Abstract
A fragment of RyR1 (amino acids 4064–4210) is predicted to fold to at least one lobe of calmodulin and to bind Ca2+. This fragment of RyR1 (R4064–4210) was subcloned, expressed, refolded, and purified. Consistent with the predicted folding pattern, R4064–4210 was found to bind two molecules of Ca2+ and undergo a structural change upon binding Ca2+ that exposes hydrophobic amino acids. R4064–4210 also binds to RyR1, the L-type Ca2+ channel (Cav1.1), and several synthetic calmodulin binding peptides. Both R4064–4210 and a peptide representing the calmodulin-binding region of RyR1 (R3614–3643) alter the Ca2+ dependence of (3H)ryanodine binding to RyR1, suggesting that they may both be interfering with an intramolecular interaction between amino acids 4064–4210 and amino acids 3614–3643 in the native RyR1 to alter or regulate the response of the channel to changes in Ca2+ concentration. The finding that a domain within RyR1 binds Ca2+ and interacts with calmodulin-binding motifs may provide insights into the mechanism for calcium- and calmodulin-dependent regulation of this channel and perhaps for its regulation by the L-type Ca2+ channel.
INTRODUCTION
Ca2+ release channels, also known as ryanodine receptors (RyRs), regulate the release of Ca2+ from sarcoplasmic reticulum (SR) stores. The released Ca2+ triggers muscle contraction and activates a cascade of Ca2+-dependent signal transduction pathways. Ca2+ and calmodulin (CaM) are important as in vivo modulators of RyRs. RyR1 activity displays a bell-shaped dependence on Ca2+ concentration, with enhancement of channel activity in the low μM range, but inhibition of the channel in the high μM to mM range (1,2). Several motifs within the primary amino-acid sequence of RyRs have been suggested to resemble Ca2+-binding E-F hand motifs in Ca2+-binding proteins (3,4). Putative E-F hands have been identified in RyR1 at amino acids 4253–4264, 4407–4416, and 4489–4499 (5), in RyR2 at amino acids 1136–1347 and 2010–2021 (6), and in RyR3 at amino acids 3934–3945 (7). Fragments of RyR1 containing some of these E-F hand motifs have been shown to bind Ca2+ (8–10). The low-affinity inhibitory Ca2+ binding site was suggested to be within the negatively charged region between residues 1872–1923 (11,12); however, the replacement of this glutamate-rich sequence with the corresponding, less-acidic sequence from RyR2 did not change low-affinity Ca2+ inactivation of RyR1 (12). Du and MacLennan suggested that the Ca2+ binding sites for both activation and inactivation of RyR1 are located at the C-terminus between residues 3726 and 5037 (13). This region has three potential E-F hands (5) and several other possible Ca2+-binding sequences (14–16). Mutation of some amino acids in this region alters Ca2+ binding and Ca2+ regulation of RyR1 (17). To determine if any of these putative E-F hands are involved in Ca2+ regulation of the channel, Fessenden et al. (18) created mutant RyRs in which the putative E-F hands were scrambled either singly or in combination with other E-F hands. These investigators found that, in intact myotubes, the mutations did not affect functional responses to depolarization, caffeine, or 4-chloro-m-cresol (4-CmC) (18). However, (3H)ryanodine binding and its Ca2+ dependence were altered by the E-F-1 mutations and abolished by the E-F-2 mutations, suggesting an involvement of these E-F hands in Ca2+ regulation of the channel. Why would this be different in the myotubes? One possibility is that a Ca2+-binding protein such as CaM, which can itself both activate and inhibit the channel in a Ca2+-dependent manner, can override the need for Ca2+ binding directly to the channel in the myotubes. This unanswered question emphasizes the need for further investigation.
CaM can function as either an activator or an inhibitor of RyR1, depending on whether it has Ca2+ bound (19). At sub-μM Ca2+ concentrations, CaM is an activator of RyR1, but at higher Ca2+ concentrations, it becomes an inhibitor (19–21). Both Ca2+-free and Ca2+-bound CaMs bind to RyR1 at a site close to amino acids 3614–3643 (22,23). This sequence is likely to represent the binding site for the C-lobe of CaM (24), whereas the N-lobe may bind to an adjacent subunit within the RyR1 tetramer between amino acids 1975 and 1999 (25). We previously have shown that a carboxyterminal tail fragment of the skeletal L-type channel (Cav1.1) α1-subunit, which contains both Ca2+ and CaM binding sites, binds to RyR1. This interaction is blocked by Ca2+CaM (26). We have also shown that a peptide representing the CaM binding site on RyR1 (amino acids 3614–3643) binds to both the Cav1.1 and an expressed fragment of the carboxyterminal tail of its α1-subunit (amino acids 1393–1527) (26). The interaction of R3614–3643 with the Cav1.1 is also blocked by Ca2+ CaM (26). Three sequences on the carboxyterminal tail of the cardiac L-type Ca2+ channel (Cav1.2), designated A (1609–1628, numbering of the human cardiac channel), C (1627–1652), and IQ (1665–1685) have been implicated in CaM binding (27–30). Synthetic peptides matching these sequences or their skeletal muscle counterparts do not interact with R3614–3643 (unpublished observation), suggesting that there is another site within Cav1.1 that can bind to R3614–3643.
Since the CaM binding domains on Cav1.1 and RyR1 do not directly interact, we began to search for other interaction sites. Using 3D-PSSM (http://www.sbg.bio.ic.ac.uk), we identified a sequence within RyR1 (amino acids 4064–4210) and one within the carboxyterminal tail of the Cav1.1 α1-subunit (amino acids 1395–1540) predicted to fold like CaM and bind Ca2+. One possibility is that these CaM-like domains can interact with the CaM binding sites of these proteins and CaM would compete for this interaction. To assess this possibility, we expressed the CaM-like domain of RyR1 and examined its ability to bind Ca2+and to interact with RYR1, the L-type channel, and several CaM-binding peptides. (See Table 1 for a list of all peptides used.)
TABLE 1.
Peptides used in this study
| R4064–4210 | MFLKLKD IVGSEAFQDYVTDPRGLISK KDFQKAMDSQKQFTGPEIQFLLSCSEADENEMINFEEFAN RFQEPARDIGFNVAV LLTNLSEHVPHDPRLRNFLELAESI LEYFRPYLGRIEIMGAS RRIERIYFEISETNRAQWEMPQV |
| R3614–3627 | KSKKAVWHKLLSKQ |
| R3614–3634 | KSKKAVWHKLLSKQRRRAVVA |
| R3614–3643 | KSKKAVWHKLLSKQRRRAVVACFRMTPLYN |
| R3625–3644 | SKQRRRAVVACFRMTPLYNL |
| Skeletal IQ | KFYATFLIQEHFRKFMKRQEE |
| C-peptide | EQANEELRAIIKKIWKRTSMKLLDQV |
| CaMKII peptide | MHRQEAVDCLKKFNARRKLKG |
EXPERIMENTAL PROCEDURES
Materials
The peptides were synthesized in the core facility at Baylor College of Medicine (Houston, TX) under the direction of Dr. Richard Cook. SR membranes were prepared from rabbit hind leg and backstrap skeletal muscle as previously described (31). Protein concentrations were determined by the Lowry method (32) using bovine serum albumin as the standard. (3H)ryanodine (70–80 Ci/mmol) was purchased from Du Pont New England Nuclear (Boston, MA); unlabeled ryanodine was purchased from Calbiochem (La Jolla, CA).
