Abstract
High shear enhances the adhesion of Escherichia coli bacteria binding to mannose coated surfaces via the adhesin FimH, raising the question as to whether FimH forms catch bonds that are stronger under tensile mechanical force. Here, we study the length of time that E. coli pause on mannosylated surfaces and report a double exponential decay in the duration of the pauses. This double exponential decay is unlike previous single molecule or whole cell data for other catch bonds, and indicates the existence of two distinct conformational states. We present a mathematical model, derived from the common notion of chemical allostery, which describes the lifetime of a catch bond in which mechanical force regulates the transitions between two conformational states that have different unbinding rates. The model explains these characteristics of the data: a double exponential decay, an increase in both the likelihood and lifetime of the high-binding state with shear stress, and a biphasic effect of force on detachment rates. The model parameters estimated from the data are consistent with the force-induced structural changes shown earlier in FimH. This strongly suggests that FimH forms allosteric catch bonds. The model advances our understanding of both catch bonds and the role of allostery in regulating protein activity.
INTRODUCTION
It has been shown that FimH-mediated binding of type 1 fimbriated Escherichia coli to mannosylated glycoproteins (1–4), selectin-mediated binding of leukocytes to sialyl-Lewis-X glycoproteins or saccharides (3,5–7), Staphylococcus aureus binding to collagen (8,9), and GP-Ib-mediated platelet adhesion to von Willebrand factor (10) are all enhanced by fluid flow. In theory, this phenomenon could be explained either by a change in the rate of bond formation under shear, by a catch-bond mechanism of receptor-ligand interaction, or both. Catch bonds are bonds that become longer lived with tensile mechanical force in contrast to slip bonds that become shorter lived (11).
Single-molecule studies have recently demonstrated that selectin bonds are catch bonds (12–14), which can explain the shear threshold for leukocyte rolling (7). However, unlike the original theoretical model of catch bonds that predicted the existence of monotonically longer-lived bonds as force increases (11), the experimental single-molecule data showed a biphasic effect of force (12–14), with the bonds strengthened by moderate force, but weakened by higher forces. To explain the selectin data, a two-pathway or harpoon model (15), where a catch pathway dominates at low force and a slip pathway dominates at high force, has been proposed (13), mathematically described (16,17), and quantitatively fit to the data (16,17). More complicated two-state models have also been described by Bartolo et al. (15), Evans et al. (14), and Barsegov et al. (18) in which there are two possible bound states and mechanical force favors the conformation with the slower unbinding pathway. These two-state models offer similar fits to selectin data as the two-path models (15,16).
Shear-enhanced FimH-mediated adhesion is also biphasic (1,19) and was also proposed to involve catch bonds between FimH and its ligand mannose (2,4,19). Although single-molecule dynamic force spectroscopy experiments have demonstrated that selectins form catch bonds (12–14), the indications that FimH forms catch bonds were derived from structure-function studies. Bacteria and beads binding through FimH switch from transient or rolling to stationary adhesion when either the flow rate or viscosity is increased (4,19), showing that mechanical force increases adhesion. Steered molecular dynamics simulations predicted that tensile force induces a conformational change in a regulatory region of FimH (1). Site-directed mutagenesis showed that a mutation that favored this conformational change caused a switch from rolling to stationary adhesion even at very low force (1,4). Together, this suggests that FimH forms catch bonds because mechanical force acts through a regulatory region to allosterically switch FimH bonds from a short- to a long-lived state. To test this hypothesis, it is necessary to measure bond lifetimes. However, FimH is a two-domain protein that is unstable unless it is integrated into the tip of fimbriae (20), hampering single-molecule approaches to studying FimH bond lifetimes.
Here, we provide novel quantitative analysis of FimH-mediated adhesion of E. coli to mannosylated surfaces. We analyze the length of time that bacteria pause on a model surface at various shear stresses and report a double exponential decay in the pause duration at each shear, with higher shear favoring the slow decay rate. We show that these results cannot be explained by differences in the number of bonds contributing to each pause, but are consistent with dramatic differences in bond lifetimes. Accordingly, we present a quantitative two-state model for how force would affect the lifetime of an allosteric catch bond that fits the pause time data with parameters that are consistent with FimH structure.
MATERIALS AND METHODS
Videos of moving bacteria in flow chambers
E. coli bacteria that express the common F18 variant of FimH were continually injected into a parallel plate flow chamber coated with 200 μg/ml mannose-BSA as described previously (4). After 2 min of injection at a shear stress of 0.5 pN/μm2, the flow chamber was washed at the same shear stress to remove unbound bacteria, leaving rolling and stationary bacteria as shown in Fig. 1. We preloaded bacteria in this manner because they fail to accumulate well at some shear rates (4). The flow rate over these bacteria was then decreased or increased to the indicated shear stress and the behavior of the bacteria at the new shear stress recorded with a charge-coupled device (CCD) camera and MetaMorph video acquisition software at 37 ms per frame for 14 s.
FIGURE 1.
Pause lifetimes of E. coli bound to mannose-BSA coated surfaces. (A) Locations of several bacteria as a function of time for a medium shear stress of 0.5 pN/μm2, showing short and long pauses. (B) Fraction of measured pauses lasting at least the indicated length of time at low (0.01 pN/μm2; ⋄) and high (2 pN/μm2; □) shear. The lines shown in this figure are a fit with a double exponential decay model and give a ∼70-fold (low shear) and ∼360-fold (high shear) difference between the fast and slow decay rates.
Analyzing pause lifetimes
The bacteria in the videos were tracked using SVision's Image Based Decision technology (SVLife, Seattle, WA), which uses structure guided processing to detect bacteria and calculate their center of mass at each time step. The image resolution was 0.8 μm per pixel, and the center of mass offered a positional resolution of slightly <1 pixel, for submicron resolution. The position trajectories were then processed to determine the length of time bacteria paused (defined as movement of the center of mass of <1 pixel, or <0.8 μm). Pauses were identified and monitored in the first portion of the video, just after the switch in shear. In the last half of the video (7.3 s), existing pauses were monitored to determine how long they lasted but new pauses were ignored because there was not sufficient time left in the video to determine how long these pauses lasted. Even so, the maximum time all pauses could be monitored was 7.3 s. The minimum pause time measurable was 74 ms (two frames), because a pause could only be detected if the bacteria remained in the same position for two frames. In this manner, all the pause lifetimes were measured to the nearest 37 ms. From this, a histogram of pause lifetimes could be obtained.
Calculating the fraction of pauses surviving as a function of time
Integrating the lifetime histogram gives the cumulative number of pauses that already ended as a function of time. Subtracting this from the total number of pauses gives the number surviving as a function of time. When these data were displayed in figures or used to fit models, redundant data points were removed. Because of the double exponential decay, there was a higher frequency of events (pause terminations) at short than at long times. This meant that many of the time points, particularly at longer times, represented no new information because there were no events since the previous time point. We filtered out these redundant data points so that the small number of long-lived pauses were not visually overweighted in the graphs nor statistically overweighted during the model fitting procedures. Because all videos had different numbers of pauses, it is desirable to normalize the data to show the fraction of pauses surviving as a function of time. When no model was applied to the data, this was done by dividing the number surviving at each time by the total number of measured pauses. However, because the first data point is at 74 ms, it is impossible to determine how many pauses were too short to be measured. Thus, in the fit to any model, the total pause number was an unknown parameter to be fit. This indicates the total number of pauses lasting at least the indicated time and corresponds directly to the probability of bond survival as a function of time.
The total distance that all bacteria rolled between pauses was also calculated from these trajectories. Each time step during the analysis in which a bacteria moved >1 pixel but less than the distance it would free float as predicted by the hydrodynamic velocity, it was determined to roll. The total distance of these rolling movements was added during the time frame in which new pauses were identified, and compared to the predicted number of bonds estimated for the same videos in Figs. 5 and 8.
FIGURE 5.
