Abstract
Tetraspanins are a superfamily of transmembrane proteins implicated in cellular development, motility, and activation through their interactions with a large range of proteins and with specific membrane microdomains. The complete three-dimensional structure of the tetraspanin CD81 has been predicted by molecular modeling and from the crystallographic structure of the EC2 large extracellular domain. Periodicity of sequence conservation, homology modeling, secondary structure prediction, and protein docking were used. The transmembrane domain appears organized as a four-stranded left-handed coiled coil directly connecting to two helices of the EC2. A smaller extracellular loop EC1 contains a small largely hydrophobic β-strand that packs in a conserved hydrophobic groove of the EC2. The palmitoylable intracellular N-terminal segment forms an amphipathic membrane-parallel helix. Structural variability occurs mainly in an hypervariable subdomain of the EC2 and in intracellular regions. Therefore, the variable interaction selectivity of tetraspanins originates both from sequence variability within structurally conserved domains and from the occurrence of small structurally variable domains. In CD81 and other tetraspanins, the numerous membrane-exposed aromatic residues are asymmetrically clustered and protrude on one side of the transmembrane domain. This may represent a functional specialization of these two sides for interactions with cholesterol, proteins, or membrane microdomains.
INTRODUCTION
Tetraspanins constitute a superfamily of transmembrane glycoproteins that are involved in the regulation of cellular development, proliferation, activation, and motility. The best characterized members include CD81, CD9, CD53, CD82, CD151, CD37, and CD63. Their role is mediated by their ability to interact with other proteins such as integrins, coreceptor molecules, major histocompatibility complex antigens, and cytoplasmic kinases. Current hypotheses view tetraspanins as “molecular facilitators” that simultaneously interact with and bring into close proximity specific proteins (for reviews see Hemler (1) and Levy and Shoham (2)). This leads to the formation of large membrane complexes that may become associated with lipid rafts and other microdomains (3–5) and the cytoskeleton (4). These complexes are involved in specific activation, transduction, or signaling processes. Tetraspanins themselves undergo homologous and heterologous associations, which may form the basis of a tetraspanin web (6,7). Several tetraspanins are also involved in binding of viruses (8–10). CD81 is among the most studied tetraspanin with implications in numerous cellular processes, among which are B- and T-cell activation (2,11). It also acts as a receptor for HCV (8).
Tetraspanins are characterized by four transmembrane segments (TM1-4) linked by one short extracellular (EC1), one short intracellular (IC), and one large extracellular (EC2) stretch (12). Tetraspanins also possess a number of conserved residues including a glycine and four to eight cysteines located on the EC2 stretch. Also partially conserved is the so-called tetraspanin signature located in the TM2/IC/TM3 region. Distant members of the tetraspanin family also exist, such as RDS/ROM or uroplakins (13,14) that share the same arrangement and EC2-conserved residues but lack the signature.
Elucidation of the three-dimensional (3D) structure of tetraspanins is essential to the understanding of their function. Some important progress has been made recently concerning the large extracellular EC2 region that bears an important part of the interaction capability of tetraspanins (12). Kitadokoro et al. (15) reported the crystallographic structure of a soluble form of the tetraspanin CD81 EC2 domain. The structure appears mushroom shaped and consists of a five-helix bundle stabilized by two disulfide bridges involving a ubiquitous CCG motif and other conserved cysteines. Seigneuret et al. (16) have used molecular modeling to predict the structure of the EC2 over the whole superfamily. The structural features of the CD81 EC2 are conserved only partially among tetraspanins. The EC2 is organized in two subdomains. The first, membrane proximal, subdomain has a structurally conserved fold among tetraspanins, involving three helices (helices A, B, and E). The second subdomain is sequentially inserted within the conserved subdomain and located on its top. It is extremely variable in size and secondary structure among tetraspanins. The occurrence of two or more key disulfide bridges and other invariant residues leads to a conserved relative topology of both subdomains.
Although, the EC2 concentrates part of the interaction potential of tetraspanins, numerous specific associations have also been mapped in the transmembrane and intracellular regions (2,12,17–20). Knowledge of the 3D structure of these regions is therefore necessary to fully understand the structure-activity relationships of tetraspanins. Recently Min et al. (14) have used cryo-EM with a resolution of 7–10 Å to visualize the uroplakin particle, a structure involving the tetraspanin uroplakin 1a/b and another protein. In this work, prediction of the complete structure of the tetraspanin CD81 has been attempted. Although the experimental crystallographic structure has been used for the EC2, molecular modeling has been used for the other regions.
EXPERIMENTAL PROCEDURES
General modeling procedures
All amino acid sequences were obtained from the SWISSPROT/TrEMBL database. Protein coordinate file modifications and structural database searches were done with Whatif (21). Homology modeling was performed with the Modeller 6.2 program (22,23). Energy minimization of structures was done with CHARMM (24). Packing quality in the modeled structures was measured as residue packing values with the OS program (25). Interior cavities were measured with Grasp (26). Residue accessibility was measured with Naccess (27). Structure quality was evaluated with Whatif and Procheck (28). Protein structure visualization and representation were done with Molmol (29).
Multiple sequence alignments and analyses of transmembrane regions
An alignment of 204 tetraspanin sequences corresponding to 107 tetraspanin types was generated with the ClustalX program (30) and improved manually. Delimitation of the CD81 transmembrane regions were performed using HMMTOP2 (31) and Perscan (32,33). Calculations of hydrophobic and variability periodicity profiles and moments were done with Perscan. For each transmembrane region or transmembrane/EC2 helix region, only sequences of the alignment having >35% identity (25% for TM4) with CD81 in the region were used.
Construction of the transmembrane coiled coil template
To build the transmembrane domain of CD81, adequate coiled coil templates were first constructed. All available structures of transmembrane proteins were searched for transmembrane coiled coil motifs using the Socket program (34). A relatively regular fully antisymmetric left-handed coiled coil arrangement of four transmembrane helices was found in cytochrome oxidase subunit III (residues 128–256 of Protein Data Bank (PDB) file 2OCC, chain C) (35). A quasiideal fully antisymmetric coiled coil was built by covalently concatenating in a head-to-tail fashion identical copies of the middle region of this four-helix domain (residues 135–142, 167–174, 205–212, 241–248), which is the most regular portion. Backbone stereochemical imperfections were corrected and all side chains were then transformed to alanine with Whatif. Because no semiantisymmetric left-handed coiled coil was found in existing transmembrane protein structures, a template corresponding to such fold was built from the fully antisymmetric template by swapping two adjacent helices. Each of the two helices was first moved perpendicularly to the coiled coil axis and rotated along an axis parallel to the coil axis so that the α-carbons of its a core residues (respectively its d core residues) were best fitted to the initial position of those of the d core residues (respectively the a core residues) of the other helix. Then the rotational position of the two swapped helices were further adjusted visually so that the positions of the α-carbons of their core residues were, mutually those found for antisymmetric four-stranded left-handed coiled coils and, with regard to the α-carbons of core residues of each other adjacent helix, those of symmetric four-stranded left-handed coiled coils as in PDB file 1GCL (36).
Initial modeling and analysis of the CD81 transmembrane domain
Homology modeling of the six candidate folds of the CD81 transmembrane domain was performed using either the fully antiparallel or the semiantiparallel coiled coil templates. For each fold an alignment between the sequence of the templates and the sequence of the CD81 transmembrane domain (i.e., of trial transmembrane helical regions) was built so that the a, d, e, and g positions of the coiled coil corresponded to those identified on CD81 by sequence conservation analysis. Registering of the four helices was unambiguous due to the necessity of having these regions at similar levels. One-hundred structures were generated for each case using the model routine of Modeller and 10 structures corresponding to the lowest values of the Modeller objective function were selected and submitted to energy minimization using progressively decreasing harmonic restraints on backbone atoms. Potential hydrogen bonds were detected interactively using the SwisspdbViewer program (37) by trying all possible combinations of rotamers of polar residue pairs located at suitable proximity.
Construction and positioning of EC1 domain
Secondary structure predictions of the EC1 were done with the Jnet method (38) using multiple sequence alignments of each tetraspanin subfamilies. A model of the β-strand region of the EC1, NLLYLE, was obtained by searching the Whatif fragment database for similar segments. A segment with the sequence NLIYLA (residues 118–123 of PDB file 1IS6), was transformed to NLLYLE using the mutate function of Whatif. An ab initio NLLYLE β-strand was also constructed with canonical dihedral angle values. Soft docking of the EC1 β-strand on the CD81 EC2 crystallographic structure (PDB file 1G8Q (15)) was performed with both fragments using the GRAMM program (39). High-resolution docking of the database β-strand fragment to the EC2 was performed with the FTDOCK program (40). Docking solutions were energy-ranked and filtered to select those corresponding to the β-strand position determined by soft docking and to its expected orientation. The five best solutions were refined with the MULTIDOCK program (41) and analyzed for packing quality and steric clashes. The best model was slightly refined manually by adjusting dihedral angles to remove remaining clashes and optimize packing and hydrogen bonding.
Complete modeling of the CD81 structure
The modeling of the CD81 complete structure was also performed by homology. A full CD81 template was constructed by linking the fully antisymmetric transmembrane coiled coil template to the docked EC1-EC2 complex with helical continuity between respectively TM3 and helix A of the EC2 and TM4 and helix E of the EC2. The first stage of modeling was performed using the model routine of Modeller with the EC2 constrained to its crystallographic conformation. Hydrogen bonds between transmembrane helices were enforced by imposing conformational and interesidue distance restraints. The best structure was selected from 500 simulations on three criteria: 1), low value of the convergence function; 2), compliance with imposed restraints; 3), minimum steric clashes and packing voids. Refinement of loops was done simultaneously for EC1 loops (residues 39–42 and 49–55) and separately for the IC loop (residues 84–87) with the loop Modeller routine. In both cases, a structure with low convergence function and corresponding to the majoritary conformation for each loop was selected from 100 simulations. The N-terminal regions was manually given a helical conformation perpendicular to the transmembrane axis such that cysteine and hydrophobic side chains face toward the extracellular side and polar side chain toward the intracellular side. The C-terminal region was given a relatively extended conformation also with such side-chain orientation. The structure was then submitted to gentle energy minimization with fixed constraints on hydrogen-bonded side chains and progressively decreasing residual harmonic constraints on all other atoms. The final coordinate file has been deposited at the PDB (entry 2AVZ). Stereochemical imperfections and steric clashes are very limited (see the header of the PDB file for details). Packing quality is such that six residues over 236 have a low packing score as measured with Whatif and two packing voids with 41 and 48 Å3 internal volumes are found.
