Abstract
Mechanical unfolding and refolding of single RNA molecules have previously been observed in optical traps as sudden changes in molecular extension. Two methods have been traditionally used: “force-ramp”, with the applied force continuously changing, and “hopping”. In hopping experiments the force is held constant and the molecule jumps spontaneously between two different states. Unfolding/refolding rates are measured directly, but only over a very narrow range of forces. We have now developed a force-jump method to measure the unfolding and refolding rates independently over a wider range of forces. In this method, the applied force is rapidly stepped to a new value and either the unfolding or refolding event is monitored through changes in the molecular extension. The force-jump technique is compared to the force-ramp and hopping methods by using a 52-nucleotide RNA hairpin with a three-nucleotide bulge, i.e., the transactivation response region RNA from the human immunodeficiency virus. We find the unfolding kinetics and Gibbs free energies obtained from all three methods to be in good agreement. The transactivation response region RNA hairpin unfolds in an all-or-none two-state reaction at any loading rate with the force-ramp method. The unfolding reaction is reversible at small loading rates, but shows hysteresis at higher loading rates. Although the RNA unfolds and refolds without detectable intermediates in constant-force conditions (hopping and force-jump), it shows partially folded intermediates in force-ramp experiments at higher unloading rates. Thus, we find that folding of RNA hairpins can be more complex than a simple single-step reaction, and that application of several methods can improve understanding of reaction mechanisms.
INTRODUCTION
Mechanical force has been used to study protein-protein interactions (1), membrane surfaces (2,3), biopolymer properties (4–7), protein folding (8,9), and RNA folding (10–12). Generally, the mechanical force is applied to individual molecules using the force-ramp (or pulling) method, in which the applied force is changed continuously, at an approximately constant loading rate (pN/s). The rates of disruption of chemical bonds or macromolecular interactions can be extracted from the distributions of the rupture forces at the chosen loading rate (13). This method assumes that the disruption of the molecular interaction follows first-order kinetics at a given force and thus the force distribution represents the integration of probability of rupture over a range of force. Some RNA molecules unfold and refold reversibly and display bistability at forces close to F1/2, where the equilibrium constant of the reaction is equal to 1 (10). The free energy, ΔG°, of the unfolding or refolding reaction at zero force can be calculated from the difference between the reversible mechanical work done at F1/2 and the energy of stretching single-stranded RNA to this force (10,14,15). When the force is held constant near F1/2, the molecule transits between unfolded and folded states, as indicated by “hopping” between the values of extension corresponding to the folded and unfolded forms of the RNA. The unfolding and refolding rates can be obtained from the lifetimes of the molecule in the two states. However, the hopping events can only be experimentally observed in a narrow range of forces close to F1/2. Beyond this range, the molecule mainly stays in one state or the other.
To directly observe the rates over a larger range of force, we have implemented a force-jump method using optical tweezers. In this method, the applied force is quickly raised or lowered to a constant value and the unfolding or refolding is monitored by changes in the end-to-end distance of the molecule. Rates of unfolding and refolding can be measured independently using this method. The force-jump (also called force-clamp) method has been used to study the unfolding of proteins (9,16) and the dissociation of nucleosomes (17). The force-jump method is analogous to temperature-jump (18), pH-jump (19), and solvent-jump (20) methods. In these bulk relaxation measurements, the observed rates are a combination of forward and reverse rate constants. However, in the single molecule force-jump experiments, each change in the extension represents an unfolding or refolding event of a single molecule. Therefore, rates of both forward and reverse reactions can be directly measured. An obvious question is whether the free energy and kinetics measured by the force-jump experiments agree with those measured by the hopping and force-ramp methods. To test this question, we have studied the folding and unfolding of the transactivation response region (TAR) RNA derived from the human immunodeficiency virus (HIV) (Fig. 1). We have applied all three methods to study the mechanical unfolding of this RNA hairpin, and we find that the free energy and unfolding kinetics agree within the error of measurement. However, the refolding kinetics depend on how the force is applied. We discuss the advantages and limitations of each method.
FIGURE 1.
The 52-basepair region of the TAR RNA hairpin is flanked by two ∼500-basepair DNA/RNA handles shown with the RNA in black and the DNA in gray. The 3′ terminus of the DNA handle A and the 5′ end of the DNA handle B (gray lines) are labeled with biotin and digoxigenin, respectively. The entire molecule is attached to two microspheres coated with either streptavidin or anti-digoxigenin antibody. The drawing is not to scale. The arrows indicate the direction of the applied force (Fy). The forces in the other two directions (Fx and Fz) are approximately zero during the experiments.
MATERIALS AND METHODS
Preparation of RNA
The RNA molecules were synthesized as previously described (10–12). The DNA sequence corresponding to HIV-1 TAR RNA (Fig. 1) was cloned into a pBR322 vector (NCBI ID “J01749”) between the EcoRI and HindIII sites. A DNA template containing an upstream T7 promoter, the TAR sequence, and ∼500 basepair regions flanking the TAR sequence that will serve as “handles” were amplified from the plasmid using the polymerase chain reaction (PCR). The handle regions correspond to the sequence of pBR322 from nucleotide 3838 to 1 and from 29 to 629, respectively. Using this DNA template, the RNA containing the TAR hairpin and flanking handles was synthesized by in vitro transcription with T7 RNA polymerase (21). DNA molecules complementary to the RNA handles were also generated by PCR. The RNA and two DNA handles were mixed in stoichiometric ratio and annealed by heating the samples to 85°C and then cooling them to room temperature slowly. The DNA annealed to the 5′-end of the RNA (handle A) was biotinylated at the 3′-end and the DNA annealed to the 3′ end (handle B) contained a digoxigenin group at the 5′-terminus. The annealed sample contains a mixture of molecules. Only the RNA molecules annealed to both DNA handles A and B can be attached to two micron-sized polystyrene beads, one type coated with streptavidin and one type coated with antidigoxigenin antibody (Fig. 1).
Optical tweezers
Dual-beam optical tweezers (7,22) were used to apply mechanical force to the RNA. In the sample chamber, the streptavidin-coated bead was held by an optical trap and the antidigoxigenin-coated bead was positioned at the tip of a micropipette through suction (7,22). The micropipette was fixed to the chamber, which in turn was mounted on a piezoelectric flexure stage (MDT-631, Thorlabs, Newton, NJ). The two beads were joined by the RNA molecule annealed to the two handles. Force was applied to the RNA by moving the piezoelectric stage. The force was measured by changes in the light momentum caused by the movement of the bead in the trap (22). The change in the extension of the molecule was measured by the movements of the two beads (22). The bead on the pipette was monitored by a “light-lever” system that records the position of the reaction chamber. Since the optical trap resembles a Hooke's law spring, the position of the trapped bead indicates the force. The spring constant of the trap was calibrated by correlating the video image of the trapped bead, collected by a CCD camera (LCL-903HS, Watec, Las Vegas, NV), and the force.