Testing predictions of the allosteric catch-bond model. (A) Behavior at higher shear stress for the allosteric model. The black diamonds show the new pause survival data at 10.9 pN/μm2. The thin black line shows the predicted model behavior at 90 pN (10.9 pN/μm2) with the parameters in Table 1, and the thin dashed and dotted lines show the prediction with mean − 1 SD decrease in k200 and x20, respectively. The thick black line shows a good fit of the data with 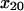 = 1.37 Å or
= 1.37 Å or 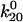 = 0.0033 s−1 and is within the predicted range. The colored lines show the model fit for the shear conditions of Fig. 3. (B) Bond number as a predictor of distance rolled for the allosteric model. The total number of bonds was estimated for each video as the normalization factor required to fit the models to the data. The distance the bacteria rolled was directly measured by tracking the bacteria in the videos. The bond number is approximately proportional to the distance rolled, (R2 = 0.8).
= 0.0033 s−1 and is within the predicted range. The colored lines show the model fit for the shear conditions of Fig. 3. (B) Bond number as a predictor of distance rolled for the allosteric model. The total number of bonds was estimated for each video as the normalization factor required to fit the models to the data. The distance the bacteria rolled was directly measured by tracking the bacteria in the videos. The bond number is approximately proportional to the distance rolled, (R2 = 0.8).
FIGURE 8.
Tests of alternative models. (A) The two-pathway model (16) (dotted line) does not describe a typical FimH data set (green diamonds; 0.01 pN/μm2) because the model requires single exponential decay. The two independent binding sites model (solid lines) requires that the high shear data (red squares; 2 pN/μm2) be shifted downward by assuming a large number of bonds broke too quickly to cause observable pauses. The resulting large number of estimated bonds can be seen as the red square outliers in panel B. (B) When the two independent binding sites model was fit to the entire data set, the total number of estimated bonds served as a poor predictor of distance rolled, unlike for the allosteric catch-bond model in Fig. 5 B.
Fitting the pause lifetime data to the model
The 7.3 s of lifetime data were fit to the model described in the results under the assumption that pauses reflected single bond lifetimes and that all the drag force was applied to one bond at a time, an assumption that we justify in the Results section. We calculated the drag force as a function of shear stress assuming an effective diameter of 1 μm, and using Goldman's approximation (21) for the drag force on a stationary sphere touching a wall in a shear field (F = 1.7 × 6πτr2, τ the shear stress, and r the particle radius.) There are some uncertainties in the force calculation that could lead to errors in the estimates of the transition state distance parameters of the model. First, the bacteria are nonspherical (∼1/2 × 1 μm) and it is unknown how the micrometer-long fimbriae contribute to the drag force. Second, the angle between a fimbria bound to the surface and the surface itself is unknown. The long length of the fimbriae suggest that the angle will be small, so that the tensile force on the bond will approach the drag force.
SAAM II software (SAAM Institute, Seattle, WA) (22) was used to obtain maximum likelihood estimates of the model parameters (23). Extended least squares (model-based, relative weighting scheme) were utilized; the measurement error in the bond survival was assumed to be zero mean and have constant fractional standard deviation. We calculated both estimates of the optimal parameter values and asymptotic standard errors—a common measure of parameter precision (24). To improve the model performance, the prior understanding that the distances involved in unbinding must be at least the distance of single atom interactions, or 1 Å, was incorporated with the use of empirical Bayesian parameters. That is, the natural logarithms of 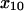 and
and 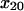 in angstroms were assumed to belong to a normal distribution with mean 2 and ± SD 0.69.
in angstroms were assumed to belong to a normal distribution with mean 2 and ± SD 0.69.
RESULTS
Duration of pauses for E. coli moving on a mannose surface
We have shown previously that E. coli bacteria binding specifically to a mannose-BSA surface through FimH exhibit a “stick-and-roll” adhesion in which they alternate between rolling and stationary adhesion at moderate shear but favor stationary adhesion at higher shear. Fig. 1 A shows stick-and-roll adhesion at a higher time resolution than shown previously (4). The rolling consists of an apparently random series of short pauses between rapid movement, whereas stationary adhesion is a much longer pause. To explore the mechanism of these pauses and thus of the stick-and-roll adhesion, we analyzed the lifetime of pauses at several shear stresses.
At each of six shear stresses, all pauses that started during the first half of the video were followed until the bacteria moved or the video ended. Fig. 1 B shows a pause survival plot calculated from the data (see Materials and Methods) for low (0.01 pN/μm2), and high (2 pN/μm2) shear stress. The data approximate a double exponential decay at each shear stress. That is, there is an inflection between two distinct decay rates in each pause survival curve, so that a fraction of pauses last very short times whereas the rest last very long times. Indeed, there are several orders of magnitude difference between the fast and slow decay rates (∼70-fold at low shear and ∼360-fold at high shear in this figure.) Raising the shear stress increases both the fraction and lifetime of long pauses. This reflects the switch from rolling to stationary adhesion reported earlier (4).
Single bonds dominate adhesive behavior
To fit a model to the pause time data it is necessary to know whether or not the pauses reflect single bonds. In particular, we asked whether the long-lived pauses were caused by multiple bonds. If two or more bonds unbind and rebind without the bacteria moving forward, this could easily explain several orders of magnitude increase in pause lifetimes. To test this, we added 5% α-methyl-mannoside, a soluble competitive inhibitor of FimH, to bacteria that were already bound to the surface. When the inhibitor entered the flow chamber, rolling bacteria immediately begin moving at the hydrodynamic velocity, whereas stationary bacteria remained stationary for some time. In either case, bacteria rarely paused again once they moved in the presence of inhibitor (Fig. 2 A). In contrast, in control experiments without inhibitor, bacteria repeatedly move and pause (Fig. 2 A). This indicates that the inhibitor did bind free FimH molecules and prevent new bonds from forming as expected. However, when we tracked the lifetime of the existing long-lived pauses, they were not affected by the presence of the inhibitor (Fig. 2 B). This demonstrates that their longevity did not depend on the formation of new bonds.
FIGURE 2.
Tests for whether single bonds cause the pauses. (A) Soluble inhibitor prevents new pauses. In the control experiment without inhibitor (▵), many new pauses began at medium shear stress (0.5 pN/μm2). In the presence of 5% α-methyl mannoside inhibitor (♦), only one new pause began. These results are normalized for the number of bacteria moving in the field of view. This indicates that the inhibitor prevented new bonds from forming. (B) Effect of soluble inhibitor on the lifetime of preexisting pauses in the same experiment as in panel A. The presence (•) or absence (▵) of 5% α-methyl mannoside inhibitor does not effect the distribution of preexisting pause lifetimes at medium shear stress (0.5 pN/μm2). No effect of inhibitor on pause lifetime is observed. (You may notice that both conditions in this experiment show a higher fraction of long pauses relative to other experiments in this article. This is because we normally counted all pauses that started in a set interval, but could not do so in this experiment because inhibitor prevents new pauses from beginning. Here we instead counted all pauses that already existed at the moment when new solution entered the chamber. Using existing pauses oversamples the long pauses. Nevertheless, this experiment is valid to compare the inhibitor with the control because both were measured the same way.) (C) Effect of changing the concentration of receptor on the surface at.0.26 pN/μm2. This was achieved by reducing the concentration of mannose-BSA in the incubation from 200 (▵) to 20 (▪) μg/ml, and resulted in ∼10-fold fewer pauses. The data here is expressed as fraction of total pauses measured, so that the difference in total pause number is not seen in the figure except as a difference in the number of nonredundant data points. Changing the receptor concentration also had no significant effect on the distribution of pause lifetimes.
The several orders of magnitude difference in pause lifetimes could in theory also result if the drag force on the bacterium is sometimes distributed over one bond and sometimes over multiple bonds. A force, f, applied across a bond has an exponential effect on the unbinding rate constant k (25,26): 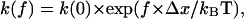 where kBT is thermal energy (41 pN Å at 25°C), and
where kBT is thermal energy (41 pN Å at 25°C), and 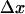 is the interaction distance, or the increase in length between the bound and transition states of the bond. It is normally assumed (25,26) that
is the interaction distance, or the increase in length between the bound and transition states of the bond. It is normally assumed (25,26) that 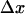 > 0 so that the unbinding rate increases with force, and the bond is a slip bond. How would the unbinding rate be affected if this force is distributed over N bonds instead of one? The ratio of unbinding rates at f and f/N is
> 0 so that the unbinding rate increases with force, and the bond is a slip bond. How would the unbinding rate be affected if this force is distributed over N bonds instead of one? The ratio of unbinding rates at f and f/N is 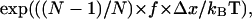 or less than
or less than 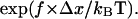 At high forces (
At high forces (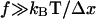 ), the bond lifetimes could increase by orders of magnitude when the mechanical load is distributed. However, we observed the double exponential decay even at low shear where there is only 0.1 pN per bacterium (Fig. 1 B). At this small force, even a fewfold increase in bond lifetime would require that
), the bond lifetimes could increase by orders of magnitude when the mechanical load is distributed. However, we observed the double exponential decay even at low shear where there is only 0.1 pN per bacterium (Fig. 1 B). At this small force, even a fewfold increase in bond lifetime would require that 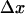 be over 400 Å, or 10-fold larger than the binding domain, which is unreasonable. Thus, the double exponential decay cannot be caused by force distribution over multiple bonds.
be over 400 Å, or 10-fold larger than the binding domain, which is unreasonable. Thus, the double exponential decay cannot be caused by force distribution over multiple bonds.
