Abstract
Sphingomyelins (SMs) are among the most common phospholipid components of plasma membranes, usually constituting a mixture of several molecular species with various fatty acyl chain moieties. In this work, we utilize atomistic molecular dynamics simulations to study the differences in structural and dynamical properties of bilayers comprised of the most common natural SM species. Keeping the sphingosine moiety unchanged, we vary the amide bonded acyl chain from 16 to 24 carbons in length and examine the effect of unsaturation by comparing lipids with saturated and monounsaturated chains. As for structural properties, we find a slight decrease in average area per lipid and a clear linear increase in bilayer thickness with increasing acyl chain length both in saturated and unsaturated systems. Increasing the acyl chain length is found to further the interdigitation across the bilayer center. This is related to the dynamics of SM molecules, as the lateral diffusion rates decrease slightly for an increasing acyl chain length. Interdigitation also plays a role in interleaflet friction, which is stronger for unsaturated chains. The effect of the cis double bond is most significant on the local order parameters and rotation rates of the chains, though unsaturation shows global effects on overall lipid packing and dynamics as well. Regarding hydrogen bonding or properties related to the lipid/water interface region, no significant effects were observed due to varying chain length or unsaturation. The significance of the findings presented is discussed.
INTRODUCTION
Sphingomyelin (SM) and phosphatidylcholine (PC) molecules are the most abundant phospholipid species encountered in eukaryotic membranes, comprising >50% of the total phospholipid content (1). Together with cholesterol, both SM and PC have been reported to be enriched in ordered but dynamic lateral domains, called lipid rafts, in various biological membranes (2–6). Despite their appealing ability to explain a wide range of cellular processes such as membrane trafficking and protein sorting, many profound characteristics of naturally occurring rafts are still unrevealed or under controversy. Notably, the existence of rafts has also been challenged (7,8). This uncertainty is partly due to limitations associated with experimental techniques, which make interpretation of experimental findings very difficult. A closely related problem, the nature of molecular interactions between the key components of lipid rafts has remained unclear. Possibly the most topical issue is the SM-cholesterol interactions, which are usually explained in terms of a “specific” interaction that may be related to intermolecular hydrogen bonding, whereas the large chain length and saturation of natural SMs might play an equally important role (9,10).
The amide-linked acyl chains in natural SMs vary from 16 to 24 carbons in length and usually contain on average only 0.1–0.35 cis double bonds per chain. Thus, the chains of SM are generally longer and more saturated than the ones in PCs. Additionally, the double bonds in the acyl chains of SM are usually located further away from the lipid/water interface (11). The long and saturated nature of SM acyl chains may have a substantial effect on its interactions with other membrane components, such as sterols and proteins. Also, the relatively large mismatch in length between the two SM chains may lead to interesting phenomena. For example, interdigitation of the long acyl chains into the opposing leaflet could provide a mechanism for transmission of information across cell membranes (12). Experimental studies of model membranes consisting of 24:0-SM (13) have revealed two distinct gel phases exhibiting qualitatively different long-chain interdigitation across the bilayer center. In gel phase 2 (lower temperature), the chains take a mixed interdigitated conformation, in which the acyl chains penetrate through the whole membrane and the ends of sphingosine (SPH) chains meet. In gel phase 1 (higher temperature), the chains are partially interdigitated, meaning that the acyl chain ends meet with the ends of SPH chains of the opposing leaflet. Even if the observations on interdigitation are usually related to single-component bilayers in the gel phase, partial interdigitation has also been predicted to be important for a fluid phase (14). In addition, different studies of long-chain glycosphingolipids in fluid-phase PC bilayers have shown evidence of chain interdigitation (15–17), although in some conditions the long chain ends of glycosphingolipids have been shown to terminate at the bilayer center (18). If chain interdigitation in biological membranes is a significant phenomenon, SMs are among the most probable candidates responsible for it.
The difficulties related to experimental studies on lipids imply that there is a great need for atomistic simulation studies to provide a more detailed insight into the properties of membrane systems. Despite a wide range of simulations carried out on different lipids (19–23), only a few recent simulation studies have concentrated on bilayer systems with SM. In most of these studies, the structure of pure SM bilayers has been analyzed and the observed differences with, e.g., PC bilayers have been mostly explained by the greater capacity of SM to form inter- and intramolecular hydrogen bonds (24–27). Also, a few studies have included mixtures of SM with other lipids, such as cholesterol and unsaturated PCs (28–30).
The aim of this study is to investigate a number of bilayers comprised of the naturally most abundant SMs and then, in a systematic manner, analyze the differences related to their structural and dynamic properties due to varying chain length and unsaturation. In addition to providing detailed insight into molecular properties of the naturally most abundant SM species, this simulation study is, to our knowledge, the first one that systematically investigates the effect of chain length and monounsaturation on fluid-phase lipid bilayers.
