Abstract
Position of the transmembrane aromatic residues of the KirBac1.1 potassium channel shifts from an even distribution in the closed state toward the membrane/solute interface in the open state model. This is the first example of an integral membrane protein making use of the observed preference for transmembrane aromatic residues to reside at the interfaces. The process of aromatic localization is proposed as a means of directing and stabilizing structural changes during conformational transitions within the transmembrane region of integral membrane proteins. All-atom molecular dynamics simulations of the open and closed conformers in a membrane environment have been carried out to take account of the interactions between the aromatic residues and the lipids, which may be involved in the conformational change, e.g., the gating of the channel.
Despite the steady increase in the number of integral membrane proteins (IMP) in the Protein Data Bank and the recent success of computational studies on biological channels (1), there is still a relatively poor understanding of the extent and nature of the interactions between IMP and the surrounding lipids, and more importantly, about the functional role these interactions might have in processes such as gating and modulation. This communication reports, to our knowledge, the first example of an IMP making use of the observed preference for transmembrane aromatic residues that reside at the interfaces. The process of aromatic localization is proposed as a means of directing and stabilizing structural changes during conformational transitions within the transmembrane region of KirBac. Multiple nanosecond molecular simulations are employed to establish a qualitative picture of the intermolecular interactions between a lipid bilayer and the aromatic residues of a membrane protein for which a high resolution x-ray closed structure (2) and an open model (3–4) are available (Fig. 1).
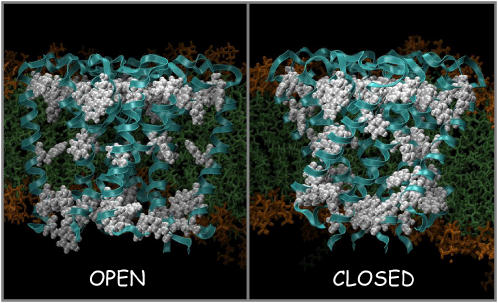
FIGURE 1 .
Schematic diagrams (drawn using VMD) of the aromatic localization at the membrane/solute interface of the open and closed state models. The protein backbone is drawn in cyan ribbons; aromatic side chains are displayed in red, space-filling format.
One feature present in all membrane proteins is the localization of their transmembrane aromatic residues at the membrane/solute interface. It has been known for a number of years that aromatic residues preferentially reside at this interface (5–7), but their functional role is unclear. These aromatic belts are suggested to anchor the protein within the flexible membrane, although several other roles related to packing, insertion, and folding (7–9) have also been attributed to them. Anchoring of the protein to the membrane is accomplished by interactions of a polar part of the aromatic amino acids of the protein with the phospholipid headgroups at the membrane-water interface, and the hydrophobic part of these amino acids with lipid acyl chains (9). Primarily, Trp and Tyr are the amino acids reported to prefer being concentrated in the lipid headgroup region near the ends of the transmembrane helices, although in this particular protein there is a high content of Phe.
The closed and the open states surprisingly show contrasting views of the location of the aromatic residues (Fig. 1). In the closed state, the aromatic residues are evenly distributed along the length of the transmembrane helices; in contrast, they show a dramatic shift toward the external and internal interfacial regions plus a band located at the central cavity section in the open state (Fig. 1). This dramatic change in the location of aromatic residues between closed and open states is the first view of an IMP using the interfacial aromatic residues as part of protein function. As these aromatic residues are highly conserved in this family of K channels (10), we propose that aromatic localization could contribute to the driving force during the conformational changes associated with gating in the Kir family.
To characterize the dynamics of these residues, >40 ns of molecular dynamics simulations were carried out in a dioleoylphosphatidylcholine lipid bilayer using NAMD. System sizes were of ∼132,000 atoms.