Without rebinding or significant force effects, the lifetime of a cluster of bonds increases as the harmonic number (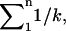 or approximately
or approximately 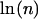 ) of n, the number of bonds (27). Increasing the lifetime of pauses several orders of magnitude (e.g., Fig. 1) would require over a billion bonds, but there are at most thousands of fimbriae per bacterium. Moreover, FimH bonds should not cluster because E. coli have only one FimH at the tip of each type I fimbria (20,28), in contrast to leukocytes, which extend long tethers with clusters of receptors at the tip. Nevertheless, we hypothesized that if multiple bonds contribute significantly to our pause data, reducing the concentration of mannose-BSA on the surface should decrease the number of bonds involved in each pause, and thus the fraction or lifetime of the long-lived pauses. When mannose-BSA was reduced 10-fold, the number of bacteria binding was greatly reduced but the pause lifetime profile of these bacteria was not changed (Fig. 2 B). In contrast, when a double exponential decay was observed for L-selectin in flow chamber assays, it was eliminated by decreasing the surface concentration twofold (29).
) of n, the number of bonds (27). Increasing the lifetime of pauses several orders of magnitude (e.g., Fig. 1) would require over a billion bonds, but there are at most thousands of fimbriae per bacterium. Moreover, FimH bonds should not cluster because E. coli have only one FimH at the tip of each type I fimbria (20,28), in contrast to leukocytes, which extend long tethers with clusters of receptors at the tip. Nevertheless, we hypothesized that if multiple bonds contribute significantly to our pause data, reducing the concentration of mannose-BSA on the surface should decrease the number of bonds involved in each pause, and thus the fraction or lifetime of the long-lived pauses. When mannose-BSA was reduced 10-fold, the number of bacteria binding was greatly reduced but the pause lifetime profile of these bacteria was not changed (Fig. 2 B). In contrast, when a double exponential decay was observed for L-selectin in flow chamber assays, it was eliminated by decreasing the surface concentration twofold (29).
Thus, all tests contradicted the notion that the pause time data, and in particular the double exponential decay, could significantly be affected by multiple bonds. We now ask whether an allosteric model could explain the FimH data quantitatively assuming the pauses reflect single bonds lifetimes as indicated by these tests.
An allosteric catch-bond model predicts a double exponential decay
A simple model of an allosteric protein has two low energy conformations separated by a single energy barrier. This barrier has height 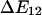 above state 1 and
above state 1 and 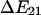 above state 2, as shown in Fig. 3 A. The transition rate from state 1 to state 2 (
above state 2, as shown in Fig. 3 A. The transition rate from state 1 to state 2 (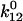 ) and the reverse rate (
) and the reverse rate (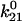 ), are related to these energy barriers, with
), are related to these energy barriers, with 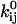 proportional to
proportional to 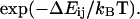 We assume that the active site can bind the ligand in both conformations, but with different strength, so there are two unbinding energies,
We assume that the active site can bind the ligand in both conformations, but with different strength, so there are two unbinding energies, 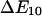 and
and  that also determine two unbinding rates
that also determine two unbinding rates 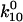 and
and 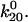 In either bound state, the complex multidimensional energy landscape thus has two escape paths—one leads to the alternate bound state and the other to unbinding. Fig. 3 A shows the one-dimensional projection of these pathways as dashed and solid lines, respectively. Because these correspond to distinct changes in the multidimensional bond structure initiating in the binding site versus the allosteric regulatory site, we assume that the receptor-ligand bond explores the two pathways independently. For this reason, the complex landscape of Fig. 3 A leads to the model of Fig. 3 B, which shows two independent transitions for each state; unbinding and conformational change.
In either bound state, the complex multidimensional energy landscape thus has two escape paths—one leads to the alternate bound state and the other to unbinding. Fig. 3 A shows the one-dimensional projection of these pathways as dashed and solid lines, respectively. Because these correspond to distinct changes in the multidimensional bond structure initiating in the binding site versus the allosteric regulatory site, we assume that the receptor-ligand bond explores the two pathways independently. For this reason, the complex landscape of Fig. 3 A leads to the model of Fig. 3 B, which shows two independent transitions for each state; unbinding and conformational change.
FIGURE 3.
Energy landscape of an allosteric catch-bond model and the associated rate constants. (A) Projections of the energy landscapes onto the direction of applied force, for the three transitions involved. The allosteric transition between state 1 (weak) and state 2 (strong) is shown as a dotted line. Unbinding transitions from states 1 or 2 are shown as solid lines. The unbound state(s) are not shown because our model doesn't assume whether there are one, two, or more unbound states, nor does our data analysis probe this part of the energy landscape. The x-dimension in this illustration can be viewed as the extension of the receptor-ligand complex. Each 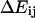 is the height of the energy barrier, whereas each
is the height of the energy barrier, whereas each 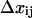 is the transition state distance (the projection of the vector from state i to the transition state to j onto the force vector). (B) The two-state model used to represent this energy landscape.
is the transition state distance (the projection of the vector from state i to the transition state to j onto the force vector). (B) The two-state model used to represent this energy landscape.
When a force f, is applied across a bond, the difference in free energy between the low-energy and transition states decreases by the amount 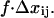 Here, as illustrated in Fig. 3,
Here, as illustrated in Fig. 3, 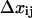 is the difference in lengths between the initial and transition states in the direction of force, and is a generalization of the interaction distance for unbinding,
is the difference in lengths between the initial and transition states in the direction of force, and is a generalization of the interaction distance for unbinding, 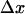 ). Thus, force affects all the transition rates exponentially, analogous to the effect of force on unbinding (25,26):
). Thus, force affects all the transition rates exponentially, analogous to the effect of force on unbinding (25,26):
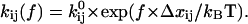 |
(1) |
This requires the common assumption (14,15,18,25) that the barriers are sharp so  are unchanged by applied force.
are unchanged by applied force.
To calculate the lifetime of the allosteric bond, we define 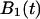 and
and  to represent the probability of occupancy of each state as a function of time. The transition rates are the four force-dependent kinetic constants,
to represent the probability of occupancy of each state as a function of time. The transition rates are the four force-dependent kinetic constants, 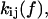 of Eq. 1, which we now abbreviate as
of Eq. 1, which we now abbreviate as 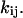 The ordinary differential equations describing this two-state model are thus:
The ordinary differential equations describing this two-state model are thus:
 |
(2) |
The initial conditions depend on experimental conditions and bond parameters. In our flow chamber experiments, the bond lifetimes are tested quickly as the bacteria move rapidly over the surface, but the bound states equilibrate slowly (see Table 1 after solution of the model). Thus, the initial conditions of the model are not 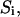 the equilibrium occupancies of the bound states (
the equilibrium occupancies of the bound states (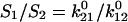 ), but rather are
), but rather are  the probability that the bond initially forms in state i. In the absence of force, the system is in thermodynamic equilibrium and the principle of detailed balance requires that the flux for initial formation equals the unbinding flux for each state:
the probability that the bond initially forms in state i. In the absence of force, the system is in thermodynamic equilibrium and the principle of detailed balance requires that the flux for initial formation equals the unbinding flux for each state: 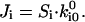 Thus:
Thus:
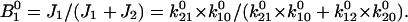 |
(3) |
TABLE 1.