Folding experiments
All unfolding/refolding experiments were done at 20–22°C in 10 mM HEPES, pH 8.0, 100 mM KCl, 1 mM EDTA, and 0.05% NaN3. Three types of experiments were done in this study: force-ramp, hopping, and force-jump. In the force-ramp experiments, the molecule was continuously stretched and relaxed by moving the piezoelectric stage at a constant rate (nm/s) in one dimension (Fig. 1, y axis) (7,10–12). The exerted force is roughly a linear function of the time (constant loading rate, pN/s). Force and extension of the molecule were recorded at a rate of 50 Hz.
In the constant force experiments (hopping and force-jump), the force was kept constant through a feedback control that employed a proportional, integrative, and differential algorithm. The average force in 5-ms intervals was compared to the desired force, and the piezoelectric stage was moved to compensate for any difference. Under the feedback mode, the standard deviation of the force varied <0.4 pN. Force and extension of the molecule were acquired at a rate of 100 Hz. In the hopping experiments, the molecule was held at constant force for times up to 2 h, while the extension of the molecule was monitored continuously. The drift in the x and z axes (Fig. 1) during the period was <1 pN. In the force-jump experiments, the force was quickly raised or lowered to a desired value by moving the piezoelectric stage at maximum speed (>200 nm/s). The time it took for the force to reach the set point was <100 ms. Once the force reached the set value, it was held constant using the feedback control until an unfolding/refolding transition occurred, as indicated by a change in extension of the molecule. After the transition, the force was increased to completely unfold the molecule or decreased to allow the molecule to refold.
RESULTS
We have investigated unfolding and refolding of an RNA hairpin structure using the force-ramp, hopping, and force-jump methods. The model system is TAR RNA derived from HIV genomic RNA. The 52-nucleotide RNA hairpin forms a stable hairpin with a three-nucleotide bulge near a six-nucleotide apical loop (Fig. 1) (23,24). We have determined the unfolding/refolding kinetics and Gibbs free energy changes using all three methods.
Hopping experiments
To directly observe unfolding and refolding events at equilibrium, we performed hopping experiments, in which the extension of the molecule was continuously monitored while the applied force was held constant. If the force is held constant in the transition range for a molecule, the RNA molecule can rapidly alternate (or hop) between two states—folded and unfolded.
We found that TAR RNA hops in a small force range near 12.4 pN. Fig. 2 A shows a 10-min trace of force (Fy) held at 12.7 pN. The histogram of force during this period is shown in Fig. 2 B. The mean value of the force was 12.7 ± 0.3 pN, consistent with the intended force (Fig. 2 A, white line). The standard deviation of force is comparable to that observed by others with laser tweezers (25). Fig. 2 C shows a trace of molecular extension under constant force condition. TAR RNA transited between the unfolded and folded states with no detectable intermediates, as shown by changes of 18 ± 2 nm in end-to-end distance of the molecule. The change in the extension, ΔX, is consistent with the transition of the TAR from hairpin form to single-stranded RNA, as estimated by the worm-like-chain interpolation formula (26):
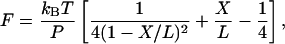 |
(1) |
where X is the extension, L is the contour length, P is the persistence length, T is the temperature, F is the force, and kB is Boltzmann's constant. Previously we estimated a persistence length of 1 nm and a contour length of 0.59 nm per nucleotide (10,12). Using the same value of the contour length and ΔX of TAR, we estimated a persistence length for single-stranded RNA ranging from 0.7 to 1.2 nm. This estimation also agrees with a recent measurement (27).
FIGURE 2.
Hopping experiment. (A) A time trace of force (Fy) under feedback control. The force was set to 12.7 pN. The white line shows the smoothed value with a 30-point sliding boxcar average. (B) Distribution of force in the y direction under feedback control. (C) A time trace of the extension of TAR RNA at 12.4 pN. The two states of the extension were ∼18 nm apart. (D) Plots of the logarithm of the rates versus force for unfolding (▪) and refolding (○). Data were fitted to Eq. 4 (solid lines). We obtained ln A of −24 ± 16 and 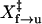 of 7 ± 5 nm for unfolding and ln A of 21 ± 8 and
of 7 ± 5 nm for unfolding and ln A of 21 ± 8 and  of 8 ± 3 nm for refolding. A is given in units of s−1.
of 8 ± 3 nm for refolding. A is given in units of s−1.
In Fig. 2 A, there are some significant deviations from the set force (>1.5 pN). These transient deviations lasted <10 ms and coincided with the unfolding/refolding events, as indicated by changes in the extension (data not shown). These datapoints reflect the lag of the force feedback during the unfolding/refolding transitions, when the position of the trapped bead moves quickly as the extension of the molecule changes.
When the force was kept constant at 12.4 pN, the ratio of the total time that the TAR hairpin stayed in the unfolded state to that in the folded state was 1.5. When force was raised to 12.7 pN, the molecule spent more time in the unfolded state (Keq = 2.4), whereas at 12.1 pN, the RNA was folded more often (Keq = 0.3). At both forces, we observed only two states of the extension of the molecule, whose extensions were ∼18 nm apart. Beyond the narrow range of forces between 12.1 and 12.7 pN, few transitions were observed; it would thus take several hours to a few days to obtain enough observations of the transitions to measure the rates with a high level of certainty. Direct observation of hopping at these forces would require significant improvement of the stability of the instrument.
The lifetimes of TAR RNA in each state are measured for a minimum of 100 hops. For a two-state reaction with first-order kinetics, the rate constant for folding is the reciprocal of the mean lifetime of the unfolded state; the rate constant for unfolding is the reciprocal of the mean lifetime of the folded state:
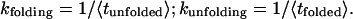 |
(2) |
The lifetimes fit a single exponential distribution for the cumulative probability, P(t), that a molecule remains unreacted for a period of time, t.
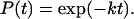 |
(3) |
Lifetimes of TAR RNA in folded or unfolded states were used to generate the probability that a reaction had not yet occurred as a function of time. The unfolding and refolding rates at each force were calculated by fitting the cumulative probabilities that the molecules were in each state to Eq. 3 (r2 > 0.95, data not shown). This treatment yielded similar rate constants to the reciprocal of mean lifetime method (Eq. 2) within the error of the measurements. The exponential behavior of this probability validates first-order kinetics for the unimolecular reaction and provides a direct measurement of the rate constants at given forces.
