Abstract
Molecular dynamics (MD) simulations have been used to unmask details of specific interactions of anionic phospholipids with intersubunit binding sites on the surface of the bacterial potassium channel KcsA. Crystallographic data on a diacyl glycerol fragment at this site were used to model phosphatidylethanolamine (PE), or phosphatidylglycerol (PG), or phosphatidic acid (PA) at the intersubunit binding sites. Each of these models of a KcsA-lipid complex was embedded in phosphatidyl choline bilayer and explored in a 20 ns MD simulation. H-bond analysis revealed that in terms of lipid-protein interactions PA > PG ≫ PE and revealed how anionic lipids (PG and PA) bind to a site provided by two key arginine residues (R64 and R89) at the interface between adjacent subunits. A 27 ns simulation was performed in which KcsA (without any lipids initially modeled at the R64/R89 sites) was embedded in a PE/PG bilayer. There was a progressive specific increase over the course of the simulation in the number of H-bonds of PG with KcsA. Furthermore, two specific PG binding events at R64/R89 sites were observed. The phosphate oxygen atoms of bound PG formed H-bonds to the guanidinium group of R89, whereas the terminal glycerol H-bonded to R64. Overall, this study suggests that simulations can help identify and characterize sites for specific lipid interactions on a membrane protein surface.
INTRODUCTION
Membrane proteins are of considerable biophysical interest. They account for ∼25% of genes (1) and for ∼50% of drug targets (2), yet they constitute only ∼0.5% of known structures (3). From a biological perspective, membrane proteins play key roles in a wide range of cellular functions, including electrical signaling across cell membranes via ion channels (4). Interactions with lipids play a key role in stabilizing the structure of membrane proteins within their lipid bilayer environment, and also may influence the function of membrane proteins (5). However, it is difficult to obtain atomic resolution information on membrane protein interactions with lipids. Some data can be extracted from lipid and/or detergent molecules present within membrane protein crystals (6–8), and also from spectroscopic studies (9). However, the crystallographic data are often incomplete (revealing only fragmentary structures of bound lipids) and may be biased toward those lipid-protein interactions which are resistant to detergent solubilization.
KcsA is a bacterial potassium channel, the structure of which is known at near atomic resolution (10). It is a relatively simple representative of the main family of membrane proteins, namely those formed by bundles of transmembrane (TM) α-helices. It is tetrameric, with the four subunits packed symmetrically around a central pore. The C-terminal TM helix (M2) faces the central pore, whereas the N-terminal TM helix (M1) faces the lipid membrane. The eight TM helices are packed as an inverted truncated cone. The wide extracellular mouth of the cone accommodates the selectivity filter, which is formed by four copies of a highly conserved sequence motif (TVGYG). This pore domain structure is conserved in the crystal structures of a number of other K channels, including inward rectifier K channels ((11), J. M. Gulbis, A. Kuo, B. Smith, D. A. Doyle, A. Edwards, C. Arrowsmith, and M. Sundstrom, unpublished)), a calcium activated K channel (13), and voltage gated K channels (14,15).
A number of experiments implicate anionic phospholipids in the structural integrity and function of KcsA (16). Electron density in the crystal structure of KcsA reveals a lipid binding site (17), but only a fragment of a lipid molecule is present in the coordinates (Protein Data Bank (PDB) code 1K4C). The lipid fragment in the crystal structure of KcsA appears to occupy a site distinct from nonspecific annular lipid interactions (18). In particular, two arginine residues (R64 and R89) lie close to the anticipated location of the polar headgroup of a lipid, and thus electrostatic interactions with a negatively charged headgroup could be mediated by these side chains (19). Significantly, the presence of negatively charged lipids is required for ion conduction through the KcsA potassium channel, suggesting that binding of lipid to KcsA is important for channel function (16,17). Fluorescence experiments also support a specific anionic lipid binding site at the interface between two monomers in the tetrameric channel structure (20). Mass spectrometry experiments indicate that KcsA binds to phosphatidylglycerol (PG) rather than to phosphatidylcholine (PC) (21). From a functional perspective, it appears specific lipids are required for optimal KcsA channel function (16,17,22). It has been suggested that these lipids may specifically promote assembly of the KcsA tetramer (23). Once correctly folded, KcsA channel activity requires the presence of negatively charged lipids (16). The exact structure of the anionic lipid headgroup is not critical as channel activity is observed in the presence of PG, phosphatidylserine (PS), or cardiolipin (16,17).
Molecular dynamics (MD) simulations (24–27) provide a useful complement to experimental investigations of membrane proteins. Whereas earlier simulations provided only limited information on membrane-lipid interactions (28) as a result of short (∼1 ns) simulation times, current simulation times are of the order of 10–100 ns and so facilitate more detailed analysis of lipid-protein interactions. Here we adopt a computational approach to unmasking details of the specific interactions of phospholipids with KcsA, revealing how anionic lipids bind to a site provided by the two key arginine residues (R64 and R89) at the interface between adjacent subunits.
METHODS
The 2 Å resolution structure of KcsA in the presence of a high concentration of K+ ions (PDB code 1K4C) was used as the basis of all simulations. An acetyl group was attached to the N-terminus of KcsA (residue 22), and the C-terminal carboxylate was protonated. The side chain of Glu71 was protonated to form a diacid hydrogen bond (H-bond) with the carboxylate group of Asp80, in agreement with the earlier simulation studies (29,30) and with structural data (10). The rest of the ionizable residues were in their default ionization state. Quanta (http://www.accelrys.com/quanta/overview.html) was used for interactive modeling of lipid molecules. Structural diagrams were prepared using VMD (31) and RasMol (32).