Parameter estimates for the allosteric catch-bond model describing FimH
| Transition | Rate constant | Distance parameter |
|---|---|---|
| 1 → off |
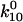 = 6.01 ± 0.24 s−1 = 6.01 ± 0.24 s−1
|
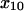 = 1.37 ± 0.30 Å = 1.37 ± 0.30 Å |
| 2 → off |
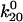 = 0.007 ± 0.006 s−1 = 0.007 ± 0.006 s−1
|
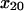 = 1.76 ± 0.63 Å = 1.76 ± 0.63 Å |
| 1 → 2 |
 = 0.210 ± 0.014 s−1 = 0.210 ± 0.014 s−1
|
 = 8.58 ± 0.25 Å = 8.58 ± 0.25 Å |
| 2 → 1 |
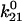 = 0.105 ± 0.013 s−1 = 0.105 ± 0.013 s−1
|
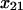 = −4.2 ± 2.1 Å = −4.2 ± 2.1 Å |
The parameters here are those used for the fit in Fig. 3. The mean ± SD is calculated by the SAAM II software as described in the Materials and Methods section.
By deriving 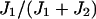 from the reverse fluxes, we do not require knowledge of the number or the energies of the unbound states. We derived this ratio earlier by specifically assuming there were two unbound states (30) and it is simple to do so for one unbound state, but the result will always be Eq. 3, as long as the assumption of thermodynamic equilibrium holds.
from the reverse fluxes, we do not require knowledge of the number or the energies of the unbound states. We derived this ratio earlier by specifically assuming there were two unbound states (30) and it is simple to do so for one unbound state, but the result will always be Eq. 3, as long as the assumption of thermodynamic equilibrium holds.
In a flow chamber experiment, force can only be applied after formation of the bond, so the initial conditions are force-independent. This assumption also requires that rebinding occurs slowly relative to transition between any unbound states, so that the distribution of unbound states is not significantly affected by how force alters previous bonds. We make this assumption because different FimH molecules should participate in the formation of new bonds as bacteria roll or move forward, and because the number of FimH binding to the surface appears to be small. We will address the impact of this assumption on model behavior after estimating the model parameters.
Solving Eq. 2 with initial conditions of Eq. 3 determines the bond survival probability 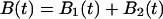 as a function of time
as a function of time 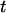 after initial formation. We consider the constant force situation, because this relates to the pause time experiments where bacteria do not move during pauses as described in the previous section. The force must ramp up from zero, but if this occurs quickly relative to bond lifetimes, this situation can be approximated with an instantaneous increase in force from zero to f, making for a simple linear model, with this solution:
after initial formation. We consider the constant force situation, because this relates to the pause time experiments where bacteria do not move during pauses as described in the previous section. The force must ramp up from zero, but if this occurs quickly relative to bond lifetimes, this situation can be approximated with an instantaneous increase in force from zero to f, making for a simple linear model, with this solution:
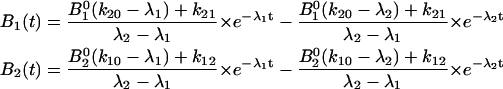 |
(4) |
where 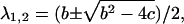
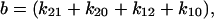 and
and 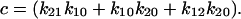
The bond survival would thus be:
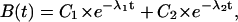 |
(5) |
where 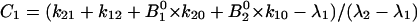 and
and 
This model predicts a double exponential decay in the number of bonds surviving as a function of time, so explains the unique behavior of FimH seen in Fig. 1 B. The mean lifetime 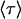 of the allosteric bond can be calculated from the probability distribution of bond lifetimes, which is the negative derivative of the survival function (
of the allosteric bond can be calculated from the probability distribution of bond lifetimes, which is the negative derivative of the survival function (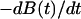 ):
):
 |
(6) |
For an allosteric bond to be a catch bond,  must increase with force. This will depend on the parameters
must increase with force. This will depend on the parameters 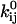 and
and 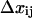 as well as the range of force in question, as
as well as the range of force in question, as 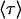 is a function of all of these.
is a function of all of these.
Fitting the allosteric catch-bond model to the pause time data
Data were collected from six different shear stresses in triplicate, and fit with a single set of parameters. Because an unknown number of pauses could be too short to be detected (<74 ms), (this problem also arises in single-molecule force probe experiments), each curve requires an adjustable normalization parameter for total number of binding events. The flow chamber allows us to check these total bond numbers as a predictive test of the model, which we do later. The data and model fit are shown in Fig. 4 A, with the parameter estimates and their errors shown in Table 1.
FIGURE 4.
The allosteric catch-bond model fit to the length of time bacteria pause. (A) The fraction of pauses surviving is graphed as a function of the time since each pause started, for a shear stress of 0.01 pN/μm2 (green diamonds), 0.5 pN/μm2 (blue triangles), 1 pN/μm2 (magenta circles), and 2 pN/μm2 (red squares). The model was fit using SAAM II software as described in the Materials and Methods section, and the parameters of this fit are given in Table 1. The results for 0.05 and 0.26 pN/μm2 are not shown in the figure to avoid cluttering. The triplicate experiments are shown as three sizes of symbols, and the model predictions as lines. At each force, the model predicts a double exponential decay in bond survival: 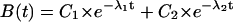 (Eq. 5). The model behavior can be understood from how force affects the two lifetimes 1/
(Eq. 5). The model behavior can be understood from how force affects the two lifetimes 1/ and 1/
and 1/ (C, dashed and dotted lines), and their coefficients C1 and C2 (B, dashed and dotted lines), because all are derived parameters of force and the eight parameters of Table 1). The overall mean lifetime
(C, dashed and dotted lines), and their coefficients C1 and C2 (B, dashed and dotted lines), because all are derived parameters of force and the eight parameters of Table 1). The overall mean lifetime 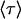 is shown in panel C by the heavy solid line.
is shown in panel C by the heavy solid line.
We can now explore the behavior of the allosteric catch-bond model in the context of these parameters. A look at Table 1 shows that in the absence of force, state 1 unbinds three orders of magnitude faster than does state 2, while the conversion rates between states are intermediate (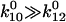 >
> 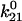 ≫
≫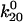 ). Thus, a bond in state 1 will usually unbind directly, whereas a bond in state 2 will usually revert to state 1 and then unbind rather than directly unbinding via
). Thus, a bond in state 1 will usually unbind directly, whereas a bond in state 2 will usually revert to state 1 and then unbind rather than directly unbinding via 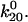 Because of this, the fast decay rate approximates
Because of this, the fast decay rate approximates 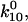 whereas the slow decay rate approximates
whereas the slow decay rate approximates 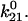 This can also be shown mathematically. In the limit that
This can also be shown mathematically. In the limit that 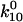 is greater than the other rate constants,
is greater than the other rate constants, 
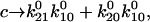 and
and 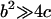 in Eq. 4 (recall that
in Eq. 4 (recall that 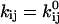 when there is no force). In this case,
when there is no force). In this case, 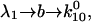 and the Taylor expansion of the square root function can be used to show that
and the Taylor expansion of the square root function can be used to show that 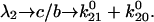 If furthermore
If furthermore 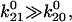 then
then 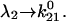 Thus, given the parameters in Table 1, there is two orders of magnitude difference between the short and long pause lifetimes.
Thus, given the parameters in Table 1, there is two orders of magnitude difference between the short and long pause lifetimes.
The initial conditions of the bonds can be calculated using Eq. 3 and the parameters are given in Table 1. 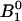 = 99.8% of the bonds form in state 1, whereas only
= 99.8% of the bonds form in state 1, whereas only 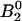 = 0.2% of the bonds form in state 2. This occurs because the rate constants for FimH dictate that the absolute height of the energy barrier between state 2 and the unbound states is much higher than that between state 1 and the unbound states (as illustrated in the cartoon in Fig. 3), and thus the flux through this state is much lower. Furthermore, the majority of bonds never reach state 2 even though it is thermodynamically favored (
= 0.2% of the bonds form in state 2. This occurs because the rate constants for FimH dictate that the absolute height of the energy barrier between state 2 and the unbound states is much higher than that between state 1 and the unbound states (as illustrated in the cartoon in Fig. 3), and thus the flux through this state is much lower. Furthermore, the majority of bonds never reach state 2 even though it is thermodynamically favored ( ) because they unbind before they can convert (
) because they unbind before they can convert (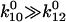 ); that is, only
); that is, only 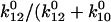 ), or 3.5%, of the bonds that form initially in state 1 will convert to state 2 before they unbind. The proportion of long lifetimes calculated by Eq. 4 with the parameters of Table 1 is ∼3.7% in the absence of applied force, and it can now be seen that this mostly reflects conversion between the states but also reflects a small fraction of bonds that start in state 2.