The apparent rate constant for the reaction at force F, k(F), is assumed to depend exponentially on force (13,14,28):
 |
(4) |
where X‡ is the distance to the transition state; kB is the Boltzmann constant; T is the temperature in Kelvin; and A is a factor that may also be written as the product of k(0) and km (10,14), in which k(0) is the rate constant at zero force and km reflects the effects of the handles and other instrumental factors (10,15). Values of ln k obtained by hopping experiments were plotted as a function of the force and fit to Eq. 4 (Fig. 2 D). We obtained 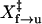 of 7 ± 5 nm and
of 7 ± 5 nm and 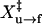 of 8 ± 3 nm from the slopes of the fits, and ln Af→u of −24 ± 16 and ln Au→f of 21 ± 8 from the intercepts (Table 1); the factor A has units of s−1.
of 8 ± 3 nm from the slopes of the fits, and ln Af→u of −24 ± 16 and ln Au→f of 21 ± 8 from the intercepts (Table 1); the factor A has units of s−1.
TABLE 1.
Parameters in the mechanical unfolding of TAR RNA
| Unfolding
|
Refolding
|
|||
|---|---|---|---|---|
| ln A* |
 (nm) (nm) |
ln A |
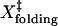 (nm) (nm) |
|
| Force-jump | −28 ± 2 | 8.2 ± 0.5 | 22 ± 3 | 8 ± 1 |
| Hopping | −24 ± 16 | 7 ± 5 | 21 ± 8 | 8 ± 3 |
| Force-ramp (0.4 pN/s) | −28.8 ± 0.9 | 8.4 ± 0.8 | 28.8 ± 0.9 | 10.9 ± 0.9 |
Factor A has the unit of s−1.
The equilibrium constant at a given force, Keq(F), can be calculated either from the ratio of the unfolding and refolding rate constants, kf→u/ku→f, or from the ratio of total time that the molecule spends in each state, tfolding/tunfolding (10,15). If both rate constants are exponential functions of force, Keq(F) is also exponentially dependent on the force:
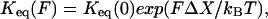 |
(5) |
where ΔX is the change in the end-to-end distance for the reaction. As expected, the two estimations of Keq(F) for TAR RNA agree. For a two-state reaction,
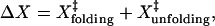 |
(6) |
where 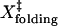 and
and  are the distances to the transition state of the folding and unfolding reactions. The sum of
are the distances to the transition state of the folding and unfolding reactions. The sum of 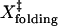 and
and  for TAR RNA is 15 ± 8 nm, roughly agrees with the observed ΔX (18 ± 2 nm). However, the large standard deviation of ΔX prevents a clear verification.
for TAR RNA is 15 ± 8 nm, roughly agrees with the observed ΔX (18 ± 2 nm). However, the large standard deviation of ΔX prevents a clear verification.
The change in free energy of the reaction is equal to the reversible work, FΔX. At Keq(F) = 1, the unfolding and refolding rates are equal, the reaction is reversible, and the free energy of unfolding the hairpin equals the work done at this force (F1/2). Using values of X‡ and ln A for folding and unfolding, we determined that F1/2 is 12.4 ± 0.3 pN (Table 2). Hence, the reversible work to unfold the hairpin at this force is 134 ± 18 kJ/mol (32 ± 11 kcal/mol). The standard free energy at zero force, ΔG°(0), is obtained by subtracting the work needed to stretch the unfolded molecule to this force, ΔG(stretch), from the reversible work done at F1/2 (assuming the effect of force on the folded form is negligible) (14,15):
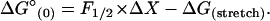 |
(7) |
We estimated ΔG(stretch) by integrating the worm-like-chain interpolation formula (Eq. 1) from 0 to F1/2 for a 52-nucleotide single-stranded RNA. The value of this adjustment is 46.8 kJ/mol (11.2 kcal/mol). Thus, ΔG°(0) for folding TAR RNA at 22°C is 88 ± 18 kJ/mol (21 ± 11 kcal/mol).
TABLE 2.
ΔG for folding TAR RNA
| F1/2 (pN) | ΔX1/2 (nm) | Work (kJ/mol) | ΔG0pN,22°C (kJ/mol) | |
|---|---|---|---|---|
| Hopping | 12.4 ± 0.3 | 18 ± 2 | 134 ± 18 | 88 ± 18 |
| Force-jump | 12.7 ± 0.2 | 18 ± 2 | 138 ± 18 | 90 ± 18 |
| Force-ramp (0.4pN/s) | 12.3 ± 0.1 | 18 ± 2 | 133 ± 16 | 86 ± 16 |
We were able to measure rate constants at three forces in the small force range (12.1–12.7 pN) that the molecule hops. Beyond this narrow range, few transitions were observed. Even at forces close to F1/2, the TAR hairpin hops slowly: the average lifetimes at F1/2 are ∼30–40 s as compared to 1–2 s for P5ab hairpin derived from Tetrahymena thermophila ribozyme (10). With the feedback control, the mean value of force in the direction of applied force (Fy) can be held constant over a long period of time. However, the drift in laser power and focus gradually changes the force in the other two directions (Fx and Fz). We stopped the experiment once the average of Fx or Fz drifted by >1 pN. The longest we were able to maintain a constant force was slightly over 2 h. Therefore, direct observation of hopping at very small (<0.1 s−1), or very large (>10 s−1) rates would require significant improvement in the stability of the laser trap.
Force-jump experiments
The hopping experiments are limited by the frequency of unfolding/refolding transitions and by the current instrumentation. To directly measure rate constants in a larger region of force, we implemented a force-jump method using the optical tweezers. Each force-jump cycle included a pair of independent measurements of lifetimes of folded and unfolded states, usually at different forces. The force-jump experiments employed the same force feedback control as the hopping experiments. A typical experiment cycle started at a low force (Fig. 3 A, top) where the RNA was fully folded. The force was then raised rapidly to the unfolding force by moving the piezoelectric stage at maximal speed (>200 nm/s) such that the bead on the micropipette moved away from the trapped bead. During this force jump, the DNA/RNA handles of the molecule were quickly stretched as shown by a sudden increase in the extension of the molecule (Fig. 3 A, bottom). Once the force reached the desired value, the feedback mechanism maintained a constant tension on the molecule. At constant tension, the end-to-end distance of the molecule remained relatively constant until the unfolding occurred, and then the extension quickly increased by ∼18 nm in a single step. After the transition, the force was raised to 20 pN at a rate of 1.5 pN/s. The extension increased as the handles and single-stranded RNA were further stretched. The force was kept at 20 pN for 3 s, then the force was quickly dropped to a refolding force; a decrease in extension by ∼18 nm signaled the refolding transition. Once the RNA refolded, the force was lowered to ∼5 pN for 3 s to insure that the new cycle began from the same initial state. We assumed that each pair of lifetimes of the folded and unfolded states was measured under similar conditions. Notably, the unfolding and refolding transitions of TAR hairpin at all measured forces were characterized by a single-step transition with ΔX of 18 ± 2 nm.
FIGURE 3.