SIMULATION DETAILS
We have carried out a series of simulations on lipid bilayer systems, each comprised of 128 sphingomyelin (SM) molecules in explicit water using the GROMACS package (31,32). The molecules studied, introduced in Fig. 1, were varied in acyl chain length and unsaturation. Ten biologically relevant species were selected, of which five were saturated (16:0, 18:0. 20:0, 22:0, and 24:0) and five were unsaturated (16:1cisΔ9, 18:1cisΔ9, 20:1cisΔ11, 22:1cisΔ13, and 24:1cisΔ15) (33,34). The most common base in mammalian SM, 18:1 sphingosine, carrying one trans double bond between the fourth and fifth carbons and exhibiting a D-erythro enantiomeric configuration (11), was chosen as the other chain of each molecule (see Fig. 1).
FIGURE 1.
Structure of 16:1cis,Δ9-SM, one of the molecules under study. In the simulation series, the length and saturation of the acyl chain was varied. The sphingosine (SPH) chain, present in each molecule, consists of 18 carbons and carries one trans double bond.
The united atom force-field parameters for 16:0-SM have been validated previously (27). The different molecular species of SM were constructed on the basis of this force field, by adding CH2 groups and cis double bonds to the acyl chains. Recently, studies on unsaturated lipids have been published, utilizing varying parameters for the cis double bond, based on either GROMOS force-field (35) or ab initio calculations (36). In our study, we utilize the parameters from a previously published simulation of palmitoyloleoylphosphatidylcholine (POPC) for the cis double bonds (35). We also consider an alternative description for the double-bond region (36) and hence study the effect of parameterization on the simulation results (see “Effect of double-bond parameters”). For water, we used the simple point charge model (37). The long-range electrostatic interactions were handled using the particle-mesh Ewald technique (38), which has been shown to be a reliable method to account for long-range interactions in lipid bilayer systems (39,40). The details of the implementation of particle-mesh Ewald have been discussed elsewhere (40). A single 1.0-nm cutoff distance was used for the Lennard-Jones interactions without shift or switch functions. All bond lengths within lipids were constrained with the LINCS algorithm (41), whereas the SETTLE algorithm (42) was used for water.
As a starting structure, we used the coordinates of a fully hydrated dipalmitoylphosphatidylcholine (DPPC) bilayer from a previously published simulation study (39), in which the corresponding atoms were replaced or added and the structure was stabilized by energy minimization. For SMs with longer chains, more space between the monolayers was created. The system was hydrated with 3655 water molecules (42 wt % H2O), which is well above the measured limit values of full hydration: 35 wt % H2O for 18:0-SM in 328 K (43) or 29.5 wt % H2O for 24:0-SM in 313 K (44). Finally, the energy of the whole system was minimized again and the water was equilibrated in a short 20-ps simulation with restrained lipid positions.
The simulations were performed in the NpT ensemble. In the beginning, the systems were equilibrated for 4.0 ns by Berendsen thermostat with a time constant τ = 0.1 ps and by Berendsen barostat with τ = 1.0 ps (45). After that, we switched to a Nosé-Hoover thermostat (46,47), with τ = 0.1 ps, and a Parrinello-Rahman barostat (48,49) with a time constant τ = 1.0 ps to reproduce the correct ensemble. In each case, the lipid bilayer and water were separately coupled to the heat bath and the semiisotropic pressure coupling was applied separately in the xy direction (bilayer plane) and the z direction (bilayer normal). The temperature used, T = 323 K, is above the main phase transition temperature of Tm = 319–321 K of 24:0-SM (33,44,50,51), which has the highest Tm of the lipids under study. For the time step, we used a value of 2.0 fs. In total, each system was simulated for 50.0 ns, of which 10.0 ns was regarded as an equilibration period and was not included in any of the analysis steps described later. Snapshots of some selected bilayers are shown in Fig. 2.
FIGURE 2.
Snapshots from the simulations of 16:0-SM (top left), 16:1-SM (top right), 24:0-SM (bottom left), and 24:1-SM (bottom right) bilayers. The water molecules have been removed for clarity. The acyl chains of opposite monolayers have been rendered with small spheres and colored differently (orange/blue), whereas the other parts of SM molecules have been rendered by bonds, coloring by atom types (black, carbon; red, oxygen; blue, nitrogen; yellow, phosphorus; white, hydrogen). A few selected SM molecules have been shown in white to better indicate interdigitation.
RESULTS AND DISCUSSION
Bilayer structure
Many important quantities, such as chain ordering and diffusion in lipid bilayers are related to the average area per lipid, 〈A〉. This quantity, calculated from the xy dimensions of the simulation box, can be used as an indicator to monitor equilibration during simulations. Fig. 3 shows the average area per lipid versus time, A(t), of the 10 simulated systems. Each simulation was first started from a loose structure characterized by A(0) = 0.65 nm2. Within a few nanoseconds, this structure spontaneously organized into a more compact one. On the basis of the curves in Fig. 3, we have chosen to cut the first 10 ns from each simulation trajectory and use the remaining 40 ns for analysis.
FIGURE 3.
Average area per lipid versus time in the studied systems. In each panel, the black curve represents saturated lipids and the shaded curve unsaturated ones.