Simulation protocols were derived from those described in previous studies of KcsA (33–35). Thus, all simulations were performed using GROMACS (www.gromacs.org) (36) . The GROMOS96 force field (37) was used as implemented in GROMACS, including development 43a2 for improved alkane dihedrals. Van der Waals interactions were modeled using a 6-12 Lennard-Jones potential with a cutoff at 1 nm. Long range electrostatic interactions were computed using the particle mesh Ewald (PME) method (38). The LINCS algorithm (39) was used to preserve the bond lengths, and the time step was set at 2 fs. Simulations were carried out at a constant pressure of 1 atm and at a temperature of 310 K, which is above the main phase transition for POPC (268 K) (40), POPE (299 K) (41), and POPG (269 K) (42) bilayers. We note that POPE is a lipid which favor the hexagaonal HII phase, and so a POPE bilayer may exhibit “curvature frustration”, which could be released by interactions with a protein (as discussed by, e.g., Lee (5)).
Both the temperature and pressure of the system were controlled by the Berendsen method (43) for the initial 2 ns equilibration, after which pressure coupling used the Parrinello-Rahman method (44). The applied pressure was controlled anisotropically, each direction being treated independently with the trace of the pressure tensor kept constant at 1 atm. Energy minimizations were performed employing a steepest descent algorithm. Whenever restraints were required, a harmonic potential of force constant 105 kJ mol−1 nm−2 was applied to all nonhydrogen atoms.
For the simulation in a mixed POPG/POPE lipid bilayer (see below), the bilayer was generated by editing lipids taken from a simulations of KcsA in POPC (34,35), i.e., starting from a configuration of POPC molecules equilibrated around a KcsA molecule. This was provided by the 15 ns frame from the KcsA/POPC simulation. Lipid headgroups were selected at random and “mutated” from PC to phosphatidyl ethanolamine (PE) or PG in the appropriate ratio. After reinsertion of KcsA, solvation, and inclusion of K+ ions, the resultant system was energy minimized. The system was then first subjected to a short (3 ns) MD run, during which the protein and crystallographic K+ ions were restrained, to relax the positions and orientations of the mixed lipid bilayer and solvent molecules. Then 7 ns of unrestrained MD was run, to equilibrate the lipids around the protein. Finally, a 20 ns production run was performed.
RESULTS
Modeling bound lipids
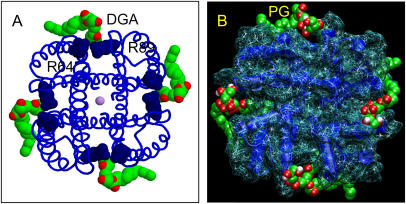
A lipid fragment was observed in the crystal structure of KcsA (10,17) (Fig. 1 A). The electron density was such that only the 1,2-sn-diacylglycerol (DGA) fragment of the lipid was built into the model, plus a small number of tail atoms (a total of 8 and 13 carbons for the sn-1 and sn-2 chains, respectively). The sn-1 tail lies in a groove between the pore helix and M2 helix of one monomer, whereas the sn-2 tail interacts less strongly with the protein. The narrowest region of the groove is formed by the side chains of residues P63 and L86 from adjacent subunits. Just “above” (i.e., extracellular to) these two side chains are those of R64 and R89, respectively, forming a potential binding site for lipid headgroups. The side chain of Trp87 is packed against the grove, in agreement with site-directed spin labeling studies (45).
FIGURE 1.
(A) Crystal structure of KcsA (PDB code 1K4C) viewed down the pore axis from the extracellular face. The K ions are shown in lilac, the R64 and R89 residues in dark blue, and the bound diacyl glycerol molecule in C = green/O = red spacefill format. (B) KcsA (shown as deep blue helices and a pale blue wireframe surface representation), with four POPG lipids modeled into each binding site shown in spacefill format (C = green/O = red).
To explore lipid-protein interactions, we constructed three models with different lipids present at the binding site defined by the crystallographic DGA fragment: palmitoyloleoyl phosphatidic acid (POPA), palmitoyloleoyl phosphatidylethanolamine (POPE), and palmitoyloleoyl phosphatidylglycerol (POPG) (Fig. 1 B). Palmitoyloleoyl lipids were used as POPG has been shown to be present in KcsA crystals (17). Phosphatidic acid (PA), PE, and PG were used to explore the headgroup specificity of the site. In each case the glycerol moiety of the DGA was used as a template onto which the headgroup was modeled interactively, and then palmitoyl and oleoyl chains were added to the sn-1 and sn-2 positions. The completed lipid was then fitted to the original crystallographic DGA conformation via a least squares method. The KcsA tetramers, each with four modeled lipids, were then docked into a POPC bilayer, obtained from previous simulations of KcsA in a POPC bilayer (34).
Progress of simulations
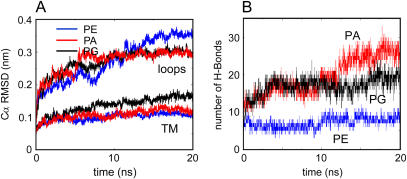
Three 20 ns duration simulations were performed, one for each of the modeled lipids (see Table 1). The “quality” of each simulation was monitored by examining the conformational drift of the protein from the initial structure, measured as the Cα atom root mean square deviation (RMSD) versus time (Fig. 2 A), and also from the root mean square fluctuations of the Cα atoms as a function of residue number (data not shown). The Cα RMSDs were all relatively low (0.1–0.15 nm after 20 ns for the TM domain, i.e., excluding surface loops), indicating little conformational drift. This range is comparable to that seen for a 15 ns simulation of KcsA in POPC without any bound lipid (an extension of the simulation described in Domene and Sansom (34)), suggesting that the presence of the modeled lipid does not influence the conformational stability of the core fold of the protein. However, there were some small differences between the three simulations. Thus, for the TM domain the final RMSD is somewhat higher for KcsA-PG than for the other two simulations. For the loops, the RMSD for KcsA-PE is significantly higher than for KcsA-PA or KcsA-PG. Thus, all three simulation systems are stable on a 20 ns timescale, but significant differences are seen.