Force-jump experiment. (A) Two cycles of the force-jump experiments. Time traces of the force and extension of the molecule are plotted in the top and bottom panels, respectively. (B) Plots of the probability of the folded hairpin as a function of the time at 12.7 pN (+, 322 observations) and at 14.2 pN (×, 144 observations). Dashed curves represent the fit of data to a single exponential (Eq. 3). (C) Plots of the logarithm of the rate constants versus force for unfolding (▪) and refolding (○). A fit of the unfolding rates versus force to Eq. 4 (solid line) yields ln A of −28 ± 2 and 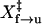 of 8.2 ± 0.5 nm. For refolding, we derived ln A of 22 ± 3 and
of 8.2 ± 0.5 nm. For refolding, we derived ln A of 22 ± 3 and 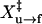 of 8 ± 1 nm.
of 8 ± 1 nm.
In the force-jump experiments, a rate constant at each force was obtained using the method described for hopping experiments. Fig. 3 B shows the cumulative probability of folded TAR hairpin as a function of time at 14.2 pN and 13.6 pN. At both forces, unfolding followed first-order kinetics and rates were extrapolated from a single exponential fitting. It is obvious that the hairpin unfolds faster at 14.2 pN. We have also compared rate constants when force is jumped or dropped by 3, 5, and 8 pN. The measured rates appear to be dependent only on the final force (data not shown). Fig. 3 C shows a plot of the values of ln k versus force; ln kf→u can be fit as a linear function of the force (Eq. 4) yielding ln A of −28 ± 2 and 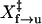 of 8.2 ± 0.5 nm (Table 1). A similar fit for refolding data gives ln A of 22 ± 3 and
of 8.2 ± 0.5 nm (Table 1). A similar fit for refolding data gives ln A of 22 ± 3 and 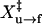 of 8 ± 1 nm.
of 8 ± 1 nm.
We further obtained F1/2 of 12.7 ± 0.2 pN (Table 2). The reversible work to unfold the hairpin at this force is 138 ± 18 kJ/mol (33 ± 4 kcal/mol). The stretching correction estimated from the worm-like-chain model is 47.7 kJ/mol (11.4 kcal/mol). Using Eq. 7, ΔG°(0),22°C for folding TAR RNA is 90 ± 18 kJ/mol (22 ± 4 kcal/mol).
The force-jump experiments are conducted under constant force/force-clamp conditions, similar to the hopping experiments. The major difference between the two methods is that in the force-jump experiments, measurements of unfolding and refolding transitions are decoupled. This feature allows us to independently measure the unfolding rates from 12.7 to 14.2 pN and refolding rates from 10.9 to 12.4 pN. With the larger force range, we were able to determine values of X‡ and ln A with higher precision, which in turn led to a better estimation of F1/2 and ΔG°(0),22°C. Notably, the values of ΔG°(0),22°C estimated using both methods are similar (Table 2). However, the standard deviation measured using force-jump methods is only ∼⅓ of that from hopping experiments.
Force-ramp experiments
Single molecules of TAR RNA hairpin flanked by DNA/RNA handles were stretched and relaxed repeatedly by moving the micropipette mounted on a piezostage at a constant rate (nm/s); this generated an almost constant loading/relaxation rate of pulling (pN/s) (7,10). On the force-extension curves, the unfolding transition of TAR hairpin is characterized by a sudden increase in the extension with a drop in the force (Fig. 4 A, gray curves). Before and after the transition, the force increased monotonically, reflecting the elasticity of the DNA/RNA handles and the elasticity of the handles plus single-stranded TAR RNA, respectively (10). Extension of the molecule increased by ∼18 nm in the unfolding transition. The increase is consistent with ΔX observed in the hopping and force-jump experiments. In response to the sudden increase in extension once the hairpin unfolds, the trapped bead moved rapidly toward the center of the optical trap (22), leading to a drop in the force. The slope of this unfolding, or ripping, transition is equal to the spring constant (pN/nm) of the laser trap. TAR RNA always unfolded in a single step at loading rates ranging from 0.4 pN/s to 5 pN/s. The force of RNA unfolding was 13.4 ± 0.5 pN at 0.4 pN/s (Fig. 4 B, top, gray bars). As expected (13), the mean ± standard deviation of the force required to unfold the hairpin increased as the loading rate was increased; at 1.7 pN/s the unfolding force was 14.3 ± 0.8 pN (Fig. 4 B, bottom, gray bars).
FIGURE 4.
Force-ramp experiment. (A) Typical force-extension curves of TAR RNA collected at loading rates of 0.4 and 1.7 pN/s. Unfolding trajectories are shown in gray and refolding in black. (B) Distribution of the unfolding (gray) and refolding (black) force at two loading rates. The unfolding force is defined as the force at which the molecule starts to rip; the refolding force is the force at which the zipping starts. (C) Distribution of the number of nucleotides in the zipping transition at two unloading rates. Measured ΔX in nm was converted to the number of single-stranded nucleotides with equivalent length at the refolding force using Eq. 1. The persistence length and the contour length of the RNA were assumed to be 1 nm and 0.59 nm, respectively. (D) Plots of the ln[r ln[1/N]] and ln[−r ln[1/U]] versus force. N and U are the folded and unfolded fractions, respectively. Solid boxes and crosses represent unfolding data collected at 0.4 and 1.7 pN/s, respectively. Open circles and asterisks represent refolding data collected at 0.4 and 1.7 pN/s, respectively. Data collected at 0.4 pN/s were fit to Eqs. 8 and 9 (solid lines). For unfolding, ln A and 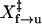 are −28.8 ± 0.9 and 8.4 ± 0.8 nm, respectively. For refolding, ln A is 28.8 ± 0.9 and
are −28.8 ± 0.9 and 8.4 ± 0.8 nm, respectively. For refolding, ln A is 28.8 ± 0.9 and 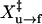 is 10.9 ± 0.9 nm. Fitting for data collected at 1.7 pN/s were not shown.
is 10.9 ± 0.9 nm. Fitting for data collected at 1.7 pN/s were not shown.