The average values of area per lipid have been plotted in Fig. 4 A as a function of chain length. It turns out that a comparison of the simulated values for 〈A〉 with experimental observations is rather challenging, since the reported systematic studies on SM are unfortunately few and varying. Maulik et al. (52) have measured the membrane thickness for a number of chain lengths and then combined their results with estimated lipid volumes to yield average areas per molecule. These values ranged from ∼0.64 nm2 for 16:0-SM to ∼0.59 nm2 for 24:0-SM (T = 323 K). This case is discussed below in more detail. More recent studies by Maulik et al. have utilized x-ray diffraction and found 0.47 nm2 for 16:0-SM (53) and 0.55 nm2 for 18:0-SM (43) at T = 328 K, together with 0.470 nm2 and 0.613 nm2 for 24:0-SM at temperatures T = 313 K and 333 K, respectively (44). The trends reproduced by our simulations were more clearly predicted in a fairly recent study, based on Langmuir film balance at a surface pressure of 30 mN/m and T = 303 K (54). In this study, the area decreased from 0.525 nm2 for 16:0-SM to 0.486 nm2 for 18:0-SM and to 0.477 nm2 for 24:0-SM. The results for unsaturated SMs were significantly higher, 0.615 nm2 for 18:1-SM and 0.607 nm2 for 24:1-SM. Although the absolute values of this study are not comparable with our simulations due to different conditions, the trends agree.
FIGURE 4.
Average structural quantities of the simulated systems as a function of acyl chain length: (A) area per lipid and (B) bilayer thickness. The bilayer thickness is the peak-to-peak distance of the electron-density plot across each system (see Fig. 8). The error bars in A are the standard deviation of mean values calculated in 4-ns blocks, whereas in B the maximum error for determining the peak-to-peak distance from the electron density plots was 0.05 nm.
The above-mentioned systematic studies (52,54) indicate a decreasing area per molecule for an increasing chain length, whereas some of the separate studies (43,44,53) have yielded results not in line with this trend. Here we put more weight on comparing our results with the systematic studies, because they produce more consistent information about the trends that we are interested in. Further, given that lipid monolayers and bilayers are known to behave somewhat differently (55,56), we feel that the most suitable study to be compared with our simulation data is the one by Maulik et al. (52).
As mentioned above, the values found by Maulik et al. ranged from 0.64 nm2 for 16:0-SM to 0.59 nm2 for 24:0-SM (T = 323 K). These values were found by dividing the estimated molecular volume by the measured bilayer thickness. By following the method applied in Maulik et al. (52) and using our simulated values for the bilayer thickness (see Fig. 4 B), we found that the approximate values for 〈A〉 were qualitatively consistent with those shown in Fig. 4 A. However, the approximate results were systematically ∼0.03 nm2 larger than those given in Fig. 4 A. That may partly explain the relatively large values reported in (52) for 〈A〉, since most of the above-discussed experimental studies are in favor of considerably smaller values for the average area per lipid. Nevertheless, the qualitative trend reported in Maulik et al. (52) is in line with our simulation data, that is, 〈A〉 decreases for an increasing chain length. This conclusion is also supported by studies of Petrache et al. (57), who found that for saturated PCs the average area per molecule decreases for an increasing chain length.
As for effects due to unsaturation, we found monounsaturation to result in a clear increase in the observed area per lipid for all systems, on average by 0.036 ± 0.007 nm2. As systematic experimental studies on the double-bond effect on surface area are lacking for SMs, we compare our results with earlier simulations carried out on PCs. The difference in average area per lipid between POPC and DPPC, structurally resembling the difference between 18:0-SM and 18:1-SM, has been reported in different studies to range from 0.043 nm2 (58) to 0.047 nm2 (59). The fact that this value is smaller for SM can be traced back to hydrogen bonding, as intermolecular hydrogen bonding between SMs has been shown to lead to smaller values for the average area per lipid (27). Also, the effect of double bonds on the main phase transition temperature has been observed to be smaller for SM than for PC (33).
The thickness of each bilayer was estimated by dp–p, the peak-to-peak distance of the calculated electron density profiles (see Fig. 8, A and B). The thickness, plotted in Fig. 4 B, increases linearly when the acyl chain length is increased. Again, it is difficult to compare the absolute values and trends with experiments because the conditions in different studies vary unsystematically, resulting in dp–p values between 4.1 and 5.6 nm for different lipids (43,44,53,60). Instead, the increasing trend of 0.12 ± 0.01 nm per CH2 group, obtained by fitting to Fig. 4 B, shows an excellent agreement with a systematic x-ray study on saturated SMs at T = 323 K (52), which reported an increase of 0.13–0.14 nm per CH2 group. The average effect of unsaturation on the bilayer thickness is 0.25 ± 0.04 nm for all systems, as estimated from Fig. 4 B. This can be compared with the reported ∼0.3 nm difference in thickness between POPC and DPPC bilayers (59).
FIGURE 8.
Electron densities of (A and B) the whole system, (C and D) the sphingosine chains, (E and F) the acyl chains, (G and H) the sphingosine and the acyl chains of opposite monolayers separately, and (I and J) the molecular number densities. Color coding follows that of Fig. 5.
In all, considering the experimental difficulties in finding accurate estimates for 〈A〉 and dp–p, together with the small number of systematic studies on different SMs in similar conditions, the values predicted by our model for these structural parameters are in reasonably good agreement with experimental findings.