RESULTS
Identification of transmembrane helical regions
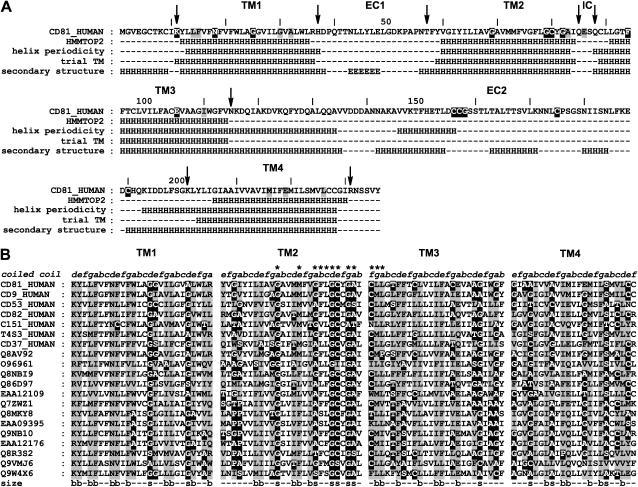
The first task of the modeling procedure of the transmembrane domain of human CD81 was to identify the limits of the four transmembrane helices in the sequence. Complete spanning of the lipid bilayer (∼40 Å) by a transmembrane helix requires an at least 26-residue length (42). The results obtained on CD81 with HMMTOP2 (30), which has recently been evaluated as one of the most efficient transmembrane segment determination methods (43,44) are shown in Fig. 1 A. A model-independent method was also used, which is valid for both transmembrane and water-exposed helices and is based on the detection of regions with helical periodicity of residue variability in a multiple sequence alignment (32,33). Both methods yielded relatively similar limits for transmembrane helices, which ranged between 24 and 27 residues. The results of the second method also indicate that for TM3 and TM4, the periodicity extends beyond transmembrane regions to the EC2. This suggests that transmembrane helices in CD81 completely span the bilayer and that TM3 and TM4 extend significantly to the extracellular aqueous phase (see below). To model the transmembrane domain of CD81, it was therefore decided to first assign trial “transmembrane helical regions”, i.e., the portions of the helices that span the bilayer, leaving the exact determination of the helix ends to later modeling stages. Each 26-residue transmembrane span was positioned taking into account the results of the two prediction methods. This corresponds to positions that are in between polar or charged residues located in the nearby aqueous regions (Fig. 1 A).
FIGURE 1.
Secondary structure and size conservation in the transmembrane regions of CD81. (A) Location of transmembrane helical regions and other regular secondary structure in the human CD81 sequence (H, helix; E, extended). (First row) Human CD81 sequence, positions corresponding to highly (>90%) or significantly (>75%) conserved residues of tetraspanin are, respectively, highlighted in black and gray; arrows indicate polar or charged residues lining the transmembrane regions. (Second row) Transmembrane helical regions obtained using HMMTOP2. (Third row) Transmembrane helical regions obtained with Perscan with an alignment of 41 tetraspanin sequences having >35% identity with CD81 in the transmembrane domain. (Fourth row) Starting trial 26-residue transmembrane helical regions used for the modeling study. (Fifth row) Final secondary structure of CD81 obtained from molecular modeling and the EC2 crystallographic structure. (B) Size conservation at the coiled coil core residue positions of the transmembrane helical regions of tetraspanins. Small residues (G, A, S, T, C) are highlighted in black and bulky residues (I, L M, V, W, Y, F, E, Q, N, K, R) are highlighted in gray. Only core residues (a, d, e, g) have been shaded for clarity. (First row) Assignment of coiled coil positions. (Last row) Size-conserved positions (s, small; b, bulky). Twelve transmembrane residues belonging to the tetraspanin signature are indicated by asterisks.
Identification of transmembrane bundle type and interior residues
In the transmembrane domain of membrane proteins, exterior residue facing the bilayer are on the average more variable and hydrophobic than residues located in the protein interior (32,33,45–47). This gives rise to a characteristic periodicity of conservation and hydrophobicity along the sequence that can be used to determine the helical pitch and to identify the interior helix side. Because tetraspanins constitute a superfamily, there are two types of conserved or homologous residue sequence positions (homologous means that the corresponding sequence position is occupied by residues of the same chemical type): 1), positions that are conserved or homologous for the whole superfamily; 2), positions that are always conserved or homologous within sequences of each tetraspanin type but different from one type to the other. In an ensemble of 204 tetraspanin sequences, transmembrane segments could be unambiguously aligned but a significant sequence heterogeneity was found. Respectively, 105, 125, 118, and 52 sequences had >30% local identity with human CD81 for TM1-4, respectively.
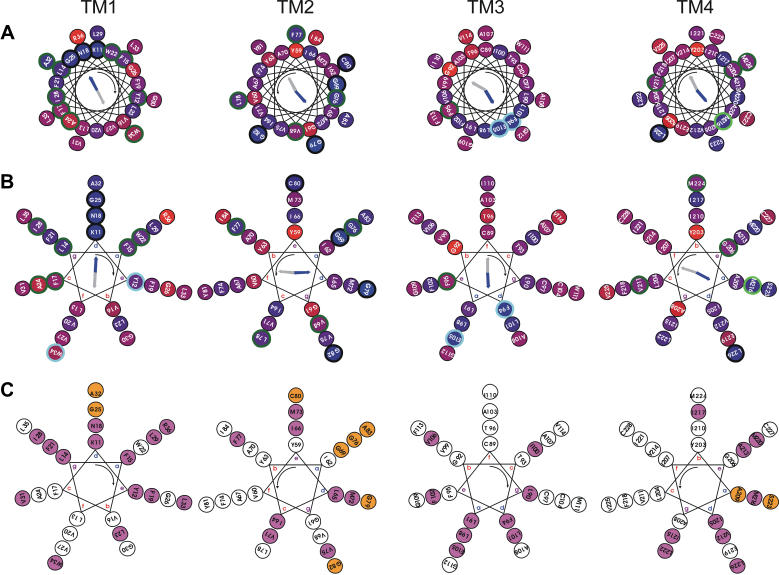
Fig. 2 shows the mapping on helical wheels of residue conservation for the four transmembrane helical regions of CD81 together with the calculated hydrophobic and conservation moments. Two values of helical pitch were considered: 3.6 (Fig. 2 A) and 3.5 (Fig. 2 B) residues per turn. This corresponds, respectively, to canonical straight α-helices and left-handed coiled coils. Examination of the latter case was suggested by numerous recent evidences of a left-handed coiled coil organization of adjacent transmembrane helices in the structures of membrane proteins (34,48–52). The packing of coiled coils involves specific core residue positions termed a and d for the more internal and e and g for the more external with repetitive spacing in the sequences (heptad repeats) (53,54). Fig. 2 shows that both helix models yield a clear periodicity of conservation: in both cases conserved and homologous residues are mainly segregated in one sector of each helix, which corresponds to the direction of the conservation moment and is opposite to that of the hydrophobic moment. This confirms that the transmembrane domain of CD81 indeed consists of four helices that are organized as an approximately square bundle. Helix interfaces correspond to the more conserved and hydrophilic sectors. In agreement with the observation of Liu et al. (52), the hydrophobic moment appears as a faithful reporter of the lipid-exposed helix sides, when calculated over a superfamily. Fig. 2 also indicates that for each helix, the coiled-coil model yields a more clearcut segregation of conserved and homologous residues in a narrower helix sector. Furthermore, for this model, there are two adjacent periodicity positions in which the most conserved residues are concentrated and that correspond to the direction of the conservation moment and are opposite to that of the hydrophobic moment. This is what is expected for the a and d core residue positions of a transmembrane left-handed coiled coil. This strongly suggests that CD81 transmembrane helical regions are organized as a bundle that corresponds or at least is close to that of a four-stranded left-handed coiled coil. Another argument is presented in Fig. 2 C. Helix interactions in soluble (53,54) or transmembrane (34,48,52,55) coiled coils are known to occur as knobs into hole interactions between side chains of residues located at helix positions a/d and e/g. The importance of residue side-chain volume conservation among homologous proteins in such packing has been emphasized (51–53). Fig. 2 C displays the helical wheel representation of side-chain volume conservation for a left-handed coiled coil. Helix positions in which small and bulky residues are conserved are indicated. The helical positions with conserved size are mainly the same, which are suggested above on the basis of residue type conservation to be the a/d and e/g positions of the proposed CD81 transmembrane coiled coil. There is a definite size conservation of small residues (mainly glycine and alanine) at positions a and d. The importance of such residues in promoting tight packing of transmembrane coiled coil helices has previously been emphasized (51,52,55,56). Such size conservation of specific core residues in transmembrane helix sequences is a further argument in favor of a four-stranded left-handed coiled coil model for the organization of the CD81 transmembrane domain. Fig. 1 B shows the pattern of core residue size conservation for representative sequences. Significantly, the so-called “tetraspanin signature”, for nine of its 13 positions, actually correspond to a size-conservation pattern of core residues in TM2 and TM3. Tetraspanin sequences with local identity in each transmembrane region lower than ∼30% were found to deviate locally from such conservation properties. This may arise from correlated mutations (57) in the superfamily in which concerted residue volume variations occur.
FIGURE 2.