Creation of the construct for expressing R4064–4210
To subclone the fragment of RyR1 cDNA coding for the region of RyR1 predicted to fold like CaM, we used PCR primers containing NdeI and HindIII restriction sites, with a stop-codon in the reverse primer. PCR reactions amplified the desired DNA fragment by selective priming of the full-length rabbit skeletal RyR1 cDNA. The amplified PCR products were agarose gel-purified and then ligated into pCR-Blunt vector (Invitrogen, San Diego, CA). The ligates were transformed into One-Shot TOP-10 Competent Cells (Invitrogen) for amplification. After confirmation by DNA sequencing, the desired DNA fragments were excised using the enzymes whose restriction sites were pre-incorporated into primers. The released DNA fragment was subcloned into pET23a(+) or pET28a(+) vectors (Novagen, Madison, WI) between NdeI and HindIII sites. The subcloned products were transformed into DH5α competent cells (Invitrogen) to amplify. DNA sequencing was performed again for verification of the desired DNA sequences in the expression vectors.
Expression, purification, and refolding of R4064–4210
The expression vectors containing the DNA construct for R4064–4210 were transformed into BL21(DE3) competent cells (Novagen). The fragment R4064–4210 was found almost exclusively in inclusion bodies when expressed either with or without a 20-amino acid His tag (MGSSHHHHHHSSGLVPRGSH) at the N-terminus of the fragment (in pET28a(+) or pET23a(+) vector). To purify this fragment from inclusion bodies, the cytoplasmic proteins were first extracted using Bacterial Protein Extraction Reagent (B-PER; Pierce Biotechnology, Rockford, IL) and then the bacterial membrane proteins were extracted by B-PER Bacterial Protein Extraction Reagent with lysozyme and nuclease. The remaining insoluble material, which is greatly enriched in R4064–4210, was washed with inclusion body wash buffer (50 mM Tris-HCl pH 7.4, 1% Triton or CHAPS, 5 mM DTT, and 5 mM EDTA). R4064–4210 was extracted with 50 mM Tris-HCl pH 7.4 and 2 M urea. The His-tagged protein was purified with a chelating sepharose column (Amersham Pharmacia Biotech, Piscataway, NJ) with a buffer containing 2 M urea. The chelating sepharose-purified R4064–4210 was dialyzed against 20-mM Tris-HCl pH 7.4 in a 10,000 MWCO cassette (Pierce). The dialysis buffer was changed twice to remove the urea and refold the protein. The urea-extracted untagged protein was refolded in a manner similar to the His-tagged protein and purified by anion exchange (HiTrap Q HP) and phenyl sepharose chromatography (Amersham Pharmacia Biotech).
Circular dichroism spectroscopy
To demonstrate the refolding of R4064–4210, circular dichroism (CD) spectroscopy was used to analyze its secondary structure. Untagged R4064–4210 (0.1 mg/ml) was incubated in 5 mM Tris-HCl pH7.9, 1 mM EGTA, or l mM Ca2+ for 10 min at room temperature. CD spectra were recorded on an Aviv CD instrument, Model #62A DS (Aviv, Lake Wood, NJ) from 188 to 250 nm with 1 nm/step, using a 2-mm quartz cell. Data were processed by subtracting the CD signal of buffer/additives and by averaging the data obtained from three independent experiments.
Analysis of 45Ca binding to R4064–4210 by equilibrium dialysis
R4064–4210 (15 μM, 100 μl) in 30 mM HEPES pH 7.0, 0.2 mM PMSF, and 5 mM β-mercaptoethanol was injected into one of two chambers of an equilibrium dialysis cell (Bel-Art Products, Pequannock, NJ), separated by a 3500 MWCO membrane. The same amount of the buffer containing different concentrations of 45Ca2+ (5–200 μM, 1 μCi/μmol) was injected into the other cell. The equilibrium dialysis cells were incubated at 20°C overnight with rotation, after which 50 μl was removed from each chamber for analysis of 45Ca2+ by liquid scintillation counting. 45Ca2+ bound to R4064–4210 was calculated from the difference in 45Ca2+ radioactivity in the two chambers. The data were fit with a four-parameter Hill equation in SigmaPlot (SPSS, Chicago, IL).
ANSA fluorescence analysis probes conformational changes of R4064–4210 induced by Ca2
To assess the ability of the RyR1 fragment to undergo Ca2+-induced conformational changes, we used 8-anilino-1-naphthalenesulfonic acid ammonium (ANSA) (Sigma-Aldrich, St. Louis, MO) fluorescence analysis. ANSA (1 μM) was incubated in 30 mM MOPS (pH 7.2) and either 5 mM EGTA (low Ca2+) or 2 mM Ca2+ (high Ca2+) for 5 min at room temperature in the presence or absence of untagged R4064–4210 (0.4 μM). Fluorescence emission spectra from 430 to 600 nm were collected with 370-nm excitation using an ISS PC1 Photon Counting Spectrofluorometer (ISS, Champaign, IL), with a 0.5-mm slit for excitation and 2-mm slit for emission, 1 nm/step. Relative fluorescence was corrected for buffer contribution. All experiments were repeated at least three times.
R4064–4210 binding to CaM-binding peptides by ANSA fluorescence analysis
R4064–4210 (0.5 μM) and ANSA (2 μM) were incubated in 30 mM MOPS (pH 7.2) containing 5 mM EGTA, 50 μM Ca2+, 500 μM Ca2+, or 5 mM Ca2+ for 5 min at room temperature; 5 μM of different peptides (R3614–3643, C-peptide (27), CaM kinase II Inhibitor (Calbiochem) or the IQ-peptide (30)) were then added. ANSA fluorescence emission data at 470 nm were collected on an SLM 8000C Spectrofluorometer (SLM Instruments, Urbana, IL) with a 370-nm excitation, 8-nm bandpass for excitation, and 16-nm bandpass for emission. Data were processed by subtracting the background of buffer and/or additives where appropriate and the data were fit with a four-parameter Hill equation in SigmaPlot (SPSS).
Identification of the amino acids interacting with R4064–4210
To determine which amino acids in R3614–3643 were involved in R4064–4210 binding, we used overlapping peptide fragments from amino acids 3614–3644 to interact with R4064–4210 in the presence of ANSA. R4064–4210 (0.5 μM) and ANSA (2 μM) were incubated in 30 mM MOPS (pH 7.2) containing 5 mM EGTA (low Ca2+) or 500 μM Ca2+ (high Ca2+) for 5 min at room temperature. Peptides (R3614–3627, R3614–3634, R3614–3643, and R3625–3644) were then added and the ANSA fluorescence data were collected on an SLM 8000C Spectrofluorometer (SLM Instruments).
The effect of R4064–4210 and synthetic peptides on (3H)ryanodine binding
SR membranes (10 μg) were incubated with (3H)ryanodine (5 nM) for 16 h at room temperature in 100 mM NaCl, 30 mM MOPS (pH7.2), 100 μg/ml BSA, and 0.1% CHAPS, containing increasing CaCl2 with 10 μM different proteins or peptides. Bound radiolabel was separated from free by filtration through Whatman GF/F filters with 5 × 3 ml washes of ice cold buffer (Whatman, Brentford, Middlesex, UK). Nonspecific binding was determined in the presence of 10-μM unlabeled ryanodine.