Chain structure
We describe the orientational order of the hydrocarbon chains by the deuterium order parameter,
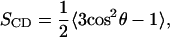 |
(1) |
where θ is the angle between a selected C-H vector and the bilayer normal. As the apolar hydrogens are not explicitly present in united-atom simulations, we reconstructed the corresponding C-H vectors using backbone chain configuration to calculate the SCD values.
The resulting order parameter values are shown in Fig. 5. The average chain order in liquid SM bilayers is higher than in bilayers of structurally matched PCs, as discussed earlier (27). This effect is related to the lower average area per lipid and the stronger intermolecular hydrogen bonding between SMs. Although the order parameter profiles of the different SM systems are similar in shape and height, some differences are yet evident.
FIGURE 5.
Deuterium order parameters as a function of carbon position along the chain (for carbon numbering, see Fig. 1). The values have been plotted separately for the sphingosine chains of (A) saturated and (B) unsaturated systems and for the acyl chains of (C) saturated and (D) unsaturated systems, the different systems being separated by gray shades as indicated in the inset of C. The error of each data point was <ΔSCD = 0.03. The error was estimated from the standard deviation of mean values, calculated in 4-ns blocks.
The most striking difference is the local minimum in order, located at the lower carbon of the cis double bond of the unsaturated acyl chains (Fig. 5 C), which is lacking for the saturated acyl chains (Fig. 5 D). This is a general phenomenon caused by double bonds and observed, for example, in NMR order parameter profiles of monounsaturated PCs, such as POPC (61). Apart from the local disorder at the double bond region, no further striking effects can be observed at other regions of the unsaturated chains, but rather a slight overall decrease in order when compared to the saturated lipids. The effect of unsaturation is roughly between 0.03 and 0.05 in units of SCD values for both acyl and sphingosine chains. The difference seems to be less significant toward the ends of the longer acyl chains, indicating the local nature of the double bond's disordering effect. We wish to stress that the parameterization of the double-bond region has some effects on the results here, as will be discussed in Effect of double-bond parameters, but that the presented general observations are not significantly affected.
According to common interpretation, the cis double bond introduces a kink into the average chain structure and thus disturbs the packing of the chains. However, in a gel-phase bilayer, the unsaturated chains are more likely to adopt “crankshaft” than “boomerang” conformations due to steric hindrance effects (62). By this notation, the term boomerang refers to a structure where the two linear segments of the unsaturated chain, separated by the double bond, are oriented at an angle of ∼130° with respect to each other. In the “crankshaft”-like conformation, the single bonds adjacent to the double bond set their torsional angles in such a way that the two chain segments before and after the double bond are essentially parallel with each other (see Li et al. (62) for details). To clarify the chain structure, we have plotted distributions of local chain orientations in a few systems in Fig. 6. The distributions clearly indicate the effect of the double bond on the chain orientation to be local. Even when the chain is bent almost parallel to the bilayer plane at the double-bond location and its close vicinity, the orientation of the ends of unsaturated chains is very similar to that found in the saturated chains. Thus, the crankshaft conformation seems to be a more valid description for the SM systems discussed here.
FIGURE 6.
Distributions for the angle between the bilayer normal and the Cn−1 to Cn+1 vector for carbon Cn. The red curves represent the two carbons at the C=C double bond and the black curves are the next adjacent carbons. The lighter curves are further away from the double bond.
Despite the local nature of the effects that unsaturation has on acyl chain orientation, some more general effects are clearly observed. For example, the acyl chain length along the bilayer normal is reduced on average by 0.17 ± 0.02 nm upon monounsaturation, as shown in Fig. 7. Notably, the unsaturation has a shortening effect on the SPH chain as well, on average by 0.11 ± 0.01 nm. This is an indication that the double bond not only affects the local orientation of the chain in which it is located, but also has an overall effect on chain packing.
FIGURE 7.
Average values for chain length, Lz, in different systems as a function of the number of carbons in the acyl chain, obtained by projecting the end-to-end vector of each chain on the z axis. The starting points of the chains, C1, were chosen as indicated in Fig. 1. The error bars are the standard deviation of mean values calculated in 4-ns blocks.
Another interesting observation from Fig. 7 is that the acyl chain length increases on average by 0.08 ± 0.04 nm per added CH2 group. This is more than half of the observed increase in bilayer thickness, which implies that the longer chains must at least partly interdigitate into the opposing monolayer.
Density profiles
To quantify interdigitation of the acyl chains across the bilayer center, we have plotted electron density profiles ρe(z) and molecular number density profiles 〈Nmol(z)〉 in Fig. 8. The molecular number density was calculated by considering the maximum extension of the molecule along the z axis, taking the van der Waals radii for the atoms into account, as introduced in a previous work (63,64).
The electron density profiles shown in Fig. 8, A and B, first illustrate that interdigitation is increased for an increasing acyl chain length, as expected. The shapes of ρe(z) curves across the whole system resemble those measured by x-ray diffraction, indicating a clear local maximum at the bilayer center for the longer chains (52). Separate electron density plots for the acyl and sphingosine chains in Fig. 8, G and H, show clearly that the long acyl chains are mostly responsible for the observed local maximum at the bilayer center. Nevertheless, instead of interdigitating to the opposing leaflet, the acyl chain ends prefer to reside on the side of their own monolayer, as indicated by the peak in the chain densities calculated separately for the monolayers (see Fig. 8, G and H).