TABLE 1.
Summary of simulations
| Simulation | Membrane composition | Bound lipid modeled | Duration (ns)* |
|---|---|---|---|
| KcsA-PA | 242 POPC | 4 × POPA | 2 + 20 |
| KcsA-PE | 242 POPC | 4 × POPE | 2 + 20 |
| KcsA-PG | 242 POPC | 4 × POPG | 2 + 20 |
| KcsA-PE/PG | 174 POPE + 68 POPG† | None | 7 + 20 |
The duration is given as equilibration time plus production time. A longer equilibration was run for KcsA-PE/PG to allow for more complete mixing of the lipids.
Approximately equal numbers of each lipid in either leaflet.
FIGURE 2.
(A) RMSD of Cα atoms from their initial coordinates for the loops and TM domains of KcsA as a function of time for simulations KcsA-PE (blue), KcsA-PG (black), and KcsA-PA (red). (B) Number of H-bonds from modeled lipid to KcsA as a function of time, for simulations KcsA-PA (red), KcsA-PG (black), and KcsA-PE (blue).
Interactions of modeled lipids
The interactions of the modeled lipids with KcsA were monitored in terms of the number of lipid-protein contacts (defined using a 0.35 nm cutoff) and number of lipid-protein H-bonds (defined using a 0.25 nm cutoff for the hydrogen-acceptor distance, and 60° for the donor-hydrogen-acceptor angle) as a function of time during each simulation. The number of interatomic contacts (data not shown) was approximately constant with respect to time for KcsA-PE, but showed a significant time-dependent increase for KcsA-PA and KcsA-PG. The two anionic lipids exhibited significantly more contacts with the protein, mainly to polar and charged side chains, whereas there was a notable absence of POPE lipid headgroup interactions. Thus for both of the anionic lipids, the “strength” of the lipid-protein interaction increased over the course of the simulation.
The numbers of H-bonds that the modeled lipids formed with the protein were analyzed (Fig. 2). Each simulation began with ∼10 H-bonds (i.e., ∼2.5 per lipid molecule). Note that the numbers of potential H-bonds acceptors/donors are 8/3 for POPE, 8/0 for POPA, and 10/2 for POPG. In simulations of KcsA-PA and KcsA-PG, the number of H-bonds per modeled lipid increases with respect to time, to ∼5 per lipid for PG and ∼6 for PA, whereas no such increase is seen for PE. During the KcsA-PA simulation the number of residues in contact with the modeled lipids remained constant, indicating that subtle side-chain rearrangements and/or lipid reorientation underlay the increase in H-bond interactions. Thus, in terms of number of H-bonds to the protein at the intersubunit binding lipid site, the simulations indicate PA > PG ≫ PE.
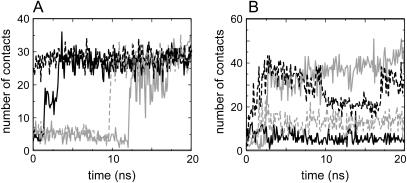
It is informative to examine the time courses of the lipid-protein contacts for the individual lipid molecules in simulations of KcsA-PA and KcsA-PG (Fig. 3). For KcsA-PA it can be seen that one by one the four lipid molecules form tighter interactions with the protein, such that by ∼15 ns all four PA molecules have maximized their interactions. For KcsA-PG the time course is a little more complex, with fluctuations throughout the simulation, such that the equilibrium situation appears to be one in which two of the four PG headgroups form tight interactions with their binding sites on KcsA. This may correlate with the absence of strong electron density for the headgroup region in the x-ray structure of KcsA.
FIGURE 3.
Atomic contacts from lipid headgroups to protein side chains for simulations: (A) KcsA-PA and (B) KcsA-PG. In each case, the four line styles correspond to contacts to each of the four lipid binding sites of KcsA, sampled every 0.1 ns.
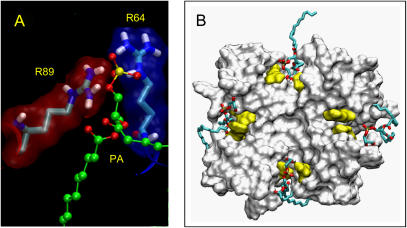
We have examined the nature of the interactions between the lipid headgroups and the protein for KcsA-PA and for KcsA-PG. The headgroup of POPA, especially the ionized oxygens of the phosphate, dominates the H-bonding of the lipid. The main headgroup contacts were with polar and charged amino acids. Hydrophobic interactions were less clear cut, but both P63 and L86 (see above) played a role. The main headgroup contacts for KcsA-PA were with the key residues R64 and R89 (Fig. 4 A). All four POPA headgroups formed contacts with both of these arginines for the entire last 10 ns of the simulation. Note that these two arginines are from neighboring KcsA monomers, one (R64) at the start of the P-helix and the other (R89) at the N-terminus of the M2 helix, and thus link the PA headgroup to a functionally key region of the channel, thus strengthening the intersubunit interaction. The phosphate groups of the POPA molecules are thus able to form strong electrostatic interactions with both arginines, and once these are formed they are not lost for the remainder of the simulation. We may therefore describe an interaction in which all three contributors (R64, R89, and a lipid headgroup) act as a “stabilizing” interaction, due to its longevity.
FIGURE 4.
