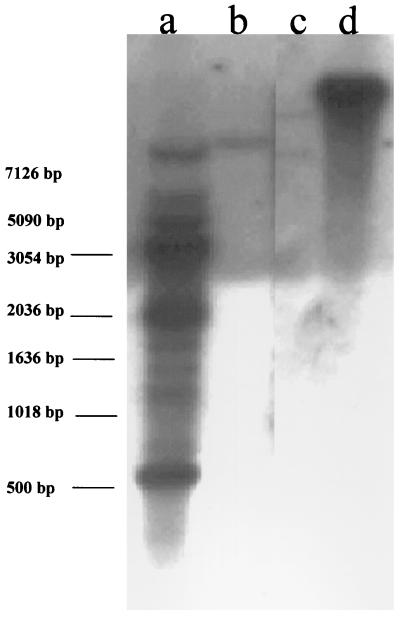
Cell line FB 9.1 was characterized further by Southern blot analysis to determine the organization of plasmid DNA in the DEN-2-resistant cell line. Cellular DNA was digested with EcoRI, an enzyme that recognizes nine sites in pIE1D2prMir, and KpnI, which has no recognition sites within the plasmid DNA. Five EcoRI sites reside in the hr5 enhancer region (bp 1 to 488) of the hr5/IE-1 baculovirus regulatory sequence, producing very small fragments upon digestion. Lane a of Fig. 5 shows results after digestion of cell line FB 9.1 DNA with EcoRI. Based on EcoRI restriction digests of pIE1D2prMir, the bands seen at approximately 650, 1,200, 2,000, and 3,000 bp were from intact plasmids. A number of additional bands also were observed, probably from junctions between plasmid and cellular DNA, suggesting that plasmids were integrated at multiple sites in the cell genome. Digestion with KpnI revealed predominantly a high-molecular-weight band greater than plasmid unit length. This suggests integration of multiple copies of the plasmid DNA into the genome but does not rule out the presence of extrachromosomal plasmid arrays similar to those seen elsewhere for other transformed C6/36 cell lines (26).
FIG. 5.
Southern blot analyses of DNA isolated from cell lines FB 9.1, H 9.1, and C6/36. Lane a, DNA from FB 9.1 digested with EcoRI; lanes b and c, DNA from C6/36 cells digested with EcoRI and KpnI, respectively; lane d, DNA from FB 9.1 digested with KpnI. Blots were hybridized with pIE1D2prMir labeled with 32P. Locations of size markers are indicated at the left.
Northern blot analysis.
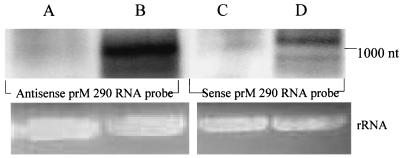
PTGS in transgenic plants is characterized by low steady-state levels of transgene transcripts. H 9.1 and FB 9.1 cells were analyzed by Northern blot hybridization for the presence of sense or antisense D2prM290 transcripts (Fig. 6). Hybridization with both prM290 sense and antisense RNA probes revealed a transcript in FB 9.1 cells of approximately 1,100 nt that was of the size predicted for the D2prMir transcript [D2prMir + 3′ untranslated region and poly(A)], indicating the probable existence of intracellular dsRNA (Fig. 6). No specific hybridization was seen with H 9.1 cellular RNA. Prior to blotting, the agarose gel used in the Northern blot analysis was stained with ethidium bromide and showed similar quantities of mosquito rRNA, indicating that similar amounts of total cellular RNA were loaded in each lane (Fig. 6).
FIG. 6.
Northern blot analysis of mRNA transcripts produced in FB 9.1 and H 9.1 transformed cell lines. Blots were probed for sense (A and B) or antisense (C and D) D2prM290 sequence. RNA was isolated from H 9.1 (A and C) and FB 9.1 (B and D) cells. Probe was strand-specific, labeled transcript from D2prM290 insert. Ethidium bromide staining of rRNA in each lane is shown below.