The refolding of the TAR hairpin depended on how fast the force was relaxed (Fig. 4 A, black curves). At a relaxing rate of 0.4 pN/s, the hairpin refolded in a single step, or so-called “zipping” transition. The zipping transition is indicated by a sudden decrease in extension with an increase in force, opposite of the unfolding transition. The decrease in extension, ΔX, of the zipping was converted to the number of the single-stranded nucleotides folded at the refolding force using Eq. 1 and the above-mentioned persistence and contour length (Fig. 4 B, top). The average number of nucleotides folded in the zipping was 44, slightly shorter than the expected 52 nucleotides. The discrepancy could result from a few nucleotides folding before the transition. The refolding force was 11.5 ± 0.8 pN at a relaxing rate of 0.4 pN/s. When the molecule was relaxed at a rate of 1.7 pN/s, the RNA refolded in multiple steps (Fig. 4 A). As the force was lowered from 20 pN to ∼12 pN, the extension decreased monotonically indicating that the molecule shortened with the relaxation of force. Then, within a narrow range of force, the extension of the molecule oscillated many times before the zipping transition was observed. After the zipping transition, the force-extension curve overlapped the stretching curve before the unfolding, indicating that the RNA hairpin was refolded. The force-extension curve of refolding at 1.7 pN/s was distinctive in three ways. First, the decrease in extension in the zipping transition (ΔXzipping) had a mean equivalent of 22 nucleotides folded (Fig. 4 C, bottom), significantly lower than 44 nucleotides observed at the slower loading rate (Fig. 4 C, top). This observation indicates that basepairs formed during the first/oscillating phase of refolding, which is also evident from the force-extension curve (Fig. 4 A). Second, the refolding trajectories in the oscillating phase were stochastic and show no clear common intermediates. The distribution of ΔXzipping was also broader and less symmetrical at the loading rate of 1.7 pN/s than at 0.4 pN/s. These observations suggest that the molecule refolds in multiple pathways with partially folded intermediates. Despite the different refolding trajectories, the subsequent unfolding curves display the usual unfolding transitions, indicating that the molecule always refolded into a similar structure. Third, unfolding also becomes less reversible at higher loading rates, as indicated by the increase in hysteresis in the force-extension curve (Fig. 4 A), and the larger difference between unfolding and refolding forces (Fig. 4 B).
The unfolding force is defined as the force at which the molecule starts to unfold. Similarly, the refolding force is defined as the force at which the molecule starts to basepair. On the force-extension curve (Fig. 4 A), the starting points of the unfolding and refolding transitions correspond to the left and right end of the ripping and zipping transitions, respectively. The subsequent changes in force and extension reflect the movement of the trapped bead in response to these transitions. Hence, if the RNA unfolds reversibly, the mean unfolding and refolding forces differ by changes of the force (ΔF ∼ 1.8 pN) during the transition, equal to the product of the change in the end-to-end distance of the molecule (16–18 nm) and the spring constant of the trap (∼0.11 pN/nm). By this criterion, also supported by the observation that the hysteresis in the unfolding and refolding force-extension curves is comparatively small (Fig. 4 A), TAR RNA unfolds almost reversibly at a loading of 0.4 pN/s (〈Funfolding〉 − 〈Frefolding〉 = 1.9 pN). In contrast, refolding of the TAR RNA at 1.7 pN/s usually starts at forces higher than the zipping forces (Fig. 4 A). Although it is hard to pinpoint the start point of refolding at 1.7 pN/s, the difference between 〈Funfolding〉 and 〈Frefolding〉 is >1.8 pN, indicating that unfolding is irreversible at this loading rate.
The kinetics of RNA unfolding can also be obtained by the force-ramp method (pulling) (10,11). For a two-state system in which k(F) is exponentially dependent on the force, the distribution of the ripping or unfolding force can be used to calculate the factor, Af→u, and the distance to the transition state,  using the following equation (10,13,28):
using the following equation (10,13,28):
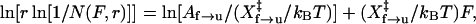 |
(8) |
where N(F,r) is the fraction of folded molecule at the force F and loading rate r. Values of A and X‡ are obtained from the slope and intercept of a plot of the left-hand side of Eq. 7 versus force. This equation is essentially the same as that derived by Evans and Ritchie (1997) to describe the ruptures of molecular adhesion bonds in atomic force microscopy (AFM) experiments (13,28). From the values of A and X‡, the rate constant at force F, k(F), can be obtained using Eq. 4.
If the molecule is refolded reversibly in a single step, a similar equation applies to the refolding:
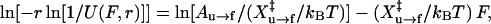 |
(9) |
where U(F,r) is the fraction of unfolded molecule at the force F and relaxation rate r; Au→f is the refolding factor; and 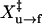 is the distance to the transition state of the refolding reaction. Eq. 9 assumes that the unloading rate is much slower than the rate of basepair formation such that refolding appears to be a single step reaction. Both Eqs. 8 and 9 assume a constant loading/unloading rate. In the real experiments, the loading/unloading rate at <2 pN may deviate from the mean value by >0.2 pN/s. Since the transition forces are 10–20 pN for RNA, we used the mean velocity from 2–20 pN as the loading/unloading rate.
is the distance to the transition state of the refolding reaction. Eq. 9 assumes that the unloading rate is much slower than the rate of basepair formation such that refolding appears to be a single step reaction. Both Eqs. 8 and 9 assume a constant loading/unloading rate. In the real experiments, the loading/unloading rate at <2 pN may deviate from the mean value by >0.2 pN/s. Since the transition forces are 10–20 pN for RNA, we used the mean velocity from 2–20 pN as the loading/unloading rate.
To obtain values of ln A and X‡ in the rate equations, we calculated the fractions of the unfolded (U) and folded (N) molecules at various forces by integrating the histogram of the force distribution (Fig. 4 B) over the corresponding range of the force. Plots of ln[r ln[1/N]] and ln[−r ln[1/U]] as a function of the force (Fig. 4 D) were fit to Eqs. 8 and 9, respectively. Unfolding data collected at the two different rates (Fig. 4 D, solid boxes and crosses) appear to overlap. We obtained ln A of −28.8 ± 0.9 and 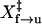 of 8.4 ± 0.8 nm from a linear fit of data collected at 0.4 pN/s (Table 1). In contrast, ln[−r ln[1/U]] versus force plots show different trends for the two rates. The plot of data from 0.4 pN/s (Fig. 4 D, open circles) is almost linear, whereas the plot of data from 1.7 pN/s (asterisks) displays curvature. When the two sets of data were fit to Eq. 9, different slopes and y intercepts were obtained. The derivation of Eq. 9 (13,28) assumes a single-step transition; however, at the force of 1.7 pN/s, the TAR RNA refolds in multiple steps that probably involve partially folded intermediates (Fig. 4 A). Clearly, Eq. 9 fails to describe refolding of TAR RNA in multiple steps. At a force of 0.4 pN/s, the hairpin to single-strand transition is two-state and the fit to Eq. 9 is linear. We obtained ln A of 28.8 ± 0.9 and
of 8.4 ± 0.8 nm from a linear fit of data collected at 0.4 pN/s (Table 1). In contrast, ln[−r ln[1/U]] versus force plots show different trends for the two rates. The plot of data from 0.4 pN/s (Fig. 4 D, open circles) is almost linear, whereas the plot of data from 1.7 pN/s (asterisks) displays curvature. When the two sets of data were fit to Eq. 9, different slopes and y intercepts were obtained. The derivation of Eq. 9 (13,28) assumes a single-step transition; however, at the force of 1.7 pN/s, the TAR RNA refolds in multiple steps that probably involve partially folded intermediates (Fig. 4 A). Clearly, Eq. 9 fails to describe refolding of TAR RNA in multiple steps. At a force of 0.4 pN/s, the hairpin to single-strand transition is two-state and the fit to Eq. 9 is linear. We obtained ln A of 28.8 ± 0.9 and 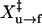 of 10.9 ± 0.9 nm for refolding (Table 1). From values of X‡ and ln A, we estimated F1/2 of 12.3 ± 0.1 pN. At this force, ΔX measured is 18 ± 2 nm. We estimated ΔG°(0),22°C for folding TAR RNA is 86 ± 16 kJ/mol (21 ± 4 kcal/mol) (Table 2).
of 10.9 ± 0.9 nm for refolding (Table 1). From values of X‡ and ln A, we estimated F1/2 of 12.3 ± 0.1 pN. At this force, ΔX measured is 18 ± 2 nm. We estimated ΔG°(0),22°C for folding TAR RNA is 86 ± 16 kJ/mol (21 ± 4 kcal/mol) (Table 2).