Assessing the interaction of R4064–4210 with Cav1.1 by pulldown assays
His-tagged R4064–4210 (3 nM) or a synthetic His tag alone were incubated with 300 μl of a Ni charged chelating sepharose bead slurry in binding buffer consisting of 50 mM MOPS (pH 7.4), 20 mM imidazole, 1 mM CaCl2 (designated high Ca2+ buffer) or 2 mM EGTA (designated low Ca2+ buffer) for 30 min. The IQ-peptide (3 nM, sequence: KFYATFLIQEHFRKFMKRQEE) from the C-terminus of the skeletal L-type Ca2+ channel (Cav1.1, α1-subunit) or digitonin-solubilized T-tubule membranes (24 μg) were added to this bead slurry. After 1-h incubation, the beads were washed twice with 300-μl binding buffer and proteins were eluted with SDS sample buffer for electrophoresis. The pulldown of the IQ-peptide by R4064–4210 was assessed by Coomassie staining of the SDS gel (in duplicate and repeated three times). Cav1.1 in the pulldown was determined by Western blotting with an anti-Cav1.1 α1s antibody, MA3-920 (Affinity BioReagents, Golden, CO), and Alexa Fluor 680 (Molecular Probes, Eugene, OR) as secondary antibody (in triplicate and repeated twice). An Odyssey Infrared Imaging System (LI-COR, Lincoln, NE) was used to analyze the Western blots. To verify the specific interaction of R4064–4210 with Cav1.1, parallel Western blotting experiments were conducted, but with an anti-sodium/potassium ATPase α-1 antibody, MA3-929 (Affinity BioReagents).
RESULTS
R4064–4210 is predicted by 3D-PSSM to fold similar to calmodulin
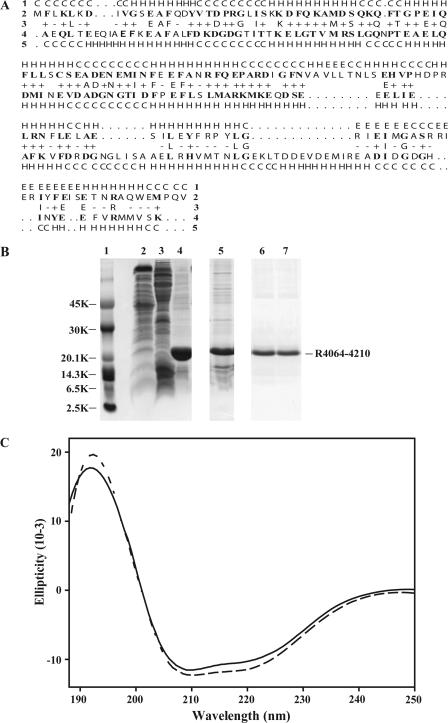
We have previously shown that a synthetic peptide matching the CaM binding site on RyR1 bound to Cav1.1, and, conversely, that a CaM binding peptide from Cav1.1 bound to RyR1 (26). Since the CaM binding peptides from these two channels do not themselves interact (unpublished observation), we began to look for sequences within RyR1 that could bind to the carboxy-terminal tail of the Cav1.1 α1-subunit. A region of Cav1.1 (amino acids 1395–1540) that contains the putative E-F hands was previously predicted by 3D-PSSM (http://www.sbg.bio.ic.ac.uk) to fold like CaM and was shown to interact with CaM binding peptides from RyR1 and Cav1.1 (26). The CaM binding peptide from RyR1 has been shown to bind both Ca2+CaM and apoCaM (22,23). We found a region of RyR1 (amino acids 4064–4210) that was also predicted to fold in a manner similar to CaM (>95% confidence level, 21% sequence identity, Fig. 1 A).
FIGURE 1.
Expression, refolding, and predicted secondary structure of R4064–4210. (A) Secondary structural comparison of R4064–4210 with calmodulin by 3D-PSSM Fold prediction. Row 2 shows the amino-acid sequence of R4064–4210. Row 4 is the sequence of Paramecium CaM, found to be the best structural match to R4064–4210 in the 3D-PSSM Fold Library. The bold letters in rows 2 and 4 are identical/similar amino acids between R4064–4210 and calmodulin. Row 1 predicts the secondary structures of R4064–4210 and row 5 shows the secondary structures of Paramecium calmodulin found in the Fold Library (H, α-helix; E, β-strand; and C, coil). Row 3 demonstrated the identity between R4064–4210 and Paramecium calmodulin. The letters mean that R4064–4210 and Paramecium calmodulin have the same amino acids in the alignment. The plus (+) versus minus (−) symbols indicate a good-versus-poor match between the sequence of R4064–4210 and Paramecium calmodulin, respectively. The R4064–4210 sequence was predicted to have >95% probability of folding like CaM. The overall sequence identity was 21%. (B) Expression, purification, and refolding of R4064–4210. The His-tagged and untagged proteins have similar expression, purity, and solubility. For simplicity, only the results for the His-tagged protein are shown. Cytoplasmic proteins were extracted by B-PER Bacterial Protein Extraction Reagent (lane 2), and membrane proteins were extracted by B-PER Bacterial Protein Extraction Reagent with lysozyme and nuclease (lane 3), and insoluble material (lane 4). Upon comparing the molecular size of His-tagged R4064–4210 to the markers (lane 1), it was found that R4064–4210 was overexpressed in the inclusion body (lane 4). 2 M urea extracted R4064–4210 from the inclusion body (lane 5). The urea-extracted materials were purified by chelating sepharose (Amersham Pharmacia Biotech) in the presence of 2 M urea for all buffers (lane 6). The purified R4064–4210 was refolded by dialyzing against 20 mM Tris-HCl pH7.4 (lane 7). (C) R4064–4210 is rich in α-helical structure as assessed by CD spectroscopy. R4064–4210 (0.1 mg/ml) was incubated in 5 mM Tris-HCl (pH7.9), 1 mM EGTA (solid line), or l mM Ca2+ (dashed line) for 10 min at room temperature. The CD spectra were recorded from 188 to 250 nm.
Extraction and refolding of R4064–4210 from inclusion bodies after expression in Escherichia coli
A fragment of RyR1 representing this putative CaM-like domain and designated R4064–4210 was expressed in BL21(DE3) cells. The protocol used for the expression generated 30–50 mg of R4064–4210 per liter of culture. The R4064–4210 was found almost exclusively in inclusion bodies but could be solubilized in 2 M urea (lane 5 of Fig. 1 B). The urea-solubilized R4064–4210 was refolded by dialysis into buffer without urea and was soluble at concentrations as high as 10 mg/ml (lane 7 of Fig. 1 B).
Assessing the α-helical content of R4064–4210
R4064–4210 appeared to be both homogenous and structured as assessed by its CD signal at both low (solid line of Fig. 1 C) and high Ca2+ concentrations (dashed line of Fig. 1 C). There is a strong positive CD maximum at 192 nm and two negative CD maxima centered at 210 and 222 nm, indicative of proteins with a high α-helical content (33). Furthermore, the high α-helical structural content is consistent with the prediction for folding obtained with 3D-PSSM (Fig. 1 A). The CD spectrum of this fragment is similar to that of CaM (24). Unlike CaM, Ca2+ produced only small changes in the α-helical content of R4064–4210, suggesting that the structural changes in this fragment upon binding Ca2+ are probably not as extensive as those in CaM.