), or 3.5%, of the bonds that form initially in state 1 will convert to state 2 before they unbind. The proportion of long lifetimes calculated by Eq. 4 with the parameters of Table 1 is ∼3.7% in the absence of applied force, and it can now be seen that this mostly reflects conversion between the states but also reflects a small fraction of bonds that start in state 2.
In the presence of force, we have assumed there is no change in the initial conditions; rather all changes occur once force is applied after initial binding. The biggest change is on the transition rate to the long-lived state 2, because 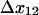 has the largest absolute distance of all transitions (8.5 Å);
has the largest absolute distance of all transitions (8.5 Å); 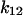 increases from 0.2 to 6.0 s−1 between 0 and 16 pN, whereas the unbinding rate
increases from 0.2 to 6.0 s−1 between 0 and 16 pN, whereas the unbinding rate 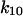 only increases from 6 to 10 s−1. Thus, by 2 pN/μm2, the fraction of long-lived pauses increases dramatically from 3.7 to 35%. (Fig. 4 B). This occurs not from a change in initial conditions, but because force causes the bonds that form in state 1 to convert more readily into the long-lived state 2. The next largest effect of force is on the rate of the return transition to state 1, because
only increases from 6 to 10 s−1. Thus, by 2 pN/μm2, the fraction of long-lived pauses increases dramatically from 3.7 to 35%. (Fig. 4 B). This occurs not from a change in initial conditions, but because force causes the bonds that form in state 1 to convert more readily into the long-lived state 2. The next largest effect of force is on the rate of the return transition to state 1, because 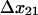 = −4.2 Å is next largest and negative;
= −4.2 Å is next largest and negative; 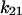 decreases from 0.1 to 0.02 s−1. At the same time,
decreases from 0.1 to 0.02 s−1. At the same time, 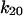 only increases from 0.007 to 0.014 s−1, and so
only increases from 0.007 to 0.014 s−1, and so 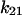 still accounts for most of the loss from state 2. Thus, force increases the lifetime of state 2 (Fig. 4, A and C), and there is now over three orders of magnitude difference in lifetime between the short and long pauses.
still accounts for most of the loss from state 2. Thus, force increases the lifetime of state 2 (Fig. 4, A and C), and there is now over three orders of magnitude difference in lifetime between the short and long pauses.
Thus, the allosteric model can describe a catch bond. The FimH parameters result in a catch bond both because force favors the long-lived state and because the lifetime of the long-lived state increases. Between 0 and 16 pN, these two effects combine to increase the mean bond lifetime 25-fold (Fig. 4 C), as calculated from the estimated parameters using Eq. 6. With other parameter sets, the allosteric model may result in a variety of slip bonds or catch bonds, but categorizing model behavior for all ranges of parameter values is beyond the scope of this work.
Testing predictions of the allosteric catch-bond model
Fig. 4, B and C, demonstrate how the bacteria should behave in higher shear conditions if the model and the assumptions used to fit the model are correct. The model predicts that above 40 pN, almost all of the bonds would convert to state 2, resulting in single exponential decay with the slower decay rate, and no further effect of force on the coefficients (Fig. 4 B). A second prediction is that, as force is increased above 24 pN, the lifetime of the long-lived state becomes shorter because the direct unbinding pathway is enhanced by force and eventually begins to dominate (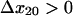 ); that is, because this state can unbind via a slip pathway (direct unbinding) or a catch pathway (reversion to a weakly bound state before unbinding), the long-lived state itself should appear to be a biphasic two-pathway catch bond, defined in Pereverzev et al. (16). The overall mean lifetime resulting from both states also describes a biphasic catch bond (Fig. 4 C).
); that is, because this state can unbind via a slip pathway (direct unbinding) or a catch pathway (reversion to a weakly bound state before unbinding), the long-lived state itself should appear to be a biphasic two-pathway catch bond, defined in Pereverzev et al. (16). The overall mean lifetime resulting from both states also describes a biphasic catch bond (Fig. 4 C).
To test the predictions above, we measured pause times of bacteria rolling at 10.9 pN/μm2, calculated to apply a force of 90 pN on each bacterium. The normalized pause survival data are shown in Fig. 5 A. The model predicts that at 90 pN force, k01 increases to 108 s−1, but k21 increases to 2 × 107 s−1, so that virtually all bonds enter state 2 without requiring any change in initial conditions. As predicted by the model, the decay is approximately single exponential. The model also predicts that the decay rate will approximate k02 = 0.13 s−1, faster than at moderate force. A faster decay is indeed observed, confirming the predicted biphasic lifetime. To quantitatively compare the prediction to the experiment at the new shear stress, the error of the estimates as well as the estimates themselves must be considered (Table 1). Only two of the eight parameters (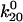 and
and 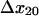 ) had a significant effect on the decay rate at 10.9 pN/μm2 when varied within their standard deviations, (Fig. 5 A). Adjusting either
) had a significant effect on the decay rate at 10.9 pN/μm2 when varied within their standard deviations, (Fig. 5 A). Adjusting either 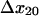 to 1.37 Å (within the estimated value of 1.73 ± 0.6 Å; Table 1) or
to 1.37 Å (within the estimated value of 1.73 ± 0.6 Å; Table 1) or 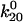 to 0.0033 s−1 (within 0.007 ± 0.006 s−1) gave a good fit to the model without significantly affecting the fit to the previous data. Thus, the very high shear stress data qualitatively and quantitatively validate the model.
to 0.0033 s−1 (within 0.007 ± 0.006 s−1) gave a good fit to the model without significantly affecting the fit to the previous data. Thus, the very high shear stress data qualitatively and quantitatively validate the model.
For a second test of the model, we checked whether the total number of binding events that were used to normalize the data made sense. This number should be proportional to the distance rolled by bacteria during the time the pauses were cataloged in each video, because rolling bacteria should move similar distances between pauses. The reason the number of binding events had to be estimated is that it could not be measured directly from the videos because pauses under 74 ms could not be detected. Fig. 5 B shows that the number of binding events is indeed proportional to the distance rolled over a wide range of shears, with an r2 value of 0.8. The allosteric catch-bond model can thus not only explain all the complex pause time data of rolling bacteria, but correctly predict both the high shear behavior and the distances rolled.
For a third test of the model, we measured the real-time response of bacteria to a switch in shear stress to see whether the transitions between short and long pauses right after the switch reflect the predicted transition rates between states 1 and 2. We bound bacteria to the mannose-BSA surface at moderate shear (0.2 pN/μm2) to have many bacteria rolling on the surface—that is, binding with short-lived pauses. When the shear stress was increased to 2 pN/μm2, these rolling bacteria quickly became stationary with long-lived pauses. When the shear stress was decreased to 0.01 pN/μm2, the stationary bacteria slowly began rolling again (Fig. 6 A) on short-lived pauses. To measure the real-time rate of conversion between short and long pauses, we measured the number of moving bacteria over time (Fig. 6 B). The moving behavior converted to stationary at a rate of 4.9 s−1. In contrast, the stationary behavior converted back to moving much more slowly, at a rate of 0.042 s−1. By combining Eq. 1 and the parameters in Table 1, the allosteric catch-bond model would predict that bonds in state 1 would convert to state 2 at a rate of 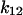 = 6 ± 1 s−1 at 16 pN (corresponding to 2 pN/μm2), and that bonds in state 2 would unbind or convert back to state 1 at a rate of
= 6 ± 1 s−1 at 16 pN (corresponding to 2 pN/μm2), and that bonds in state 2 would unbind or convert back to state 1 at a rate of 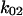 +
+ 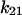 = 0.11 ± 0.2 s−1 at 0.1 pN (at 0.01 pN/μm2). Thus the allosteric catch-bond model predicts values close to the measured values, and in particular explains the two orders of magnitude difference between the real-time responses of bacteria to increases versus decreases in shear stress.
= 0.11 ± 0.2 s−1 at 0.1 pN (at 0.01 pN/μm2). Thus the allosteric catch-bond model predicts values close to the measured values, and in particular explains the two orders of magnitude difference between the real-time responses of bacteria to increases versus decreases in shear stress.
FIGURE 6.