Hence, interdigitation in SM systems is not only due to acyl chains; the role of SPH is also important. Fig. 8, C and D, shows that interdigitation due to the sphingosine chain is rather considerably more pronounced in unsaturated SMs, and the difference is particularly evident in the case of short acyl chains. Fig. 8, E and F, in turn, demonstrates that interdigitation of the acyl chains in saturated and unsaturated SMs is largely similar, since the electron density profiles for the acyl chains are almost identical (see also discussion below).
To better characterize the significance of unsaturation on interdigitation, let us consider Fig. 8, G and H. They show that the density maximum of acyl chains is a bit higher for saturated systems, indicating a stronger tendency of the longer saturated chains to bend before the center of the bilayer (see the maximum at z ≈ 0.4 nm). In unsaturated SMs, this effect is also present, but it is somewhat weaker. The molecular number densities shown in Fig. 8, I and J, further indicate that interdigitation in unsaturated SMs is slightly more prominent than in saturated lipids. In particular, although the number of interdigitated molecules shows no clear difference when saturated and unsaturated acyl chains of lengths 22 and 24 carbons are compared, there is a notable difference in the case of short-chained lipids (16 and 18 carbons), indicating more preferential interdigitation for unsaturated lipids. This can be understood on the basis of the chain-length analysis in Fig. 7, which shows that the sphingosine chain is actually longer than (or as long as) the shorter acyl chains. Now, as the double bond shortens the acyl chain, this leads to stronger interference between sphingosine chains of the opposite monolayers and thus stronger molecular interdigitation. Summarizing, although the differences are minor, our results propose that interdigitation in unsaturated SMs is slightly more prominent than in saturated sphingomyelins. The possible biological significance of this finding and related consequences are discussed at the end of this article.
Lateral diffusion
To investigate lateral diffusion of the lipids, their center of mass (CM) coordinates were projected onto the xy plane and the two-dimensional mean-squared displacements 〈[r(t)]2〉 were plotted in Fig. 9. The tracer diffusion coefficient characterizing the lateral diffusion of individual molecules is ideally obtained from the slope of these graphs at long times by:
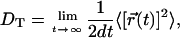 |
(2) |
where d = 2 is the dimensionality of the dynamic process. Here, we have accounted for the fact that the CM positions of the two lipid monolayers may fluctuate in time, and hence the above calculation for SM molecules was performed with respect to the CM position of the corresponding monolayer.
FIGURE 9.
Mean-squared displacements of the CM positions of the lipids in the bilayer plane in (A) saturated and (B) unsaturated systems.
The diffusion coefficients, obtained from the slope of the mean-squared displacement graphs (Fig. 9) between 5 and 15 ns, using all the data for analysis (40 ns), are shown in Fig. 10. It is clear from the figure that increasing the chain length of saturated SMs slows down the lateral diffusion rate. The trend is not monotonic for unsaturated SMs, which is likely due to the relatively short simulation time and the resulting scatter in the data, as seen from the graphs of Fig. 9, especially for 24:1-SM and 20:1-SM.
FIGURE 10.
Lateral diffusion coefficients of the lipids as a function of acyl chain length, obtained from the slope of the graphs in Fig. 9 from the period between 5 and 15 ns. The error bars have been estimated by considering each of the two monolayers in the studied systems separately.
Overall, the simulation results are in favor of an idea that an increasing acyl chain length leads to a reduction in the lateral diffusion rate. This is in line with experiments. Vaz et al. (65) measured the lateral diffusion of NBD-PE (N-(7-nitro-2,1,3-benzoxadiazol-4-yl)phosphatidylethanolamine) in POPC and dilauroylphosphatidylcholine bilayers (at 298 and 293 K, respectively) and found that the lateral diffusion coefficient decreased monotonically with increasing chain length of the diffusing particle. Very recently, Dustman et al. (66) have also concluded that for monounsaturated lipids the lateral diffusion rates decrease with increasing chain length. As for actual quantitative values, recent pulsed-field gradient NMR measurements by Filippov et al. have resulted in a value of 0.6 × 10−7 cm2/s for 16:0-SM at 323 K (67). This is in full agreement with our simulation data.
As the differences in DT values between saturated and unsaturated lipids are expected to be related to the average area per lipid and the degree of chain interdigitation, it is interesting to find that lateral diffusion is faster in unsaturated SM bilayers. Since increasing interdigitation is expected to slow down diffusion (see Fig. 8), whereas increasing area per molecule enhances diffusion (Fig. 3), it seems evident that here the role of interdigitation for lateral diffusion is not as important as the role of area per molecule, or, more precisely, the free area available for diffusion. The diffusion of the monolayers with respect to each other, however, should mainly depend on the interleaflet friction (viscosity) and thus be an indicator of the degree of interdigitation. To analyze this, we plotted the mean-squared displacements of the CM positions of the two monolayers with respect to each other (data not shown). Although the statistics is quite poor, one clear observation can be made. The root mean-squared velocity, deduced from the relative displacements of the monolayers on a 10-ps timescale is 9.4 ± 0.1 nm/ns for saturated systems and 8.8 ± 0.1 nm/ns for unsaturated systems, the difference being observable for all of the chain lengths. The effect can also be observed from the total relative displacements of the monolayers after 40 ns, which are ∼2–3 times larger for saturated than for unsaturated systems. The picture which emerges from these findings is consistent with the above results for interdigitation: unsaturated SM bilayers are characterized by slightly stronger interdigitation compared to saturated SMs, and hence interleaflet friction is more notable in unsaturated SM bilayers.