Mechanism of DEN-2 resistance.
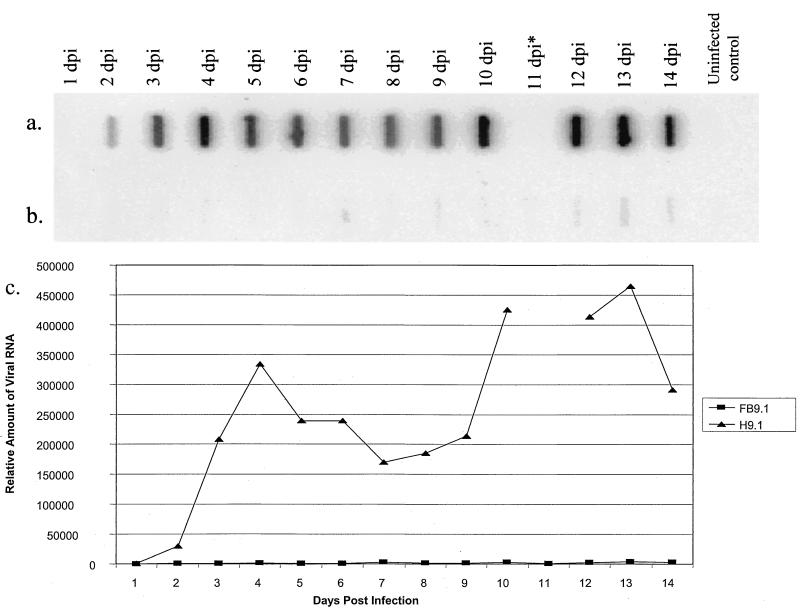
RNA silencing results in sequence-specific degradation of target mRNA. To determine the fate of DEN-2 genome RNA in resistant cell lines, FB 9.1 cell cultures and H 9.1 cell controls were infected with DEN-2 (Jamaica 1409) at an MOI of 0.01, and total RNA was extracted every 24 h for 14 days and analyzed for accumulation of DEN-2 RNA by blot hybridization. DEN-2 RNA concentration increased daily in C6/36 cells (data not shown) and, after a 24-h delay, accumulated over the entire time course in H 9.1 cells (Fig. 7). However, only minimal DEN-2 RNA accumulated in FB 9.1 cells, even at 14 days postinfection.
FIG. 7.
Slot blot analysis comparing DEN-2 RNA accumulation in FB 9.1 and H 9.1 cells after DEN-2 challenge. Total cellular RNA from each cell line on days 1 to 14 following DEN-2 challenge was blotted and probed for DEN-2 RNA, with a 32P-labeled probe complementary to the NS2B-NS3 gene region of DEN-2 RNA. (a) RNA from H 9.1 cells. (b) RNA from FB 9.1 cells. (c) Relative amounts of DEN-2 RNA determined by phosphorimager analysis. dpi, days postinfection. *, data from 11-day-postinfection time point for H 9.1 cells were unavailable.
Detection of siRNA.
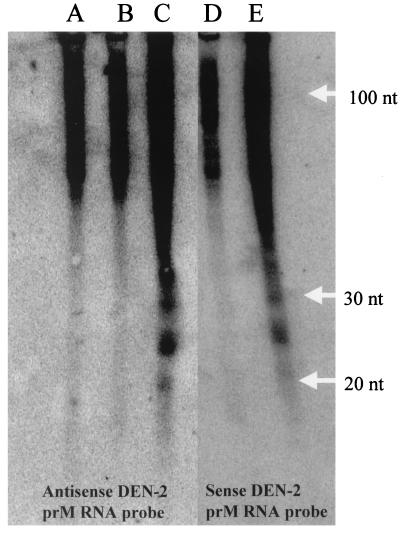
RNA silencing in plants resulted in the accumulation of a class of small RNAs that were approximately 21 to 25 nt in length (14). We reasoned that, if RNA silencing occurs in mosquito cells, the resistant cell lines should produce siRNAs that are readily detectable by Northern blot analysis with a probe specific for DEN-2 prM RNA. In addition, since the irRNA is constitutively transcribed in the FB 9.1 cell line, both sense and antisense DEN-2-specific siRNAs should be detected in these cells. However, the control cell line H 9.1 should not contain these RNAs. Indeed, we were able to detect RNAs of 21 to 25 nt in length that hybridized to both sense and antisense RNA probes derived from the prM region of DEN-2. In contrast, no small RNAs of this size were detected in H 9.1 cells (Fig. 8).