Real-time response to changes in shear stress. Bacteria were bound at 0.2 pN/μm2, unbound bacteria were washed away, and then bacteria were switched from moderate (0.2 pN/μm2) to high (2 pN/μm2) and then back to low (0.01 pN/μm2) shear stress at times indicated by the dotted vertical lines. (A) Sample trajectories of individual bacteria are shown. The bacteria stopped moving almost immediately when the shear was turned up. However, each bacterium waited a longer time before beginning to move again when the shear was turned back down. (B) The total number of moving bacteria is reported at each time point. Bacteria are defined as moving if they move at any time over the next 1 s, because the short-lived pauses generally lasted <1 s. At each change in shear, the data is fit to get a transition rate between moving and stationary behavior (solid gray line). The rates obtained in this fit are 4.9 s−1 for the switch to stationary adhesion and 0.042 s−1 for the reversion from stationary to moving. When the flow rate is switched from low to high, the fluid movement was observed to increase in speed in less than one frame (37 ms), but the switch from high to low took place over several seconds, probably due to a slow decrease in pressure in the tubing between pump and chamber. This may explain why the response is a little slow at first for this transition.
Evaluating the initial conditions
Our two assumptions about the initial conditions should be evaluated now that the model parameters and behavior are known for these experiments. First, we assumed that transition rates between states 1 and 2 were slow relative to the wait time before bonds were probed. This assumption means that the initial conditions reflect on-rates and not the equilibrium between the two states. This can now be validated: states 1 and 2 equilibrate at a rate of 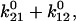 or 0.3 s−1, while bonds were tested at the video frame rate of 1/37 ms = 27 s−1, (and force added even more quickly.) The relative on-rates into the two states can be calculated with Eq. 3 and reflect the difference in absolute energies of the two unbinding transition states (peaks of solid lines in Fig. 3.) Thus, with the estimated parameters for FimH, most of the bonds form in state 1 (98.8%) even though state 2 is thermodynamically favored after equilibration of the complex even in the absence of force (
or 0.3 s−1, while bonds were tested at the video frame rate of 1/37 ms = 27 s−1, (and force added even more quickly.) The relative on-rates into the two states can be calculated with Eq. 3 and reflect the difference in absolute energies of the two unbinding transition states (peaks of solid lines in Fig. 3.) Thus, with the estimated parameters for FimH, most of the bonds form in state 1 (98.8%) even though state 2 is thermodynamically favored after equilibration of the complex even in the absence of force (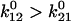 ). This assumption is important to model behavior. If the bonds were allowed to equilibrate before being probed, which would happen with a half-lifetime of 3 s, then
). This assumption is important to model behavior. If the bonds were allowed to equilibrate before being probed, which would happen with a half-lifetime of 3 s, then  or 67%, would already be in state 2 even in the absence of force. This explains why the majority of bonds in Fig. 2 B are long-lived even though the force is relatively low, because in this inhibitor experiment we were forced to probe the fate of all preexisting pauses after several minutes of equilibration. This demonstrates the history dependence that can arise with a two-state model and the importance of considering how experimental protocols affect initial conditions.
or 67%, would already be in state 2 even in the absence of force. This explains why the majority of bonds in Fig. 2 B are long-lived even though the force is relatively low, because in this inhibitor experiment we were forced to probe the fate of all preexisting pauses after several minutes of equilibration. This demonstrates the history dependence that can arise with a two-state model and the importance of considering how experimental protocols affect initial conditions.
Second, we assumed that bond on-rates were slow relative to transition rates between any unbound states so that early bonds do not influence the initial conditions of later bonds. This cannot be directly tested with the estimated parameters, because we do not know the energy levels of the unbound states or even how many such states there are. If there is one unbound state, it is of necessity in equilibrium with itself and the assumption holds. If there are two unbound states with transition rates comparable to those between the two bound states, then the rate of equilibration between them will be also ∼0.3 s−1. This is fast relative to the rate of bond formation, which we calculate at 0.01–0.1 s−1 per FimH (moving bacteria were calculated to pause at a rate of 10 s−1, and there are hundreds to thousands of FimH per bacterium). In the future, experiments may be performed so that on-rates are faster than transition rates, for example, in repeated atomic force microscopy pulls. In this case, force would increase the probability that a bond rebinds while still in state 2 from the last bond, and the fraction of long lifetimes would increase with repeated pulls at high force. The precise effect of force in such a case is beyond the scope of this article. However, it is worth noting that the vast majority of FimH bonds that reach state 2 are estimated to do so by conversion (3.5–100% depending on force) rather than by forming directly into state 2 (0.2%). Thus, if the number that form into state 2 directly increases even by many fold, it will hardly affect model behavior. For this reason, it would have minimal effect if this assumption is wrong for our experiments.
Alternative models do not explain the data
Although the allosteric catch-bond model is conceptually simple (arising from a single assumption of allostery) it has eight parameters that must be fit to the data. To further evaluate the validity of the model, we tested whether the data could be explained by a mathematically simpler model.
Previous models used for catch bonds cannot explain all the FimH data. The four-parameter two-pathway catch-bond model (see cartoon, Fig. 7 A) that fits all published selectin data (16) can partially describe the data presented here for FimH. If one could only detect pauses that lasted >2 s, then simple single-exponential decay would be observed at all shears (view only time points after 2 s in Figs. 4 A and 5). There is a biphasic effect of shear stress on this single decay rate, so that the slowest decay rate is observed at moderate shear stress (slopes of line and data with red squares after 2 s), as predicted for a two-pathway catch bond. However, the two-pathway model cannot explain the full range of FimH data. Because this model assumes only a single bound state, it doesn't explain the double-exponential decay observed even at a single shear stress when short pauses can be detected as in our experiments (Fig. 8 A). Previously published two-state models also do not describe the double exponential decay observed in the FimH data, because they use assumptions suited to explain the single-exponential decay selectin data (14,18).
FIGURE 7.
Alternative models. (A) Two-pathway bond model. (B) Two independent binding sites model. In each model, each rate constant 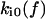 reflects an unstressed rate constant
reflects an unstressed rate constant  and a transition state distance
and a transition state distance 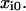
Alternatively, one might ask whether the double exponential decay might be caused by two independent binding sites (see cartoon, Fig. 7 B). This could reflect two mannose-binding proteins on the bacteria or two distinct mannose-binding sites or structures within FimH. This model requires five parameters: unbinding rates 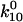 and
and 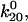 transition distances
transition distances 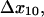 and
and 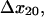 and B2, the fraction of long-lived bonds. This model can indeed describe the double exponential decay at any single shear stress (Fig. 8 A). The model was fit to the entire data set as before, and the parameters were estimated as:
and B2, the fraction of long-lived bonds. This model can indeed describe the double exponential decay at any single shear stress (Fig. 8 A). The model was fit to the entire data set as before, and the parameters were estimated as: 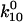 = 4.4 ± 0.2 s−1,
= 4.4 ± 0.2 s−1,  = 0.11 ± .01 s−1,
= 0.11 ± .01 s−1,  = 6.2 ± 0.1 Å,
= 6.2 ± 0.1 Å, 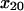 = −2.7 ± 0.4 Å, and B2 = 6 ± 0.3%. However, because the proportion of long-lived bonds does not change with force, this model does not explain why a higher proportion of measured pauses are long-lived as shear stress increases. Instead, it must assume that more and more pauses are too short to be detected at high force (Fig. 8 A). As a result, the normalization factors required to fit the model to the entire data set fail to predict the distances bacteria move (Fig. 8 B); that is, the two-independent bond-type model contradicts the observation that the bacteria stop moving at high shear. Moreover, this model does not predict the biphasic effect of shear that is observed on the lifetime of the long-lived pauses, instead predicting that the lifetime of the long-lived pauses should get longer at 10.9 pN/μm2 (not shown) whereas the data show much shorter pauses (Fig. 5 A).
= −2.7 ± 0.4 Å, and B2 = 6 ± 0.3%. However, because the proportion of long-lived bonds does not change with force, this model does not explain why a higher proportion of measured pauses are long-lived as shear stress increases. Instead, it must assume that more and more pauses are too short to be detected at high force (Fig. 8 A). As a result, the normalization factors required to fit the model to the entire data set fail to predict the distances bacteria move (Fig. 8 B); that is, the two-independent bond-type model contradicts the observation that the bacteria stop moving at high shear. Moreover, this model does not predict the biphasic effect of shear that is observed on the lifetime of the long-lived pauses, instead predicting that the lifetime of the long-lived pauses should get longer at 10.9 pN/μm2 (not shown) whereas the data show much shorter pauses (Fig. 5 A).