Helical wheel representation of residue types and size conservation in CD81 transmembrane regions. (A) Representation of residue type conservation using a 3.6 residue-per-turn helical pitch (α-helix). The conservation of the residue position is indicated by the inner color of the circles in a linear red to blue scale with blue meaning 100% conserved. Residue positions that are conserved or homologous in all sequences are, respectively, circled in black and light blue. Residues positions that are variable among all sequences but conserved or homologous in all groups of sequences corresponding to a single tetraspanin species are, respectively, circled in dark and light green. The threshold for residue conservation or homology is 90%. Considered homologous residue families are: positive (H, K, R), negative-amidated (D, E, N, Q), hydroxylated (S, T), and aromatic (F, Y, W). The calculated conservation and hydrophobic moments are indicated as blue and gray arrows, respectively. (B) Same as panel A but using a 3.5 residue-per-turn helical pitch (coiled coil). (C) Representation of residue size conservation using a 3.5 residue-per-turn helical pitch (coiled coil). Residue positions that are conservatively occupied by a bulky (I, L M, V, W, Y, F, E, Q, N, K, R) and a small (G, A, S, T, C) side chain are, respectively, filled in magenta and orange. The threshold for size conservation is 95%. For calculation of conservation properties of TM1-4, local alignments of 81, 81, 88, and 64 sequences having 35, 35, 35, and 25% local identity with human CD81 were, respectively, used for each TM region. All helices are viewed from the intracellular side and the helix rotation sense is indicated by the curved arrow. For coiled coils (3.5 residues per turn) the canonical residue positions a–d are indicated.
Relative topography of the transmembrane and EC2 domains
In CD81, two helices of the transmembrane domain, namely TM3 and TM4 are sequentially adjacent to two helices of the EC2 domain, respectively, helices A and E (15), which are roughly parallel in the EC2 structure. This raises the question of whether there is a continuity of the helical conformation or a nonregular loop conformation between the transmembrane and EC2 helices. A strong indication of helical continuity comes from the examination of the helical periodicity of hydrophobicity and conservation in this region. Such an approach has already been performed in photosynthetic reaction centers (58). Both soluble and transmembrane surface helices are expected to display a sequence periodicity of conservation and hydrophobicity. If a transmembrane and a soluble helix are separated by a nonordered region, a drop of both periodicities in the sequence is expected. If there is a continuous helix, when it emerges from the bilayer, conserved residues remain on the same side toward the protein interior whereas the more hydrophobic side in contact with lipids is continued by a more hydrophilic side in contact with the aqueous phase (Fig. 3 A). One therefore expects a continuous conservation periodicity profile and a drop of hydrophobicity periodicity profile. As shown in Fig. 3 B, this is the latter situation that is observed both in the TM3-helix A, and helix E-TM4 regions. This strongly suggests the presence of continuous helices between the transmembrane domain and the EC2.
FIGURE 3.
Characterization of the continuity between transmembrane and EC2 helices. (A) Principle of the method (58). A transmembrane bundle is depicted in which a transmembrane helix in continuity with a soluble helix is highlighted. The helix is in contact with the protein interior and the external medium in both environments. When the helix emerges from the bilayer, conserved (nonhatched) and variable (hatched) residues, respectively, remain on the interior and exterior side of the helix whereas the more hydrophobic residues (dark gray) and the more hydrophilic (light gray) switch to opposite sides. This yields a continuous conservation periodicity profile and a drop of hydrophobicity periodicity profile. (B) Variation in the index of the helical (3.6 residue-per-turn) periodicity for sequence conservation (solid line) and sequence hydrophobicity (dotted line). (Top) Region connecting TM3 to EC2 helix A. (Bottom) Region connecting EC2 helix E to TM4. The periodicity analysis was done with the Perscan software with alignments of, respectively, 47 and 39 sequences with >35% homology with CD81 in the corresponding regions. The horizontal dotted line corresponds to the limit of significant periodicity. Identical results were obtained when a pitch of 3.5 residue-per-turn was used.
Arrangement of helices in the CD81 transmembrane bundle
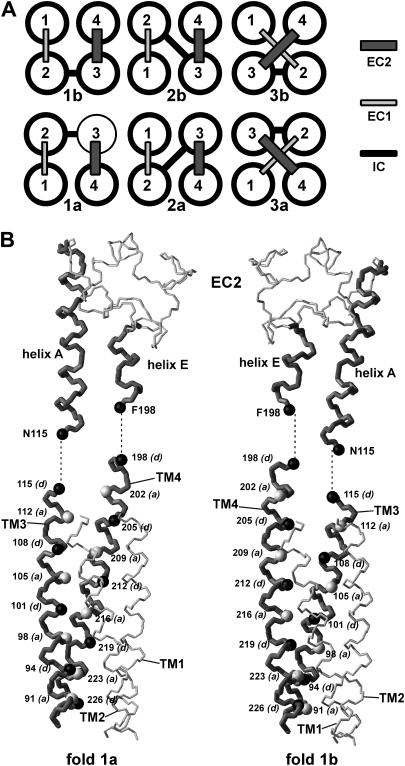
The above results suggest that the transmembrane domain of CD81 can be modeled as a square four-stranded left-handed coiled coil. There are six possible such square arrangements corresponding to six permutations of the four transmembrane helices and therefore to six configurations of the soluble segments IC, EC1, and EC2 (Fig. 4 A). These constitute three pairs of pseudomirror images. The first pair (1a and 1b) corresponds to a fully antiparallel left-handed coiled coil (i.e., all adjacent helices are antiparallel) whereas the two others (2a/b and 3a/b) correspond to a semiantiparallel left-handed coiled coil (i.e., two sets of adjacent helices are parallel and two sets are antiparallel).
FIGURE 4.
Possible folds of a square four-helix bundle and discrimination between folds 1a and 1b. (A) The six possible folds obtained by permutation of helices. (B) Backbone representation of the EC2 and its connectivity with the modeled transmembrane domain for folds 1a (left) and 1b (right). Transmembrane helices TM3 and TM4 and EC2 helices A and E are drawn in dark gray and thicker bonds. The Cα atoms of the internal coiled coil core residues a and d are, respectively, represented in light gray and black and labeled.
To discriminate between the different folds, CD81 transmembrane domain structures corresponding to each fold were modeled by homology, using fully antiparallel or semiantiparallel transmembrane left-handed coiled coil templates (see “Experimental Procedures”). A first criteria to select the correct fold was to investigate the possibilities of direct connectivity between transmembrane and EC2 helices. This made it possible to eliminate readily transmembrane folds 3a and 3b. Indeed, in these two folds TM3 and TM4 helices are not adjacent. The measured minimum distance between helix axes of TM3 and TM4 in the modeled 3a and 3b folds is 13.8 Å. On the other hand, in the EC2, helices A and E are in contact and have a 10.2-Å interaxis distance. It is therefore not possible to connect the EC2 helices to TM3 and TM4 in the framework of transmembrane folds 3a and 3b.
In all four other folds, helices TM3 and TM4 are adjacent and in close contact. The relative position of TM3 and TM4 is similar in folds 1a and 2a, as in folds 1b and 2b. A more detailed analysis of the connectivity with the EC2 can be used to discriminate between these two positions. Fig. 4 B compares the connectivity of the EC2 helices A and E with TM3 and TM4 in the framework of folds 1a and 1b. In fold 1a, the TM3 and TM4 interface mainly involves the a core residues with more distant d residues, whereas the opposite occurs in fold 1b. With fold 1a, TM3 and TM4 can be readily connected to helices A and E of EC2 while matching both the assignment of core a and d residues and the interhelix residue distance (the distance between the α-carbons of N115 and F198 is, respectively, 10.8 and 11.4 Å in the EC2 and in the transmembrane fold 1a). On the other hand, there is no connectivity between TM3, TM4, and the EC2 and the coiled coil corresponding to configuration 1b that satisfies at the same time distance criteria between connecting residues (the N115-F198 distance is 8.6 Å), the assignment of core a and d residues, and a continuous helical conformation. A similar argument can be used to discard fold 2b. The existence of transmembrane coiled coil folds (1a and 2a), which can be connected to the EC2 domain with a helical continuity and a consistent succession of helical residues is a further argument in favor of such continuity.
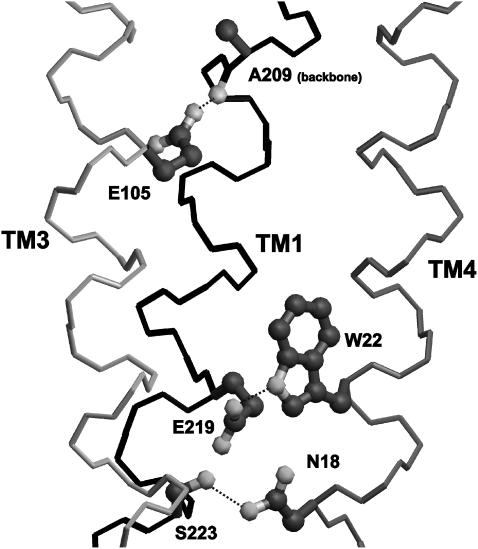
The two remaining transmembrane folds 1a and 2a have identical positioning of TM3 and TM4 and swapped positioning of TM1 and TM2. These, respectively, correspond to a fully antiparallel and a semiantiparallel left-handed coiled coil. The second arrangement is a priori less probable because it is not found among natural or artificial soluble four-stranded coiled coils nor in transmembrane proteins. Furthermore, in transmembrane proteins, sequentially adjacent helices usually directly interact (59), which is the case in fold 1a but not in fold 2a. To further discriminate between these two folds, helix-helix interactions were examined. Stability of transmembrane helical bundles mainly results from: 1), surface complementarity between adjacent helices resulting in close packing and efficient Van der Waals interactions; and 2), hydrogen bonding between polar residues located at helix-helix interfaces (60–62). Helix packing in the modeled CD81 transmembrane domains corresponding to the two folds was examined by calculating the packing values (25,52) of helix interior residues a, d, e, and g, as well as the volume of internal cavities, representative of packing defects. As shown in Table 1, there is a significantly better packing in the fully antiparallel fold 1a. This is more marked for TM1 and TM2, which are swapped in the two folds. This is due to a better size complementarity of interior core residues in fold 1a than in fold 1b. There are several small size-conserved residues in TM2 that interact with also mostly size-conserved very bulky residues of TM1 in fold 1a (see below). In fold 2a, TM1 and TM2 are in contact on the opposite sides of their interior surfaces so that this size complementarity is mostly lost. Both models were also evaluated with regards to hydrogen bonding of interior residues of adjacent helices. There are five interior hydrogen-bonding side chains, namely N18 and W22 in TM1, E105 in TM3, as well as E219 and S223 in TM4 (glutamic acids are expected to be protonated). As indicated in Table 1, fold 1a affords the formation of two hydrogen bonds between side chains of adjacent helices, namely N18-S223 and W22-E219. Oppositely, no side-chain/side-chain hydrogen bonds are possible with fold 1b due to the fact that TM1 and TM4 are not adjacent. In none of the two folds is a side chain available for hydrogen bonding with E105 of TM3. However, in membrane protein structures, 50% of interior glutamic acid side-chain hydrogen-bond with backbone atoms of adjacent helices (62). Here, it was indeed possible to form a hydrogen-bond between the protonated carboxyl of E105 and the peptide carbonyl of A209 of TM2 (in both folds because the TM3-TM4 relation is the same). Interestingly, Liu et al. (52) recently reported that alanine carbonyls constitute a preferential site for interhelix hydrogen bonds of aspartic acid and asparagine side chains. In both models, TM2 has no interhelix hydrogen bond, being devoid of suitable interior residues. In all, these data indicate that fold 1a not only affords a better packing of the transmembrane domain of CD81 but also a higher interhelix hydrogen bonding potential. It is therefore likely that this is this fold that is adopted by CD81.