FIG. 8.
Northern blot hybridization to detect siRNA in mosquito cell lines. Total RNA was extracted from untransformed C6/36 cells (A), the H 9.1 cell line (B and D), and the FB 9.1 cell line (C and E) and enriched for small RNA (Materials and Methods). Lanes A to C were probed with 32P-labeled strand-specific antisense RNA complementary to the DEN-2 prM gene; lanes D and E were probed with sense RNA from the DEN-2 prM gene.
DISCUSSION
In this study, we present evidence that an RNAi-like defense against DEN virus infection can be efficiently triggered in mosquito cells by dsRNA corresponding to a portion of the DEN-2 genome. PTGS in plants and RNAi in invertebrates are triggered by intracellular dsRNA and result in degradation of mRNA with cognate sequences. Both are thought to represent an evolutionarily ancient system that has as one function protection of organisms from virus infection and transposon invasion.
The hr5 enhancer and immediate-early 1 (IE-1) promoter region of Autographa californica nuclear polyhedrosis virus has been shown elsewhere to allow efficient gene expression in C6/36 cells (17, 18) and was selected for transcription of the sense and antisense RNAs and irRNAs. Transformation with a plasmid designed to express irRNA resulted in a much higher proportion of resistant cell lines (44%) than did transformation with plasmids that expressed either positive-sense (14%) or antisense (7%) RNA alone. These results mirror those reported by Waterhouse et al. (39), who showed that 44 to 54% of plants were resistant to potato virus Y after transformation with an inverted-repeat DNA containing viral genome sequences, whereas less than 15% of plants were resistant to potato virus Y after transformation with sense or antisense DNA constructs. Other researchers have reported similar increases in the frequency of PTGS when inverted-repeat DNA constructs were used in place of sense or antisense DNA constructs (7, 35, 36, 38). irRNAs also have been transcribed from transgenes in Drosophila to initiate stable, heritable gene silencing, further suggesting that this strategy may be very efficient for interfering with RNA virus replication in mosquitoes (9, 19, 20).
Our previous work (1, 29) demonstrated that resistance to DEN-2 in mosquito cells and adult mosquitoes could be induced by expression of the D2prM290 sequence from a SIN virus expression vector and that expression of RNA with antisense polarity and that of RNA with sense polarity were equally effective. Virus resistance had many of the characteristics of RNA silencing, including the presence of D2prM290-specific siRNA, and we assumed that the trigger was production of DEN-2 dsRNA as part of the partially double-stranded SIN virus replicative intermediate. In the work reported here we showed that expression of a nuclear transcript that is capable of forming dsRNA provides a more efficient trigger of viral interference than does expression of transcripts with either sense or antisense polarity, giving further evidence that the presence of intracellular dsRNA triggers RNA silencing in mosquito cells.
pIE1D2prMir, the plasmid used in transformation to produce the FB 9.1 cell line, was constructed with DEN-2 (New Guinea C) prM sequences, and cells transformed by this plasmid were challenged with the DEN-2 (Jamaica 1409) strain. The sequences of the two virus genomes in the 290-nt prM region differ by approximately 4%, and this should not have disrupted complementary sequence recognition thought to be required for an RNA silencing mechanism. Resistance persisted over as many as 50 cell passages due to stable integration of the transgene. Lack of resistance to West Nile virus challenge indicated that the specificity of interference was determined by expression of DEN-2 sequences from the transforming plasmid.