(A) Snapshot of a POPA molecule interacting with R64 and R89 from the KcsA-PA simulation. (B) Snapshot (t = 15 ns) from the KcsA-PG simulation showing the four bound POPG molecules (in bonds format) with the KcsA solvent accessible surface (seen from the extracellular face) with the R64 and R89 residues in yellow.
Inspection of the KcsA-PG simulation revealed that when a “stabilizing” interaction occurred, this involved not the phosphate group (as seen for POPA), but rather the (headgroup) glycerol moiety of POPG. Both arginines (R64 and R89) are able to form a close association with the headgroup of POPG. Only two of the four POPG lipid headgroups formed stabilizing interactions with the two arginines. Of the other two POPGs, one (marked with an asterisk in Fig. 4 B) has the glycerol headgroup directed away from the protein toward the surrounding solvent, whereas the other (marked with a dagger symbol in Fig. 4 B) interacted for part of the simulation with R89, at other times forming short-lived (10–100 ps) H-bonds with the backbone oxygen atom of A55. Thus, the headgroup of bound PG shows some degree of motion relative to KcsA.
Simulations in a mixed lipid bilayer
The plasma membrane of Escherichia coli, in which KcsA was expressed before crystallization, is composed of ∼30% POPG and ∼70% POPE. However, it is not known if the distribution of POPG between the two leaflets of the bilayer is uniform or asymmetric (46). A number of experimental studies of KcsA have used bilayers with PE and PG in a ∼3:1 ratio (16,17,47). Therefore, we simulated KcsA in a mixed lipid bilayer containing POPE and POPG in a ∼3:1 ratio, the distribution of each lipid being approximately equal in either leaflet. Thus, the lower (intracellular) leaflet of the simulation system contained 93 POPE and 40 POPG molecules; the upper leaflet contained 81 POPE and 28 POPG molecules. In this, the first simulation of KcsA in a mixed lipid bilayer, lipid molecules were not modeled into the four binding sites before the start of the simulation (Table 1). Instead, the interaction of lipid molecules with these sites was monitored as the simulation progressed.
The progress of the simulation was monitored via evaluation of the time-dependent Cα RMSD from the initial structure. The RMSD was somewhat lower than that for the equivalent simulation in a POPC bilayer (34) (data not shown), rising to ∼0.2 nm after 7 + 20 ns (see Methods). Thus KcsA appears to be to some extent stabilized by the mixed (anionic and neutral) lipid bilayer. As we will see, this may be explained by interactions at the specific lipid binding sites discussed above.
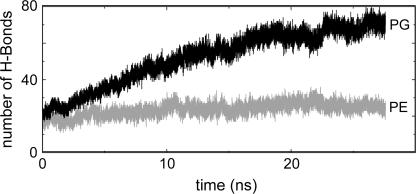
At the end of the KcsA-PE/PG simulation, an average of ∼35 lipids form interactions (cutoff 0.35 nm) between their headgroups and the protein surface. However, if one monitors the number of such interactions with respect to time, it rises steadily from ∼25 at the start of the simulation to ∼35 at the end, having reached an apparent equilibrium for the last 5 ns of the simulation. This rise is due to an enrichment of the layer of lipid in contact with KcsA with PG at the expense of PE. Thus, the overall composition of the bilayer (Table 1) is 174 PE and 68 PG, i.e., a PE/PG ratio of 2.6. If one averages over the final 20 ns of the KcsA-PE/PG simulation, the ratio for those lipids whose headgroups are within 0.35 nm of the lipid is PE/PG = 1.4. This implies that the overall affinity of PG for annular sites on KcsA is about twice that of PE, although substantially longer simulations would be needed to obtain an accurate estimate of relative affinities.
The enrichment of the layer of lipids around KcsA in contact with POPG is seen if one counts the number of lipid-protein H-bonds (Fig. 5). The glycerol oxygens of the POPG headgroups dominate, with an increase in the number of H-bonds occurring over the first ∼25 ns of the simulation, until a plateau of ∼90 H-bonds (∼70 to PG, ∼20 to PE) is reached. This compares with ∼55 H-bonds in a KcsA/POPC simulation (34). If we normalize these by division by the number of lipid headgroups within 0.35 nm of the protein, we obtain averages of 3.9, 1.5, and 2.6 H-bonds formed by a bound (i.e., annular) PG, PE, and PC, respectively. Thus, despite the presence of three H-bond donors on the PE headgroup it does not form as many H-bonds to KcsA as PG. This indicates a degree of overall selectivity in KcsA-lipid interactions.
FIGURE 5.
Simulation KcsA-PE/PG. Number of H-bonds between phospholipids and protein as a function of time for the entire simulation (7 + 20 ns). The number of H-bonds that POPG (solid) and POPE (shaded) form with KcsA are shown separately.
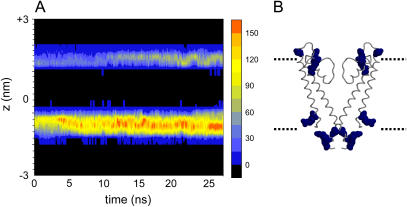
Previous analyses of simulations of KcsA in POPC have emphasized interactions with amphipathic aromatic (Trp, Tyr) and basic (Arg, Lys) side chains (35). In the KcsA-PE/PG simulation, interactions of lipid headgroups with Arg side chains of KcsA are of particular importance. This is especially true of the interactions with the PG headgroups, where Arg residues form the main interaction partners. Aromatic and other hydrophobic residues formed about the same number of contacts in the KcsA-PE/PG simulation as they did in earlier KcsA-POPC simulations (35). A spatial and temporal breakdown of the pattern of lipid headgroup-arginine interactions during the KcsA-PE/PG simulation reveals a steady increase in such interactions at both the intracellular and extracellular membrane interfaces (Fig. 6).