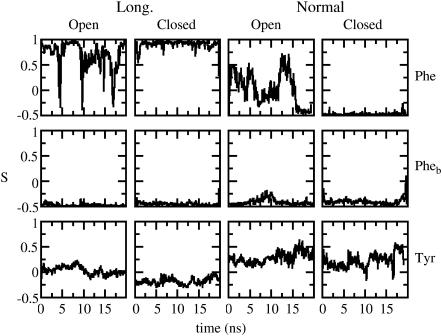
Previous simulation studies have probed the role of aromatics in anchoring proteins to lipid bilayers solely (11,12). Our simulation aims to shed light on the role of the aromatic residues in the conformational changes implicated in the gating mechanism. We have analyzed the nature of the interactions between the aromatic residues of the protein in the open and closed states and the lipid membrane. In KirBac1.1, there are 24 Tyr, 12 Trp, and 44 Phe residues in the transmembrane domain, including the slide helices (there are 460 residues in total in the transmembrane domain). When the channel is open, the distribution of these residues is in three bands: 32 (12 Tyr + 20 Phe) residues in the upper (periplasmic) belt, 40 (12 Tyr + 12 Trp + 16 Phe) residues in the lower (cytoplasmic) belt, and 8 Phe residues in the central belt. Phe appears to have no preference for the interface over the core of the membrane (13). During the simulation time, the number of contacts between the lipid bilayer and the open conformer are greater than for the closed conformer, not surprisingly as the surface-accessible area of the open form is greater. It is also observed that the number of contacts between each type of aromatic residues and the lipids is also greater in the open conformer due to the movement of some of the originally buried residues toward the lipids. Two order parameters, SL and SN, were calculated to characterize the orientation of these residues in both conformers in the bilayer. SN (½(3cos2 − 1)) is the angle between the axis normal to the bilayer and a vector normal to the plane of the aromatic ring, and SL is the angle between the axis normal to the bilayer and a vector from Cβ to Cα in Tyr and Phe. The time evolution trends in the orientation of Tyr and Trp residues remain unchanged between the closed and open conformers unlike Phe residues. SL and SN values fluctuate by ∼16° and ∼35°, respectively, on a picosecond timescale in both Tyr and Trp. However, Trp residues change their orientations from parallel to perpendicular on a nanosecond timescale. The picture that emerges from the analysis of the SN and SL values for Phe residues is somehow more complex. Those Phe residues located in the pore helix and buried within the protein (Phe102 and Phe103) do not move at all in either the closed or the open conformers. In contrast, the rest of Phe residues behave very differently; SN and SL values globally change on a nanosecond timescale in the open conformer, whereas they remain constant in the closed conformer with fluctuations on a picosecond timescale of ∼45° and ∼16° in the open and closed conformations, respectively. Representative cases are plotted in Fig. 2. In summary, the orientation of the Phe residues dramatically changes in the open conformation, whereas it remains uniform in the closed state, except for the residues that are buried in the protein and are totally immobilized in both the closed and open conformations.
FIGURE 2 .
Orientation of representative cases of aromatic rings in the open and closed conformations as a function of time. The first row corresponds to any nonburied Phe residues; the second row, labeled Pheb, corresponds to Phe112 or Phe113; and the third row corresponds to any of the Tyr residues. Data are smoothed in windows of 75 ps. Data corresponding to only one of the residues of one of the four chains of the tetramer are shown for clarity as they all behave in a similar manner.
The interactions have been analyzed further in terms of H-bonding between lipid and aromatic residues. A hydrogen bond was counted if the donor-acceptor distance was <0.35 nm and the angle between hydrogen-donor-acceptor was <60°. Interactions between headgroups and aromatic residues of the closed conformer are appreciable with contributions coming from Trp>Tyr>Phe; they become negligible for the open conformer with the exception of Tyr (but these are the residues located in the slide helices). It might appear contradictory that there is a decrement in hydrogen bonds as the surface exposure of aromatic residues to lipids increases in going from closed to open. This observation may be accounted by the fluctuations of the aromatic residues discussed above, and it should also be noted that it may be advantageous to have fewer lipid/aromatic contacts when the channel is in the open state to prevent the stabilization of the open state of the channel for a long time.
The results presented in this communication provide computational and direct structural evidence of the nature of interaction between a lipid bilayer and IMPs. Many membrane proteins are involved in the transport of ions and small molecules, and these are likely to undergo significant conformational changes within their transmembrane section as the protein moves from one state to another. The process of aromatic localization may be part of the driving force as the protein cycles between the different conformational states. In this model, the aromatic residues can be thought of as a buoy in the sea, directing the movement of any attached item to the correct level. Even if the time spent in one conformational state is not preferred over another, it can be envisioned that the location of the transmembrane aromatic residues is optimized to prevent the formation of an energetic well during transitions between states. Therefore, the process of aromatic localization may be a general method used by membrane proteins as the transmembrane sections move between various conformational states and aromatic residues are not exclusively used in order that a membrane protein is stably inserted in a bilayer. Though aromatic belts seem to facilitate the anchoring of the protein within the membrane through their interactions with the polar heads of membrane lipids (Trp and Tyr), we also suggest that in this particular family of IMPs, the interfacial aromatic residues (Phe) play a role in guiding conformation changes as the channel cycles between various gating states.
