Abstract
The availability of seven different structures of cytochrome f (cyt f) from Chlamydomonas reinhardtii allowed us, using Brownian dynamics simulations, to model interactions between these molecules and their redox partners, plastocyanin (PC) and cytochrome c6 (cyt c6) in the same species to study the effect of cyt f structure on its function. Our results showed that different cyt f structures, which are very similar, produced different reaction rates in interactions with PC and cyt c6. We were able to attribute this to structural differences among these molecules, particularly to a small flexible loop between A-184 and G-191 (which has some of the highest crystallographic temperature factors in all of the cyt f structures) on the cyt f small domain. We also showed that deletion of the cyt f small domain affected cyt c6 more than PC, due to their different binding positions on cyt f. One function of the small domain in cyt f may be to guide PC or cyt c6 to a uniform dock with cyt f, especially due to electrostatic interactions with K-188 and K-189 on this domain. Our results could serve as a good guide for future experimental work on these proteins to understand better the electron transfer process between them. Also, these results demonstrated the sensitivity and the power of the Brownian dynamics simulations in the study of molecular interactions.
INTRODUCTION
The cytochrome b6f (cyt b6f) complex is an oligomeric membrane protein complex, which is one of the three major redox complexes residing in the thylakoid membrane (1,2). Electrons pass through this complex from Photosystem II to Photosystem I. Cytochrome f (cyt f), which is one of the main subunits of this complex, transfers electrons to one of two mobile electron carriers, namely, plastocyanin (PC) or cytochrome c6 (cyt c6), which transfers the electron to the Photosystem I complex (3). The availability of Chlamydomonas reinhardtii, cyt f, PC, and cyt c6 structures allowed us to model electron transfer interactions between these redox partners in the same species.
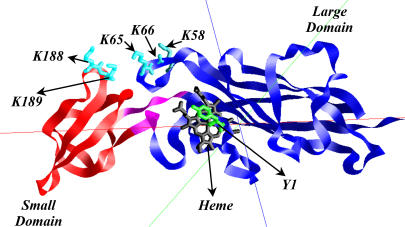
The luminal portion of cyt f is a ∼28 kDa β-sheet protein consisting of two domains (4–6). As can be seen in Fig. 1, the larger of the two domains, residues 1–170 and 230–251 in C. reinhardtii, binds the heme. The residues of the large domain are highly conserved. In contrast, the residues of the small domain (residues 171–229) are less conserved among different organisms (7). Five important lysine residues in C. reinhardtii cyt f contribute to a positive electrostatic field that it is believed to attract negative charges on PC or cyt c6. Of these, K-58, K-65, and K-66 are located on the large domain, and K-188 and K-189 are found on the small domain (4,8–11).
FIGURE 1.
C. reinhardtii cyt f, structure C of the PDB code1CFM, with its large and small domains colored in blue and red, respectively. The bridge connecting the two domains is shown in magenta. Heme and some of the key basic residues on cyt f are also shown. Tyr-1, the ligand to the heme, is shown as green.
PC is an 11 kDa “blue” copper, β-sheet protein that has two clusters of negatively charged residues called the upper and lower cluster, respectively (7,10,12–14). The upper cluster consists of residue Nos. 59–61, but it should be noted that PCs from all algae and some species of higher plants have a two-residue deletion in this region (13). For example, C. reinhardtii PC has only two negatively charged residues at positions No. 59 and No. 61, but there is a third negatively charged residue at position No. 85. The lower cluster consists of residue Nos. 42–44 and either No. 45 or No. 79, which are conserved in all higher plants and green algal PCs. Close to these clusters, there is also another negative group, D-53. These eight anionic residues produce a large negative electrostatic field surrounding PC (15).
Cyt c6 is only present in some algae and cyanobacteria (16), although a cyt c6-like protein in a higher plant (Arabidopsis) has been recently reported (17). Cyt c6 from C. reinhardtii is a 10 kDa α-helical heme protein with no sequence homology to PC (10,18,19). Nonetheless, both of these proteins have a similar pattern of negatively charged residues on their surfaces, resulting in very similar electrostatic potentials. According to Ullmann et al. (20), residues E-47, D-41, and E-54 on cyt c6 correspond to D-53, D-59–D-61, and E-85 on PC (the upper cluster), respectively. Glutamate Nos. 69–71 on cyt c6 correspond to the negative residues of the lower negative cluster of PC (D-42, E-43, D-44 (20)).
Experimental data support the electrostatic nature of the interactions of cyt f with PC. Some of these in vitro studies include cross-linking (21); chemical replacement of positively charged groups on cyt f and negatively charged groups on PC (22–25); increasing the salt concentrations (26–28); mutations of negatively charged residues on PC (29,30) and positively charged residues on cyt f (31,32); and cyt f-PC complexes studied by NMR (33–36). Computational modeling has also pointed toward the electrostatic interactions of PC or cyt c6 with cyt f (8–11,37–40). However, experimental work by Soriano et al. (41) and Zhou et al. (42) showed much smaller electrostatic interactions between cyt f and PC in vivo in C. reinhardtii.
Gong et al. (43) used a C. reinhardtii mutant of cyt f, which lacked the small domain, and studied its interactions with PC in vivo. They concluded that deleting the small domain did not affect the electron transfer rate to PC and, therefore, this domain was not involved in the interactions. Their observation matched with in vivo mutation of the two basic residues K-188 and K-189 on the small domain of cyt f (41). Note that in all of these experiments, PC would have been the electron acceptor for cyt f. However the in vitro electron transfer studies (31,32) plus the NMR complexes between the cyt f and PC (33–36) indicated that these lysine residues are involved in the interactions with PC. Computational modeling (8–11,37–40) also pointed toward the importance of the positively charged residues on cyt f on its interactions with PC.
In this work, we used Brownian dynamics (BD) simulations to examine the role of C. reinhardtii cyt f structure on its ability to interact with both C. reinhardtii PC and cyt c6. To do this, we carried out simulations using the seven available C. reinhardtii cyt f structures, which show small differences in conformation, particularly in the small domain. Three of these structures are found in the unit cell of the crystal structure of the truncated cyt f (i.e., cyt f lacking the transmembrane tail) obtained by Chi et al. (6); another three are found in the crystal structure solved by Sainz et al. (44), also of the truncated cyt f; and the seventh is the extramembrane domain of the cyt f subunit of the cyt b6f complex (2). We modeled the interaction of PC and cyt c6 with all seven intact cyt f structures and their respective mutants containing only the large domain (i.e., small domain deletion mutants). We were interested in understanding the role of the cyt f structure, both the small and the large domains, in interacting with its redox partners at the molecular level.
In BD simulations (45–48), a mobile diffusing molecule, such as PC or cyt c6, is allowed to dock with a target molecule, such as cyt f, under the influence of an electrostatic field plus random Brownian motions. BD simulations provide two important pieces of information (among many others): 1), the rate at which two proteins interact with each other, and 2), the structure of the complexes formed.
Our studies showed that different C. reinhardtii cyt f structures, which are very similar, produced different reaction rates when interacting with PC and cyt c6. We were able to attribute this to structural differences between these molecules, particularly to a small flexible loop on the small domain. We also showed that deletion of the small domains affected cyt c6 more than PC, due to its different binding position on cyt f. Our results could serve as a good guide for future experimental work on these proteins to understand better the process of electron transfer between them. These results demonstrate the sensitivity and the power of the BD simulations in the study of the molecular interactions.
METHODS
Molecular structures
The crystal structures for C. reinhardtii cyt f, PC, and cyt c6 were obtained from the Protein Data Bank (PDB; http://www.rcsb.org/pdb/; (49)). The seven cyt f structures used were three structures in the unit cell of the PDB code of 1CFM (6), three structures in the unit cell of the PDB code of 1EWH (44), and the extramembrane domain of cyt f subunit from the cyt b6f complex with the PDB code of 1Q90 (2). The PC structure used was that of PDB code 2PLT (12), and the cyt c6 used was that of PDB code 1CYJ (19). The residue numbering that we used in this work is that of the PDB files. We generated the cyt f small domain deletion mutant structures by removing residues 171–229 from the intact structures (no energy minimization was performed, keeping the orientation of the side chains unchanged).