Thus, simpler models with fewer parameters may describe some aspects of the data, but do not explain the complexity of data for the pause lifetimes in FimH-mediated bacterial adhesion.
DISCUSSION
The allosteric catch-bond model explains and even predicts the phenomena we report here and elsewhere for E. coli bacteria rolling on mannose-BSA. This model is based on a single hypothesis that was suggested by steered molecular dynamics simulations and site-directed mutagenesis (1). We assumed that FimH is an allosteric protein in which the native conformation of a regulatory region stabilizes a weakly bound state, whereas a force-induced conformation of the regulatory region converts the bond to a strongly bound state. This quantitatively explains the coexistence of short and long pauses in the pause time data, and thus the stochastic switching between rolling and stationary behavior reported earlier (4). It also quantitatively explains why there is an increasingly higher fraction of long-lived pauses as flow increases and thus why the bacteria transition into the stationary state at high shear (4). Moreover, this model correctly predicted that the effect of shear stress on the lifetime of the long-lived pauses would be biphasic, so that very high shear shortens the pauses and causes the bacteria to move again. The estimated parameters for the model are consistent with the original structural hypothesis; the extension of 8.5 Å for 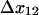 is comparable to the three-amino-acid extension observed in the steered molecular dynamics simulations (1). Finally, the allosteric catch-bond model explains why a large number of nonactive site mutations increase the binding strength of FimH (1,31). Although mutations in general cause a loss of function, nonactive site mutations can easily increase the activity of an allosteric protein by destabilizing the low-affinity state; that is, by causing a loss of inhibitory function.
is comparable to the three-amino-acid extension observed in the steered molecular dynamics simulations (1). Finally, the allosteric catch-bond model explains why a large number of nonactive site mutations increase the binding strength of FimH (1,31). Although mutations in general cause a loss of function, nonactive site mutations can easily increase the activity of an allosteric protein by destabilizing the low-affinity state; that is, by causing a loss of inhibitory function.
In contrast, alternative models do not explain the data. This includes the two-pathway model (16) (cannot explain double exponential decay), the assumption that there are two independent bond structures (cannot explain the increased fraction of long-lived pauses and stationary behavior at high shear), and the notion that long-lived pauses reflect multiple bonds (cannot explain the inability of inhibitors or changes in receptor concentration to significantly affect the pause lifetimes). This strongly suggests that FimH forms allosteric catch bonds with mannose. Table 1 gives an estimation of all the rate constants and the structural distances. Because they are based on a complicated system of whole bacteria, the parameters presented here have to be refined in the future using methods such as the atomic force microscopy, optical tweezers, or cell-free bead tethering assays that can more accurately measure the effect of force on single bond lifetimes.
Nevertheless, these data provide the first experimental demonstration that catch bonds can involve force regulation of double exponential decays in bond survival. Such a double exponential decay was not seen in selectin single-molecule experiments. When two-state models were recently applied to selectin catch bonds, they had to be applied in such a way that the model reduced to single exponential decay to fit the data. Evans et al. (14) assumed that the two states are in rapid equilibrium. Barsegov et al. (18) primarily calculated average lifetimes and did not address the absence of the double exponential decay in the Marshall selectin data being described. Finally, we (16) showed that the selectin data (5–7) can also be fit with a two-pathway, single-state model. This model can be understood as a limiting case of the allosteric model when one lifetime is too short to be resolved and only the long lifetime is observed in the experiments. Thus, published selectin experiments show single exponential decay in bond lifetimes, and because there are multiple models that can explain those data it remains to be determined whether the selectin bonds are also allosteric two-state catch bonds. Our work is thus the first experimental demonstration of the two-state model, first suggested by Bartolo et al. (15).
The assumptions about the initial conditions made in this article are different from those made previously to explain selectin catch bonds with two-state models. Here we assumed that transition rates were slow relative to the rate at which bonds were probed, so that initial conditions reflected the relative probability of forming into the two states. These assumptions were validated by the estimated parameters. In the model of Evans et al. (14), however, it is assumed that transition rates are rapid relative to all other rates so that initial conditions do not affect bond lifetimes. This is the same assumption made earlier by Bartolo et al. (15) and is consistent with the single exponential decay observed for P-selectin in constant force experiments (12). In the model of Barsegov et al. (18) it is assumed that initial conditions reflect the equilibrium distributions of the two bound states. This is consistent with the long time spent in contact with the surface relative to the estimated transition rates in their model fit. Thus, unlike one state models (25,26), the allosteric model is history dependent, and different experimental methods and bond parameters require different assumptions about the initial conditions. History dependence has been observed for P-selectin (32), but it remains to be determined whether the data can be explained with an allosteric model.
The allosteric catch-bond model presented here illustrates the type of experiments that need to be conducted for the future characterization of catch bonds. It is necessary that bond lifetimes be measured over a wide range of timescales (from the millisecond to the minute timescale for FimH, and in the microsecond to second timescale for the shorter-lived selectins). It should also be possible to test for allosteric conformational changes by testing structural variants of receptors or ligands to probe the effects of potential regulatory regions. Crystal structures and molecular dynamics simulations can be used to predict relevant mutants for these studies (1).
In the context of mechanical regulation, the concept of allostery is not merely a mathematical formalism, but has practical meaning. First, proteins that are known to undergo allosteric conformational changes may be able to form catch bonds. For example, the activated (longer-lived) state of the integrin-fibronectin bond has recently been shown to have a longer structure than the inactive state in both experiments (33) and simulations (34), but it remains unclear whether it forms an allosteric catch bond. Mechanical regulation through allostery has also been shown for motor proteins; the affinity of kinesin for microtubules and of myosin for actin is allosterically regulated by nucleotides ATP and ADP, and it has been proposed that the binding of these are in turn regulated by mechanical force due to internal strain (35,36). It now appears that actomyosin bonds also display catch-bond behavior in optical tweezers studies (William Guilford, University of Virginia, personal communication, 2005). Second, inhibition of medically relevant catch bonds with competitive inhibitors can be difficult (1) because soluble ligands create no significant drag force and thus do not bind strongly. If a catch bond is allosteric, however, it should be possible to design allosteric inhibitors that stabilize the weak state and prevent strong binding. Third, the allosteric model suggests design principles for engineering catch bonds for technological applications (19). A well-understood allosteric catch bond could be engineered to alter the force sensitivity and the specificity through mutations in the regulatory region and active site, respectively; or, a novel catch bond can be engineered from an allosteric protein by mechanically coupling it to its allosteric modulator. Because allostery is the dominant mechanism explaining most protein regulation in biology, designing mechanosensitive bonds or components from allosteric proteins offers enormous flexibility for nature or engineers.
Acknowledgments
We thank H. Qian, O. Prezhdo, Y. Pereverzev, and J. Glomset for valuable discussions and comments on the manuscript.
We gratefully acknowledge support by the National Institutes of Health, R01 AI50940 (E.V.S.) and P41 EB-001975 (P.V.), and the Resource Facility for Population Kinetics at the University of Washington.