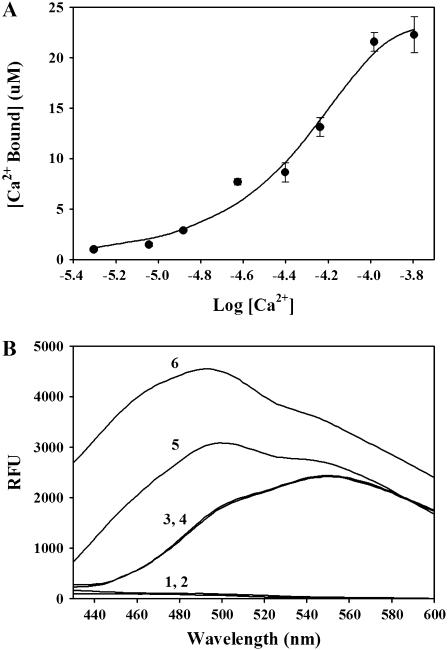
The ability of R4064–4210 to bind Ca2+
To assess the ability of R4064–4210 to bind Ca2+ we used micro-equilibrium dialysis with 45Ca2+. We found that two molecules of 45Ca2+ bound per mol of R4064–4210 (1.9 ± 0.3 pmol 45Ca2+ bound per pmol of R4064–4210, with an apparent affinity of 60 ± 12 μM, n = 3, and a Hill coefficient of 1.6 ± 0.4; see Fig. 2 A). To confirm the ability of R4064–4210 to bind Ca2+ we also used terbium fluorescence. We found that terbium fluorescence was increased in the presence of R4064–4210 and that the increase is inhibited by Ca2+ (data not shown), suggesting that Ca2+ competes with terbium for a common binding site on this fragment of RyR1.
FIGURE 2.
R4064–4210 binds Ca2+. (A) Analysis of 45Ca2+ binding to R4064–4210 by equilibrium dialysis. R4064–4210 (15 μM, 100 μl) in 30 mM HEPES pH 7.0 was injected into one of two chambers of an Equilibrium Dialysis cell, separated by a 3500-MWCO membrane. The same amount of the buffer containing different concentrations of 45Ca2+ was injected into the other chamber. After overnight incubation, 50-μl aliquots were removed from each chamber for determination of 45Ca2+ by liquid scintillation counting. 45Ca2+ bound to R4064–4210 was determined from the difference in 45Ca2+ radioactivity in the two chambers. By a four-parameter Hill fitting, R4064–4210 apparent affinity for Ca2+ (EC50) = 60 ± 12 μM, maximum binding (Bmax) = 1.9 ± 0.3 pmol 45Ca2+ bound per pmol of R4064–4210, and the Hill coefficient = 1.6 ± 0.4. (B) Ca2+ binding to R4064–4210 alters ANSA fluorescence. R4064–4210 (0.4 μM) was incubated with ANSA (1 μM) in 30 mM MOPS (pH 7.2), with either 5 mM EGTA (low Ca2+) or 2 mM Ca2+ (high Ca2+) for 5 min at room temperature. Fluorescence emission spectra from 430 to 600 nm were collected with a 370-nm excitation wavelength. 1, R4064–4210 (low Ca2+); 2, R4064–4210 (high Ca2+); 3, ANSA (low Ca2+); 4, ANSA (high Ca2+); 5, R4064–4210 + ANSA (low Ca2+); and 6, R4064–4210 + ANSA (high Ca2+).
Ca2+-induced changes in R4064–4210 assessed by ANSA fluorescence probe analysis
To determine if R4064–4210 undergoes a structural change upon binding Ca2+, we used ANSA, a probe that increases its fluorescence when bound to the hydrophobic sites on proteins (34). We found that Ca2+ increased ANSA fluorescence in the presence of R4064–4210 (Fig. 2 B), suggesting that Ca2+ binds to R4064–4210 and induces exposure of hydrophobic regions.
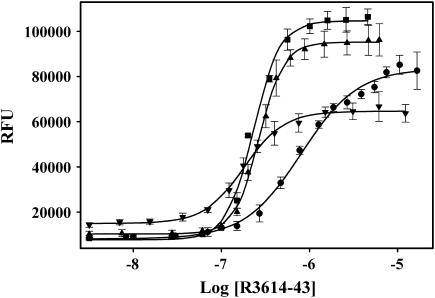
R4064–4210 binds to the CaM binding motif (R3614–3643) of RyR1
R4064–4210 binds directly to a peptide (R3614–3643) representing the CaM binding site on RyR1. We found a very dramatic increase in ANSA fluorescence with increasing concentrations of R3614–3643 (Fig. 3). If the R4064–4210 region of RyR1 is important for Ca2+-dependent regulation of RyR1 activity, the interactions should be in some way regulated by the binding of Ca2+. To assess this, we examined the interaction of R3614–3643 with R4064–4210 at different Ca2+ concentrations (Fig. 3). We found that R3614–3643 apparently interacts with R4064–4210 at the all of the Ca2+ concentrations tested, but that the affinity and the apparent magnitude of the ANSA fluorescence changes were different at different Ca2+ concentrations. The apparent affinities of R3614–3643 for R4064–4210 at <10 nM, 50 μM Ca2+, 500 μM Ca2+, and 5 mM Ca2+ were 803 ± 47 nM, 230 ± 7 nM, 268 ± 8 nM, and 181 ± 10 nM, respectively. The magnitude of the ANSA fluorescence change was also different at the different Ca2+ concentrations, with the smallest change occurring at 5 mM and <10 nM Ca2+ (concentrations where RyR1 is less active). An interaction of the CaM-like domain (R4064–4210) at both high and low Ca2+ concentrations is consistent with R3614–3643 being a binding site for both apoCaM and Ca2+CaM (22,23).
FIGURE 3.
R4064–4210 interaction with R3614–3643 at different Ca2+ concentrations by ANSA fluorescence probe. 0.5μM R4064–4210 and 2 μM ANSA was incubated in 30 mM MOPS (pH 7.2) containing 5 mM EGTA, •; 50 μM Ca2+, ▪; 500 μM Ca2+, ▴; or 5 mM Ca2+, ▾ for 5 min at room temperature. Then the incubated materials were titrated with R3614–3643 and the ANSA fluorescence data were collected. Ca2+ concentrations affect significantly the interaction of R4064–4210 with R3614–3643 (EC50 = 803 ± 47, 230 ± 7, 268 ± 8, and 181 ± 10 nM; and the Hill coefficient = 1.4 ± 0.1, 2.8 ± 0.2, 3.1 ± 0.2, and 1.9 ± 0.2 for 5 mM EGTA, 50 μM Ca2+, 500 μM Ca2+, and 5 mM Ca2+, respectively).
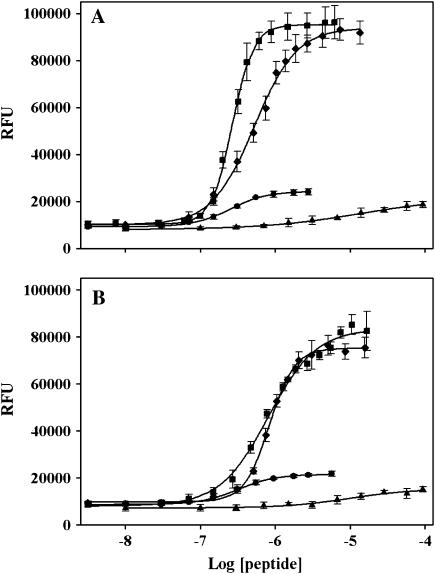
Amino-acid determinants in R3614–3643 required for R4064–4210 binding
To determine which amino acids within the CaM binding site of RyR1 were required for the interaction with R4064–4210, we tested the ability of a series of overlapping peptides from this region for the ability to interact with R4064–4210 using ANSA fluorescence to assess the interactions. We found that a peptide corresponding to amino acids 3614–3634 (solid circles in Fig. 4 A) had an apparent affinity (EC50 = 251 ± 17 nM) that was similar to R3614–3643 (268 ± 8 nM) (solid squares in Fig. 4 A) at high Ca2+; however, the magnitude of the fluorescence change induced by R3614–3634 was much less than that induced by R3614–3643. This finding suggests that determinants between amino acids 3614–3634 were important for affinity, but that determinants between amino acids 3635 and 3643 are needed to induce the conformational change at high Ca2+. The R3614–3634 peptide had a higher apparent affinity for R4064–4210 (320 ± 21 nM, n = 3) (solid circles in Fig. 4 B) than R3614–3643 (803 ± 47 nM, n = 3) (solid squares in Fig. 4 B) at low Ca2+, but again the maximal fluorescence change was much less than that produced by R3614–3643. These data suggest that the amino acids between 3635 and 3643 did not contribute greatly to the affinity but were responsible for the greater change in ANSA fluorescence with R4064–4210. A peptide corresponding to amino acids 3625–3644 produced a maximal fluorescence change comparable to R3614–3643 but with a twofold decrease in affinity (EC50 = 525 ± 27 nM) at high Ca2+ (solid diamonds in Fig. 4 A). The affinities of the two peptides were similar at low Ca2+ (835 ± 23 vs. 803 ± 47 nM for 3614–3643) (solid diamonds in Fig. 4 B). A peptide corresponding to amino acids 3614–3627 (solid up-triangles in Fig. 4, A and B) had little effect on the ANSA fluorescence at either high or low Ca2+ concentrations.