Molecular representations
All molecular representations were made using the program GRASP (50). Root mean-square (RMS) distance deviations were calculated using the program Deep View (Swiss-PDB Viewer (51); http://www.expasy.org/spdbv/). Fig. 7 was also generated by this program.
FIGURE 7.
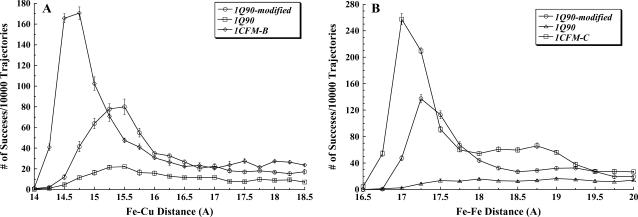
Interactions of the extramembrane domain of the cyt f subunit from the cyt b6f complex (PDB code 1Q90) with PC and cyt c6 at 10 mM ionic strength and pH 7.0. The conditions are the same as for Fig. 3. (A) The 1Q90 cyt f and its modified form in which the loop of residues A-184-G-191 is replaced with the corresponding loop from the 1CFM-B cyt f structure (see Methods section) interacting with PC. (B) The 1Q90 cyt f and its modified form interacting with cyt c6. The interactions of 1CFM-B with PC and 1CFM-C with cyt c6 are shown for comparison.
Brownian dynamics simulations
The simulations were carried out using program MacroDox v. 3.2.1 (http://pirn.chem.tntech.edu/macrodox.html) exactly as described in detail by Gross and Pearson (10) and Haddadian and Gross (11). Typically, five sets of 10,000 trajectories at 10 mM ionic strength and pH 7.0 were carried out (to obtain and minimize the error values in the simulations).
The equation of the motion used in the BD algorithm for each trajectory is the Ermak-McCammon equation (52):
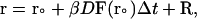 |
(1) |
where  and
and 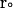 are the final and initial distances, respectively, between the center of mass of the mobile molecule (PC or cyt c6) and the center of mass of the target molecule (cyt f) before and after a time step of
are the final and initial distances, respectively, between the center of mass of the mobile molecule (PC or cyt c6) and the center of mass of the target molecule (cyt f) before and after a time step of 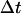 ;
; 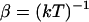 ;
; 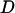 is the relative diffusion coefficient of the two molecules;
is the relative diffusion coefficient of the two molecules; 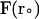 is the external force on the mobile molecule at
is the external force on the mobile molecule at 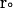 ; and R is a random (Brownian) vector with the following properties (53,54):
; and R is a random (Brownian) vector with the following properties (53,54):
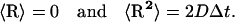 |
(2) |
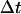 should be sufficiently small so that there is a minimal change in the external force (i.e.,
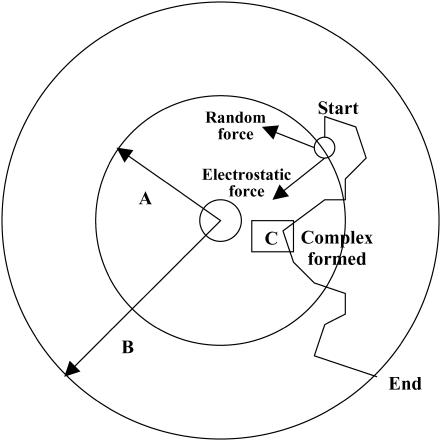
should be sufficiently small so that there is a minimal change in the external force (i.e., 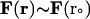 ). For each trajectory, the center of mass of the mobile molecule is positioned on the surface of a sphere of radius 90 Å, centered at the center of the mass of the target molecule as shown in Fig. 2 (sphere A). At the start of each trajectory, the program determines the position and the orientation of the mobile molecule randomly on this sphere. The mobile molecule is subjected to a force,
). For each trajectory, the center of mass of the mobile molecule is positioned on the surface of a sphere of radius 90 Å, centered at the center of the mass of the target molecule as shown in Fig. 2 (sphere A). At the start of each trajectory, the program determines the position and the orientation of the mobile molecule randomly on this sphere. The mobile molecule is subjected to a force, 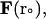 in our case an electrostatic force, and moves accordingly, after which
in our case an electrostatic force, and moves accordingly, after which 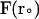 and R are recalculated. When the mobile molecule leaves a sphere of 200 Å radius from the center of the mass of the target molecule, the trajectory is concluded (sphere B in Fig. 2). In addition, at each step of the trajectory, the overlaps between all of the atoms of the mobile molecule and the target molecule are checked and prevented. In MacroDox, both of the molecules are treated as rigid rotating bodies. An equation similar to Eq. 1 describes the rotation of the mobile molecule due to the torque exerted on it by the target molecule (55).
and R are recalculated. When the mobile molecule leaves a sphere of 200 Å radius from the center of the mass of the target molecule, the trajectory is concluded (sphere B in Fig. 2). In addition, at each step of the trajectory, the overlaps between all of the atoms of the mobile molecule and the target molecule are checked and prevented. In MacroDox, both of the molecules are treated as rigid rotating bodies. An equation similar to Eq. 1 describes the rotation of the mobile molecule due to the torque exerted on it by the target molecule (55).
FIGURE 2.
BD methodology. PC or cyt c6 is randomly placed on a sphere A, 90 Å distance from the center of mass of cyt f. It is allowed to move one step at a time under the influence of an electrostatic field and a random Brownian factor. Many such steps form a trajectory, which is terminated when the mobile molecule exits sphere B (200 Å). The smallest Fe-Cu or Fe-Fe distance is recorded for each trajectory (point C).
MacroDox determines the closest approach of the two molecules based on a set of preselected reaction criteria. In our simulations, these reaction criteria were chosen as metal-to-metal distances to select for the electron transfer-active complexes. The shorter the distance between metal centers, the higher the chance of electron transfer (Moser et al. (56,57)). In the cyt c6-cyt f interactions, we used iron-to-iron, Fe-Fe, distance as the reaction criterion, and for cyt f-PC interactions, iron-to-copper, Fe-Cu, distance was used.
The smallest value of the reaction criterion for the trajectory shown in Fig. 2 is at point C. For each successful trajectory, MacroDox records the distance of C, the structure of the complex formed in the form of a PDB file, the 15 closest electrostatic contacts in the complex, and the electrostatic interaction energy for the complex. After all of the trajectories have been concluded, the number of successes is determined and plotted as a function of their metal-to-metal distances. Interaction rates were calculated as a function of metal-to-metal distances using equations derived by Northrup et al. (45–50,55). These equations calculate the diffusion-controlled second order rate constants, k2, from the fraction of trajectories that met the preset metal-to-metal distances. In this study, a value of 16 Å cutoff for Fe-Cu and a value of 18 Å for Fe-Fe distances were used (this will be discussed further in the results section).
BD methodology and the program MacroDox have been used successfully to study the cyt f-PC and cyt f-cyt c6 interactions (9–11,38,40) plus the interactions between other proteins (58,59). For a description of the MacroDox strengths and weaknesses, see Gross and Pearson (10), Haddadian and Gross (11), and Gross (40).
Electrostatic calculations
MacroDox uses a modified Tanford-Kirkwood pK algorithm (60) to assign charges on the molecules. In addition, the charge on H-37 and H-87 on PC and one of the histidine residues on cyt f (H-25) were set to zero, because they are ligated to the metal centers (the other histidine residue on cyt f is far away from the metal center). The charge on the single histidine on cyt c6 was also set to zero, due to the fact that it is a heme ligand. C-84 is a ligand to the Cu atom on PC; its sulfur atom was assigned a net charge of −1 (15), and the Cu atom was given a charge of +2. The heme charges for both cyt f and cyt c6 were Fe (+2): two ring nitrogen atoms (−1 each) and the two propionic acid side chains (−1 each).
The electrostatic potentials were calculated using the Warwicker/Watson finite difference method to solve for the linearized Poisson-Boltzmann equation (61). MacroDox uses a 61 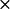 61
61 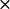 61 cubic grid, with its center positioned at the center of the mass of the protein, to solve for the electrostatic potential. We used a grid spacing of 3.6 Å, followed by a smaller spacing of 1.2 Å for the electrostatic potential calculations.