FIGURE 6.
(A) Interactions (cutoff 0.35 nm) between lipid headgroups and arginine side chains for simulation KcsA-PE/PG. The number of interactions is shown on a blue to red scale (black = no interactions) as a function of time and position along the z axis. The intracellular interface is at z ∼ −1 nm; the extracellular interface is at z ∼ +1 nm. (B) X-ray structure of KcsA (two subunits only) with arginine side chains in spacefill format (blue), aligned on z to match the diagram in A.
At the extracellular interface, a number of the Arg-lipid interactions appear to take place at the specific lipid binding sites discussed above. Thus, the KcsA-PE/PG simulation was examined to identify any lipids which adopted a position on the surface of the protein at the potential binding sites discussed above. We defined the KcsA lipid binding site by including only those residues which were identified by the previous simulation studies to constantly remain in contact while a modeled lipid was occupying the tight crevice between each monomer. Thus, the contacts between the residues R64, R89, L86, and W87 at each binding site and neighboring lipids were classified as potential binding events.
Over the course of the simulation, two of the four binding sites formed tight interactions with PG headgroups. The first of these sites was occupied by a POPG molecule right from the start of the simulation, in part due to the random placement of a PG molecule close to this site during the initial setup of the simulation. Both arginines R64 and R89 formed long-lived interactions with a single POPG molecule at this site throughout the simulation. As in the KcsA-PG simulation, R64 interacted with the glycerol moiety of the lipid, whereas Thr61 and R89 both interacted with the headgroup moiety.
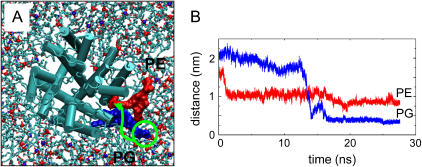
In contrast, for the first ∼12 ns of the simulation the second site did not form a tight interaction with a lipid molecule. However, just before midway through the simulation, a PG molecule binds to the site (even though there was a PE molecule that was initially closer to the site). Thus, a selective binding event was observed during the simulation (Fig. 7). Once bound to the site, the PG molecule remained there for the rest of the simulation. In addition to the tight interaction with PG, there was also a looser interaction with an adjacent PE molecule at a binding crevice. However, this involved only the glycerol backbone and acyl tail moieties of POPE, in contrast to the POPG lipid, which only formed a significant number of contacts at any binding site via its polar headgroup.
FIGURE 7.
(A) Snapshot from the KcsA-PE/PG simulation at 10 ns, seen looking down on the extracellular face of the channel (shown as cylinders for helices). The lipid molecules are shown in bonds format. Two lipid molecules (one PG in blue, one PE in red) close to KcsA are highlighted. The green arrow indicates schematically the way in which the PG headgroup forms a close interaction with KcsA over the course of the simulation. (B) Distance between centers of mass of R64 and the polar headgroups of lipids PG596 (blue) and PE449 (red). These are the same two lipid molecules shown in spacefill format in A.
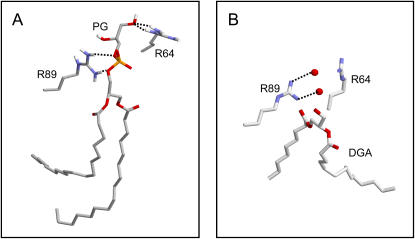
It is informative to examine the nature of the interactions between the POPG molecule, which enters the binding site at ∼12 ns, and to compare this with the bound lipid fragment in the crystal structure of KcsA (Fig. 8). In the simulation, the PG molecule binds between the two adjacent subunits. The hydrophobic tails form weak and somewhat flexible interactions with the hydrophobic surface of the channel protein. The phosphate of the headgroup forms two H-bonds to R89 of one subunit, and the glycerol of the headgroup forms two H-bonds to the R64 of the adjacent subunit. If one compares this with the crystal structure it can be seen that the approximate location of the phosphate oxygens in the simulation snapshot is replaced by two water molecules. Thus, it seems that a spontaneous POPG binding event in the simulation is able to reveal the nature of the interactions of KcsA with an anionic headgroup phospholipid molecule. It is possible that in the crystal partial occupancy of the anionic headgroup region occurs.
FIGURE 8.
KcsA interaction with a POPG molecule from simulation KcsA-PE/PG (at the end of the simulation). A shows the H-bonding interactions of the bound POPG molecule (PG596; see Fig. 7) with the two arginines residues (R64 and R89 of the subunits A and B) that form the binding site. B shows approximately the same view of the corresponding two arginines in the crystal structure of KcsA, along with two water molecules (red spheres) that interact with the side chain of R89.
DISCUSSION
It is useful to note the different nomenclatures used to discuss lipid-protein interactions. For example, Lee distinguishes between annular and nonannular sites (7), whereas Palsdottir and Hunte define three classes of bound lipid: annular, nonannular, and integral. This study is concerned primarily with binding of anionic lipids to specific, nonannular sites at the interfaces between adjacent subunits of KcsA. Previous simulation studies (35,48) have focused on nonspecific interactions of annular lipids. The results of the current simulations agree with and extend experimental studies of the specificity of KcsA lipid interactions (17–19,21). In particular, recent studies (49) indicate that the nonannular binding sites of KcsA are specific for anionic phospholipids. Detailed examination of our simulations provides some indication that there is a degree of modulation of KcsA flexibility by bound lipids (especially decreased flexibility of loops in the presence of anionic lipids). This may be of functional relevance. X-ray diffraction reveals that the conformation of the selectivity filter in KcsA can switch between an “active” and “inactive” conformation (10,50). The latter conformation is seen in crystals of KcsA at low [K+] or in the presence of tetraethylammonium (51). In parallel with these structural studies, simulations have indicated that filter flexibility may play a key role in K channel function (52–55). One may therefore speculate that the role of bound anionic lipids may be to stabilize the “active” conformation of the selectivity filter (i.e., the conformation favored by a high concentration of [K+] in the crystallographic experiments). However, more systematic studies of, e.g., the influence of the species of bound lipid on the free energy of filter distortion are needed to be certain about this mechanism.