TABLE 1.
Helix-helix interactions in CD81 transmembrane folding models
| Fully antiparallel (1a) | Semiantiparallel (1b) | ||
|---|---|---|---|
| TM1 | 0.477 ± 0.009 | 0.421 ± 0.012 | |
| Average packing value of core residues | TM2 | 0.463 ± 0.010 | 0.420 ± 0.013 |
| TM3 | 0.449 ± 0.009 | 0.431 ± 0.011 | |
| TM4 | 0.498 ± 0.010 | 0.475 ± 0.009 | |
| Total internal cavity volume (Å3) | 145 ± 56 | 293 ± 67 | |
| Potential interhelix side-chain H-bonds | 2 | 0 |
Packing values are averages for each transmembrane helix of values measured for a, d, e, and d type residues and on 10 distinct homology modeling simulations. Internal cavity volumes are averages on 10 distinct homology modeling simulations.
Conformation and positioning of the EC1 domain
The first extracellular segment EC1 of tetraspanins is variable in size and sequence among tetraspanin species. The secondary structure of the EC1 of CD81 and other tetraspanins was predicted by methods designed for soluble proteins (38). Despite high sequence divergence, the EC1 of most tetraspanins has a conserved secondary structure, namely a small central β-strand (extended structure) flanked by nonregular structure stretches. Among 37 tetraspanin species (corresponding to 110 sequences) for which this prediction was carried out, 26 contained such a β-strand. This may reflect the nontotal accuracy of the method (38) or a limited structural heterogeneity among tetraspanins. The secondary structure predictions of the EC1 of representative tetraspanins are shown in the left side of the alignment of Fig. 5 A. The length of the predicted EC1 β-strand is variable ranging from three to seven residues. Again, this may represent an actual variable length of the β-strand or an accuracy limitation of the method.
FIGURE 5.
Characterization of the conformation of the CD81 small extracellular domain EC1 and its interaction with the large extracellular domain EC2. (A) Sequence alignment of tetraspanins emphasizing the conservation of secondary structure and residue polarity in the EC1 region and in the conserved subdomain of the EC2. The predicted regular secondary structure regions of the EC1 as well as the experimental secondary structure regions of the EC2 obtained by crystallography for CD81 are indicated on top, respectively, in green and red (E, extended; H, helix). The limits of the predicted EC1 β-strand are boxed in green on each tetraspanin sequence. Hydrophobic residues are in blue and polar residues are in pink. EC2 residues involved in contact with the EC1 are indicated by an asterisk. (B) Prediction of EC1 β-strand/EC2 interactions. (Left) Identification of the EC1 β-strand interaction region on the EC2 by blind soft docking. The EC1 β-strand is shown as backbone-only bond representation and colored in green. The molecular surface of the EC2 is colored to emphasize hydrophobic (blue) and polar (pink) residues in the EC1 interaction region. Results of 30 simulations of docking are shown. (Right) Final modeling of the EC1 β-strand/EC2 interaction by high-resolution docking followed by manual adjustment. EC1 side chains are shown. Two modeled EC1-EC2 hydrogen bonds are indicated in red.
As shown in Fig. 5 A, for all sequences, the EC1 β-strand is enriched in hydrophobic residues. In CD81, it has the sequence ELLYLQ. Considering the predicted continuity of TM3 and TM4 with EC2 helices and the predicted relative topology of TM1-4 in fold 1a, the EC1 is expected to be in contact with a region made of residues belonging to helices A, E, and B (15) of the conserved subdomain of the EC2 (16). The right side of the alignment in Fig. 5 A highlights the polar and nonpolar EC2 residues corresponding to this region in several tetraspanin sequences. In the CD81 EC2 structure this region forms a groove and its center corresponds to a conserved hydrophobic surface (15,16). This raises the possibility that in the CD81 structure, the EC1 β-strand packs in this groove, mainly through hydrophobic contact. To obtain a model-independent evaluation of this hypothesis, “blind” docking simulations were performed. The EC1 β-strand was considered as a ligand and its interaction site on the EC2 was predicted. A soft-docking approach was used, which, at the expense of accuracy of the docked complex, allows for uncertainties in the conformation of the ligand and the target (39). The left side of Fig. 5 B shows the result of such a simulation using a six-residue NLLYLE strand that was modeled from a homologous strand region obtained by structural database search (see “Experimental Procedures”). Similar results were obtained with a six-residue strand modeled ab initio. Almost all docking results aim to a location of the β-strand at the expected position. Although this approach indicates that the considered EC2 region constitutes a preferential site for a six-residue β-strand, its resolution is limited. However, once the approximate position of the EC1 strand relative to the EC2 was delimited, it was possible to use a higher resolution docking method (40,41) to predict the most likely arrangement. As shown in the right side of Fig. 5 B, this method indicates that the EC1 strand packs inside the EC2 groove with its hydrophobic residues interacting with the hydrophobic surface. Two hydrogen bonds with the EC2 were tentatively formed by slightly adjusting the conformation of two side chains.
Conformation of the N- and C-terminal regions
The N- and C-terminal regions of CD81 are close to the intracellular membrane surface. Secondary structure prediction suggests a nonordered conformation for these segments. However, the N-terminal region contains cysteine residues known to be palmitoylated in situ (12,63,64). When this segment is placed on a helical wheel, hydrophobic residues and palmitoylable cysteines are located on one side of the helix and hydrophilic residues on the other side (not shown). Such amphipathic helices are common in membrane protein structures and adopt a membrane-parallel orientation with hydrophobic and hydrophilic residues, respectively, facing inward and outward (65). It is proposed that the N-terminal segment once palmitoylated is structured at the membrane interface and adopts such an amphipathic helix conformation. The C-terminal region of CD81 probably adopts a random conformation although hydrophobic and hydrophilic side chains may on the average also orient, respectively, toward and away from the membrane.
Complete molecular modeling and architecture of the 3D CD81 structure
To obtain a complete 3D model of the CD81 structure, a homology modeling procedure was used with a full template, built by assembling a fully antiparallel transmembrane left-handed coiled coil template with the predicted EC1 β-strand/EC2 complex. Exact ends of transmembrane helices were fixed by trial and error. The IC end of TM1 was fixed to Y12 because a two-residue loop was necessary to orient the C-terminal amphipathic helix perpendicularly to the transmembrane domain. The IC ends of TM2 and TM3 were fixed to A83 and Q88. This is necessary to have the palmitoylable C89 exposed to the bilayer. This also affords similar level ends to the two helices and a four-residue IC loop sufficient for closure. The IC end of TM4 was fixed to C228 to end at a similar level relative to the bilayer as the other helices. On the EC side, extending TM1 and TM2, respectively, by two and three residues was necessary to avoid packing voids in the EC1 region of the final 3D structure. This locates the EC ends of TM1 and TM2 next to two proline residues. Prolines have been demonstrated to be efficient breakers at the end of transmembrane helices (66). Finally, the N-terminal and C-terminal segments were, respectively, given a helical and roughly extended conformation orienting hydrophobic residues and cysteines toward the transmembrane domain and hydrophilic residues toward the IC side. The final limits of the secondary structure of CD81 are indicated in Fig. 1 A.
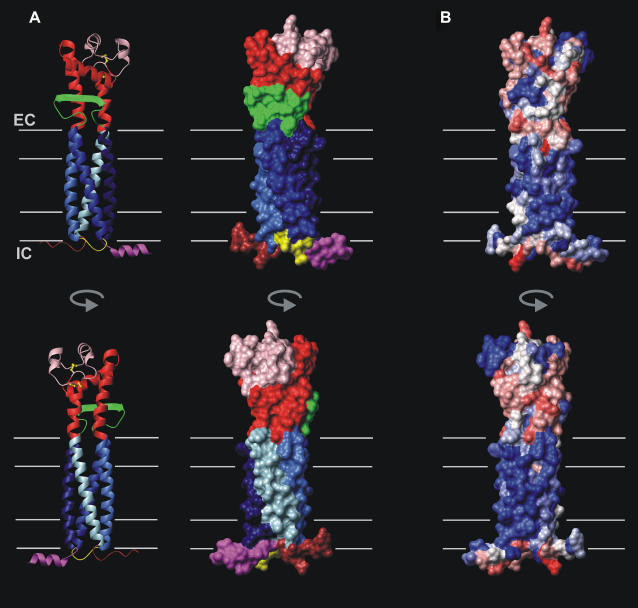
The resulting 3D structure of CD81 is shown in Fig. 6 A. The structure is cylindrical and very compact. After emerging from the bilayer, TM3 and TM4 become helices A and E of the EC2 and remain packed, although departing from their coiled coil geometry due to the constraints of the EC2 tertiary structure. The EC1 packs against the conserved subdomain of the EC2 mainly on helices A and E with its β-strand running roughly antiparallel to helix B. Due to this tight packing of the EC1 in the EC2 groove, the whole extracellular domain more or less retains its mushroom shape. It protrudes out of the bilayer by 33 Å. The EC2 variable subdomain (16) is located on the side opposite to the EC1 and forms one half of the mushroom head. In CD81, it contains two small helices (C and D) (15). It is linked to the conserved subdomain by two disulfide bridges. On the intracellular side, the N-terminal amphipathic helix, the IC loop, and the C-terminal disordered domain emerge in the aqueous phase at similar levels, presumably interacting with the membrane surface.