61 cubic grid, with its center positioned at the center of the mass of the protein, to solve for the electrostatic potential. We used a grid spacing of 3.6 Å, followed by a smaller spacing of 1.2 Å for the electrostatic potential calculations.
All of the simulations were carried out on a Silicon Graphics O2 workstation (IRIX 6.5) (Mountain View, CA).
RESULTS
The interactions of the six different structures of truncated cyt f with PC
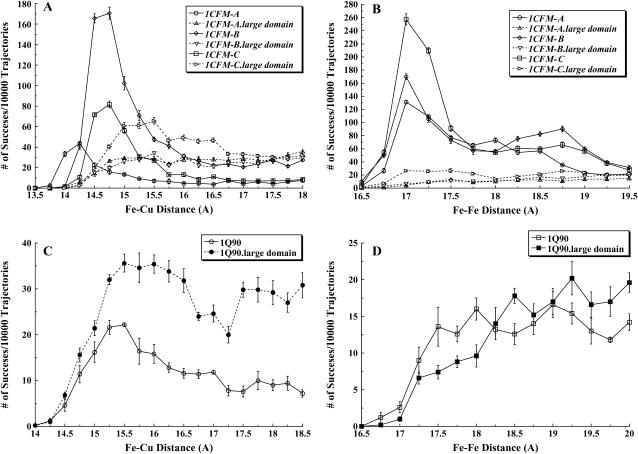
We modeled the interactions between different cyt f structures and their large domains (small domain deletion mutants) with PC in the C. reinhardtii system using the BD simulation program MacroDox at 10 mM ionic strength and pH 7.0. A plot of the number of successful complexes formed versus the reaction criterion coordinate distance is shown in Fig. 3 A for PC interacting with each of the three cyt f structures in the unit cell of the PDB code 1CFM. The reaction criterion for cyt f-PC interactions was the Fe-Cu distance.
FIGURE 3.
C. reinhardtii cyt f-PC and cyt f-cyt c6 interactions. The intact cyt fs and their corresponding large domains were interacted with PC and cyt c6 at 10 mM ionic strength and pH 7.0. Five sets of 10,000 trajectories each were carried out, after which the average of the number of successful complexes per 10,000 trajectories were plotted as a function of Fe-Cu (for cyt f-PC interactions) and Fe-Fe (for cyt f-cyt c6 interactions) distance at the closest approach. The number of complexes with closest metal-to-metal distances between 15 and 15.25 Å are indicated by the point at 15 Å, etc. (A) Three cyt f structures in the PDB code of 1CFM and their corresponding large domains interacting with PC. (B) Three cyt f structures in the PDB code of 1CFM and their corresponding large domains interacting with cyt c6. (C) The extramembrane domain of the cyt f subunit from the cyt b6f complex (PDB code 1Q90) and its large domain interacting with PC. (D) The extramembrane domain of the cyt f subunit from the cyt b6f complex (PDB code 1Q90) and its large domain interacting with cyt c6.
Of the three different cyt f molecules in the unit cell of the crystal structure of 1CFM, structure 1CFM-B produced the largest peak, with a maximum number of complexes at 14.75 Å; structure 1CFM-C produced an intermediate number of complexes, with a maximum also at 14.75 Å; and structure A produced the smallest number of complexes, with a peak at 14.25 Å.
Table 1 compares the total number of complexes formed with a Fe-Cu distance of ≤16 Å (the complexes within the peaks in Fig. 3 A) for cyt f structures 1CFM-A, 1CFM-B, and 1CFM-C. These are defined as “close-distance” complexes and are considered electron-transfer active. The cutoff value of 16 Å was chosen to classify these complexes in cyt f-PC interactions for two reasons. First, essentially all complexes formed by electrostatic forces are included, whereas those formed by random Brownian motions alone are excluded. The number of complexes formed due to Brownian motion alone can be determined by turning off the electrostatic field. No complexes are formed with Fe-Cu distances ≤16 Å in the absence of the electrostatic field (data not shown). Second, rates calculated using this criterion closely resemble experimental values (for a discussion of these points see Gross and Pearson (10), Haddadian and Gross (11), and Gross (40)). However, selection of the cutoff distances affects only the magnitude of the reaction rates but not the relative effectiveness order of the rates.
TABLE 1.
Calculated interaction rates, k2, and the number of close-distance complexes formed for truncated intact cyt f and its large domain interacting with PC
| Intact Cyt f
|
Large domain of Cyt f
|
|||
|---|---|---|---|---|
| No. of complexes /10,000 trajectories |
k2* (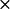 108) M−1 S−1 108) M−1 S−1
|
No. of complexes /10,000 trajectories |
k2* (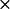 108) M−1 S−1 108) M−1 S−1
|
|
| 1CFM-A | 155±2 | 5.0±0.5 | 160±4 | 5.0±0.4 |
| 1CFM-B | 641±18 | 20.2±0.7 | 148±3 | 4.7±0.5 |
| 1CFM-C | 292±4 | 9.4±0.6 | 297±7 | 9.3±0.5 |
| 1EWH-A | 516±9 | 16.4±0.7 | 198±2 | 6.2±0.5 |
| 1EWH-B | 87±3 | 2.8±0.3 | 2±0.2 | 0.05±0.04 |
| 1EWH-C | 933±7 | 28.4±1.2 | 329±6 | 10.3±0.7 |
| 1Q90 | 94±5 | 3.1±0.3 | 147±5 | 4.6±0.4 |
| 1Q90-modified† | 334±5 | 10.8±0.6 | – | – |
| 1CFM-B-large‡ domain+K-188 and K-189 | 2060±2 | 56.1±3.5 | – | – |
The close-distance complexes were defined as those with Fe-Cu distances 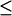 16 Å, which are considered electron transfer active. The second order diffusion-controlled rate constants, k2, were calculated for the formation of these complexes, as described in the Methods section. Five sets of 10,000 trajectories each were carried out to obtain the error values.
16 Å, which are considered electron transfer active. The second order diffusion-controlled rate constants, k2, were calculated for the formation of these complexes, as described in the Methods section. Five sets of 10,000 trajectories each were carried out to obtain the error values.
The structure of 1Q90 cyt f was altered in its small domain, which does not affect the interactions of its large domain with PC.
Only the large domain of 1CFM-B plus residues K-188 and K-189, but without the rest of the small domain, was used. Since a large number of close-distance complexes were formed, we carried only 1000 trajectories.
It can be seen that the greatest number of close-distance complexes is formed for 1CFM-B, an intermediate number for 1CFM-C, and the smallest number for 1CFM-A. The 16 Å cutoff distance was also used to calculate second order rate constants for the interactions (k2), which are also shown in Table 1.
Three cyt f structures in the PDB file 1EWH also showed different interactions with PC (Table 1). Structure 1EWH-C produced the greatest number of close-distance complexes of any of the seven cyt f structures examined. Structure 1EWH-A produced an intermediate number of close-distance complexes, whereas structure 1EWH-B had the smallest number of close-distance complexes formed. Thus, both sets of crystal structures showed one structure that formed a large number of complexes, one an intermediate number, and one a small number. However, the exact number of complexes formed and the corresponding interactions rates are not the same for the members of both sets. For example, 1EWH-C produced 933 ± 7 complexes corresponding to an interaction rate of 28.4 ± 1.2 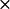 108 M−1s−1, whereas the values of the number of complexes formed and the interactions rates were 641 + 18 and 20.2 ± 0.7
108 M−1s−1, whereas the values of the number of complexes formed and the interactions rates were 641 + 18 and 20.2 ± 0.7 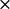 108 M−1s−1 for 1CFM-B. Differences in complex formation and interaction rates were also observed when comparing 1CFM-C with 1EWH-A and 1CFM-A with 1EWH-B.
108 M−1s−1 for 1CFM-B. Differences in complex formation and interaction rates were also observed when comparing 1CFM-C with 1EWH-A and 1CFM-A with 1EWH-B.
The interactions of the six different structures of truncated cyt f with cyt c6.