It is also important to consider the methodological limitations of the simulations. Although of reasonable duration, ∼20 ns simulations do not fully sample protein (56) or lipid motions. To some extent we have attempted to offset this by comparing the results of multiple simulations, but longer simulations would clearly be beneficial. These studies are restricted to the closed state conformation of KcsA. Although the x-ray structure of the open state is not known, it would be possible to extend the analysis to open state models (57). We should also recall that the x-ray structure of KcsA has a short N-terminal helix and a more extended C-terminal domain omitted (58) and these may also play a role in lipid-protein interactions.
Despite these limitations, the use of comparative simulations to study KcsA-lipid interactions provides a paradigm for simulation studies of specific lipid interactions of other membrane proteins. Such interactions may play an important role in the regulation of some K channels, such as the inward rectifier K channels, which have been shown to be modulated by phosphatidylinositol 4,5-bisphosphate (59,60). More generally, there is accumulating structural evidence of specific lipid-protein interactions of functional importance for a wide variety of membrane proteins (5,61). For example, in the Rhodobacter sphaeroides photosynthetic reactions center, mutation of a key arginine residue involved in interactions with cardiolipin results in a decrease in thermal stability (61). Similar effects of mutation of phospholipid binding site residues have been shown for the yeast cytochrome bc1 complex (8). Studies on, e.g., bacterial outer membrane proteins (62) suggest that simulations may be able to play an important role in identifying “hotpots” for lipid interactions on a membrane protein surface, reinforcing the general implications of this study. It would therefore seem that by combining comparative analysis of simulations of a wide range of membrane protein along with structural bioinformatics studies of the water-membrane interface region of membrane proteins (63), we may be able to arrive at a more general model of the nature of lipid binding sites in membrane proteins and of their role in protein stability and function.
Acknowledgments
Our thanks to our colleagues for discussions concerning this work.
This work was supported by grants from the Wellcome Trust and the Biotechnology and Biological Sciences Research Council. S.S.D. was an Engineering and Physical Sciences Research Council funded research student. C.D. is a Royal Society University Research Fellow.
References
- 1.Wallin, E., and G. von Heijne. 1998. Genome-wide analysis of integral membrane proteins from eubacterial, Archean, and eukaryotic organisms. Protein Sci. 7:1029–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Terstappen, G. C., and A. Reggiani. 2001. In silico research in drug discovery. Trends Pharmacol. Sci. 22:23–26. [DOI] [PubMed] [Google Scholar]
- 3.White, S. H. 2004. The progress of membrane protein structure determination. Protein Sci. 13:1948–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hille, B. 2001. Ionic Channels of Excitable Membranes. Sinauer Associates, Sunderland, MA.
- 5.Lee, A. G. 2004. How lipids affect the activities of integral membrane proteins. Biochim. Biophys. Acta. 1666:62–87. [DOI] [PubMed] [Google Scholar]
- 6.Fyfe, P. K., K. E. McAuley, A. W. Roszak, N. W. Isaacs, R. J. Codgell, and M. R. Jones. 2001. Probing the interface between membrane proteins and membrane lipids by x-ray crystallography. Trends Biochem. Sci. 26:106–112. [DOI] [PubMed] [Google Scholar]
- 7.Lee, A. G. 2003. Lipid-protein interactions in biological membranes: a structural perspective. Biochim. Biophys. Acta. 1612:1–40. [DOI] [PubMed] [Google Scholar]
- 8.Palsdottir, H., and C. Hunte. 2004. Lipids in membrane protein structures. Biochim. Biophys. Acta. 1666:2–18. [DOI] [PubMed] [Google Scholar]
- 9.Marsh, D., and T. Pali. 2004. The protein-lipid interface: perspectives from magnetic resonance and crystal structures. Biochim. Biophys. Acta. 1666:118–141. [DOI] [PubMed] [Google Scholar]
- 10.Zhou, Y., J. H. Morais-Cabral, A. Kaufman, and R. MacKinnon. 2001. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 Å resolution. Nature. 414:43–48. [DOI] [PubMed] [Google Scholar]
- 11.Kuo, A., J. M. Gulbis, J. F. Antcliff, T. Rahman, E. D. Lowe, J. Zimmer, J. Cuthbertson, F. M. Ashcroft, T. Ezaki, and D. A. Doyle. 2003. Crystal structure of the potassium channel KirBac1.1 in the closed state. Science. 300:1922–1926. [DOI] [PubMed] [Google Scholar]
- 12.Reference deleted in proof.