Rotational motions
The rotational motions of different parts of the lipids can be examined by utilizing the second rank reorientational autocorrelation functions C2(t):
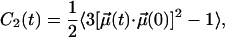 |
(3) |
where 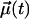 is a unit vector that defines the chosen rotational mode. Three different rotational modes were analyzed: the headgroup, the interfacial region, and the C-H bond vectors along the acyl chains.
is a unit vector that defines the chosen rotational mode. Three different rotational modes were analyzed: the headgroup, the interfacial region, and the C-H bond vectors along the acyl chains.
For the interfacial region, we chose a vector from sphingosine C3 to C1, and for the headgroup, a vector from headgroup phosphorus (P) to nitrogen (N). Fig. 11 shows the decay half-times, t1/2, of the reorientational C2(t) functions for these vectors. One can see that the headgroups are much more mobile than the interfacial regions in each system. Also, the rotational motions tend to get slower when the acyl chain length is increased, which might be due to decreased area per lipid. The effect of unsaturation is also quite significant: the decay time of each rotational motion is two to three times slower for the saturated than for the unsaturated version of a lipid.
FIGURE 11.
Decay half-times of selected rotational autocorrelation functions versus acyl chain length, plotted for saturated and unsaturated systems separately. The half-time, t1/2, is defined as the time where the autocorrelation function drops to half of its initial value. The error bars have been obtained by splitting the analyzed trajectory into two 20-ns parts and considering these parts separately.
For the acyl chains, we have calculated the average C2(t) functions separately for each of the previously constructed C-H vectors. The characteristic time τ is used to quantify the decay of the autocorrelation functions, as introduced in our previous study (27). In the same study, we indicated that these parameters, calculated from the simulation of a DPPC bilayer, are roughly comparable to experimental NMR relaxation times. As the NMR relaxation time measurements are lacking for SM chains, we concentrate here on comparing the simulated systems with each other.
The rotational motions of the chains, indicated by τ of the individual C-H autocorrelation functions, are increasing in roughly exponential fashion when going from chain ends toward the interfacial region. It can be noted that both chains of 24:0-SM are significantly slower than in the other systems, which may be a reflection of the main phase transition temperature of this particular lipid, which is already close to the simulation temperature. The effect of cis double bonds on chain dynamics can be clearly seen in Fig. 12, which indicates that the lower carbon of the C=C bond is always slowed down. However, the ends of the saturated chains are just as fast as those of the unsaturated chains. This is mainly enabled by the next adjacent dihedral bonds to the double bond, which are faster and thus compensate the slowing down (68). The details of the dynamics near the double bond are a consequence of the utilized force-field parameters. In this study, the general observations are not significantly affected by the different parameters available, as reported and discussed in “Effect of double-bond parameters”, below, and in the Supplementary Material.
FIGURE 12.
Characteristic decay time, τ, of the rotational autocorrelation functions of C-H bonds as a function of the carbon position along the chain. The τ-values have been plotted separately for each system, with the values for the acyl chain indicated in black and those for the sphingosine chain in gray. For numbering of carbons, see Fig. 1.
The fast decay of the rotational autocorrelation functions, identified by t1/2, behaves similarly to τ in a qualitative manner. The values of t1/2 increase exponentially from subpicosecond values to tens of picoseconds for acyl chains and up to nanoseconds for sphingosine chains (data not shown).
Hydrogen bonding and related characteristics
The tendency of sphingomyelin to form intra- and intermolecular hydrogen bonds is related to many of its characteristic structural and dynamic properties in bilayers. It is thus interesting to study the possible differences in hydrogen bonding caused by variations in chain length and saturation. We have used the following geometrical criteria to find hydrogen bonds from the simulation trajectory: the acceptor-hydrogen distance dah ≤ 0.25 nm and the donor-hydrogen-acceptor angle θdha ≤ 90°. More detailed discussion on the utilized method can be found elsewhere (27).
The average numbers of detected hydrogen bonds in the studied systems are presented in Table 1. The trends in our results agree with earlier simulation studies comparing the effects of cis versus trans unsaturation in PCs, where the average number of hydrogen bonds changed with increasing area per lipid (69). As the average lipid-lipid distance increases upon unsaturation, the number of intermolecular hydrogen bonds between lipids tends to decrease, whereas the number of intramolecular bonds increases. The same logic can be applied to the number of lipid-solvent bonds, which increases with greater area per lipid, as there is more space for water molecules. As there are no great differences in the average area per lipid values between the sphingomyelin systems studied here, the absence of striking differences in the hydrogen-bonding properties is not surprising. However, our results do not disagree with the above-mentioned earlier study for PCs.