As can be seen in Fig. 3 B, similar to cyt f-PC interactions, three cyt f molecules in the unit cell of 1CFM showed different numbers of complexes formed with cyt c6. However, in the case of cyt cc, the largest peak of complexes formed occurred with cyt f 1CFM-C rather than 1CFM-B. An intermediate number of complexes was formed for 1CFM-B, and the smallest number of complexes was observed for 1CFM-A as was the case for PC. In all three interactions, the greatest number of complexes was observed at 17 Å. Thus, the peak for complex formation occurred at a larger distance for cyt c6 than for PC due to the fact that the Fe atom in cyt c6 is buried ∼1 Å deeper in the molecule than is the Cu atom in PC. This is also indicated by the fact that the smallest Fe-Fe distance for cyt f-cyt c6 interactions occurred at 16.5 Å compared to 13.75 Å for cyt f-PC interactions.
The number of complexes formed with a Fe-Fe distance of ≤18 Å (the complexes within the peaks in Fig. 3 B) for cyt f interacting with cyt c6 as well as the corresponding interaction rates are shown in Table 2. Similar to the PC interactions, these complexes are defined as “close-distance” and are considered electron-transfer active.
TABLE 2.
Calculated interaction rates, k2, and the number of close-distance complexes formed for truncated intact cyt f and its large domain interacting with cyt c6
| Intact Cyt f
|
Large domain of Cyt f
|
|||
|---|---|---|---|---|
| No. of complexes /10,000 trajectories |
k2* (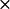 108) M−1 S−1 108) M−1 S−1
|
No. of complexes /10,000 trajectories |
k2* (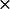 108) M−1 S−1 108) M−1 S−1
|
|
| 1CFM-A | 413±3 | 13.5±0.8 | 37±3 | 1.2±0.2 |
| 1CFM-B | 467±4 | 15.1±0.6 | 41±2 | 1.3±0.2 |
| 1CFM-C | 677±10 | 21.4±0.8 | 110±2 | 3.5±0.3 |
| 1EWH-A | 476±5 | 15.4±0.6 | 69±4 | 2.2±0.2 |
| 1EWH-B | 61±3 | 2.0±0.3 | 1±0.4 | 0.02±0.01 |
| 1EWH-C | 1022±11 | 31.2±0.8 | 163±7 | 5.2±0.5 |
| 1Q90 | 39±3 | 1.3±0.3 | 24±1 | 0.8±0.2 |
| 1Q90-modified† | 366±4 | 12.0±0.6 | – | – |
| 1CFM-C-large‡ domain+K-188 and K-189 | 1220±4 | 35.9±3.1 | – | – |
The close-distance complexes were defined as those with Fe-Fe distances 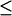 18Å, which are considered electron transfer-active. The second order diffusion-controlled rate constants, k2, were calculated for the formation of these complexes, as described in the Methods section. Five sets of 10,000 trajectories each were carried out to obtain the error values.
18Å, which are considered electron transfer-active. The second order diffusion-controlled rate constants, k2, were calculated for the formation of these complexes, as described in the Methods section. Five sets of 10,000 trajectories each were carried out to obtain the error values.
The structure of 1Q90 cyt f was altered in its small domain, which does not affect the interactions of its large domain with PC.
Only the large domain of 1CFM-C plus residues K-188 and K-189, but without the rest of the small domain, was used. Since a large number of close-distance complexes were formed, we carried only 1000 trajectories.
For three cyt f molecules in the unit cell of 1EWH interacting with cyt c6, unlike 1CFM cyt f structures, the greatest number of close-distance complexes formed was for 1EWH-C, which was the same 1EWH cyt f structure that showed the greatest number of complexes formed interacting with PC. 1EWH-A cyt f showed an intermediate number of complexes formed, and 1EWH-B structure showed the least, both in agreement with the PC results (Table 2).
Thus, considering the number of close-distance complexes formed and the interaction rates with PC and cyt c6, there are real differences between the six structures for the truncated cyt f.
The interactions of the cyt f large domains with PC
Gong et al. (43) showed that removing the small domain from C. reinhardtii cyt f had no effect on its interactions with PC in vivo. Note that no experiments were carried out under conditions in which cyt c6 was the electron acceptor. In our simulations, when cyt f 1CFM-A was used, there was a small increase in the number of complexes formed between 14.25 and 15.0 Å after which the number of complexes formed remained constant (Fig. 3 A). 1CFM-B showed identical results to within the limit of error. Studies with the large domain of 1CFM-C, however, showed a larger number of complexes formed with a broad peak at 15–15.5 Å.
In all three 1CFM cyt f structures, removal of the small domain decreased the number of complexes formed at the smallest Fe-Cu distances (i.e., those ≤14.75 Å, Fig. 3 A). However, the total number of close-distance complexes formed by electrostatic forces (i.e., those with Fe-Cu distances of ≤16 Å, Table 1) was 23% of that observed for the intact truncated cyt f for 1CFM-B and ∼100% for 1CFM-C and 1CFM-A intact cyt f structures. The same is true for the corresponding interaction rates. The large domains of 1EWH-A and 1EWH-C cyt f showed 38% and 35%, respectively, of the number of complexes formed for the corresponding intact truncated cyt f structures. However, 1EWH-B cyt f showed only 2% of the complexes formed for the corresponding intact truncated cyt f. In conclusion, except for 1EWH-B cyt f, the large domain alone showed a significant percentage of the number of complexes formed with the intact cyt f structures to explain the results of Gong et al. (43).
The interactions of the cyt f large domains with cyt c6.
In contrast to the results with PC, when the large domain was interacted with cyt c6, very few close-distance complexes were formed (Table 2). Maximum complex formation was observed for the large domains of 1CFM-C and 1EHW-C, which showed 16% of the number of complexes formed with the corresponding intact cyt fs. Thus, the small domain appears to be more important for the interaction of cyt f with cyt c6 than with PC. These results may reflect the difference in the binding sites on cyt f for PC and cyt c6 as was discussed by Haddadian and Gross (11).
The interactions of the cyt f subunit from the crystal structure of the cyt b6f complex
The interactions of the extramembrane domain of the cyt f subunit from the crystal structure of C. reinhardtii cyt b6f complex (PDB code 1Q90; (2)) with both PC and cyt c6 were also studied. Fig. 3 C shows that intact 1Q90 cyt f produced a peak of complexes at 15.5 Å. Interestingly, the large domain of 1Q90 cyt f produced a greater number of close-distance complexes than did the intact truncated cyt f (Fig. 3 C and Table 1). These results suggest that there is something about the small domain of 1Q90cyt f that inhibits complex formation. In contrast to PC, removing the small domain decreased the number of close-distance complexes formed and the corresponding interaction rate for 1Q90 cyt f interacting with cyt c6 (Fig. 3 D and Table 2).
Complex formation
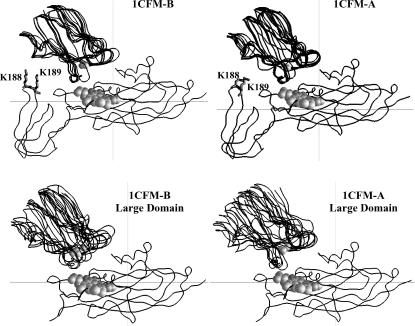
The question arises as to whether the complexes observed in BD simulations for different cyt f structures and their large domains interacting with PC and cyt c6 have unique orientations or not. Fig. 4 depicts the peptide backbone overlays of five complexes each for the cyt f structures 1CFM-A and 1CFM-B and their corresponding large domains interacting with PC. Among the three structures in the unit cell of the 1CFM cyt f crystal, these structures resulted in the highest and the lowest interaction rates with PC, respectively (Table 1). These structures were chosen randomly from the complexes with the Fe-Cu distances less than the peak distances in the plots of the complexes formed (Fig. 3 A). As can be seen, the overall orientation of PC with respect to cyt f did not change in any of the complexes of either intact truncated cyt f or in their corresponding large domains. In the presence and the absence of the small domain, PC docks on cyt f with an inclination toward the small domain, which matches well with the NMR structures (33–36). However, the lack of the small domain caused more heterogeneity in the structure of the complexes formed.
FIGURE 4.