- 13.Jiang, Y., A. Lee, J. Chen, M. Cadene, B. T. Chait, and R. MacKinnon. 2002. Crystal structure and mechanism of a calcium-gated potassium channel. Nature. 417:515–522. [DOI] [PubMed] [Google Scholar]
- 14.Jiang, Y., A. Lee, J. Chen, V. Ruta, M. Cadene, B. T. Chait, and R. Mackinnon. 2003. X-ray structure of a voltage-dependent K+ channel. Nature. 423:33–41. [DOI] [PubMed] [Google Scholar]
- 15.Long, S. B., E. B. Campbell, and R. MacKinnon. 2005. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 309:897–902. [DOI] [PubMed] [Google Scholar]
- 16.Heginbotham, L., L. Kolmakova-Partensky, and C. Miller. 1998. Functional reconstitution of a prokaryotic K+ channel. J. Gen. Physiol. 111:741–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valiyaveetil, F. I., Y. Zhou, and R. MacKinnon. 2002. Lipids in the structure, folding and function of the KcsA channel. Biochemistry. 41:10771–10777. [DOI] [PubMed] [Google Scholar]
- 18.Williamson, I. M., S. J. Alvis, J. M. East, and A. Lee. 2002. Interactions of phospholipids with the potassium channel KcsA. Biophys. J. 83:2026–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williamson, I. M., S. J. Alvis, J. M. East, and A. G. Lee. 2003. The potassium channel KcsA and its interaction with the lipid bilayer. Cell. Mol. Life Sci. 60:1581–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alvis, S. J., I. M. Williamson, J. M. East, and A. G. Lee. 2003. Interactions of anionic phospholipids and phosphatidylethanolamine with the potassium channel KcsA. Biophys. J. 85:3828–3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Demmers, J. A. A., A. van Dalen, B. de Kruijiff, A. J. R. Heck, and J. A. Killian. 2003. Interaction of K channel KcsA with membrane phospholipids as studied by ESI mass spectrometry. FEBS Lett. 541:28–32. [DOI] [PubMed] [Google Scholar]
- 22.van Dalen, A., M. van der Laan, A. J. M. Driessen, A. Killian, and B. de Kruijff. 2002. Components required for membrane assembly of newly synthesized K+ channel KcsA. FEBS Lett. 511:51–58. [DOI] [PubMed] [Google Scholar]
- 23.van Dalen, A., S. Hegger, A. Killian, and B. Kruijff. 2002. Influence of lipids on membrane assembly and stability of the potassium channel KcsA. FEBS Lett. 525:33–38. [DOI] [PubMed] [Google Scholar]
- 24.Forrest, L. R., and M. S. P. Sansom. 2000. Membrane simulations: bigger and better? Curr. Opin. Struct. Biol. 10:174–181. [DOI] [PubMed] [Google Scholar]
- 25.Ash, W. L., M. R. Zlomislic, E. O. Oloo, and D. P. Tieleman. 2004. Computer simulations of membrane proteins. Biochim. Biophys. Acta. 1666:158–189. [DOI] [PubMed] [Google Scholar]
- 26.Bond, P. J., and M. S. P. Sansom. 2004. The simulation approach to bacterial outer membrane proteins. Mol. Memb. Biol. 21:151–162. [DOI] [PubMed] [Google Scholar]
- 27.Roux, B. 2005. Ion conduction and selectivity in K+ channels. Annu. Rev. Biophys. Biomolec. Struct. 34:153–171. [DOI] [PubMed] [Google Scholar]
- 28.Tieleman, D. P., L. R. Forrest, H. J. C. Berendsen, and M. S. P. Sansom. 1999. Lipid properties and the orientation of aromatic residues in OmpF, influenza M2 and alamethicin systems: molecular dynamics simulations. Biochemistry. 37:17554–17561. [DOI] [PubMed] [Google Scholar]
- 29.Ranatunga, K. M., I. H. Shrivastava, G. R. Smith, and M. S. P. Sansom. 2001. Side-chain ionization states in a potassium channel. Biophys. J. 80:1210–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bernèche, S., and B. Roux. 2002. The ionization state and the conformation of Glu-71 in the KcsA K+ channel. Biophys. J. 82:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33–38. [DOI] [PubMed] [Google Scholar]
- 32.Sayle, R. A., and E. J. Milner-White. 1995. RasMol: biomolecular graphics for all. Trends Biochem. Sci. 20:374–376. [DOI] [PubMed] [Google Scholar]
- 33.Shrivastava, I. H., and M. S. P. Sansom. 2000. Simulations of ion permeation through a potassium channel: molecular dynamics of KcsA in a phospholipid bilayer. Biophys. J. 78:557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Domene, C., and M. S. P. Sansom. 2003. A potassium channel, ions and water: simulation studies based on the high resolution x-ray structure of KcsA. Biophys. J. 85:2787–2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deol, S. S., P. J. Bond, C. Domene, and M. S. P. Sansom. 2004. Lipid-protein interactions of integral membrane proteins: a comparative simulation study. Biophys. J. 87:3737–3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindahl, E., B. Hess, and D. van der Spoel. 2001. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J. Mol. Model. (Online). 7:306–317. [Google Scholar]
- 37.van Gunsteren, W. F., P. Kruger, S. R. Billeter, A. E. Mark, A. A. Eising, W. R. P. Scott, P. H. Huneberger, and I. G. Tironi. 1996. Biomolecular Simulation: The GROMOS96 Manual and User Guide. Biomos & Hochschulverlag AG an der ETH Zurich, Groningen and Zurich.