The plasmid integration pattern in FB 9.1 cells was complex and appeared to be a combination of fragments or single copies of the plasmid integrated at multiple sites and unit-length inserts, possibly tandemly aligned. Although we have not determined the plasmid copy number of the FB 9.1 cell line, previous studies in this laboratory have shown that stably transformed C6/36 cells can contain high copy numbers of unit-length plasmid, sometimes organized in large arrays (26). A transcript of the predicted size from the D2prMir insert was detected by using both sense and antisense DEN-2 prM RNA probes, indicating that transcription from the transgene could form dsRNA. No attempt was made to distinguish between nuclear and cytoplasmic transcripts.
FB 9.1 cells challenged with DEN-2 failed to accumulate DEN-2 genome RNA. Infection of untransformed C6/36 cells with DEN-2 resulted in production of genomic RNA that was readily detectable by blot hybridization by 24 h postinfection and increased linearly at least until day 5 (1). Infection of transformed cells containing no DEN sequence (null cell line H 9.1) also resulted in accumulation of DEN-2 genome RNA, although RNA production appeared to be delayed by 24 h. In contrast, DEN-2-resistant cell line FB 9.1 contained barely detectable viral genome even 14 days after challenge, suggesting that DEN-2 RNA was degraded. Even though the FB 9.1 cell line produced no DEN-2 E antigen detectable by IFA (Fig. 4) and little virus RNA (Fig. 7) over a 14-day period after DEN-2 infection, FB 9.1 cells released infectious DEN-2, although with at least a 100-fold-lower titer than in either C6/36 or H 9.1 cells. This observation most likely reflects the relative sensitivity of the IFA and slot blot hybridization assays and the virus plaque assay.
Most importantly, 21- to 25-nt RNAs derived from the triggering DEN-2 sequences, the hallmark of RNA silencing, were present in resistant cells. Biochemical studies in Drosophila embryo lysates and Schneider S2 cells have suggested a mechanism for RNA silencing involving an enzyme called Dicer with RNase III and helicase domains that degrades dsRNA to 21- to 23-bp fragments. These are denatured to produce siRNA effectors (3, 41) that associate with an RNA-induced silencing complex to act as guides for sequence-specific degradation of cognate mRNA (15). Dicer-like genes occur in phylogenetically diverse organisms (3), and our stable cell lines can be used to examine whether similar mechanisms implement the mosquito RNA silencing system.
If mosquito cells mount an innate antiviral response triggered by the presence of dsRNA such as that found in both SIN and DEN virus replicative intermediates, how are arboviruses able to sustain lifelong persistent infections in their mosquito vectors? Plant viruses have evolved counterdefenses against PTGS (2), and recent studies by Ding and colleagues have shown that an insect virus also encodes a protein that suppresses RNA silencing (22). We will use these cell lines to investigate the possible existence of DEN viral mechanisms to counter RNA silencing that might have resulted in this evolutionary balance between virus and host.
Acknowledgments
We thank Tanya Allen-Miura for her help with the cloning of the pIE1-null plasmid.
This work was supported by the National Institute of Allergy and Infectious Diseases, grants AI34014, AI48740, and TW1130.