Orientations of the complexes formed for PC interacting with cyt f. The peptide backbones are shown for complexes formed with 1CFM-B (which showed the highest interaction rates) and the 1CFM-A (which had the smallest interaction rates). For each cyt f, five complexes, with cutoff distances less than the peak distances (Fig. 3 A) , were randomly chosen. The overlays were done using program GRASP (50). The cyt f heme and the cu atoms in PCs are in gray and shown as space-filling models.
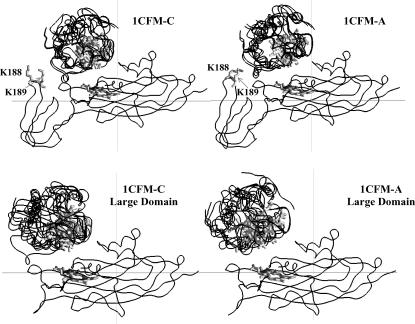
Fig. 5 shows the peptide backbone overlays of five complexes each, for the cyt f structures 1CFM-A and 1CFM-C and their corresponding large domains interacting with cyt c6. Among three cyt f structures in the PDB code of 1CFM, these structures resulted in the highest and the lowest interaction rates, respectively (Table 2). These were chosen randomly from the complexes with the Fe-Fe distances less than the peak distances in the plots of the complexes formed (Fig. 3 B). As in the case of the PC interactions, the overall orientation of cyt c6 with respect to cyt f did not change in any of the intact truncated cyt f complexes or in their corresponding large domains. Also, the absence of the small domain caused more heterogeneity in the structure of the complexes formed.
FIGURE 5.
Orientation of the complexes formed for cyt c6 interacting with cyt f. The peptide backbones are shown for complexes formed with 1CFM-C (which showed the highest interaction rates) and the 1CFM-A (which had the smallest interaction rates). For each cyt f complex, five complexes, with cutoff distances less than the peak distances (Fig. 3 B) were randomly chosen. The overlays were done using the program GRASP (50). The cyt f and cyt c6 hemes are in gray and shown as ribbon models.
The same results as above were also observed for intact and large domains of cyt f structures from the PDB codes 1EWH and 1Q90 interacting with both PC and cyt c6 (data not shown).
It should be mentioned that the complexes formed in our simulations are not the final electron transfer-active state. However, they have both the involvement of electrostatic plus the hydrophobic interactions and are close to the final complex (Haddadian and Gross (11) ). Schlarb-Ridley et al. (62) suggested a mechanism for the cyt f-PC interactions considering a rearrangement of the initial complex to form the final electron-active complex. We believe that the complexes we obtained are not the initial docking complex due to the fact that they are not electrostatically optimized. The electrostatic complexes would involve maximum interactions of the positive residues on the spine of cyt f with the residues on the negative patches of PC and cyt c6. However in these complexes, H87 on PC and the heme of cyt c6 would lie far from the cyt f heme (data not shown). The complexes obtained from the simulations are not also the final electron-active complex due to the lack of effects such as desolvation, packing, etc. in the simulations. However, our complexes have the hydrophobic surfaces surrounding the H-87 on PC and heme of cyt c6 facing the hydrophobic residues surrounding the heme of cyt f (Haddadian and Gross (11)). Therefore they are very close to the final complex. It should also be mentioned that the complexes we obtained resemble very well the NMR complexes obtained by Ubbink et al. (33), Crowley et al. (35), and Lange et al. (36).
DISCUSSION
Structural differences in the seven cyt f structures
The observation that in both cyt f-PC and cyt f-cyt c6 interactions the reaction rates were different for the seven C. reinhardtii cyt fs can be explained by differences in their structures, which is evident from their superimpositions. Table 3 shows the RMS distance deviation of the atoms in cyt f structures of the PDB codes 1CFM and 1EWH from their corresponding atoms in the extramembrane domain of cyt f subunit from the cyt b6f complex, PDB code 1Q90. The RMS was calculated using backbone atoms only for intact truncated cyt f, as well as for the large and small domain structures. As can be seen, among all of the intact, truncated cyt f structures, the 1CFM-A and B and the 1EWH-B structures had the lowest and the highest deviations from the 1Q90 cyt f structure, respectively. The large difference between 1EWH-B cyt f and its large domain interaction rates with PC and cyt c6 compared to the rest of cyt f structures can be attributed to its overall different structure from the rest of cyt f molecules (Table 3, its large and small domains also have some of the highest RMS deviations).
TABLE 3.
RMS distance deviation of the backbone atoms in cyt f structures of the PDB codes 1CFM and 1EWH from the corresponding backbone atoms in the extramembrane domain of Cyt f subunit from the Cyt b6f complex (PDB code 1Q90)
| Intact (Å) | Large domain (Å) | Small domain (Å) | |
|---|---|---|---|
| 1CFM.A | 0.6 | 0.9 | 0.7 |
| 1CFM.B | 0.6 | 0.8 | 1.1 |
| 1CFM.C | 0.8 | 0.8 | 1.1 |
| 1EWH.A | 0.7 | 0.9 | 0.8 |
| 1EWH.B | 1.0 | 0.9 | 1.1 |
| 1EWH.C | 0.8 | 0.6 | 1.0 |
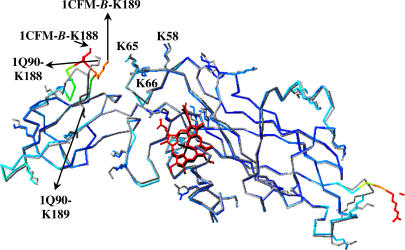
The superimposition of only the large domains showed that they were very similar, which is demonstrated by their very similar RMS values (Table 3). Also, all of the side chains of the cyt f basic residues located on the large domain, particularly K-58, K-65, and K-66 that are important in electrostatic interactions with both PC and cyt c6 (4,9–11,32) have more or less the same orientations (Fig. 6; this is the case for all of the seven cyt f structures). However, when we used only the small domains to calculate the RMS values, we obtained some of the largest deviations, which were not expected, considering the smaller number of atoms involved. Four of the six cyt f small domain structures resulted in RMS deviations of ≤1 Å. In contrast, only one cyt f structure and none for the cyt f large domain had duration ≥1 Å. A closer study of the structures revealed that in the small domain the loop formed by residues A-184-G-191 had the most variances (i.e., >2 Å RMS deviations) in all of the cyt f structures. This loop contains the important basic residues K-188 and K-189 that are involved in the electrostatic interaction with both PC and cyt c6 (4,9–11,32). The side chains of these residues have different orientations in all of the seven cyt f structures (Figs. 4 and 5). In all seven cyt f structures, this loop also had some of the highest crystallographic temperature factors (β-factors), which implied its flexibility (Fig. 6 (2,6,44)).
FIGURE 6.
Overlay of the cyt f structure B from the PDB code 1CFM on the extramembrane domain of the cyt f subunit from the cyt b6f complex (PDB code 1Q90). The 1CFM-B cyt f backbone and all of its basic residues were colored by their temperature factors (β-factors), in which the molecule is colored from dark blue for low β-factors to red for high β-factors (the heme is colored red for visualization purpose). The backbone and the all of the basic residues of the 1Q90 cyt f are shown in gray and its heme in black. As can be seen, the loop of residues A-184-G-191 of the 1CFM-B cyt f has some of the highest β-factors compared to the rest of the molecule and is oriented very differently than the 1Q90 loop. The key basic residues in both cyt fs are labeled. This figure was generated by the program Deep View (51).
The effect of replacing the A-184-G-191 loop
To check for the importance of the A-184-G-191 loop, we removed this loop from 1Q90 cyt f and replaced it with the corresponding loop in the 1CFM-B structure using the program Deep View (51). The loop from 1CFM-B was chosen because of its high rate of interaction with both PC and cyt c6 and its small overall deviation from the 1Q90 structure (RMS = 0.61 Å). We then interacted the modified 1Q90 cyt f with both PC and cyt c6, which resulted in a large increase in the number of close-distance complexes formed (Fig. 7). We obtained a 3.5-fold increase in the cyt f-PC interaction rate and a ninefold rise in the cyt f-cyt c6 interaction rate (Tables 1 and 2). Therefore, the position of this loop and the orientation of the basic residues K-188 and K-189 within this loop are important factors in both cyt f-PC and cyt f-cyt c6 interactions. In fact, the difference in the orientation of K-188 and K-189 side chains in intact 1Q90 cyt f from that of the 1CFM-B structure (Fig. 6) may explain the observation that the 1Q90 cyt f had the smallest cyt f-PC and cyt f-cyt c6 interaction rates (Tables 1 and 2, within simulation error).