FIGURE 4.
Mapping the amino acids in R3614–3643 that bind to R4064–4210. R4064–4210 (0.5 μM) and ANSA (2 μM) were incubated in 30 mM MOPS (pH 7.2) containing 500 μM Ca2+ (A, high Ca2+) or 5 mM EGTA (B, low Ca2+) for 5 min at room temperature. Peptides R3614–3627, R3614–3634, R3614–3643, and R3625–3644 were added at the indicated concentrations, and the ANSA fluorescence were collected. ANSA fluorescence data in the presence of R4064–4210 per peptide were processed by subtracting the effect of R4064–4210 and peptide on the fluorescence of ANSA. (•, R3614–3634 with EC50 = 251 ± 17 nM, the Hill coefficient = 1.8 ± 0.2 for high Ca2+ and 320 ± 21 nM, 1.7 ± 0.2 for low Ca2+; ▪, R3614–3643 with EC50 = 268 ± 8 nM, the Hill coefficient = 3.1 ± 0.2 for high Ca2+ and 803 ± 47 nM, 1.4 ± 0.1 for low Ca2+; ♦, R3625–3644 with EC50 = 525 ± 27 nM, the Hill coefficient = 1.5 ± 0.1 for high Ca2+ and 835 ± 23 nM, 2.6 ± 0.2 for low Ca2+; and ▴, R3614–3627.)
Effects of R4064–4210 on (3H)ryanodine binding to RyR1
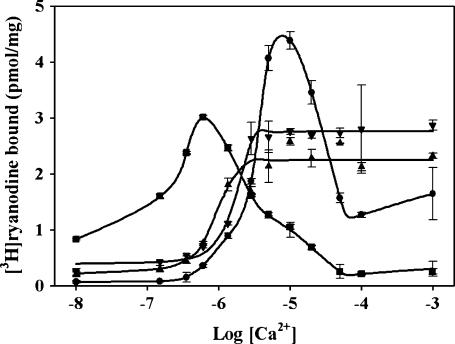
If the interaction between amino acids 3614–3643 and 4064–4210 occurs within the native RyR1, and if this interaction in some way regulates the functional state of RyR1, both R4064–4210 and R3614–3643 would be predicted to modulate RyR1 activity by interfering with interactions within RyR1. To test this possibility, we examined the effects of R4064–4210 and R3614–3643 on the (3H)ryanodine binding to RyR1 at different Ca2+ concentrations. For comparison, the effects of CaM on (3H)ryanodine binding are also shown. (3H)Ryanodine binding is frequently used to assess functional effects on RyR1 since ryanodine preferentially binds to the open state of the channel (35). Ca2+ titration of (3H)ryanodine binding to SR membranes in the presence and absence of R4064–4210, R3614–3643, and CaM is shown in Fig. 5. (3H)Ryanodine binding to SR membranes shows a bell-shaped dependence on Ca2+ concentration similar to the Ca2+ dependence of channel activity: binding is low at nM Ca2+ concentrations, increases in sub-μM Ca2+, and reaches maximum at ∼10-μM Ca2+, and then decreases (Fig. 5, curve with solid circles). Our surprising finding were that both R4064–4210 and R3614–3643 (Fig. 5, curves with solid up-triangles and solid down-triangles, respectively) slightly enhanced (3H)ryanodine binding at <10 nM Ca2+, inhibited (3H)ryanodine at intermediate Ca2+ and prevented Ca2+ inhibition of (3H)ryanodine binding at high Ca2+. For comparison, CaM activates the channel at low Ca2+ but inhibits the channel at high Ca2+ (Fig. 5, curve with solid squares). Since these peptides and fragments would be expected to compete with intramolecular interactions of these same sequences within RyR1, these findings suggest that an interaction between amino acids 3614–3643 and 4064–4210 within RyR1: 1), somewhat inhibits activity in low Ca2+ (perhaps stabilizing a resting state of the channel); and 2), enhances channel activity at intermediate Ca2+ and inhibits channel activity at high Ca2+. CaM might activate the channel at low Ca2+ and inhibit the channel at intermediate Ca2+ by competing with these intramolecular interactions.
FIGURE 5.
Peptides alter (3H)ryanodine binding to SR membranes. SR membranes (10 μg) were incubated with 5 nM (3H)ryanodine in 100 mM NaCl, 30 mM MOPS (pH 7.2) containing increasing CaCl2 concentration with or without 10 μM CaM, R4064–4210, or R3614–3643 for 16 h at room temperature. The samples were filtered through Whatman GF/F filters with five washes and the radioactivity determined by liquid scintillation counting. Control, •; CaM, ▪; R4064–4210, ▴; and R3614–3643, ▾.
R4064–4210 binds to CaM binding motifs
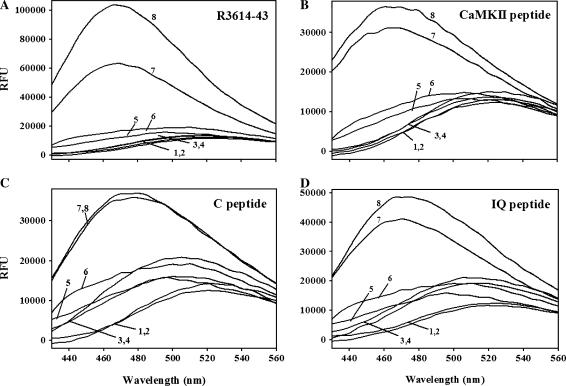
Since R4064–4210 is predicted to bind Ca2+ and interact with CaM binding motifs within RyR1, we asked whether it could bind to other CaM binding motifs. The CaM binding sites on voltage-dependent calcium channels and RyR1 are complex sites. In both types of protein, more than one motif has been implicated in CaM binding (22,25,27–30). We examined the ability of R4064–4210 to interact with synthetic peptides representing CaM-binding motifs from these channels. Also, to determine if this is a more general protein-protein interaction motif, we assessed the ability of R4064–4210 to bind to the CaM binding peptide from CaM kinase II (aa 281–309, Calbiochem). We used changes in ANSA fluorescence in the presence of R4064–4210 to assess the interactions. We found that ANSA fluorescence in the presence of R4064–4210 increased in the presence of CaM binding peptides, R3614–3643, calmodulin kinase II inhibitor peptide (CaMKII peptide), the CaV1.2 C-peptide, and the CaV1.2 IQ-peptide (Fig. 6), suggesting that all of these peptides could bind to the CaM-like region of RyR1 and the interactions again occurs at both high and low Ca2+ concentrations. Peptides that did not bind CaM had no effects on the ANSA fluorescence in the presence of R4064–4210 (data not shown). These interactions were confirmed by tryptophan fluorescence changes and pulldown assays with His-tagged R4064–4210 (data not shown).