References
- 1.Thomas, W. E., E. Trintchina, M. Forero, V. Vogel, and E. V. Sokurenko. 2002. Bacterial adhesion to target cells enhanced by shear force. Cell. 109:913–923. [DOI] [PubMed] [Google Scholar]
- 2.Isberg, R. R., and P. Barnes. 2002. Dancing with the host; flow-dependent bacterial adhesion. Cell. 110:1–4. [DOI] [PubMed] [Google Scholar]
- 3.Konstantopoulos, K., W. D. Hanley, and D. Wirtz. 2003. Receptor-ligand binding: ‘catch’ bonds finally caught. Curr. Biol. 13:R611–R613. [DOI] [PubMed] [Google Scholar]
- 4.Thomas, W. E., L. Nilsson, M. Forero, E. V. Sokurenko, and V. Vogel. 2004. ‘Stick-and-roll’ bacterial adhesion mediated by catch-bonds. Mol. Microbiol. 53:1545–1557. [DOI] [PubMed] [Google Scholar]
- 5.Finger, E. B., K. D. Puri, R. Alon, M. B. Lawrence, U. H. von Andrian, and T. A. Springer. 1996. Adhesion through L-selectin requires a threshold hydrodynamic shear. Nature. 379:266–269. [DOI] [PubMed] [Google Scholar]
- 6.Alon, R., S. Chen, K. D. Puri, E. B. Finger, and T. A. Springer. 1997. The kinetics of L-selectin tethers and the mechanics of selectin-mediated rolling. J. Cell Biol. 138:1169–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yago, T., J. Wu, C. D. Wey, A. G. Klopocki, C. Zhu, and R. P. McEver. 2004. Catch bonds govern adhesion through L-selectin at threshold shear. J. Cell Biol. 166:913–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li, Z. J., N. Mohamed, and J. M. Ross. 2000. Shear stress affects the kinetics of Staphylococcus aureus adhesion to collagen. Biotechnol. Prog. 16:1086–1090. [DOI] [PubMed] [Google Scholar]
- 9.Mohamed, N., T. R. Rainier, and J. M. Ross. 2000. Novel experimental study of receptor-mediated bacterial adhesion under the influence of fluid shear. Biotechnol. Bioeng. 68:628–636. [PubMed] [Google Scholar]
- 10.Doggett, T. A., G. Girdhar, A. Lawshe, D. W. Schmidtke, I. J. Laurenzi, S. L. Diamond, and T. G. Diacovo. 2002. Selectin-like kinetics and biomechanics promote rapid platelet adhesion in flow: the GPIb(alpha)-vWF tether bond. Biophys. J. 83:194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dembo, M., D. C. Torney, K. Saxman, and D. Hammer. 1988. The reaction-limited kinetics of membrane-to-surface adhesion and detachment. Proc. R. Soc. Lond. B Biol. Sci. 234:55–83. [DOI] [PubMed] [Google Scholar]
- 12.Marshall, B. T., M. Long, J. W. Piper, T. Yago, R. P. McEver, and C. Zhu. 2003. Direct observation of catch bonds involving cell-adhesion molecules. Nature. 423:190–193. [DOI] [PubMed] [Google Scholar]
- 13.Sarangapani, K. K., T. Yago, A. G. Klopocki, M. B. Lawrence, C. B. Fieger, S. D. Rosen, R. P. McEver, and C. Zhu. 2004. Low force decelerates L-selectin dissociation from P-selectin glycoprotein ligand-1 and endoglycan. J. Biol. Chem. 279:2291–2298. [DOI] [PubMed] [Google Scholar]
- 14.Evans, E., A. Leung, V. Heinrich, and C. Zhu. 2004. Mechanical switching and coupling between two dissociation pathways in a P-selectin adhesion bond. Proc. Natl. Acad. Sci. USA. 101:11281–11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartolo, D., I. Derenyi, and A. Ajdari. 2002. Dynamic response of adhesion complexes: beyond the single-path picture. Phys. Rev. E. 65:051910. [DOI] [PubMed] [Google Scholar]
- 16.Pereverzev, Y., O. V. Prezhdo, M. Forero, E. Sokurenko, and W. Thomas. 2005. The two-pathway model for the catch-slip transition in biological adhesion. Biophys. J. 89:1446–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pereverzev, Y. V., O. V. Prezhdo, W. E. Thomas, and E. V. Sokurenko. 2005. Distinctive features of the biological catch bond in the jump-ramp force regime predicted by the two-pathway model. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 72:010903. [DOI] [PubMed] [Google Scholar]
- 18.Barsegov, V., and D. Thirumalai. 2005. Dynamics of unbinding of cell adhesion molecules: transition from catch to slip bonds. Proc. Natl. Acad. Sci. USA. 102:1835–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forero, M., W. E. Thomas, C. Bland, L. Nilsson, E. V. Sokurenko, and V. Vogel. 2004. A catch-bond based smart nano-adhesive sensitive to shear stress. Nano Lett. 4:1593–1597. [Google Scholar]
- 20.Choudhury, D., A. Thompson, V. Stojanoff, S. Langermann, J. Pinkner, S. J. Hultgren, and S. D. Knight. 1999. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science. 285:1061–1066. [DOI] [PubMed] [Google Scholar]
- 21.Goldman, A. J., R. G. Cox, and H. Brenner. 1967. Slow viscous motion of a sphere parallel to a plane wall. II. Couette flow. Chem. Eng. Sci. 22:653–660. [Google Scholar]
- 22.Barrett, P. H., B. M. Bell, C. Cobelli, H. Golde, A. Schumitzky, P. Vicini, and D. M. Foster. 1998. SAAM. II. Simulation, Analysis, and Modeling Software for tracer and pharmacokinetic studies. Metabolism. 47:484–492. [DOI] [PubMed] [Google Scholar]
- 23.Sheiner, L. B., and S. L. Beal. 1985. Pharmacokinetic parameter estimates from several least squares procedures: superiority of extended least squares. J. Pharmacokinet. Biopharm. 13:185–201. [DOI] [PubMed] [Google Scholar]
- 24.Landaw, E. M., and J. J. DiStefano III. 1984. Multiexponential, multicompartmental, and noncompartmental modeling. II. Data analysis and statistical considerations. Am. J. Physiol. 246:R665–R677. [DOI] [PubMed] [Google Scholar]
- 25.Evans, E. 2001. Probing the relation between force—lifetime—and chemistry in single molecular bonds. Annu. Rev. Biophys. Biomol. Struct. 30:105–128. [DOI] [PubMed] [Google Scholar]
- 26.Bell, G. I. 1978. Models for the specific adhesion of cells to cells. Science. 200:618–627. [DOI] [PubMed] [Google Scholar]
- 27.Tees, D. F. J., J. T. Woodward, and D. A. Hammer. 2001. Reliability theory for receptor-ligand bond dissociation. J. Chem. Phys. 114:7483–7496. [Google Scholar]
- 28.Hahn, E., P. Wild, U. Hermanns, P. Sebbel, R. Glockshuber, M. Haner, N. Taschner, P. Burkhard, U. Aebi, and S. A. Muller. 2002. Exploring the 3D molecular architecture of Escherichia coli type 1 pili. J. Mol. Biol. 323:845–857. [DOI] [PubMed] [Google Scholar]
- 29.Dwir, O., A. Solomon, S. Mangan, G. S. Kansas, U. S. Schwarz, and R. Alon. 2003. Avidity enhancement of L-selectin bonds by flow: shear-promoted rotation of leukocytes turn labile bonds into functional tethers. J. Cell Biol. 163:649–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas, W. E. 2003. Shear stress enhances bacterial adhesion. PhD thesis. University of Washington, Seattle, WA.
- 31.Sokurenko, E. V., V. Chesnokova, D. E. Dykhuizen, I. Ofek, X. R. Wu, K. A. Krogfelt, C. Struve, M. A. Schembri, and D. L. Hasty. 1998. Pathogenic adaptation of Escherichia coli by natural variation of the FimH adhesin. Proc. Natl. Acad. Sci. USA. 95:8922–8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marshall, B. T., K. K. Sarangapani, J. Lou, R. P. McEver, and C. Zhu. 2005. Force history dependence of receptor-ligand dissociation. Biophys. J. 88:1458–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chigaev, A., T. Buranda, D. C. Dwyer, E. R. Prossnitz, and L. A. Sklar. 2003. FRET detection of cellular alpha(4)-integrin conformational activation. Biophys. J. 85:3951–3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin, M., I. Andricioaei, and T. A. Springer. 2004. Conversion between three conformational states of integrin I domains with a C-terminal pull spring studied with molecular dynamics. Structure (Camb). 12:2137–2147. [DOI] [PubMed] [Google Scholar]
- 35.Rosenfeld, S. S., and H. L. Sweeney. 2004. A model of myosin V processivity. J. Biol. Chem. 279:40100–40111. [DOI] [PubMed] [Google Scholar]
- 36.Rosenfeld, S. S., P. M. Fordyce, G. M. Jefferson, P. H. King, and S. M. Block. 2003. Stepping and stretching. How kinesin uses internal strain to walk processively. J. Biol. Chem. 278:18550–18556. [DOI] [PMC free article] [PubMed] [Google Scholar]