FIGURE 6.
Architecture and polarity of the modeled CD81 3D structure. (A) Ribbon (left) and surface (right) representation of the CD81 tertiary structure and topology. TM1-TM4, the conserved and variable subdomains of the EC2, the EC1, the IC loop, and the N-terminal and C-terminal regions are, respectively, represented in marine blue, blue, royal blue, light blue, red, pink, green, yellow, magenta, and brown. Disulfide bridges are in yellow. (B) Surface representation of residue polarity calculated according to Eisenberg et al. (67). The surface color range linearly from red to blue, corresponding, respectively, to polar and hydrophobic. The expected limits of the lipid bilayer (40-Å thickness) and of the hydrophobic region (30-Å thickness) are indicated as thin gray lines.
Fig. 6 B shows the distribution of polarity (67) on the molecule. The transmembrane domain surface is remarkably hydrophobic. Although moderately polar residues are often found on the transmembrane surface of intrinsic proteins (46), this is hardly the case for CD81 with only two threonines, which are in a situation where they might hydrogen bond. Positively or negatively charged residues are only present in the region where the transmembrane domain is expected to emerge in the extracellular aqueous phase. There are no charged residues at intracellular transmembrane helix ends, although several occur at the nearby aqueous stretches. All these factors contribute to provide to CD81 a relatively long hydrophobic portion of the transmembrane domain (∼35-Å length). Although the IC loop is highly hydrophilic, the N-terminal membrane parallel helix and the C-terminal disordered region are amphipathic (the hydrophobicity brought by palmitoylation of cysteine residues is not represented in Fig. 6 B). The extracellular region is mainly hydrophilic but is also characterized by the presence of hydrophobic surface regions. Although the hydrophobic patch initially found on the crystallographic structure of the EC2 in the conserved subdomain (15,16) is now partially hidden by the EC1, the region of the extracellular domain located below helix E remains largely hydrophobic due to the presence of four apolar residues in the EC1 strand.
The experimental data available to evaluate the modeled CD81 structure consists first of residues known to be accessible in CD81 and other tetraspanins. The structure was found to be in agreement with this data (Table 2). CD81 contains six palmitoylable cysteines (12,63,64) located in portions of the protein close to the intracellular headgroup region of the membrane. In the case of C6, C9, and C89, such accessibility was explicitly used in the modeling procedure. C80, C227, and C228 are found to be also accessible. Furthermore, CD81 residues that correspond sequentially to palmitoylable cysteines located at other positions in the tetraspanin CD151 (12) are also accessible. Unlike CD81, most tetraspanins are also glycosylated. CD9 is expected to be glycosylated at two asparagine side chains of the EC1 (68) that correspond to EC1 residues largely solvent-accessible in the CD81 model. Besides, in the CD81 modeled structure, no EC2 residue known to be accessible is masked by the EC1. In particular, CD81 residues involved in binding of the E2 glycoprotein of HCV (12,15,69) remain accessible. This is also the case for CD81 residues located in the structurally conserved subdomain of the EC2 and corresponding to N-glycosylated positions in other tetraspanins (12,16) and for a residue that corresponds to a cysteine involved in disulfide-mediated dimer formation in RDS/peripherin (13).
TABLE 2.
Surface accessibility in the modeled CD81 structure of tetraspanin residues involved in posttranslational modification and ligand binding
| CD81 residue | Corresponding residue in other tetraspanin | Accessible surface (Å2) | Accessibility criteria | Reference |
|---|---|---|---|---|
| C6 | – | 52.2 | Palmitoylation | 12 |
| C9 | – | 49.8 | Palmitoylation | 12 |
| K52* | N51 (CD9) | 96.5 | N-glycosylation | 79 |
| P53* | N52 (CD9) | 27.3 | N-glycosylation | 79 |
| C80 | – | 23.0 | Palmitoylation | 12 |
| Y81* | C80 (CD151) | 139.3 | Palmitoylation | 12 |
| C89 | – | 102.5 | Palmitoylation | 12 |
| D138* | N129 (CD53, CD63) | 106.0 | N-glycosylation | 16 |
| N141* | C151(peripherin) | 97.8 | Intermolecular disulfide | 13 |
| L162 | – | 28.5 | HCV glycoprotein E2 binding | 80 |
| T163 | – | 79.5 | HCV glycoprotein E2 binding | 15 |
| I182 | – | 114.5 | HCV glycoprotein E2 binding | 80 |
| N184 | – | 17.6 | HCV glycoprotein E2 binding | 80 |
| F186 | – | 140.5 | HCV glycoprotein E2 binding | 15 |
| D196 | – | 50.8 | HCV glycoprotein E2 binding | 15 |
| M224* | C242 (CD151) | 88.2 | Palmitoylation | 12 |
| V225* | C243 (CD151) | 64.0 | Palmitoylation | 12 |
| C227 | – | 28.3 | Palmitoylation | 12 |
| C228 | – | 74.7 | Palmitoylation | 12 |
In the first column, residues without asterisks are known to be accessible in CD81 whereas those with an asterisk are CD81 residues corresponding sequentially to residues known to be accessible in other tetraspanins and listed in the second column. Accessible surface includes side chain and Cα.
Another type of data that can be used to evaluate the CD81 structural model is the cryo-EM work of Min et al. (14) on the uroplakin particle. This evaluation is complicated by the fact that the uroplakin particle building blocks are heterodimers of the tetraspanin uroplakin 1a or 1b with the nontetraspanin single span transmembrane protein uroplakin 2 or 3. The current resolution does not allow one to distinguish the two proteins. Each uroplakin particle heterodimer appears as a relatively cylindrical transmembrane structure with a small (5-Å length) intracellular and a prominent (65-Å length) extracellular domain. According to the authors, the long length of this extracellular domain is mainly due to the nontetraspanin uroplakin 2 or 3. On the other hand, Min et al. also show that the transmembrane zone of the heterodimer can accommodate five transmembrane helices with four forming a square bundle and the fifth located on the side of this square bundle. This is consistent with the modeling work if one assumes that the square bundle corresponds to the tetraspanin component of the heterodimer. Therefore, it appears that the CD81 structural model is compatible with the cryo-EM data.
Interactions between transmembrane helices in the CD81 modeled structure
Two types of interactions are responsible for stabilizing the transmembrane domain of CD81 and other tetraspanins, namely Van der Waals interactions and hydrogen bonds. Due to the accuracy limits of molecular modeling, only a brief and qualitative description of these is given here. First, there appears to be a size complementarity of many small and bulky core residues in contact belonging to adjacent transmembrane helices. In all there are 26 proximities between such residue pairs, more than half occurring between size-conserved residues. In particular, near the intracellular end of TM2, there is a cluster of small residues belonging to the tetraspanin signature (G69, G76, G79, C80, G82, and A83) that mainly interacts with another cluster of very bulky residues of TM1 (Y12, F15, N18, F19, and W22) in a size-complementary manner (not shown). Such observations suggest that size complementarity does indeed play a role in the stability of the CD81 transmembrane domain. Fig. 7 shows the modeled (and therefore stereochemically approximative) network of hydrogen bonding between transmembrane helices in CD81. There are two side-chain hydrogen bonds between TM1 and TM4, namely N18-S223 and W22-E219. TM4 is also suggested to be connected with TM3 through an H-bond between the side chain of E105 and the backbone carbonyl of A209. On the other hand, TM2, which is the less polar of all transmembrane helices, contains no interior hydrogen bonding groups.
FIGURE 7.
Hydrogen bonds between transmembrane helices in the modeled CD81 3D structure. The backbones of TM1, TM3, and TM4 are drawn in bond representation and, respectively, colored in black, light gray, and gray. Side chains and backbone involved in H-bonds are shown in ball-and-stick and colored in dark gray except polar atoms that are drawn in light gray. H-bonds are depicted as dotted lines.
Topography of aromatic residues at the CD81 transmembrane domain surface
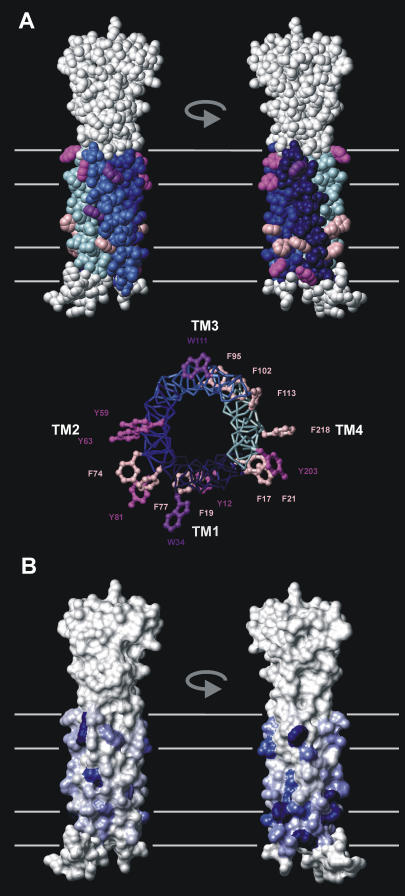
As found in transmembrane protein experimental structures (42), the modeled CD81 structure possesses a number of aromatic residues located on the surface of the membrane-embedded domain (Fig. 8 A). Trp and Tyr side chains are on the average more superficial than Phe side chains, as is also observed experimentally (70). Considering the size of the CD81 transmembrane domain, the number of such aromatic residues is high, i.e., 16. This is mainly due to a high number of Phe residues. Another uncommon property is that the distribution and orientation of aromatic residues is nonuniform. There are more aromatic residues (mainly Phe) in one side of the transmembrane domain located toward TM1 and TM2. In addition, the aromatic residues on this side appear to have a more protruding orientation (Fig. 8 A). This latter property is due to differences in the coiled coil positions occupied by the exposed aromatic residues in each TM helix (b, c, and f positions have more protruding side chains than e and g position). Despite a limited residue to residue conservation, the general trend of asymmetry of distribution of such aromatic amino acids is conserved. Among 63 tetraspanin species with transmembrane domains having more than 25% sequence identity with that of CD81, 49 contain between 10 and 19 exposed transmembrane aromatic residues. Fig. 8 B shows a mapping of the frequency of aromatic residues in these sequences on the transmembrane surface of the CD81 structure. There is a definite accumulation of aromatic residues on the side of the transmembrane domain located toward TM1 and TM2. Tetraspanins that correspond to this pattern include the most common members, CD9, CD53, CD82, CD151, CD37, CD151 except for CD63.