FIGURE 6.
R4064–4210 binds to CaM binding motifs. R4064–4210 (0.5 μM) and ANSA (2 μM) were incubated in 30 mM MOPS (pH 7.2), 5 mM EGTA (low Ca2+), or 500 μM Ca2+ (high Ca2+) for 5 min at room temperature. CaM-binding peptides (5 μM, R3614–3643 peptide, A; CaMKII peptide, B; C-peptide, C; and IQ-peptide, D) were added and ANSA fluorescence emission spectra were collected from 430 nm to 560 nm with a 370-nm excitation wavelength. (Curve 1, ANSA + EGTA; curve 2, ANSA + Ca2+; curve 3, ANSA + peptide + EGTA; curve 4, ANSA + peptide + Ca2+; curve 5, ANSA + R4064–4210 + EGTA; curve 6, ANSA + R4064–4210 + Ca2+; curve 7, ANSA + R4064–4210 + peptide + EGTA; and curve 8, ANSA + R4064–4210 + peptide + Ca2+.)
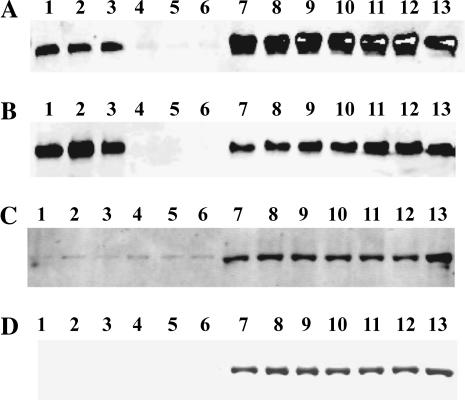
R4064–4210 binds to the Cav1.1 by pulldown analysis
If the R4064–4210 sequence can bind to the CaM binding site on Cav1.1, it should be able to pull down native Cav1.1. We tested this using His-tagged R4064–4210 and digitonin-solubilized T-tubule membranes containing the Cav1.1. Fig. 7 shows that R4064–4210 pulls down the Cav1.1 protein. Consistent with our previous findings, the pulldown of Cav1.1 by R4064–4210 does not require Ca2+ binding to either R4064–4210 or Cav1.1 (Fig. 7 A for low Ca2+ and Fig. 7 B for high Ca2+). To confirm that the pulldown is specific for Cav1.1, pulldown data of Na/K ATPase by R4064–4210 are also shown in Fig. 7. At either low Ca2+ (Fig. 7 C) or high Ca2+ condition (Fig. 7 D), R4064–4210 does not pulldown Na/K ATPase.
FIGURE 7.
R4064–4210 binds to the native Cav1.1. His-tagged R4064–4210 or the synthetic His-tag alone were incubated with Ni-charged chelating sepharose beads and digitonin solubilized T-tubule membranes in either low, <10 nM Ca2+ (A), or high, 1 mM Ca2+ (B). After centrifugation, the pellets and supernatants were electrophoresced on 10% SDS polyacrylamide gels. After transfer to Immobilon (Millipore, Billerica, MA), the Western blots were developed with an anti-α1 antibody from Affinity BioReagents. For comparison, Western blots were also developed with an anti-Na/K ATPase α1 antibody, with C for low Ca2+ and D for high Ca2+. (Lanes 1–3, pulldown with His-tagged R4064–4210; lanes 4–6, pulldown with His tag only; lanes 7–9, supernatant from the pulldown with His-tagged R4064–4210; lanes 10–12, supernatants from the pulldowns with His-tag only; and lane 13, starting T-tubule preparation for the pulldown assay.)
DISCUSSION
RyRs are activated by the binding of Ca2+ at μM concentrations, but are inhibited by higher concentrations (1,2). Our data suggest that an expressed fragment containing two putative E-F hands (R4064–4210) and a synthetic peptide (R3614–3643) that represents the CaM binding motif of RyR1 interfere with an interaction that regulates the response of RyR1 to Ca2+. These findings raise questions as to whether Ca2+ binding to sites between amino acids 4064 and 4210 is involved in the Ca2+ regulation of the channel and, if so, how? One possibility is that Ca2+ binding to R4064–4210 establishes which determinants within both the 3614–3643 CaM binding site and amino acids 4064–4210 are actually interacting, and as a result, the functional outcomes of the interaction. We have previously shown that Ca2+ binding to CaM causes an N-terminal shift in its interaction within amino acids 3614–3643 and converts CaM from an activator to an inhibitor of the channel (20,21). It is conceivable that a similar shift in the interaction of amino acids 4064–4210 with amino acids 3614–3643 could occur when Ca2+ binds to the two sites between amino acids 4064 and 4210. It is, however, also possible that Ca2+ binding to other sites within RYR1 alters the aa4064–4210 to aa3614–3643 interaction. The effects of R4064–4210 and R3614–3643 on the Ca2+ dependence of (3H)ryanodine binding supports the possibility that this intramolecular interaction is important for both Ca2+ activation and inhibition of the channel. The location of the Ca2+ activation and inhibition sites remain to be determined, but could conceivably be the two sites within amino acids 4064–4210. Scrambling the sequence of putative E-F hand 1 and E-F hand 2 within aa4064–4210 does not alter the functional responses to either depolarization or activators (caffeine or 4-CmC) when compared with wtRyR1 expressed in myotubes (19). The question is whether 4-CmC or caffeine can accurately assess alterations in the affinity of Ca2+ activation or inhibition sites. Another question is whether the presence of endogenous CaM (which we propose competes for the interaction of aa3614–3643 with aa4064–4210) can obscure apparent changes in Ca2+ activation or inhibition of RyR1. Fessenden et al. (36) found that RyR1 expressed in dyspedic 1B5 myotubes was activated by 4-CmC, whereas RyR3 was not. Given that RyR1 and RyR3 are both activated by Ca2+ with only an approximately twofold difference in Ca2+ affinity (37), but have the same apparent affinity for caffeine, one has to question whether altered responses to caffeine and 4-CmC can be used to indirectly assess RyR1 responses to Ca2+. In addition, caffeine and 4-CmC not only increase Ca2+ affinity of RyR1 but also alter the maximal activity. Additional data obtained by Fessenden et al. (18) with SR membranes argues that these two E-F hands play an important role in Ca2+ regulation of the channel. Scrambling of the sequence of EF1 altered the Ca2+ dependence of (3H)ryanodine binding, with respect to both enhancement and inhibition. Even more surprisingly, scrambling of EF2 abolished high-affinity (3H)ryanodine binding to membranes. Without a direct analysis of the binding of Ca2+ to these E-F hand mutants, these studies do not rule out E-F hands 1 and 2 as sites of Ca2+ regulation of the channel. The sequence from 4064 to 4210 binds Ca2+ and modulates the Ca2+ dependence of channel activity and we cannot, therefore eliminate the possibility that the Ca2+ sites within this region are regulatory sites.
If the region containing E-F-1 and E-F-2 represents a site at which Ca2+ regulates channel activity, the interaction of the CaM binding motif (R3614–3643) with this sequence suggests that CaM alters channel activity by interfering with these interactions in a Ca2+-dependent fashion, activating when it is in its apoCaM form and inhibiting in its Ca2+ bound form, both of which bind to R3614–3643. We suggest that CaM activates the channel at low Ca2+ and inhibits the channel at intermediate Ca2+ by competing with the intramolecular interactions between amino acids 4064–4210 and 3614–3643.