Acknowledgments
C.D. thanks the Royal Society and the Engineering and Physical Sciences Research Council National Service for Computational Chemistry Software. S.V. and M.L.K. thank the National Institutes of Health and the National Center for Supercomputing Applications Supercomputing facilities. C.V.-B. is grateful to the Wellcome Trust and D.A.D. to the Structural Genomics Consortium.
References
- 1.Roux, B. 2005. Ion conduction and selectivity in K(+) channels. Annu. Rev. Biophys. Biomol. Struct. 34:153–171. [DOI] [PubMed] [Google Scholar]
- 2.Kuo, A., J. M. Gulbis, J. F. Antcliff, T. Rahman, E. D. Lowe, Z. Jochen, J. Cuthbertson, F. M. Ashcroft, T. Ezaki, and D. A. Doyle. 2003. Crystal structure of the potassium channel KirBac1.1 in the closed state. Science. 300:1922–1926. [DOI] [PubMed] [Google Scholar]
- 3.Domene, C., D. A. Doyle, and C. Venien-Bryan. 2005. Modeling of an ion channel in its open conformation. Biophys. J. 89:L01–L03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuo, A., C. Domene, L. N. Johnson, D. A. Doyle, and C. Venien-Bryan. 2005. Two different conformational states of KirBac3.1 potassium channel revealed by electron crystallography. Structure. 13:1463–1472. [DOI] [PubMed] [Google Scholar]
- 5.Arkin, I., and A. Brunger. 1998. Statistical analysis of predicted transmembrane alpha-helices. Biochim. Biophys. Acta. 1429:113–128. [DOI] [PubMed] [Google Scholar]
- 6.Persson, S., J. A. Killian, and G. Lindblom. 1998. Molecular ordering of interfacially localized tryptophan analogs in ester- and ether-lipid bilayers studied by H-2-NMR. Biophys. J. 75:1365–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braun, P., and G. von Heijne. 1999. The aromatic residues Trp and Phe have different effects on the positioning of a transmembrane helix in the microsomal membrane. Biochemistry. 38:9778–9782. [DOI] [PubMed] [Google Scholar]
- 8.Hu, W., K. C. Lee, and T. A. Cross. 1993. Tryptophans in membrane-proteins—indole ring orientations and functional implications in the gramicidin channel. Biochemistry. 32:7035–7047. [DOI] [PubMed] [Google Scholar]
- 9.Gaede, H., W. Yau, and K. Gawrisch. 2005. Electrostatic contributions to indole-lipid interactions. J. Phys. Chem. B. 109:13014–13023. [DOI] [PubMed] [Google Scholar]
- 10.Shealy, R. T., A. D. Murphy, R. Ramarathnam, E. Jakobsson, and S. Subramaniam. 2003. Sequence-function analysis of the K-selective family of ion channels using a comprehensive alignment and the KcsA channel structure. Biophys. J. 84:2929–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tieleman, D. P., L. R. Forrest, M. S. P. Sansom, and H. J. C. Berendsen. 1998. Lipid properties and the orientation of aromatic residues in OmpF, influenza M2, and alamethicin systems: molecular dynamics simulations. Biochemistry. 37:17554–17561. [DOI] [PubMed] [Google Scholar]
- 12.Domene, C., P. J. Bond, S. S. Deol, and M. S. P. Sansom. 2003. Lipid/protein interactions and the membrane/water interfacial region. J. Am. Chem. Soc. 125:14966–14967. [DOI] [PubMed] [Google Scholar]
- 13.Ulmschneider, M. B., and M. S. P. Sansom. 2001. Amino acid distributions in integral membrane protein structures. Biochim. Biophys. Acta. 1512:1–14. [DOI] [PubMed] [Google Scholar]