FIGURE 8.
Asymmetric distribution of exterior transmembrane domain aromatic residues in CD81 and tetraspanins. (A) (Top) CPK representation of the modeled CD81 structure highlighting exterior aromatic residues of the transmembrane domain. Helices TM1-4 are colored as in Fig. 6 A except for Phe, Tyr, and Trp residues, which are colored pink, magenta, and purple, respectively. The rest of the molecule is in gray. Two opposite orientations corresponding to the aromatic poor (left) and aromatic rich (right) sides are shown. (Bottom) Bond representation of the CD81 transmembrane domain viewed from the extracellular side. Only exterior aromatic side chains are shown to illustrate their protruding or nonprotruding character. (B) Surface representation of the modeled CD81 structure colored according to the frequency of occurrence of transmembrane domain aromatic residues in tetraspanins at the corresponding accessible residue position. Frequencies were obtained from an alignment of 49 distinct tetraspanin species with both >25% homology to CD81 in the transmembrane domain and >10 exterior aromatic residues (one sequence per species). The surface color ranges linearly from white to marine blue corresponding, respectively, to 0 and 0.77 (maximum occurrence found) aromatic residue frequency. Orientations of the molecule are identical to panel A.
DISCUSSION
In this work the complete structure of the tetraspanin CD81 has been modeled using various prediction and modeling methods as well as the crystallographic structure of the EC2 domain. These results are consistent with current knowledge of transmembrane protein structure. Furthermore, these predictions, although mostly derived independently from each other, constitute a self-consistent ensemble. The accuracy of the modeling is related to the current status of computational methods. The best methods of transmembrane helix prediction are in error on average by one helix turn at each end (46). The secondary structure prediction method used to predict the EC1 β-strand have an accuracy of only ∼75% (38). Also, here, the transmembrane domain of CD81 has been modeled using a low computational cost homology modeling procedure that has allowed to test the influence of several parameters. Computationally intensive modeling strategies of transmembrane domains exist that provide more global searches of conformational space and have been recently applied to transmembrane bundles with four different helices (47,71). Because the final modeled CD81 structure has a suitable packing quality and is consistent with experimental data, it appears to be a plausible working model that might be improved in the future both experimentally and computationally.
Some features of the CD81 structure suggest a new interpretation of one aspect of the EC2 structural data. In the EC2 crystal structure, a surface hydrophobic patch contributed by residues from helices A, B, and E is apparent and corresponds to crystallographic contact between adjacent EC2 molecules. This suggested that the hydrophobic patch might be involved in tetraspanin interactions with themselves (15,16). Here, in the modeled CD81 structure, these residues are in part masked by their interaction with the EC1. This suggests that such masked residues are actually conserved internal hydrophobic residues of the extracellular domain that become unmasked in the soluble EC2. This is consistent with the reports that the removal of the EC2 by mutagenesis of CD151 does not affect its association with itself and other tetraspanins (72) and that mutations of two residues involved in the CD81 EC2 hydrophobic patch (F150 and V146), although decreasing EC2 oligomerization in solution, have no effect on CD81 homodimerization in situ (73). Other structural factors are likely involved in tetraspanin-tetraspanin interactions.
The availability of the CD81 modeled structure raises the question of which aspects of the structure are common to all tetraspanins. It seems likely that the arrangement of the transmembrane domain as a four-stranded fully antisymmetric left-handed coiled coil is common to most if not all tetraspanins. This provides an explanation for the conservation of many small and bulky residues in the transmembrane domain of tetraspanins (including the tetraspanin signature). The situation is more complex with regard to the conservation of hydrogen bonding between transmembrane helices. Only two of the transmembrane hydrogen bonding residues of CD81 are highly conserved among tetraspanins, namely N18 and E105 (occasionally replaced by Q). Most tetraspanins possess a hydroxylated residue corresponding to S223 or at a nearby interior position of TM4 that may provide hydrogen bonding to the conserved asparagine of TM1. The position of A209 always corresponds to a small-size residue, a feature that may facilitate hydrogen bonding of the TM3 conserved glutamine to the backbone carbonyl. On the other hand, most tetraspanins do not possess a polar residue corresponding to E219 of CD81. Rather a conserved E or Q residue is found in TM4 at a position corresponding to interior residue M216 of CD81. A polar residue located at such position would still be able to hydrogen bond to the TM1 tryptophan corresponding to W22 (conserved in ∼50% of tetraspanins). Alternatively, in other tetraspanins, unlike for CD81, this TM4 E/Q residue would be at a suitable distance to the E/Q of TM3 for H-bonding. Finally, in most tetraspanins, TM2 is poor in hydrogen bonding residues. This suggests that, whereas the exact interhelix hydrogen-bonding network may be variable in tetraspanins, it mostly involves TM1-TM4 and TM3-TM4 interaction. It seems also likely that the continuity of EC2 helices with TM2 and TM3 is shared by most, if not all tetraspanins.
The majority of tetraspanins, including all well-characterized members, appears to possess a β-strand region in the EC1. The β-strand is always enriched in hydrophobic residues, which interact with conserved hydrophobic residues of the EC2 groove. In a previous study, it was found that, despite limited sequence similarity, helices A, B, and E of the EC2 form a structurally conserved subdomain among tetraspanins (16). These data suggest that, for the majority of tetraspanins, the EC1 shares a similar pattern, i.e., it has a largely conserved structure despite significant sequence divergence. Because the conserved EC2 subdomain and the EC1 are packed together, these appear to constitute a structurally conserved extracellular subdomain. This conserved subdomain is topped by a smaller structurally variable subdomain (16). For CD81 and most tetraspanins, the region of the conserved extracellular subdomain located below helix B and opposite to the EC2 variable side is highly hydrophobic. This is due to the occurrence of apolar residues in the EC1 β-strand as well as to conserved EC2 hydrophobic residues not masked by the EC1. Therefore, this region constitutes a potential site for protein-protein interactions.
Structural conservation on the intracellular side of tetraspanin appears to be more contrasted. The small IC loop connecting TM2 and TM3 is four-residue long in the CD81 structural model. It corresponds to a sequence pattern that is found in ∼60% of tetraspanins and therefore likely adopts a similar conformation. The N-terminal intracellular stretch of CD81, which contains two palmitoylable cysteines, has been modeled as a membrane-parallel amphipathic helix; 40% of tetraspanin types contain a comparable amphipathic pattern. Although many tetraspanins lack one cysteine in this region, it is often replaced by a hydrophobic residue so that the amphipathic character is retained. The idea that this amphipathic helix is formed upon palmitoylation suggests a possible mechanism for regulation of tetraspanin interactions. Recently, the heterologous interactions between tetraspanins have been shown to depend upon palmitoylation (63). Interactions between the amphipathic N-terminal helices may be involved in such interactions. Finally, the intracellular C-terminal stretch is among the most divergent region in tetraspanins. Although it is suggested to be disordered in CD81, it may adopt specific conformations in other members, especially because it is often involved in very specific functions (12).
The relatively long hydrophobic transmembrane region found for CD81 may be significant in view of the partial association of tetraspanins with lipid rafts (3,4). In several occasions (1,3), palmitoylation of tetraspanins was found to have no effect on their raft location. Rafts are characterized by a larger thickness of the hydrophobic bilayer portion and the length of the transmembrane domain has been suggested as a sorting criteria for intrinsic proteins (74). A possible role of Trp and Tyr side chains located near the headgroup region must also be considered because these may adapt to hydrophobic mismatches through transversal changes in their position (75). It may be the combined properties of the hydrophobic region and of the aromatic belt that allow tetraspanins to reside both in raft and nonraft regions. A related structural pattern that appears to be conserved, at least in ∼50 tetraspanin species is constituted by clusters of protruding aromatic residues asymmetrically located on one side of the transmembrane domain. Such lateral asymmetry may reflect a specialization of the two sides of the transmembrane domain for distinct protein interactions, e.g., homologous or heterologous tetraspanin interactions or interactions with tetraspanins and with other partners. Alternatively, because recent data by Charrin et al. (76) suggests a physical interaction between cholesterol and tetraspanins, it is possible that the transmembrane surface aromatic residues are involved in one or several cholesterol-binding sites. Indeed, recently proposed putative cholesterol sites of other proteins seem to invariably involve aromatic residues (77–79). This may be also significant for tetraspanin-raft interaction. In a more speculative manner, this lateral asymmetry of aromatic residue distribution in the transmembrane domain of tetraspanin may also favor their location at the boundary of two distinct types of membrane microdomains, e.g., raft and nonraft. The idea that proteins with asymmetric lipid interactions might stabilize lipid regions with differing compositions has been proposed (80).
One particularity of tetraspanin interactions is their various levels of specificity. Several tetraspanins share identical molecular partners (e.g., CD4, CD8, EWI-1, EWI-F) or partners from the same type (e.g., integrins, MHC molecules, other tetraspanins). On the other hand, each tetraspanin also has very specific partners (1,2,12,17–20). This work suggests that the selectivity of tetraspanins originates from two simultaneous properties: i), sequence variability within structurally conserved domains; ii), occurrence of limited-size structurally variable domains.