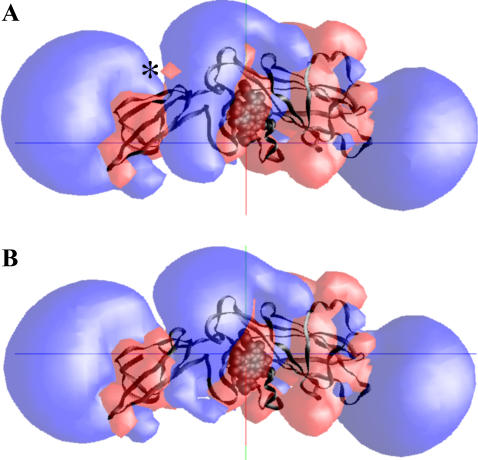
Fig. 8 shows the electrostatic fields simulation of reduced 1Q90 cyt f and its A-184-G-191 loop-modified structure. As can be seen, the loop-modified cyt f structure has a larger positive field in the vicinity of the heme and lacks the small negative field on top of the small domain (marked with an asterisk in Fig. 8). This enhancement of stronger positive and weaker negative fields assist cyt f in better attracting the negative fields of PC and cyt c6, resulting in higher rates of interactions.
FIGURE 8.
Electrostatic fields of the extramembrane domain of the cyt f subunit from the crystal structure of the C. reinhardtii cyt b6f complex: (A) reduced cyt f and (B) reduced cyt f, whose loop of residues A-184-G-191 is replaced with the corresponding loop from the 1CFM-B cyt f structure (see Methods section). The electrostatic field contours at +1 kT/e (blue) and −1 kT/e (red) were calculated at 10 mM ionic strength and pH 7.0. The small negative field of wild-type cyt f on top of its small domain is marked with an asterisk. The heme is shown as a space-filling model, and the backbone of cyt f is colored black.
Also, different conformations of the cyt f molecules provide different steric hindrances for PC or cyt c6 upon docking. As was mentioned in the Methods section, at each step of the trajectory, the overlaps between all of the atoms of the mobile molecule and the target molecule are checked and prevented. Thus, the surface features of the two molecules are considered in the simulations, and at close approach PC or cyt c6 would feel different steric hindrances due to different cyt f structures. Therefore, besides the different electrostatic forces that bring the molecules close, these steric hindrances also affect the number of close-distance complexes formed and the interaction rates.
The role of the small domain of cyt f
The deletion of the small domain caused a greater degree of heterogeneity in the structures of the complexes formed (Figs. 4 and 5). Also, in contrast to the intact cyt f interactions, the cyt f large domains, when interacting with either PC or cyt c6, produce no significant peaks in the plots of the number of complexes formed versus distance (Fig. 3). These observations suggest that the cyt f small domain plays a role in guiding PC and cyt c6 to a uniform dock with cyt f, involving electrostatic interactions with K-188 and K-189 on this domain.
Gong et al. (43) used a C. reinhardtii mutant of cyt f, which lacked the small domain, and studied its interactions with PC in vivo. They concluded that deleting the small domain did not affect the electron transfer rate to PC and, therefore, this domain was not involved in the interactions in vivo. Their observations agreed with in vivo mutation of the two basic residues K-188 and K-189 on the small domain of cyt f (41). However the in vitro electron transfer studies (31,32) plus the NMR complexes between the cyt f and PC (33–36) indicated that these lysine residues are involved in the interactions with PC. Computational modeling (8–11,37–40) also pointed toward importance of these residues in the interactions.
In this study, among all of the cyt f structures used, only the 1EWH-B structure resulted in a significant difference between the reaction rates of intact cyt f structures and their corresponding large domains with PC (Table 1). Two of the cyt f structures, 1CFM-A and 1CFM-C, resulted in identical reaction rates with PC for the intact and their corresponding large domain structures. However, when cyt c6 interacted with the cyt f large domain, close-distance complex formation was inhibited compared to the intact cyt f in all of the seven cyt f structures examined.
The different binding positions of PC and cyt c6 on cyt f may explain the observation that the deletion of the small domain affected the cyt f-cyt c6 interactions more than those for cyt f-PC. As can be seen in Figs. 4 and 5, the overall orientation of PC or cyt c6 with respect to different cyt f molecules, within the limit of heterogeneity of the complexes formed, did not change in any of the complexes formed with intact cyt f or in their corresponding large domains. In the presence and the absence of the cyt f small domain, PC docks vertically on cyt f with its Cu atom close to the heme with an inclination toward the small domain. This position of PC agreed well with the NMR structures (33–36). In contrast, as we previously reported (10,11), cyt c6 docks horizontally on top of the cyt f farther away from the small domain, covering a larger area of the cyt f molecule.
Deleting the small domain has two effects on cyt f interactions. First, it weakens the positive electrostatic field of cyt f, particularly by removing the residues K-188 and K-189. Second, it reduces steric hindrance due to the presence of the small domain itself. These two opposing factors affect PC because of its inclination toward the small domain, more than cyt c6. The reduction in steric hindrance partially balances the weakening of the positive electrostatic field, resulting in a small effect in complex formation and interaction rates in cyt f-PC interactions. In contrast to PC, cyt c6 (with its binding position farther away from the small domain) is not affected as much by the steric relaxation resulting from the deletion of this domain and is mostly affected by the removal of positive charges on this domain (particularly K-188 and K-189). Therefore, cyt f-cyt c6 reactions were inhibited to a greater degree by the deletion of the small domain.
To check for the effectiveness of these two opposing factors, we carried out simulations with the large domain of 1CFM-B plus residues K-188 and K-189 (but without the rest of the small domain) interacting with PC, and the large domain of 1CFM-C plus residues K-188 and K-189 (but without the rest of the small domain) interacting with cyt c6. As can be seen in Tables 1 and 2, the presence of positive electrostatic fields of these two residues resulted in an interaction rate that is 2.8 times larger than the intact 1CFM-B interacting with PC, whereas the rate for 1CFM-C large domain plus K-188 and K-189 interacting with cyt c6 increased only by 1.7-fold compared to the intact 1CFM-C-cyt c6 interactions. Thus, when K-188 and K-189 were added to the large domain, the positive electrostatic field increased enhancing interactins with PC.
CONCLUSIONS
Our studies showed that different C. reinhardtii cyt f structures, which are very similar, produced different reaction rates in interactions with PC and cyt c6. We were able to attribute this to structural differences between the seven cyt f structures, particularly to a small flexible loop on the cyt f small domain. In all of the seven cyt f structures, this loop had some of the highest crystallographic temperature factors (β-factors), which implied its flexibility (Fig. 6 (2,6,44)). We also showed that deletion of the small domains affected cyt c6 to a greater extent than PC, due to its different binding position on cyt f. The function of the small domain in cyt f may be to guide PC or cyt c6 to a uniform dock with cyt f, especially due to electrostatic interactions with K-188 and K-189 on this domain. The question of which of these seven cyt f structures resembles the physiological molecule is difficult to answer; it may be possible that the cyt f structure, especially in the loop of residues A-184-G-191, undergoes structural changes during the interactions with its redox partners—a case of induced local fit.
One of our purposes in doing this study was to lay the groundwork for future experiments on these proteins to understand better the electron transfer process between them. Experimental work such as changing the size of the flexible loop, fixing it in position, or modifying it with a probe to check for its motion are required to verify its effects on the interactions. Also, these results signified the sensitivity and the power of the BD simulations in the study of the molecular interactions.