To better understand the differences in folding at the two unloading rates we studied refolding at an intermediate unloading rate of 1 pN/s. Under these conditions, the TAR hairpin mostly refolded in a single step, but nearly every force-extension curve displayed some fluctuations just before the folding transition (Fig. 5 A). Similarly, the force-extension curves become noisier immediately after the unfolding transition. The change in the extension of the molecule is calculated by the relative movement of the two beads: the movement of the bead on the micropipette and the motion of the trapped bead. The bead on the micropipette was moved by the piezoelectric stage at a constant rate (nm/s) and its position is a linear function of time with little noise (data not shown). The motion of the trapped bead is affected by the tension of the molecule, given that the trap resembles a Hooke's law spring. Therefore, the fluctuation on the force-extension curve is mainly affected by the noise in the force. On the time trace of force (Fig. 5 B), fluctuation of force increased significantly right after the unfolding transition and just before the refolding transition. We determined the force noise of over 150 pulling curves with the unfolding and refolding transitions excluded. During the unfolding transition (Fig. 5 C), the standard deviation of force (σF) in 0.2-pN bins was ∼0.025 pN before the transition (asterisks) and rose to 0.1 pN after the unfolding (open circles). The σF then decreased to <0.03 pN as force increased to ∼16 pN. Above 16 pN, σF was at a level comparable to that of the hairpin, ∼0.025 pN. The overlapping force region between σF before (asterisks) and after (open circles) the unfolding transition reflects the range of the unfolding force (Fig. 3 B). A similar trend was observed in the refolding (Fig. 5 D). σF was∼0.025 pN between 16–18 pN. As force was lowered below 16 pN, σF increased to a maximum of 0.11 pN. After the folding transition, σF immediately dropped to a level of between 0.02 and 0.03 pN. Clearly, force fluctuation increased significantly only when single-stranded RNA was in the force range of the transition. This suggests that under these conditions, partial folding/unfolding events occurred before the zipping transition.
FIGURE 5.
Force-ramp experiment at intermediate loading rate. (A) Force-extension curves of TAR RNA collected at a loading rate of 1.0 pN/s. (B) A time trace of force in the same force-ramp cycle. (C and D) Noise of the force (σF) in 0.2 pN bins as a function of force. Neither unfolding nor refolding transitions are included. Open circles and asterisks represent the noise due to single-strand and hairpin RNA, respectively. The overlap force region between the forms of the RNA reflects the distribution of the transition forces.
DISCUSSION
Folding pathways
Only one stable structure was predicted by the Mfold program (29) (http://www.bioinfo.rpi.edu/applications/mfold/old/rna/form1.cgi) for the TAR RNA (Fig. 1) at room temperature. The prediction is consistent with all available structures of TAR RNA under various conditions (30–33). When the hairpin unfolds into single-stranded RNA, the end-to-end distance of the entire molecule including the DNA/RNA handles will increase roughly by the length of the single-stranded RNA. In all experiments, a change in the extension of ∼18 nm was observed upon unfolding, which is consistent with the estimation of 52-nucleotide single-stranded RNA by the worm-like-chain model.
Although the TAR hairpin appeared to unfold in a single step no matter how force was applied, it refolded through different pathways depending on how force was relaxed. Under constant force conditions, the single-stranded RNA refolded into a hairpin without detectable intermediate states (Figs. 2 C and 3 A). When the molecule was relaxed at 0.4 pN/s, the RNA also refolded in a single step (Fig. 4 A). However, refolding involved multiple intermediates at an unloading rate of 1.7 pN/s (Fig. 4 A). Before the zipping transition, the force-extension curves oscillate back and forth suggesting some partial folding/unfolding events. During these fluctuations, the curves also significantly deviate from the worm-like-chain model for single-stranded RNA with handles indicating that some basepairs are formed. The stochastic trajectories suggest that refolding pathways are heterogeneous. The broad distribution of the zipping distance (Fig. 4 C) indicates that the RNA structure does not reach a common intermediate before the zipping transition occurs.
Interestingly, at an intermediate unloading rate of 1.0 pN/s, TAR RNA mostly refolds in a single step, whereas the force-extension curves show significant fluctuations right before the zipping transition (Fig. 5). The fluctuations in the force-extension curves likely represent many successive partial folding and unfolding events. These fluctuations are similar to those at 1.7 pN/s (Fig. 4 A), although the amplitude is much smaller.
It was at first surprising that the folding pathway of TAR RNA changes with the unloading rate. There must be a slow step in the folding of TAR RNA that is sensitive to the mechanical perturbation. A zipper model was proposed for folding RNA hairpins (34,35), in which a few basepairs closing the apical loop are formed first (nucleation) and then the formation of the helix quickly proceeds (zipping). When force is held near F1/2, the extension varies little for a few seconds followed by a fast zipping (Fig. 2 C). It is clear that the nucleation is the slow step and that few basepairs are formed during the nucleation. There are many possible ways for a 52-nucleotide single-stranded RNA to close a loop by forming two successive basepairs. If a nonnative basepair is formed, it has to be disrupted before the correct structure can be reached. Under constant force conditions, the molecule has sufficient time to try different combinations of basepairing until the correct one is formed. The lifetime of the unfolded state reflects a conformational search for the correct first basepairs. However, at faster unloading rates, the force may drop significantly before the misnucleated structures can rearrange. Instead, these structures are stabilized and can even proceed to form additional basepairs at lower force. As shown in Fig. 4 A, the force-extension curves at an unloading rate of 1.7 pN/s fluctuate many times before a small zipping transition occurs (Fig. 4 C). The fluctuation reflects successive partial folding and unfolding to reach a state with all basepairs native, at which the zipping occurs. The fluctuations thus also represent a conformational search. However, the amplitudes are visibly larger than the nucleation process under constant force, indicating that many basepairs are formed during the former process. Apparently, nucleation is perturbed by the relaxation of the force.