Acknowledgments
Note added in proof: Recently, Kovalenko et al. (81) reported the occurrence of heptad repeats in three transmembrane helices (TM1-3) of 28 tetraspanin sequences and emphasized the role of contacts between small and bulky residues in TM1-TM2 interactions in the tetraspanin CD9.
I thank Drs. Hélène Conjeaud and Miglena Angelova for critically reading the manuscript and Josette Barreau for correcting the manuscript and for her continuous support.
This work was supported by grants from the Centre National de la Recherche Scientifique, The Institut National de la Santé et de la Recherche Médicale, and the Agence Nationale de la Recherche contre le SIDA.
Abbreviations used: TM1-4, first to fourth transmembrane segment; EC1-2, first and second extracellular stretch; IC, intracellular loop; HCV, hepatitis C virus.
References
- 1.Hemler, M. E. 2003. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu. Rev. Cell Dev. Biol. 19:397–422. [DOI] [PubMed] [Google Scholar]
- 2.Levy, S., and T. Shoham. 2005. The tetraspanin web modulates immune-signalling complexes. Nature Rev. Immunol. 5:136–148. [DOI] [PubMed] [Google Scholar]
- 3.Claas, C., C. S. Stipp, and M. E. Hemler. 2001. Evaluation of prototype transmembrane 4 superfamily protein complexes and their relation to lipid rafts. J. Biol. Chem. 276:7974–7984. [DOI] [PubMed] [Google Scholar]
- 4.Delaguillaumie, A., J. Harriague, S. Kohanna, G. Bismuth, E. Rubinstein, M. Seigneuret, and H. Conjeaud. 2004. Tetraspanin CD82 controls the association of cholesterol-dependent microdomains with the actin cytoskeleton in T lymphocytes: relevance to co-stimulation. J. Cell Sci. 117:5269–5282. [DOI] [PubMed] [Google Scholar]
- 5.Vogt, A. B., S. Spindeldreher, and H. Kropshofer. 2002. Clustering of MHC-peptide complexes prior to their engagement in the immunological synapse: lipid raft and tetraspan microdomains. Immunol. Rev. 189:136–151. [DOI] [PubMed] [Google Scholar]
- 6.Charrin, S., S. Manie, M. Billard, L. Ashman, D. Gerlier, C. Boucheix, and E. Rubinstein. 2003. Multiple levels of interactions within the tetraspanin web. Biochem. Biophys. Res. Commun. 304:107–112. [DOI] [PubMed] [Google Scholar]
- 7.Kovalenko, O. V., X. Yang, T. V. Kolesnikova, and M. E. Hemler. 2004. Evidence for specific tetraspanin homodimers: inhibition of palmitoylation makes cysteine residues available for cross-linking. Biochem. J. 377:407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cormier, E. G., F. Tsamis, F. Kajumo, R. J. Durso, J. P. Gardner, and T. Dragic. 2004. CD81 is an entry coreceptor for hepatitis C virus. Proc. Natl. Acad. Sci. USA. 101:7270–7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pique, C., C. Lagaudriere-Gesbert, L. Delamarre, A. R. Rosenberg, H. Conjeaud, and M. C. Dokhelar. 2000. Interaction of CD82 tetraspanin proteins with HTLV-1 envelope glycoproteins inhibits cell-to-cell fusion and virus transmission. Virology. 276:455–461. [DOI] [PubMed] [Google Scholar]
- 10.Hosie, M. J., B. J. Willet, T. H. Dunsford, O. Jarret, and J. C. Neil. 1993. Upregulation of surface feline CXCR4 expression following ectopic expression of CCR5: implications for studies of the cell tropism of feline immunodeficiency virus. J. Virol. 67:1667–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levy, S., S. C. Todd, and H. T. Maecker. 1998. CD81 TAPA-1: a molecule involved in signal transduction and cell adhesion in the immune system. Annu. Rev. Immunol. 16:89–109. [DOI] [PubMed] [Google Scholar]
- 12.Stipp, C. S., T. V. Kolesnikova, and M. E. Hemler. 2003. Functional domains in tetraspanin proteins. Trends Biochem. Sci. 28:106–112. [DOI] [PubMed] [Google Scholar]
- 13.Goldberg, A. F., C. J. Loewen, and R. S. Molday. 1998. Cysteine residues of photoreceptor peripherin/rds: role in subunit assembly and autosomal dominant retinitis pigmentosa. Biochemistry. 37:680–685. [DOI] [PubMed] [Google Scholar]
- 14.Min, G., G. Zhou, M. Schapira, T. T. Sun, and X. P. Kong. 2003. Structural basis of urothelial permeability barrier function as revealed by cryo-EM studies of the 16 nm uroplakin particle. J. Cell Sci. 116:4087–4094. [DOI] [PubMed] [Google Scholar]
- 15.Kitadokoro, K., D. Bordo, G. Galli, R. Petracca, F. Falugi, S. Abrignani, G. Grandi, and M. Bolognesi. 2001. CD81 extracellular domain 3D structure: insight into the tetraspanin superfamily structural motifs. EMBO J. 20:12–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seigneuret, M., A. Delaguillaumie, C. Lagaudriere-Giesberg, and H. Conjeaud. 2001. Structure of the tetraspanin main extracellular domain. A partially conserved fold with a structurally variable domain insertion. J. Biol. Chem. 276:40055–40064. [DOI] [PubMed] [Google Scholar]
- 17.Imai, T., M. Kakizaki, M. Nishimura, and O. Yoshie. 1995. Molecular analyses of the association of CD4 with two members of the transmembrane 4 superfamily, CD81 and CD82. J. Immunol. 155:1229–1239. [PubMed] [Google Scholar]
- 18.Charrin, S., F. Le Naour, V. Labas, M. Billard, J. P. Le Caer, J. F. Emile, M. A. Petit, C. Boucheix, and C. Rubinstein. 2003. EWI-2 is a new component of the tetraspanin web in hepatocytes and lymphoid cells. Biochem. J. 373:409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Charrin, S., F. Le Naour, M. Oualid, M. Billard, G. Faure, S. M. Hanash, C. Boucheix, and E. Rubinstein. 2001. The major CD9 and CD81 molecular partner. Identification and characterization of the complexes. J. Biol. Chem. 276:14329–14337. [DOI] [PubMed] [Google Scholar]
- 20.Toyo-oka, K., Y. Yashiro-Ohtani, C. S. Park, X. G. Tai, K. Miyake, T. Hamaoka, and H. Fujiwara. 1999. Association of a tetraspanin CD9 with CD5 on the T cell surface: role of particular transmembrane domains in the association. Int. Immunol. 164:2043–2052. [DOI] [PubMed] [Google Scholar]
- 21.Vriend, G. 1990. WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 8:52–56. [DOI] [PubMed] [Google Scholar]
- 22.Sali, A., and T. L. Blundell. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779–815. [DOI] [PubMed] [Google Scholar]
- 23.Fiser, A., R. K. Do, and A. Sali. 2000. Modeling of loops in protein structures. Protein Sci. 9:1753–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brooks, B. R., R. E. Bruccoleri, B. D. Olafson, D. J. States, S. Swaminathan, and M. Karplus. 1983. CHARMM: a program for macromolecular energy, minimisation and dynamics calculations. J. Comput. Chem. 4:187–217. [Google Scholar]
- 25.Pattabiraman, N., K. B. Ward, and P. J. Fleming. 1995. Occluded molecular surface: analysis of protein packing. J. Mol. Recognit. 8:334–344. [DOI] [PubMed] [Google Scholar]
- 26.Nicholls, A., K. Sharp, and B. Honig. 1991. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 11:281–292. [DOI] [PubMed] [Google Scholar]
- 27.Hubbard, S. J., and J. M. Thornton. 1993. Naccess Version 2.1.1. Department of Biochemistry and Molecular Biology, University College, London, UK.