References
- 1.Kurisu, G., H. Zhang, J. L. Smith, and W. A. Cramer. 2003. Structure of the cytochrome b6f complex of oxygenic photosynthesis: tuning the cavity. Science. 302:1009–1014. [DOI] [PubMed] [Google Scholar]
- 2.Stroebel, D., Y. Choquet, J. L. Popot, and D. Picot. 2003. An atypical heme in the cytochrome b6f complex. Nature. 426:413–418. [DOI] [PubMed] [Google Scholar]
- 3.Hope, A. B. 2000. Electron transfers amongst cytochrome f, plastocyanin and photosystem I: kinetics and mechanisms. Biochim. Biophys. Acta. 1456:5–26. [DOI] [PubMed] [Google Scholar]
- 4.Martinez, S. E., D. Huang, A. Szczepaniak, W. A. Cramer, and J. L. Smith. 1994. Crystal structure of the chloroplast cytochrome f reveals a novel cytochrome fold and unexpected heme ligation. Structure. 2:95–105. [DOI] [PubMed] [Google Scholar]
- 5.Martinez, S. E., D. Huang, M. Ponomarev, W. A. Cramer, and J. L. Smith. 1996. The heme redox center of chloroplast cytochrome f is linked to a buried five-water chain. Protein Sci. 5:1081–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chi, Y. I., L. S. Huang, Z. Zhang, J. G. Fernandez-Velasco, and E. A. Berry. 2000. X-ray structure of a truncated form of cytochrome f from Chlamydomonas reinhardtii. Biochemistry. 39:7689–7701. [DOI] [PubMed] [Google Scholar]
- 7.Gray, J. C. 1992. Cytochrome f: structure, function and biosynthesis. Photosynth. Res. 34:359–374. [DOI] [PubMed] [Google Scholar]
- 8.Pearson, D. C. Jr., E. L. Gross, and E. S. David. 1996. The electrostatic properties of cytochrome f: implications for docking with plastocyanin. Biophys. J. 71:64–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pearson, D. C. Jr., and E. L. Gross. 1998. Brownian dynamics study of the interaction between plastocyanin and cytochrome f. Biophys. J. 75:2698–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gross, E. L., and D. C. Pearson Jr. 2003. Brownian dynamics simulations of the interaction of Chlamydomonas cytochrome f with plastocyanin and cytochrome c6. Biophys. J. 85:2055–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haddadian, E. J., and E. L. Gross. 2005. Brownian dynamics study of cytochrome f interactions with cytochrome c6 and plastocyanin in Chlamydomonas reinhardtii, plastocyanin and cytochrome c6 mutants. Biophys. J. 88:2323–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Redinbo, M. R., D. Cascio, M. K. Choukair, D. Rice, S. Merchant, and T. O. Yeates. 1993. The 1.5 Å crystal structure of plastocyanin from the green alga Chlamydomonas reinhardtii. Biochemistry. 32:10560–10567. [DOI] [PubMed] [Google Scholar]
- 13.Gross, E. L. 1996. Plastocyanin: structure, location, diffusion, and electron transfer mechanisms. In Oxygenic Photosynthesis: The Light Reactions. D. Ort and C. Yocum, editors. Kluwer Academic Publishers, Dordrecht, The Netherlands. 413–429.
- 14.Sigfridsson, K. 1998. Plastocyanin, an electron-transfer protein. Photosynth. Res. 57:1–28. [Google Scholar]
- 15.Durell, S. R., J. K. Labanowski, and E. L. Gross. 1990. Modeling the electrostatic potential field of plastocyanin. Arch. Biochem. Biophys. 277:241–254. [DOI] [PubMed] [Google Scholar]
- 16.Kerfeld, C. A., and D. W. Krogmann. 1998. Photosynthetic cytochromes c in cyanobacteria, algae, and plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49:397–425. [DOI] [PubMed] [Google Scholar]
- 17.Gupta, R., Z. He, and S. Luan. 2002. Functional relationship of cytochrome c6 and plastocyanin in Arabidopsis. Nature. 417:567–571. [DOI] [PubMed] [Google Scholar]
- 18.Merchant, S., and L. Bogorad. 1986. Regulation by copper of the expression of plastocyanin and cytochrome c552 in Chlamydomonas reinhardtii. Mol. Cell. Biol. 6:462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kerfeld, C. A., H. P. Anwar, R. Interrante, S. Merchant, and T. O. Yeates. 1995. The structure of chloroplast cytochrome c6 at 1.9 Å resolution: evidence for functional oligomerization. J. Mol. Biol. 250:627–647. [DOI] [PubMed] [Google Scholar]
- 20.Ullmann, G. M., M. Hauswald, A. Jensen, N. M. Kostic, and E. W. Knapp. 1997. Comparison of the physiologically equivalent proteins cytochrome c6 and plastocyanin on the basis of their electrostatic potentials. Tryptophan 63 in cytochrome c6 may be isofunctional with tyrosine 83 in plastocyanin. Biochemistry. 36:16187–16196. [DOI] [PubMed] [Google Scholar]
- 21.Morand, L. Z., M. K. Frame, K. K. Colvert, D. A. Johnson, D. W. Krogmann, and D. J. Davis. 1989. Plastocyanin cytochrome f interaction. Biochemistry. 28:8039–8047. [DOI] [PubMed] [Google Scholar]
- 22.Takenaka, K., and T. Takabe. 1984. Importance of local positive charges on cytochrome f for electron transfer to plastocyanin and potassium ferricyanide. J. Biochem. (Tokyo). 96:1813–1821. [DOI] [PubMed] [Google Scholar]
- 23.Takabe, T., K. Takenaka, H. Kawamura, and Y. Beppu. 1986. Charges on proteins and distances of electron of electron transfer in metalloprotein redox reactions. J. Biochem. (Tokyo). 99:833–840. [DOI] [PubMed] [Google Scholar]
- 24.Takabe, T., H. Ishikawa, S. Niwa, and Y. Tanaka. 1984. Electron transfer reactions of chemically modified plastocyanin with P700 and cytochrome f. Importance of local charges. J. Biochem. (Tokyo). 96:385–393. [DOI] [PubMed] [Google Scholar]
- 25.Anderson, G. P., D. G. Sanderson, C. H. Lee, S. Durell, L. B. Anderson, and E. L. Gross. 1987. The effect of ethylene diamine chemical modification of plastocyanin on the rate of cytochrome f oxidation and P-700+ reduction. Biochim. Biophys. Acta. 894:386–398. [DOI] [PubMed] [Google Scholar]
- 26.Niwa, S., H. Ishikawa, and S. Nikai. 1980. Electron transfer reactions between Cytochrome f and plastocyanin from Brassica komatsuna. J. Biochem. (Tokyo). 88:1177–1183. [DOI] [PubMed] [Google Scholar]
- 27.Qin, L., and N. M. Kostic. 1993. Importance of protein rearrangement in the electron-transfer reaction between the physiological partners cytochrome f and plastocyanin. Biochemistry. 32:6073–6080. [DOI] [PubMed] [Google Scholar]
- 28.Meyer, T. E., Z. G. Zhao, M. A. Cusanovich, and G. Tollin. 1993. Transient kinetics of electron transfer from a variety of c-type cytochromes to plastocyanin. Biochemistry. 32:4552–4559. [DOI] [PubMed] [Google Scholar]
- 29.Lee, B. H., T. Hibino, T. Takabe, P. J. Weisbeek, and T. Takabe. 1995. Site-directed mutagenetic study on the role of negative patches on silene plastocyanin in the interactions with cytochrome f and photosystem I. J. Biochem. (Tokyo). 117:1209–1217. [DOI] [PubMed] [Google Scholar]
- 30.Kannt, A., S. Young, and D. S. Bendall. 1996. The role of acidic residues of plastocyanin in its interaction with cytochrome f. Biochim. Biophys. Acta. 1277:115–126. [PubMed] [Google Scholar]
- 31.Gong, X. S., J. Q. Wen, N. E. Fisher, S. Young, C. J. Howe, D. S. Bendall, and J. C. Gray. 2000. The role of individual lysine residues in the basic patch on turnip cytochrome f for the electrostatic interactions with plastocyanin in vitro. Eur. J. Biochem. 267:3461–3468. [DOI] [PubMed] [Google Scholar]
- 32.Soriano, G. M., M. V. Pomamarev, R. A. Piskorowski, and W. A. Cramer. 1998. Identification of the basic residues of cytochrome f responsible for electrostatic docking interactions with plastocyanin in vitro: relevance to the electron transfer reaction in vivo. Biochemistry. 37:15120–15128. [DOI] [PubMed] [Google Scholar]
- 33.Ubbink, M., M. Ejdeback, B. G. Karlsson, and D. S. Bendall. 1998. The structure of the complex of plastocyanin and cytochrome f, determined by paramagnetic NMR and restrained rigid-body molecular dynamics. Structure. 6:323–335. [DOI] [PubMed] [Google Scholar]
- 34.Ejdeback, M., A. Bergkvist, B. G. Karlsson, and M. Ubbink. 2000. Side-chain interactions in the plastocyanin-cytochrome f complex. Biochemistry. 39:5022–5027. [DOI] [PubMed] [Google Scholar]
- 35.Crowley, P. B., D. M. Hunter, K. Sato, W. McFarlane, and K. Dennison. 2004. The parsley plastocyanin-turnip cytochrome f complex: a structurally distorted but kinetically functional acidic patch. Biochem. J. 378:45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lange, C., T. Cornvik, I. Diaz-Moreno, and M. Ubbink. 2005. The transient complex of poplar plastocyanin with cytochrome f: effects of ionic strength and pH. Biochim. Biophys. Acta. 1707:179–188. [DOI] [PubMed] [Google Scholar]
- 37.Ullmann, G. M., E. W. Knapp, and N. M. Kostic. 1997b. Computational simulation and analysis of dynamic association between plastocyanin and cytochrome f. Consequences for the electron-transfer reaction. J. Am. Chem. Soc. 119:42–52. [Google Scholar]
- 38.Nelson, N., D. C. Pearson Jr., and E. L. Gross. 1999. The interaction of plastocyanin with cytochrome f: a Brownian dynamics study. In Photosynthesis: Mechanisms and Effects, Vol. 3. G. Garab, editor. Kluwer Academic Publishers, Dordrecht, The Netherlands. 1493–1498.