Under constant force conditions or slow force relaxation, the folding of TAR RNA acts like a two-state Markovian process. However, when force is relaxed fast (Fig. 4 A), folding shows many different trajectories and various intermediates with distinctive extensions. The initial state formed affects the formation of the next intermediates. Such a process is certainly not Markovian. It is reminiscent of the folding of ubiquitin polyproteins (9,36). Single-molecule folding trajectories of the ubiquitin display large fluctuation in the extension followed by a rapid final contraction into the fully folded structure (9). These similar observations in protein and RNA folding suggest that folding of macromolecules can follow various pathways with continuous intermediates rather than staying in a single path with well-defined discrete states.
Rate constants
When the molecule behaves like a two-state system, rate constants can be obtained assuming first-order kinetics. The unfolding rate constants, k(F)f→u, directly measured from both the hopping (Fig. 2 D) and the force-jumping experiments (Fig. 3 C) appear to be exponentially dependent on force. We have also extrapolated k(F)f→u using 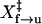 and ln Af→u obtained from the force-ramp experiments (Fig. 4 D). Results from all three measurements were pooled together (Fig. 6 A) and were fit to a linear form of Eq. 4. All experiments apparently measured the same kinetic barrier of the unfolding reaction. We obtained
and ln Af→u obtained from the force-ramp experiments (Fig. 4 D). Results from all three measurements were pooled together (Fig. 6 A) and were fit to a linear form of Eq. 4. All experiments apparently measured the same kinetic barrier of the unfolding reaction. We obtained 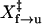 of 8.3 ± 0.1 nm and ln A of −28.3 ± 0.4.
of 8.3 ± 0.1 nm and ln A of −28.3 ± 0.4.
FIGURE 6.
Comparison of rates measured from different methods. The logarithm of the rates from force-ramp (▴), hopping (▪), and force-jump (•) are plotted as a function of the force. (A) Unfolding. Rates from all experiments are pooled together and fitted to Eq. 4. We obtained 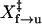 of 8.3 ± 0.1 nm and ln A of −28.3 ± 0.4. (B) Refolding. Data collected by each method were fitted to Eq. 4 independently: solid line, force-ramp; dotted line, hopping; and dashed line, force-jump.
of 8.3 ± 0.1 nm and ln A of −28.3 ± 0.4. (B) Refolding. Data collected by each method were fitted to Eq. 4 independently: solid line, force-ramp; dotted line, hopping; and dashed line, force-jump.
Refolding rates obtained from different methods show some differences. Force-jump (force-drop) measurements give a higher value of refolding rates than estimations by force-ramp (Fig. 6 B). Rates measured by hopping experiments fall between the other two. Clearly, refolding of TAR hairpin is complex as compared to unfolding. It is surprising that rates measured by force-drop (Fig. 6 B) are ∼3 times higher than those determined by hopping experiments, although the same force feedback was used in both measurements. The major difference between the two types of the experiments is the starting state of the molecule. In the hopping experiments, the RNA could form some small nonnative structures. To form the native hairpin, the RNA needs to evolve out of these local energy minima, which might slow down the overall folding. However, it is not clear to us why rates extrapolated from force-ramp experiments are slowest.
Free energy calculations
We calculated ΔG°(0),22°C from three types of measurements and the results agree well (Table 2). Values of F1/2 obtained by all three methods are close. F1/2 by force-jump is slightly higher than those by other methods because of the faster refolding rates obtained. The confidence level of ΔG°(0),22°C is mostly limited by the spatial resolution, which causes an uncertainty of ΔX at ∼2 nm. Our value was obtained in 100 mM KCl, but to compare with a nearest-neighbor free energy estimated by Mfold (29), we measured a value in 1 M salt: ΔG°(0 pN,20°C) = 124 kJ/mol (data unpublished). The calculated Mfold value is 157 kJ/mol in 1 M NaCl. Given the uncertainties in both methods for obtaining free energies and the correction of stretching energy, we consider the agreement reasonable.
Comparison of the three methods of force application
For simple two-state systems, such as the unfolding of TAR RNA, hopping, force-jump, and force-ramp methods yield similar results (Fig. 6 A). For complex reactions, such as the refolding of TAR hairpin, several methods may be needed to unravel the mechanism.
Constant force measurements allow direct measurement of the reaction rates; the effect of force on rates can be determined. In hopping experiments, the kinetics of the molecule is observed in real time under equilibrium conditions, allowing measurements of rate constants and equilibrium free energies. However, hopping experiments are limited to the narrow range of forces where both unfolding and refolding occur often enough to be observed. However, this drawback is circumvented by the force-jump or force-drop method, in which different forces are used to measure unfolding and refolding. The increase in the force range that can be used to measure the kinetics also provides better measures of the positions of kinetic barrier. The force-jump method can also be employed to measure the rate constants of individual reaction steps in complicated reactions, such as the unfolding of T. thermophila ribozyme (12).
The force-ramp method has been applied to several RNA molecules (10–12). It is a useful tool to survey heterogeneous folding/unfolding pathways and intermediates, especially for complex RNAs (12). However, quantitative interpretation of force-ramp results for complex systems is not straightforward.
It is surprising that the folding of even a relatively simple RNA hairpin, such as TAR, depends on how force is applied. Under constant force conditions, the hairpin folds without detectable intermediates (Figs. 2 C and 3 A). Even the folding kinetics appears to follow the first-order kinetics (Fig. 3 B). However, by changing the relaxation rates, we have found that the molecule instead folds in multiple pathways involving many intermediates (Fig. 5). A single method is obviously not enough to solve the mystery of RNA folding. To study complicated systems, we can use constant-force experiments to measure the overall reaction rate, or the rates of individual reaction steps; and we can apply the force-ramp method to perturb the hidden steps.
Acknowledgments
We thank Ramin Khayatpoor for technical assistance in developing codes for force-jump and Dr. Jin-Der Wen, Ms. Sophie Dumont, and Mr. Jeffrey Vieregg for suggestions and discussion.
This research was supported by National Institutes of Health grants GM-10840 (I.T.) and GM-32543 (C.B.), and Department of Energy grant DOE-AC03-76SF00098 (I.T. and C.B.).
Pan T. X. Li and Delphine Collin contributed equally to this work.