Other laboratories have also suggested that CaM might be altering RyR1 activity by interfering with an intramolecular interaction or by inducing a conformational change in the CaM binding site. Zhu et al. (38) demonstrated that a peptide representing the CaM binding site (amino acids 3614–3643) could either activate or inhibit RyR1, depending on the dose and the Ca2+ concentration. They proposed that the CaM binding site was an important modulatory site within RyR1. Consistent with this, Rodney et al. (39) showed that the R3614–3643 peptide injected in frog skeletal muscle fibers increased the occurrence of Ca2+ sparks in a dose-dependent manner, but not by displacing endogenous CaM. These studies also support the existence of an important intramolecular interaction. Gangopadhyay et al. (40) used an environment-sensitive fluorescent probe, 6-bromoacetyl-2-dimethylaminonaphthalene (badan), to study the interaction between badan-labeled calmodulin (CaM) and the CaM-binding peptide of the ryanodine receptor. They found that the interaction interface and the global conformation of the CaM-CaM binding peptide were altered by the binding of Ca2+ to CaM.
Other amino acids are also likely to contribute to the ability of CaM to regulate RyR1 activity. Yamaguchi et al. (41) created chimeras of RyR1 and RyR2 and found that five nonconserved amino acid residues (RyR1 aa3680 and 3682–3685, and RyR2 aa3647 and 3649–3652), by differentially affecting RyR helical probability, played a key role in the ability of CaM to inhibit RyR1.
We have previously proposed that the CaM binding sequence on RyR1 can interact with determinants on the Cav1.1 and that the CaM binding sequence on the Cav1.1 can bind to determinants on RyR1 (26). We now show that the expressed fragment of RyR1 from amino acids 4064–4210 can bind to the intact Cav1.1 and to peptides representing the CaM binding sites of both RyR1 and the Cav1.1 (both the C-peptide and the IQ-peptide). We propose that the CaM binding sites on RyR1 and the Cav1.1 are more general protein-protein interaction motifs that bind to regions that contain E-F hands. The significance of this for excitation-contraction coupling is that it may provides a unique mechanism for interactions between these two ion channels: the CaM binding site on the Cav1.1 binding to the RyR1 CaM-like domain and the RyR1 CaM binding site interacting with a CaM-like domain on the Cav1.1. Since R4064–4210 of RyR1 also interacts with the CaM binding site (R3614–3643) on RyR1 to stabilize the closed channel, the interaction with the carboxyterminal tail of the Cav1.1 α1-subunit might disrupt an inter- or intramolecular interaction, thereby regulating RyR1 activity.
Acknowledgments
The studies were supported in part by a grant from the Muscular Dystrophy Association, and National Institutes of Health grants No. AR41802 and No. AR44864 (to S.L.H.).
References
- 1.Liu, W., D. A. Pasek, and G. Meissner. 1998. Modulation of Ca2+-gated cardiac muscle Ca2+-release channel (ryanodine receptor) by mono- and divalent ions. Am. J. Physiol. 274:C120–C128. [DOI] [PubMed] [Google Scholar]
- 2.Meissner, G., E. Rios, A. Tripathy, and D. A. Pasek. 1997. Regulation of skeletal muscle Ca2+ release channel (ryanodine receptor) by Ca2+ and monovalent cations and anions. J. Biol. Chem. 272:1628–1638. [DOI] [PubMed] [Google Scholar]
- 3.Kretsinger, R. H. 1976. Calcium-binding proteins. Annu. Rev. Biochem. 45:239–266. [DOI] [PubMed] [Google Scholar]
- 4.Kretsinger, R. H. 1976. Evolution and function of calcium-binding proteins. Int. Rev. Cytol. 46:323–393. [DOI] [PubMed] [Google Scholar]
- 5.Takeshima, H., S. Nishimura, T. Matsumoto, H. Ishida, K. Kangawa, N. Minamino, H. Matsuo, M. Ueda, M. Hanoaka, T. Hirose, and S. Numa. 1989. Primary structure and expression from complementary DNA of skeletal muscle ryanodine receptor. Nature. 339:439–445. [DOI] [PubMed] [Google Scholar]
- 6.Nakai, J., T. Imagawa, Y. Hakamat, M. Shigekawa, H. Takeshima, and S. Numa. 1990. Primary structure and functional expression from cDNA of the cardiac ryanodine receptor/calcium release channel. FEBS Lett. 271:169–177. [DOI] [PubMed] [Google Scholar]
- 7.Hakamata, Y., J. Nakai, H. Takeshima, and K. Imoto. 1992. Primary structure and distribution of a novel ryanodine receptor/calcium release channel from rabbit brain. FEBS Lett. 312:229–235. [DOI] [PubMed] [Google Scholar]
- 8.Treves, S., P. Chiozzi, and F. Zorzato. 1993. Identification of the domain recognized by anti-(ryanodine receptor) antibodies which affect Ca2+-induced Ca2+ release. Biochem. J. 291:757–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, S. R., L. Zhang, and D. H. MacLennan. 1992. Characterization of a Ca2+ binding and regulatory site in the Ca2+ release channel (ryanodine receptor) of rabbit skeletal muscle sarcoplasmic reticulum. J. Biol. Chem. 267:23318–23326. [PubMed] [Google Scholar]
- 10.Chen, S. R., L. Zhang, and D. H. MacLennan. 1993. Antibodies as probes for Ca2+ activation sites in the Ca2+ release channel (ryanodine receptor) of rabbit skeletal muscle sarcoplasmic reticulum. J. Biol. Chem. 268:13414–13421. [PubMed] [Google Scholar]
- 11.Zorzato, F., J. Fujii, K. Otsu, M. Phillips, N. M. Green, F. A. Lai, G. Meissner, and D. H. MacLennan. 1990. Molecular cloning of cDNA encoding human and rabbit forms of the Ca2+ release channel (ryanodine receptor) of skeletal muscle sarcoplasmic reticulum. J. Biol. Chem. 265:2244–2256. [PubMed] [Google Scholar]
- 12.Hayek, S. M., X. Zhu, M. B. Bhat, J. Zhao, H. Takeshima, H. H. Valdivia, and J. Ma. 2000. Characterization of a calcium-regulation domain of the skeletal-muscle ryanodine receptor. Biochem. J. 351:57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du, G. G. and D. H. MacLennan. 1999. Ca2+ inactivation sites are located in the COOH-terminal quarter of recombinant rabbit skeletal muscle Ca(2+) release channels (ryanodine receptors). J. Biol. Chem. 274:26120–26126. [DOI] [PubMed] [Google Scholar]
- 14.Chen, S. R. W., and D. H. MacLennan. 1994. Identification of calmodulin, Ca2+, and ruthenium red-binding domains in the Ca2+ release channel (ryanodine receptor) of rabbit skeletal muscle sarcoplasmic reticulum. J. Biol. Chem. 269:22698–22704. [PubMed] [Google Scholar]
- 15.Xiong, H., X. Feng, L. Gao, L. Xu, D. A. Pasek, J. H. Seok, and G. Meissner. 1998. Identification of a two EF-hand Ca2+ binding domain in lobster skeletal muscle ryanodine receptor/Ca2+ release channel. Biochemistry. 37:4804–4814. [DOI] [PubMed] [Google Scholar]