REFERENCES
- 1.Adelman, Z. N., C. D. Blair, J. O. Carlson, B. J. Beaty, and K. E. Olson. 2001. Sindbis virus-induced silencing of dengue viruses in mosquitoes. Insect Mol. Biol. 10:265-273. [DOI] [PubMed] [Google Scholar]
- 2.Anandalakshmi, R., G. J. Pruss, X. Ge, R. Marathe, A. C. Mallory, T. H. Smith, and V. B. Vance. 1998. A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. USA 95:13079-13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernstein, E., A. A. Caudy, S. M. Hammond, and G. J. Hannon. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363-366. [DOI] [PubMed] [Google Scholar]
- 4.Caplen, N. J., Z. Zheng, B. Falgout, and R. A. Morgan. 2002. Inhibition of viral gene expression and replication in mosquito cells by dsRNA-triggered RNA interference. Mol. Ther. 6:243-251. [DOI] [PubMed] [Google Scholar]
- 5.Carlson, J. O., K. E. Olson, S. Higgs, and B. J. Beaty. 1995. Molecular manipulation of mosquitoes. Annu. Rev. Entomol. 40:359-388. [DOI] [PubMed] [Google Scholar]
- 6.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Ann. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 7.Chuang, C. F., and E. M. Meyerowitz. 2000. Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97:4985-4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. A. Driver, and C. C. Mello. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806-811. [DOI] [PubMed] [Google Scholar]
- 9.Fortier, E., and J. M. Belote. 2000. Temperature-dependent gene silencing by an expressed inverted repeat in Drosophila. Genesis 26:240-244. [DOI] [PubMed] [Google Scholar]
- 10.Gaines, P. J., K. E. Olson, S. Higgs, A. M. Powers, B. J. Beaty, and C. D. Blair. 1996. Pathogen-derived resistance to dengue type 2 virus in mosquito cells by expression of the premembrane coding region of the viral genome. J. Virol. 70:2132-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signaling, immune modulation, antiviral responses and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 12.Gould, E. A., A. Buckley, N. Cammack, A. D. Barrett, J. C. Clegg, R. Ishak, and M. G. Varma. 1985. Examination of the immunological relationships between flaviviruses using yellow fever virus monoclonal antibodies. J. Gen. Virol. 66:1369-1382. [DOI] [PubMed] [Google Scholar]
- 13.Gubler, D. J. 1998. Epidemic dengue and dengue hemorrhagic fever: a global public health problem in the 21st century, p. 1-14. In W. M. Scheld, D. Armstrong, and J. M. Hughes (ed.), Emerging infections, vol. 1. American Society for Microbiology, Washington, D.C.
- 14.Hamilton, A. J., and D. C. Baulcombe. 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286:950-952. [DOI] [PubMed] [Google Scholar]
- 15.Hammond, S. M., E. Bernstein, D. Beach, and G. J. Hannon. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404:293-296. [DOI] [PubMed] [Google Scholar]
- 16.Henchal, E. A., M. K. Gentry, J. M. McCown, and W. E. Brandt. 1982. Dengue virus-specific and flavivirus group determinants identified with monoclonal antibodies by indirect immunofluorescence. Am. J. Trop. Med. Hyg. 31:830-836. [DOI] [PubMed] [Google Scholar]
- 17.Huynh, C. Q., and H. Zieler. 1999. Construction of modular and versatile plasmid vectors for the high-level expression of single or multiple genes in insects and insect cell lines. J. Mol. Biol. 288:13-20. [DOI] [PubMed] [Google Scholar]
- 18.Jarvis, D. L., C. Weinkauf, and L. A. Guarino. 1996. Immediate-early baculovirus vectors for foreign gene expression in transformed or infected insect cells. Protein Expr. Purif. 8:191-203. [DOI] [PubMed] [Google Scholar]
- 19.Kennerdell, J. R., and R. W. Carthew. 1998. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell 95:1017-1026. [DOI] [PubMed] [Google Scholar]
- 20.Kennerdell, J. R., and R. W. Carthew. 2000. Heritable gene silencing in Drosophila using double-stranded RNA. Nat. Biotechnol. 18:896-898. [DOI] [PubMed] [Google Scholar]
- 21.Kumagai, M. H., J. Donson, G. della-Cioppa, D. Harvey, K. Hanley, and L. K. Grill. 1995. Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA. Proc. Natl. Acad. Sci. USA 92:1679-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, H., W. X. Li, and S. W. Ding. 2002. Induction and suppression of RNA silencing by an animal virus. Science 296:1319-1321. [DOI] [PubMed] [Google Scholar]