TABLE 1.
Average numbers of intra- and intermolecular hydrogen bonds in the simulated bilayer systems
| System | No. (intra) | No. (inter) | No. (solvent) |
|---|---|---|---|
| 16:0-SM | 138.6 ± 3.0 | 53.8 ± 5.3 | 710.5 ± 20.9 |
| 18:0-SM | 140.1 ± 3.3 | 65.5 ± 4.6 | 642.1 ± 20.3 |
| 20:0-SM | 138.6 ± 3.1 | 63.9 ± 5.2 | 657.8 ± 20.0 |
| 22:0-SM | 140.1 ± 3.1 | 64.8 ± 6.0 | 651.3 ± 19.5 |
| 24:0-SM | 140.1 ± 3.1 | 59.3 ± 4.5 | 644.8 ± 20.4 |
| 16:1-SM | 140.8 ± 3.2 | 59.9 ± 5.3 | 693.6 ± 17.5 |
| 18:1-SM | 140.9 ± 3.2 | 56.3 ± 4.8 | 699.5 ± 19.1 |
| 20:1-SM | 140.2 ± 3.2 | 59.4 ± 4.8 | 691.5 ± 16.1 |
| 22:1-SM | 138.9 ± 2.9 | 62.4 ± 4.7 | 689.0 ± 16.6 |
| 24:1-SM | 138.4 ± 3.0 | 56.9 ± 4.7 | 683.0 ± 18.9 |
To summarize observations made about the individual hydrogen-bond types, the intramolecular bonds are dominated by the OH group, making bonds with phosphoryl oxygens Oa (∼90%) and Ob (∼10%), whereas the intermolecular bonds are dominated by the NH groups, bonding mainly with the hydroxyl and carboxyl oxygens OOH and OC1, but also, to a varying extent, with the phosphoryl oxygens Oa–Od. In all systems, water is mainly (∼70%) hydrogen-bonded with phosphoryl oxygens Oc and Od, but also with all other polar groups of the lipids. Some systematic trends can be found from the average numbers of hydrogen bonds when comparing different systems. Unsaturation tends to redistribute the lipid-lipid hydrogen bonds in such a way that the NH group forms fewer bonds with hydroxyl oxygen OOH, but sometimes even more bonds with the carboxyl and phosphoryl oxygens. This refers to a conformational restriction, such as a need for a close proximity of the molecules to form an intermolecular bond between the NH group and OOH atom. The hydrogen bonds between lipids and water, however, show no notable redistribution but rather generally increase in number upon unsaturation.
Significant differences could not be observed in structural properties related to hydrogen bonding. The angular distribution of the P-N vectors with respect to the bilayer normal were similar for each system, as well as the average orientational profiles of water dipoles. Thus, also, the values for electrostatic potential between the bilayer center and bulk water are close to each other: ΔV = (–0.64 ± 0.04)V for saturated systems and ΔV = (−0.65 ± 0.02)V for unsaturated systems. Also, the orientational distribution of a selected interfacial vector (sphingosine C1 to C3) showed very little difference among the studied systems, reflecting similarity in the structure of the interfacial region as well.
Effect of double-bond parameters
In recent computational studies on unsaturated lipids, a number of different parameterizations for the cis double bond have been utilized (35,36,68,70). In our work, we adapted these parameters from a previously published POPC simulation, based on the GROMOS force field (35). To investigate the effect of double-bond description, we ran a short 4-ns simulation on the 16:1-SM system starting from the last configuration at 50 ns with another parameter set that is available for a united-atom model in GROMACS, originally developed for polyunsaturated lipids (36).
The most significant difference observed after changing the double-bond description was related to the local ordering and dynamics of the acyl chain around the double bond region (see Supplementary Material). This is understandable, as the major difference between the different united atom parameterizations for acyl chains with double bonds is not in how they treat the double bonds themselves, but in how the neighboring single bonds are treated. We note that the effect of the double bond reaches a few bond lengths in both directions along the acyl chain. The acyl chains appear less ordered and more mobile around the double bond, but the effect, especially in ordering, vanishes out toward either of the chain ends. In other quantities studied in this work, the effects due to double-bond description were considerably weaker.
On the basis of the above information, we have, hereby, a clear qualitative idea of how different parameterizations affect the results presented here. The changes appear in spatially local quantities and are hence quite easily predictable. The description of the double-bond region is hence not expected to lead to any significant differences in the conclusions or in the trends predicted by the simulations discussed in this work. Nevertheless, we wish to stress that care should be taken, since it is not always clear how the double-bond description affects the overall behavior of the system. It is worthwhile to test a few of the commonly employed descriptions and gauge their influence on simulation results. For obvious reasons, this is particularly the case in polyunsaturated systems: some of the previously suggested parameterizations seem not to take into account the so-called “skew” nature of the single bonds that are next to the double bond. It has been proposed that this is responsible for the extraordinary flexibility of polyunsaturated acyl chains (71).