- 28.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283–291. [Google Scholar]
- 29.Koradi, R., M. Billeter, and K. Wüthrich. 1996. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14:51–55. [DOI] [PubMed] [Google Scholar]
- 30.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. Multiple sequence alignment with Clustal X. Nucleic Acids Res. 24:4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tusnády, G. E., and I. Simon. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics. 17:849–850. [DOI] [PubMed] [Google Scholar]
- 32.Donnelly, D., J. P. Overington, and T. L. Blundell. 1994. The prediction and orientation of alpha-helices from sequence alignments: the combined use of environment-dependent substitution tables, Fourier transform methods and helix capping rules. Protein Eng. 7:645–653. [DOI] [PubMed] [Google Scholar]
- 33.Donnelly, D., J. P. Overington, S. V. Ruffle, J. H. Nugent, and T. L. Blundell. 1993. Modeling alpha-helical transmembrane domains: the calculation and use of substitution tables for lipid-facing residues. Protein Sci. 2:55–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walshaw, J., and D. N. Woolfson. 2001. Socket: a program for identifying and analysing coiled-coil motifs within protein structures. J. Mol. Biol. 307:1427–1450. [DOI] [PubMed] [Google Scholar]
- 35.Yoshikawa, S., K. Shinzawa-Itoh, R. Nakashima, R. Yaono, E. Yamashita, N. Inoue, M. Yao, M. J. Fei, C. P. Libeu, T. Mizushima, H. Yamaguchi, T. Tomizaki, et al. 1998. Redox-coupled crystal structural changes in bovine heart cytochrome c oxidase. Science. 280:1723–1729. [DOI] [PubMed] [Google Scholar]
- 36.Harbury, P. B., T. Zhang, P. S. Kim, and T. Alber. 1993. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 262:1401–1407. [DOI] [PubMed] [Google Scholar]
- 37.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 18:2714–2723. [DOI] [PubMed] [Google Scholar]
- 38.Cuff, J. A., and G. J. Barton. 2000. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins. 40:502–511. [DOI] [PubMed] [Google Scholar]
- 39.Vakser, I. A. 1996. Low-resolution docking: prediction of complexes for underdetermined structures. Biopolymers. 39:455–464. [DOI] [PubMed] [Google Scholar]
- 40.Gabb, H. A., R. M. Jackson, and M. J. Sternberg. 1997. Modelling protein docking using shape complementarity, electrostatics and biochemical information. J. Mol. Biol. 272:106–120. [DOI] [PubMed] [Google Scholar]
- 41.Jackson, R. M., H. A. Gabb, and M. J. Sternberg. 1998. Rapid refinement of protein interfaces incorporating solvation: application to the docking problem. J. Mol. Biol. 276:265–285. [DOI] [PubMed] [Google Scholar]
- 42.Lee, A. G. 2003. Lipid-protein interactions in biological membranes: a structural perspective. Biochim. Biophys. Acta. 1612:1–40. [DOI] [PubMed] [Google Scholar]
- 43.Chen, C. P., A. Kernytsky, and B. Rost. 2002. Transmembrane helix predictions revisited. Protein Sci. 11:2774–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cuthbertson, J. M., D. A. Doyle, and M. S. Sansom. 2005. Transmembrane helix prediction: a comparative evaluation and analysis. Protein Eng. Des. Sel. 18:295–308. [DOI] [PubMed] [Google Scholar]
- 45.Stevens, T. J., and I. T. Arkin. 2001. Substitution rates in alpha-helical transmembrane proteins. Protein Sci. 10:2507–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eyre, T. A., L. Partridge, and J. M. Thornton. 2004. Computational analysis of alpha-helical membrane protein structure: implications for the prediction of 3D structural models. Protein Eng. 17:613–624. [DOI] [PubMed] [Google Scholar]
- 47.Torres, J., and I. T. Arkin. 2000. Recursive use of evolutionary conservation data in molecular modeling of membrane proteins. A model of the multidrug H+ antiporter emrE. Eur. J. Biochem. 267:3422–3431. [DOI] [PubMed] [Google Scholar]
- 48.Langosch, D., and J. Heringa. 1998. Interaction of transmembrane helices by a knobs-into-holes packing characteristic of soluble coiled coils. Proteins. 31:150–160. [DOI] [PubMed] [Google Scholar]
- 49.Langosch, D., E. Lindner, and R. Gurezka. 2002. In vitro selection of self-interacting transmembrane segments: membrane proteins approached from a different perspective. IUBMB Life. 54:109–113. [DOI] [PubMed] [Google Scholar]
- 50.Ruan, W., V. Becker, U. Klingmuller, and D. Langosch. 2004. The interface between self-assembling erythropoietin receptor transmembrane segments corresponds to a membrane-spanning leucine zipper. J. Biol. Chem. 279:3273–3279. [DOI] [PubMed] [Google Scholar]
- 51.Eilers, M., A. B. Patel, W. Liu, and S. O. Smith. 2002. Comparison of helix interactions in membrane and soluble alpha-bundle proteins. Biophys. J. 82:2720–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu, W., M. Eilers, A. B. Patel, and S. O. Smith. 2004. Helix packing moments reveal diversity and conservation in membrane protein structure. J. Mol. Biol. 337:713–729. [DOI] [PubMed] [Google Scholar]
- 53.Betz, S. F., J. W. Bryson, and W. F. DeGrado. 1995. Native-like and structurally characterized designed alpha-helical bundles. Curr. Opin. Struct. Biol. 5:457–463. [DOI] [PubMed] [Google Scholar]
- 54.Lupas, A. 1996. Coiled coils: new structures and new functions. Trends Biochem. Sci. 21:375–382. [PubMed] [Google Scholar]
- 55.Javadpour, M. M., M. Eilers, M. Groesbeek, and S. O. Smith. 1999. Helix packing in polytopic membrane proteins: role of glycine in transmembrane helix association. Biophys. J. 77:1609–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Curran, A. R., and D. M. Engelman. 2003. Sequence motifs, polar interactions and conformational changes in helical membrane proteins. Curr. Opin. Struct. Biol. 13:412–417. [DOI] [PubMed] [Google Scholar]
- 57.Gobel, U., C. Sander, R. Schneider, and A. Valencia. 1994. Correlated mutations and residue contacts in proteins. Proteins. 18:309–317. [DOI] [PubMed] [Google Scholar]
- 58.Donnelly, D., and R. J. Cogdell. 1993. Predicting the point at which transmembrane helices protrude from the bilayer: a model of the antenna complexes from photosynthetic bacteria. Protein Eng. 6:629–635. [DOI] [PubMed] [Google Scholar]
- 59.Bowie, J. U. 1997. Helix packing in membrane proteins. J. Mol. Biol. 272:780–789. [DOI] [PubMed] [Google Scholar]
- 60.Liang, J. 2002. Experimental and computational studies of determinants of membrane-protein folding. Curr. Opin. Chem. Biol. 6:878–884. [DOI] [PubMed] [Google Scholar]
- 61.Schneider, D. 2004. Rendezvous in a membrane: close packing, hydrogen bonding, and the formation of transmembrane helix oligomers. FEBS Lett. 577:5–8. [DOI] [PubMed] [Google Scholar]
- 62.Adamian, L., and J. Liang. 2002. Interhelical hydrogen bonds and spatial motifs in membrane proteins: polar clamps and serine zippers. Proteins. 47:209–218. [DOI] [PubMed] [Google Scholar]
- 63.Charrin, S., S. Manie, M. Oualid, M. Billard, C. Boucheix, and E. Rubinstein. 2002. Differential stability of tetraspanin/tetraspanin interactions: role of palmitoylation. FEBS Lett. 516:139–144. [DOI] [PubMed] [Google Scholar]
- 64.Clark, K. L., A. Oelke, M. E. Johnson, K. D. Eilert, P. C. Simpson, and S. C. Todd. 2004. CD81 associates with 14–3-3 in a redox-regulated palmitoylation-dependent manner. J. Biol. Chem. 279:19401–19406. [DOI] [PubMed] [Google Scholar]
- 65.Granseth, E., G. Von Heijne, and A. Elofsson. 2005. A study of the membrane-water interface region of membrane proteins. J. Mol. Biol. 346:377–385. [DOI] [PubMed] [Google Scholar]
- 66.Nilsson, I., and G. Von Heijne. 1998. Breaking the camel's back: proline-induced turns in a model transmembrane helix. J. Mol. Biol. 293:807–814. [DOI] [PubMed] [Google Scholar]
- 67.Eisenberg, D., E. Schwarz, M. Komaromy, and R. Wall. 1984. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J. Mol. Biol. 179:125–142. [DOI] [PubMed] [Google Scholar]
- 68.Seehafer, J. G., S. C. Tang, J. R. Slupsky, and A. R. Shaw. 1988. The functional glycoprotein CD9 is variably acylated: localization of the variably acylated region to a membrane-associated peptide containing the binding site for the agonistic monoclonal antibody 50H. 19. Biochim. Biophys. Acta. 957:399–410. [DOI] [PubMed] [Google Scholar]
- 69.Drummer, H. E., K. A. Wilson, and P. Poumbourios. 2002. Identification of the hepatitis C virus E2 glycoprotein binding site on the large extracellular loop of CD81. J. Virol. 76:11143–11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ulmschneider, M. B., and M. S. Sansom. 2001. Amino acid distributions in integral membrane protein structures. Biochim. Biophys. Acta. 1512:1–14. [DOI] [PubMed] [Google Scholar]
- 71.Gottschalk, K. E., M. Soskine, S. Schuldiner, and H. Kessler. 2004. A structural model of emrE, a multi-drug transporter from Escherichia coli. Biophys. J. 86:3335–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Berditchevski, F., E. Gilbert, M. R. Griffiths, S. Fitter, L. Ashman, and S. J. Jenner. 2001. Analysis of the CD151-alpha3beta1 integrin and CD151-tetraspanin interactions by mutagenesis. J. Biol. Chem. 276:41165–41174. [DOI] [PubMed] [Google Scholar]
- 73.Drummer, H. E., K. A. Wilson, and P. Poumbourios. 2005. Determinants of CD81 dimerization and interaction with hepatitis C virus glycoprotein E2. Biochem. Biophys. Res. Commun. 328:251–257. [DOI] [PubMed] [Google Scholar]
- 74.Munro, S. 1995. An investigation of the role of transmembrane domains in Golgi protein retention. EMBO J. 14:4695–4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.De Planque, M. R., and J. A. Killian. 2003. Protein-lipid interactions studied with designed transmembrane peptides: role of hydrophobic matching and interfacial anchoring. Mol. Membr. Biol. 20:271–284. [DOI] [PubMed] [Google Scholar]
- 76.Charrin, S., S. Manie, C. Thiele, M. Billard, D. Gerlier, C. Boucheix, and E. Rubinstein. 2003. A physical and functional link between cholesterol and tetraspanins. Eur. J. Immunol. 33:2479–2489. [DOI] [PubMed] [Google Scholar]
- 77.Jamin, N., J. M. Neumann, M. A. Ostuni, T. K. Vu, Z. X. Yao, S. Murail, J. C. Robert, C. Giatzakis, V. Papadopoulos, and J. Lacapere. 2005. Characterization of the cholesterol recognition amino acid consensus sequence of the peripheral-type benzodiazepine receptor. J. Mol. Endocrinol. 19:588–594. [DOI] [PubMed] [Google Scholar]
- 78.Epand, R. M., B. G. Sayer, and R. F. Epand. 2005. Caveolin scaffolding region and cholesterol-rich domains in membranes. J. Mol. Biol. 345:339–350. [DOI] [PubMed] [Google Scholar]
- 79.Epand, R. M., B. G. Sayer, and R. F. Epand. 2005. The tryptophan-rich region of HIV gp41 and the promotion of cholesterol-rich domains. Biochemistry. 44:5525–5531. [DOI] [PubMed] [Google Scholar]
- 80.McConnell, H. M., and A. Radhakrishnan. 2003. Condensed complexes of cholesterol and phospholipids. Biochim. Biophys. Acta. 1610:159–173. [DOI] [PubMed] [Google Scholar]
- 81.Kovalenko, O. V., D. G. Metcalf, W. F. DeGrado, and M. E. Hemler. 2005. Structural organization and interactions of transmembrane domains in tetraspanin proteins. BMC Struct. Biol. 5:11. [DOI] [PMC free article] [PubMed] [Google Scholar]