- 23.Lindbo, J. A., and W. G. Dougherty. 1992. Untranslatable transcripts of the tobacco etch virus coat protein gene sequence can interfere with tobacco etch virus replication in transgenic plants and protoplasts. Virology 189:725-733. [DOI] [PubMed] [Google Scholar]
- 24.Maine, E. M. 2001. RNAi as a tool for understanding germline development in Caenorhabditis elegans: uses and cautions. Dev. Biol. 239:177-189. [DOI] [PubMed] [Google Scholar]
- 25.Miller, B. R., and C. J. Mitchell. 1986. Passage of yellow fever virus: its effect on infection and transmission rates in Aedes aegypti. Am. J. Trop. Med. Hyg. 35:1302-1309. [DOI] [PubMed] [Google Scholar]
- 26.Monroe, T. J., M. C. Muhlmann-Diaz, M. J. Kovach, J. O. Carlson, J. S. Bedford, and B. J. Beaty. 1992. Stable transformation of a mosquito cell line results in extraordinarily high copy numbers of the plasmid. Proc. Natl. Acad. Sci. USA 89:5725-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montgomery, M. K., S. Xu, and A. Fire. 1998. RNA as a target of double-stranded RNA-mediated genetic interference in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 95:15502-15507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olson, K. E., S. Higgs, P. J. Gaines, A. M. Powers, B. S. Davis, K. I. Kamrud, J. O. Carlson, C. D. Blair, and B. J. Beaty. 1996. Genetically engineered resistance to dengue-2 virus transmission in mosquitoes. Science 272:884-886. [DOI] [PubMed] [Google Scholar]
- 29.Olson, K. E., Z. N. Adelman, E. A. Travanty, I. Sanchez-Vargas, B. J. Beaty, and C. D. Blair. 2002. Developing arbovirus resistance in mosquitoes. Insect Biochem. Mol. Biol. 32:1333-1343. [DOI] [PubMed] [Google Scholar]
- 30.Ratcliff, F., B. D. Harrison, and D. C. Baulcombe. 1997. A similarity between viral defense and gene silencing in plants. Science 276:1558-1560. [DOI] [PubMed] [Google Scholar]
- 31.Ratcliff, F. G., S. A. MacFarlane, and D. C. Baulcombe. 1999. Gene silencing without DNA. RNA-mediated cross-protection between viruses. Plant Cell 11:1207-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruiz, M. T., O. Voinnet, and D. C. Baulcombe. 1998. Initiation and maintenance of virus-induced gene silencing. Plant Cell 10:937-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Sharp, P. A. 1999. RNAi and double-strand RNA. Genes Dev. 13:139-141. [PubMed] [Google Scholar]
- 35.Stam, M., R. de Bruin, S. Kenter, R. A. van der Hoorn, R. van Blokland, J. N. Mol, and J. M. Kooter. 1997. Post-transcriptional silencing of chalcone synthase in Petunia by inverted transgene repeats. Plant J. 12:63-82. [Google Scholar]
- 36.Stam, M., R. de Bruin, R. van Blokland, R. A. van der Hoorn, J. N. Mol, and J. M. Kooter. 2000. Distinct features of post-transcriptional gene silencing by antisense transgenes in single copy and inverted T-DNA repeat loci. Plant J. 21:27-42. [DOI] [PubMed] [Google Scholar]
- 37.Tenllado, F., and J. R. Diaz-Ruiz. 2001. Double-stranded RNA-mediated interference with plant virus infection. J. Virol. 75:12288-12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, M. B., and P. M. Waterhouse. 2000. High-efficiency silencing of a beta-glucuronidase gene in rice is correlated with repetitive transgene structure but is independent of DNA methylation. Plant Mol. Biol. 43:67-82. [DOI] [PubMed] [Google Scholar]
- 39.Waterhouse, P. M., M. W. Graham, and M. B. Wang. 1998. Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc. Natl. Acad. Sci. USA 95:13959-13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Westaway, E. G., J. M. Mackenzie, M. T. Kenney, M. K. Jones, and A. A. Kromykh. 1997. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA and of NS2B with NS3 in virus-induced membrane structures. J. Virol. 71:6650-6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zamore, P. D., T. Tuschl, P. A. Sharp, and D. P. Bartel. 2000. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101:25-33. [DOI] [PubMed] [Google Scholar]