- 16.Hamada, T., Y. Sakube, J. Ahnn, H. Kim, and H. Kagawa. 2002. Molecular dissection, tissue localization and Ca2+ binding of the ryanodine receptor of Caenorhabditis elegans. J. Mol. Biol. 324:123–135. [DOI] [PubMed] [Google Scholar]
- 17.Fessenden, J. D., L. Chen, Y. Wang, C. Paolini, C. Franzini-Armstrong, P. D. Allen, and I. N. Pessah. 2001. Ryanodine receptor point mutant E4032A reveals an allosteric interaction with ryanodine. Proc. Natl. Acad. Sci. USA. 98:2865–2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fessenden, J. D., W. Feng, I. N. Pessah, and P. D. Allen. 2004. Mutational analysis of putative calcium binding motifs within the skeletal ryanodine receptor isoform, RyR1. J. Biol. Chem. 279:53028–53035. [DOI] [PubMed] [Google Scholar]
- 19.Tripathy, A., L. Xu, G. Mann, and G. Meissner. 1995. Calmodulin activation and inhibition of skeletal muscle Ca2+ release channel (ryanodine receptor). Biophys. J. 69:106–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodney, G. G., B. Y. Williams, G. M. Strasburg, K. Beckingham, and S. L. Hamilton. 2000. Regulation of RyR1 activity by Ca2+ and calmodulin. Biochemistry. 39:7807–7812. [DOI] [PubMed] [Google Scholar]
- 21.Rodney, G. G., C. P. Moore, B. Y. Williams, J. Z. Zhang, J. Krol, S. E. Pedersen, and S. L. Hamilton. 2001. Calcium binding to calmodulin leads to an N-terminal shift in its binding site on the ryanodine receptor. J. Biol. Chem. 276:2069–2074. [DOI] [PubMed] [Google Scholar]
- 22.Moore, C. P., J.-Z. Zhang, and S. L. Hamilton. 1999. A role for Cysteine 3635 of RyR1 in redox modulation and calmodulin binding. J. Biol. Chem. 274:36831–36834. [DOI] [PubMed] [Google Scholar]
- 23.Rodney, G. G., J. Krol, B. Williams, K. Beckingham, and S. L. Hamilton. 2001. The carboxy-terminal calcium binding sites of calmodulin control calmodulin's switch from an activator to an inhibitor of RyR1. Biochemistry. 40:12430–12435. [DOI] [PubMed] [Google Scholar]
- 24.Xiong, L. W., R. A. Newman, G. G. Rodney, O. Thomas, J. Z. Zhang, A. Persechini, M. A. Shea, and S. L. Hamilton. 2002. Lobe-dependent regulation of ryanodine receptor Type 1 by calmodulin. J. Biol. Chem. 277:40862–40870. [DOI] [PubMed] [Google Scholar]
- 25.Zhang, H., J. Z. Zhang, C. I. Danila, S. L. Hamilton, and A. Noncontiguous. 2003. Inter-subunit binding site for calmodulin on the skeletal muscle Ca2+ release channel. J. Biol. Chem. 278:8348–8355. [DOI] [PubMed] [Google Scholar]
- 26.Sencer, S., R. V. Papineni, D. B. Halling, P. Pate, J. Krol, J. Z. Zhang, and S. L. Hamilton. 2001. Coupling of RyR1 and L-type calcium channels via calmodulin binding domains. J. Biol. Chem. 276:38237–38241. [DOI] [PubMed] [Google Scholar]
- 27.Pate, P., J. Mochca-Morales, Y. Wu, J. Z. Zhang, G. G. Rodney, I. I. Serysheva, B. Y. Williams, M. E. Anderson, and S. L. Hamilton. 2000. Determinants for calmodulin binding on voltage-dependent Ca2+ channels. J. Biol. Chem. 275:39786–39792. [DOI] [PubMed] [Google Scholar]
- 28.Pitt, G. S., R. D. Zuhlke, A. Hudmon, H. Schulman, H. Reuter, and R. W. Tsien. 2001. Molecular basis of calmodulin tethering and Ca2+-dependent inactivation of L-type Ca2+ channels. J. Biol. Chem. 276:30794–30802. [DOI] [PubMed] [Google Scholar]
- 29.Soldatov, N. M., M. Oz, K. A. O'Brien, D. R. Abernethy, and M. Morad. 1998. Molecular determinants of L-type Ca2+ channel inactivation. segment exchange analysis of the carboxyl-terminal cytoplasmic motif encoded by exons 40–42 of the human α1C-subunit gene. J. Biol. Chem. 273:957–963. [DOI] [PubMed] [Google Scholar]
- 30.Zühlke, R. D., and H. Reuter. 1998. Ca2+-sensitive inactivation of L-type Ca2+ channels depends on multiple cytoplasmic amino acid sequences of the α1C-subunit. Proc. Natl. Acad. Sci. USA. 95:3287–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hawkes, M. J., M. Díaz-Muñoz, and S. L. Hamilton. 1989. A procedure for the purification of the ryanodine receptor from skeletal muscle. Membr. Biochem. 8:133–145. [DOI] [PubMed] [Google Scholar]
- 32.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the folin phenol reagent. J. Biol. Chem. 193:265–275. [PubMed] [Google Scholar]
- 33.Venyaminov, S. Y., and J. T. Yang. 1996. Circular Dichroism and the Conformational Analysis of Biomolecules. G. D. Fasman, editor. Plenum Publishing, New York. 69–108.
- 34.Muesing, R. A., and T. Nishida. 1971. Disruption of low- and high-density human plasma lipoproteins and phospholipid dispersions by 1-anilinonaphthalene-8-sulfonate. Biochemistry. 10:2952–2962. [DOI] [PubMed] [Google Scholar]
- 35.Chu, A., M. Diaz-Munoz, M. J. Hawkes, K. Brush, and S. L. Hamilton. 1990. Ryanodine as a probe for the functional state of the skeletal muscle sarcoplasmic reticulum Ca2+ release channel. Mol. Pharmacol. 37:735–741. [PubMed] [Google Scholar]
- 36.Fessenden, J. D., C. F. Perez, S. Goth, I. N. Pessah, and P. D. Allen. 2003. Identification of a key determinant of ryanodine receptor Type 1 required for activation by 4-chloro-m-cresol. J. Biol. Chem. 278:28727–28735. [DOI] [PubMed] [Google Scholar]
- 37.Murayama, T., and Y. Ogawa. 2004. RyR1 exhibits lower gain of CICR activity than RyR3 in the SR: evidence for selective stabilization of RyR1 channel. Am. J. Physiol. Cell Physiol. 287:C36–C45. [DOI] [PubMed] [Google Scholar]
- 38.Zhu, X., J. Ghanta, J. W. Walker, P. D. Allen, and H. H. Valdivia. 2004. The calmodulin binding region of the skeletal ryanodine receptor acts as a self-modulatory domain. Cell Calcium. 35:165–177. [DOI] [PubMed] [Google Scholar]
- 39.Rodney, G. G., G. M. Wilson, and M. F. Schneider. 2005. A calmodulin binding domain of RyR increases activation of spontaneous Ca2+ sparks in frog skeletal muscle. J. Biol. Chem. 280:11713–11722. [DOI] [PubMed] [Google Scholar]
- 40.Gangopadhyay, J. P., Z. Grabarek, and N. Ikemoto. 2004. Fluorescence probe study of Ca2+-dependent interactions of calmodulin with calmodulin-binding peptides of the ryanodine receptor. Biochem. Biophys. Res. Commun. 323:760–768. [DOI] [PubMed] [Google Scholar]
- 41.Yamaguchi, N., L. Xu, K. E. Evans, D. A. Pasek, and G. Meissner. 2004. Different regions in skeletal and cardiac muscle ryanodine receptors are involved in transducing the functional effects of calmodulin. J. Biol. Chem. 279:36433–36439. [DOI] [PubMed] [Google Scholar]