- 38.Darden, T., D. York, and L. Pedersen. 1993. Particle mesh Ewald—an N.log(N) method for Ewald sums in large systems. J. Chem. Phys. 98:10089–10092. [Google Scholar]
- 39.Hess, B., H. Bekker, H. J. C. Berendsen, and J. G. E. M. Fraaije. 1997. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 18:1463–1472. [Google Scholar]
- 40.Seelig, J., and N. Waespe-Sarcevic. 1978. Molecular order in cis and trans unsaturated phospholipid bilayers. Biochemistry. 17:3310–3315. [DOI] [PubMed] [Google Scholar]
- 41.Huang, C. H., and S. S. Li. 1999. Calormetric and molecular mechanics studies of the thermotrophic phase of membrane phospholipids. Biochim. Biophys. Acta. 1422:273–307. [DOI] [PubMed] [Google Scholar]
- 42.Boggs, J. M., and B. Tummler. 1993. Interdigitated gel phase bilayers formed by unsaturated synthetic and bacterial glycerolipids in the presence of polymyxin B and glycerol. Biochim. Biophys. Acta. 1145:42–50. [DOI] [PubMed] [Google Scholar]
- 43.Berendsen, H. J. C., J. P. M. Postma, W. F. van Gunsteren, A. DiNola, and J. R. Haak. 1984. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81:3684–3690. [Google Scholar]
- 44.Parrinello, M., and A. Rahman. 1981. Polymorphic transitions in single-crystals—a new molecular-dynamics method. J. Appl. Phys. 52:7182–7190. [Google Scholar]
- 45.Gross, A., and W. L. Hubbell. 2002. Identification of protein sidechains near the membrane-aqueous interface: a site-directed spin labelling study of KcsA. Biochemistry. 41:1123–1128. [DOI] [PubMed] [Google Scholar]
- 46.Huijbregts, R. P., A. I. de Kroon, and B. de Kruijff. 2000. Topology and transport of membrane lipids in bacteria. Biochim. Biophys. Acta. 1469:43–61. [DOI] [PubMed] [Google Scholar]
- 47.Zakharian, E., and R. N. Reusch. 2004. Functional evidence for a supramolecular structure for the Streptomyces livdans potassium channel KcsA. Biochim. Biophys Res. Commun. 322:1059–1065. [DOI] [PubMed] [Google Scholar]
- 48.Domene, C., P. J. Bond, S. S. Deol, and M. S. P. Sansom. 2003. Lipid-protein interactions and the membrane/water interfacial region. J. Am. Chem. Soc. 125:14966–14967. [DOI] [PubMed] [Google Scholar]
- 49.Marius, P., S. J. Alvis, J. M. East, and A. G. Lee. 2005. The interfacial lipid binding site on the potassium channel KcsA is specific for anionic phospholipids. Biophys. J. 89:4081–4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou, Y., and R. MacKinnon. 2003. The occupancy of ions in the K+ selectivity filter: charge balance and coupling of ion binding to a protein conformational change underlie high conduction rates. J. Mol. Biol. 333:965–975. [DOI] [PubMed] [Google Scholar]
- 51.Lenaeus, M. J., M. Vamvouka, P. J. Focia, and A. Gross. 2005. Structural basis of TEA blockade in a model potassium channel. Nat. Struct. Mol. Biol. 12:454–459. [DOI] [PubMed] [Google Scholar]
- 52.Capener, C. E., P. Proks, F. M. Ashcroft, and M. S. P. Sansom. 2003. Filter flexibility in a mammalian K channel: models and simulations of Kir6.2 mutants. Biophys. J. 84:2345–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Domene, C., A. Grottesi, and M. S. P. Sansom. 2004. Filter flexibility and distortion in a bacterial inward rectifier K+ channel: simulation studies of KirBac1.1. Biophys. J. 87:256–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Noskov, S. Y., S. Bernèche, and B. Roux. 2004. Control of ion selectivity in potassium channels by electrostatic and dynamic properties of carbonyl ligands. Nature. 431:830–834. [DOI] [PubMed] [Google Scholar]
- 55.Allen, T. W., O. S. Andersen, and B. Roux. 2004. On the importance of atomic fluctuations, protein flexibility, and solvent in ion permeation. J. Gen. Physiol. 124:679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Faraldo-Gómez, J. D., L. R. Forrest, M. Baaden, P. J. Bond, C. Domene, G. Patargias, J. Cuthbertson, and M. S. P. Sansom. 2004. Conformational sampling and dynamics of membrane proteins from 10-nanosecond computer simulations. Proteins. 57:783–791. [DOI] [PubMed] [Google Scholar]
- 57.Holyoake, J., C. Domene, J. N. Bright, and M. S. P. Sansom. 2003. KcsA closed and open: modelling and simulation studies. Eur. Biophys. J. 33:238–246. [DOI] [PubMed] [Google Scholar]
- 58.Cortes, D. M., L. G. Cuello, and E. Perozo. 2001. Molecular architecture of full-length KcsA: role of cytoplasmic domains in ion permeation and activation gating. J. Gen. Physiol. 117:165–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lopes, C. M. B., H. L. Zhang, T. Rohacs, T. H. Jin, J. Yang, and D. E. Logothetis. 2002. Alterations in conserved Kir channel-PIP2 interactions underlie channelopathies. Neuron. 34:933–944. [DOI] [PubMed] [Google Scholar]
- 60.Du, X. O., H. L. Zhang, C. Lopes, T. Mirshahi, T. Rohacs, and D. E. Logothetis. 2004. Characteristic interactions with phosphatidylinositol 4,5-bisphosphate determine regulation of Kir channels by diverse modulators. J. Biol. Chem. 279:37271–37281. [DOI] [PubMed] [Google Scholar]
- 61.Fyfe, P. K., N. W. Isaacs, R. J. Cogdell, and M. R. Jones. 2004. Disruption of a specific molecular interaction with a bound lipid affects the thermal stability of the purple bacterial reaction centre. Biochim. Biophys. Acta. 1608:11–22. [DOI] [PubMed] [Google Scholar]
- 62.Baaden, M., and M. S. P. Sansom. 2004. OmpT: molecular dynamics simulations of an outer membrane enzyme. Biophys. J. 87:2942–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Granseth, E., G. von Heijne, and A. Elofsson. 2005. A study of the membrane–water interface region of membrane proteins. J. Mol. Biol. 346:377–385. [DOI] [PubMed] [Google Scholar]