References
- 1.Leckband, D. 2000. Measuring the forces that control protein interactions. Annu. Rev. Biophys. Biomol. Struct. 9:1–26. [DOI] [PubMed] [Google Scholar]
- 2.Leckband, D., W. Muller, F. Schmitt, and H. Ringsdorf. 1995. Molecular mechanisms determining the strength of receptor-mediated intermembrane adhesion. Biophys. J. 69:1162–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muller, D., J. Heymann, F. Oesterhelt, C. Moller, H. Gaub, G. Buldt, and A. Engel. 2000. Atomic force microscopy of native purple membrane. Biochim. Biophys. Acta. 1460:27–38. [DOI] [PubMed] [Google Scholar]
- 4.Hugel, T., N. Holland, A. Cattani, L. Moroder, M. Seitz, and H. Gaub. 2002. Single-molecule optomechanical cycle. Science. 296:1103–1106. [DOI] [PubMed] [Google Scholar]
- 5.Marszalek, P., H. Li, A. Oberhauser, and J. Fernandez. 2002. Chair-boat transitions in single polysaccharide molecules observed with force-ramp AFM. Proc. Natl. Acad. Sci. USA. 99:4278–4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith, S., L. Finzi, and C. Bustamante. 1992. Direct mechanical measurements of the elasticity of single DNA molecules by using magnetic beads. Science. 258:1122–1126. [DOI] [PubMed] [Google Scholar]
- 7.Smith, S., Y. Cui, and C. Bustamante. 1996. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 271:795–799. [DOI] [PubMed] [Google Scholar]
- 8.Rief, M., M. Gautel, F. Oesterhelt, J. M. Fernandez, and H. E. Gaub. 1997. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 276:1109–1112. [DOI] [PubMed] [Google Scholar]
- 9.Fernandez, J. M., and H. Li. 2004. Force-clamp spectroscopy monitors the folding trajectory of a single protein. Science. 303:1674–1678. [DOI] [PubMed] [Google Scholar]
- 10.Liphardt, J., B. Onoa, S. Smith, I. Tinoco, Jr., and C. Bustamante. 2001. Reversible unfolding of single RNA molecules by mechanical force. Science. 292:733–737. [DOI] [PubMed] [Google Scholar]
- 11.Liphardt, J., S. Dumont, S. Smith, I. Tinoco, Jr., and C. Bustamante. 2002. Equilibrium information from nonequilibrium measurements in an experimental test of Jarzynski's equality. Science. 296:1832–1835. [DOI] [PubMed] [Google Scholar]
- 12.Onoa, B., S. Dumont, J. Liphardt, S. Smith, I. Tinoco, Jr., and C. Bustamante. 2003. Identifying kinetic barriers to mechanical unfolding of the T. thermophila ribozyme. Science. 299:1892–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans, E., and K. Ritchie. 1997. Dynamic strength of molecular adhesion bonds. Biophys. J. 72:1541–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tinoco, I., Jr., and C. Bustamante. 2002. The effect of force on thermodynamics and kinetics of single molecule reactions. Biophys. Chem. 101–102:513–533. [DOI] [PubMed] [Google Scholar]
- 15.Tinoco, I., Jr. 2004. Force as a useful variable in reactions: unfolding RNA. Annu. Rev. Biophys. Biomol. Struct. 33:363–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oberhauser, A., P. Hansma, M. Carrion-Vazquez, and J. Fernandez. 2001. Stepwise unfolding of titin under force-clamp atomic force microscopy. Proc. Natl. Acad. Sci. USA. 98:468–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brower-Toland, B., C. Smith, R. Yeh, J. Lis, C. Peterson, and M. Wang. 2002. Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proc. Natl. Acad. Sci. USA. 99:1960–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turner, D., G. Flynn, S. Lundberg, L. Faller, and N. Sutin. 1972. Dimerization of proflavin by the laser raman temperature-jump method. Nature. 239:215–217. [DOI] [PubMed] [Google Scholar]
- 19.O'Leary, M., and W. J. Brummund. 1974. pH jump studies of glutamate decarboxylase. Evidence for a pH-dependent conformation change. J. Biol. Chem. 249:3737–3745. [PubMed] [Google Scholar]
- 20.Acuna, M., F. Mingot, and C. Davila. 1976. Reversibility of partial denaturation of DNA. Biochim. Biophys. Acta. 454:45–50. [DOI] [PubMed] [Google Scholar]
- 21.Milligan, J., and O. Uhlenbeck. 1989. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 180:51–62. [DOI] [PubMed] [Google Scholar]
- 22.Smith, S., Y. Cui, and C. Bustamante. 2003. Optical-trap force transducer that operates by direct measurement of light momentum. Methods Enzymol. 361:134–162. [DOI] [PubMed] [Google Scholar]
- 23.Muesing, M., D. Smith, and D. Capon. 1987. Regulation of mRNA accumulation by a human immunodeficiency virus trans-activator protein. Cell. 48:691–701. [DOI] [PubMed] [Google Scholar]
- 24.Rosen, C., J. Sodroski, and W. Haseltine. 1985. The location of cis-acting regulatory sequences in the human T cell lymphotropic virus type III (HTLV-III/LAV) long terminal repeat. Cell. 41:813–823. [DOI] [PubMed] [Google Scholar]
- 25.Lang, M., C. Asbury, J. Shaevitz, and S. Block. 2002. An automated two-dimensional optical force clamp for single molecule studies. Biophys. J. 83:491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bustamante, C., J. Marko, E. Siggia, and S. Smith. 1994. Entropic elasticity of lambda-phage DNA. Science. 265:1599–1600. [DOI] [PubMed] [Google Scholar]
- 27.Seol, Y., G. Skinner, and K. Visscher. 2004. Elastic properties of a single-stranded charged homopolymeric ribonucleotide. Phys. Rev. Lett. 93:118102. [DOI] [PubMed] [Google Scholar]
- 28.Evans, E. 2001. Probing the relation between force-lifetime-and chemistry in single molecular bonds. Annu. Rev. Biophys. Biomol. Struct. 30:105–128. [DOI] [PubMed] [Google Scholar]
- 29.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brodsky, A., and J. Williamson. 1997. Solution structure of the HIV-2 TAR-argininamide complex. J. Mol. Biol. 267:624–639. [DOI] [PubMed] [Google Scholar]
- 31.Aboul-ela, F., J. Karn, and G. Varani. 1995. The structure of the human immunodeficiency virus type-1 TAR RNA reveals principles of RNA recognition by Tat protein. J. Mol. Biol. 253:313–332. [DOI] [PubMed] [Google Scholar]
- 32.Aboul-ela, F., J. Karn, and G. Varani. 1996. Structure of HIV-1 TAR RNA in the absence of ligands reveals a novel conformation of the trinucleotide bulge. Nucleic Acids Res. 24:3974–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang, K., and I. Tinoco, Jr. 1997. The structure of an RNA “kissing” hairpin complex of the HIV TAR hairpin loop and its complement. J. Mol. Biol. 269:52–66. [DOI] [PubMed] [Google Scholar]
- 34.Cantor, C., and P. Schimmel. (1980) Biophysical Chemistry. Freeman, San Francisco.
- 35.Bloomfield, V., D. Crothers, and I. Tinoco, Jr. (1974) Physical Chemistry of Nucleic Acids. Harper and Row, New York
- 36.Schlierf, M., H. Li, and J. M. Fernandez. 2004. The unfolding kinetics of ubiquitin captured with single-molecule force-clamp techniques. Proc. Natl. Acad. Sci. USA. 01:7299–7304. [DOI] [PMC free article] [PubMed] [Google Scholar]