- 39.De Rienzo, F., R. R. Gabdoulline, M. C. Menziani, P. G. De Benedetti, and R. C. Wade. 2001. Electrostatic analysis and Brownian dynamics simulation of the association of plastocyanin and cytochrome f. Biophys. J. 81:3090–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gross, E. L. 2004. A Brownian dynamics study of the interaction of Phormidium laminosum plastocyanin with Phormidium laminosum cytochrome f. Biophys. J. 87:2043–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soriano, G. M., M. V. Ponomarev, G. S. Tae, and W. A. Cramer. 1996. Effect of the interdomain basic region of cytochrome f on its redox reactions in vivo. Biochemistry. 35:14590–14598. [DOI] [PubMed] [Google Scholar]
- 42.Zhou, J., J. G. Fernandez-Velasco, and R. Malkin. 1996. N-terminal mutants of chloroplast cytochrome f: effect on redox reactions and growth in Chlamydomonas reinhardtii. J. Biol. Chem. 271:6225–6232. [DOI] [PubMed] [Google Scholar]
- 43.Gong, X. S., S. Chung, and J. G. Fernandez-Velaso. 2001. Electron transfer and stability of the cytochrome b6f complex in a small domain deletion mutant of cytochrome f. J. Biol. Chem. 276:24365–24371. [DOI] [PubMed] [Google Scholar]
- 44.Sainz, G., C. J. Carrell, M. V. Ponamarev, G. M. Soriano, W. A. Cramer, and J. L. Smith. 2000. Interruption of the internal water chain of cytochrome f impairs photosynthetic function. Biochemistry. 39:9164–9173. [DOI] [PubMed] [Google Scholar]
- 45.Northrup, S. H., J. A. Luton, J. O. Boles, and J. C. Reynolds. 1987. Brownian dynamics simulation of protein association. J. Comput. Aided Mol. Des. 1:291–311. [DOI] [PubMed] [Google Scholar]
- 46.Northrup, S. H., J. O. Boles, and J. C. Reynolds. 1987. Electrostatic effects in the Brownian dynamics of association and orientation of heme proteins. J. Phys. Chem. 91:5991–5998. [Google Scholar]
- 47.Northrup, S. H., J. O. Boles, and J. C. Reynolds. 1988. Brownian dynamics of cytochrome c and cytochrome c peroxidase association. Science. 241:67–70. [DOI] [PubMed] [Google Scholar]
- 48.Northrup, S. H., K. A. Thomasson, C. M. Miller, P. D. Barker, L. D. Eltis, J. G. Guillemette, S. C. Inglis, and A. G. Mauk. 1993. Effect of charged amino acid mutations on the bimolecular kinetics of reduction of yeast iso-1-ferricytochrome c by bovine ferrocytochrome b5. Biochemistry. 32:6613–6623. [DOI] [PubMed] [Google Scholar]
- 49.Berman, H. M., J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, L. N. Shindyalov, and P. E. Bourne. 2000. The protein data bank. Nucleic Acids Res. 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nicholls, A., and B. Honig. 1991. A rapid finite-difference algorithm, utilizing successive over-relaxation to solve the Poisson-Boltzmann equation. J. Comput. Chem. 12:435–445. [Google Scholar]
- 51.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-pdbviewer: an environment for comparative protein modeling. Electrophoresis. 18:2714–2723. [DOI] [PubMed] [Google Scholar]
- 52.Ermark, D. L., and J. C. McCammon. 1978. Brownian dynamics with hydrodynamic interactions. J. Phys. Chem. 69:1352–1360. [Google Scholar]
- 53.Harvey, S. C. 1989. Treatment of electrostatic effects in macromolecular modeling. Proteins. 5:78–92. [DOI] [PubMed] [Google Scholar]
- 54.McCammon, J. A., and S. C. Harvey. 1987. Dynamics of Proteins and Nucleic Acids. Cambridge University Press, Cambridge.
- 55.Northrup, S. H. 1996. Theoretical simulation of protein-protein interactions. In Cytochrome c: A Multidisciplinary Approach. R. A. Scott and A. G. Mauk, editors. University Science Publishers, Sausalito, CA. 543–570.
- 56.Moser, C. C., C. C. Page, R. Farid, and P. L. Dutton. 1995. Biological electron transfer. J. Bioenerg. Biomembr. 27:263–274. [DOI] [PubMed] [Google Scholar]
- 57.Moser, C. C., J. M. Keske, K. Warncke, R. S. Farid, and P. L. Dutton. 1992. Nature of biological electron transfer. Nature. 355:796–802. [DOI] [PubMed] [Google Scholar]
- 58.Waingeh, V. F., S. L. Lowe, and K. A. Thomasson. 2004. Brownian dynamics of interactions between glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mutants and F-actin. Biopolymers. 73:533–541. [DOI] [PubMed] [Google Scholar]
- 59.Cui, M., J. Shen, J. M. Briggs, W. Fu, J. Wu, Y. Zhang, X. Luo, Z. Chi, R. Ji, H. Jiang, and K. Chen. 2002. Brownian dynamics simulations of the recognition of the scorpion toxin P05 with small–conductance calcium-activated potassium channels. J. Mol. Biol. 318:417–428. [DOI] [PubMed] [Google Scholar]
- 60.Matthew, J. B. 1985. Electrostatic effects in proteins. Annu. Rev. Biophys. Chem. 14:387–417. [DOI] [PubMed] [Google Scholar]
- 61.Warwicker, J., and H. C. Watson. 1982. Calculation of the electric potential in the active site cleft due to alpha-helix dipoles. J. Mol. Biol. 157:671–679. [DOI] [PubMed] [Google Scholar]
- 62.Schlarb-Ridley, B. G., M. Hualing, W. D. Teale, V. S. Meyer, C. J. Howe, and D. S. Bendall. 2005. Implications of the effects of viscosity, macromolecular crowding, and temperature for the transient interaction between cytochrome f and plastocyanin from the cyanobacterium Phormidium laminosum. Biochemistry. 44:6232–6238. [DOI] [PubMed] [Google Scholar]