CONCLUDING REMARKS
There is a large body of work devoted to the role of sphingomyelin in various cellular processes, including signaling events and the formation of highly ordered dynamic domains known as lipid rafts. Like many other lipids, SMs are characterized by a wide population of molecules that differ in subtle but notable ways. In particular, the acyl chain linked to the amide group of sphingosine varies in length as well as unsaturation, most common SMs being 16:0, 18:0, 22:0, 24:0, and 24:1cis,Δ15 (11,34). Although the mixed population of different acyl chains seems to be an inherent feature of sphingomyelins, its influence on the physical properties of SMs is not fully understood. There are indications that a mismatch in length between the acyl chain and the sphingosine backbone has an influence on thermotropic behavior of SM bilayers, and it has been suggested that this is due to potential interdigitation (11). In particular, the long fatty acid which is amide-bonded to the sphingosine base has been suggested to interdigitate with the cytoplasmic leaflet of a cell membrane (6). Acknowledging further the role of ceramide as a messenger in processes such as apoptosis, the biological significance of SM is remarkable.
In this work, we have employed atomic-scale molecular dynamics simulations to study bilayers with a number of structurally different sphingomyelin molecules, ranging from 16:0/16:1 to 24:0/24:1. The objective has been to elucidate the effects of chain-length mismatch and monounsaturation on bilayer structure and dynamics. In particular, we have addressed the extent of interdigitation in these systems and its implications for membrane properties.
The observed linear increase in bilayer thickness per added CH2 group is in excellent agreement with experiments, as is also the rate of lateral diffusion in a 16:0-SM bilayer. Further, the electron density profiles calculated from our simulation data for long-chain SMs reproduce the secondary peak in the middle of the bilayer, which was previously observed for 22:0-SM and 24:0-SM bilayers by x-ray diffraction (52). These findings, and in particular the agreement of our electron-density profiles with experimental data allows us to be confident that the description of our model system is on a solid ground.
Apart from increased bilayer thickness, an increasing acyl chain length has various other effects on SM bilayers. It leads to a slight reduction in the average area per lipid and consequently to a minor enhancement in the ordering of SPH and acyl chains. SMs with saturated acyl chains are more tightly packed and ordered than the monounsaturated counterparts. Further, the decreasing area per molecule (and enhanced ordering) with an increasing acyl chain length is likely the reason for a decrease in lateral diffusion rates, and for the slowing down of rotational motions at the interfacial and headgroup regions.
One of the most interesting phenomena related to lipids with a large chain-length disparity is the interdigitation of the longer chains across the bilayer center. It has been shown that interdigitation significantly affects the properties of gel-phase lipid bilayers, and it has been proposed that this might be important for bilayers in the fluid phase as well (14). Our results support this idea. We have found that in the fluid phase above Tm, there is rather significant interdigitation for all acyl chain lengths considered. The interdigitation is emphasized for an increasing chain length, and is slightly more pronounced in monounsaturated SMs. The latter finding seems to imply that in monounsaturated SM bilayers the intermonolayer friction is somewhat stronger than in bilayers comprised of saturated SMs, indicating a stronger coupling of the two leaflets in unsaturated SM systems. There is reason to acknowledge, however, that the mentioned effects are relatively minor.
As we are here dealing with the fluid phase, it is tempting to ask what is the biological significance of chain interdigitation at lower temperatures in the gel phase or in ordered domains rich in cholesterol and SM. In plasma membranes, these lipids are mainly located at the extracellular leaflet, whereas the composition of the other leaflet is highly different. This renders the question on the biological significance of interdigitation in bilayers more complex than can be explained in terms of single-component studies. Finally, the role of acyl chain length and unsaturation on SM interaction with cholesterol remains one of the problems calling for more detailed quantification. These issues are to be discussed elsewhere.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Acknowledgments
The authors thank David J. Siminovitch, Robert V. Law, Samuli Ollila, and Emma Falck for fruitful discussions. We also thank Arvi Rauk and Michal Bachar for correspondence. We acknowledge the Finnish IT Center for Science and the HorseShoe (DCSC) supercluster computing facility at the University of Southern Denmark for computer resources.
This work has, in part, been supported by the Academy of Finland through its Center of Excellence Program (P.S.N. and I.V.), the Academy of Finland grant Nos. 202598 (P.S.N), 80246 (I.V.), and 80851 (M.T.H.), the Jenny and Antti Wihuri Foundation (M.T.H.), and the Finnish Academy of Science and Letters (P.S.N.).
References
- 1.Barenholz, Y., and T. E. Thompson. 1999. Sphingomyelin: biophysical aspects. Chem. Phys. Lipids. 102:29–34. [DOI] [PubMed] [Google Scholar]
- 2.Brown, D. A., and E. London. 2000. Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J. Biol. Chem. 275:17221–17224. [DOI] [PubMed] [Google Scholar]
- 3.Edidin, M. 2003. The state of lipid rafts: From model membranes to cells. Annu. Rev. Biophys. Biomol. Struct. 32:257–283. [DOI] [PubMed] [Google Scholar]
- 4.Mayor, S., and M. Rao. 2004. Rafts: scale-dependent, active lipid organization at the cell surface. Traffic. 5:231–240. [DOI] [PubMed] [Google Scholar]
- 5.Pike, L. J. 2004. Lipid rafts: heterogeneity on the high seas. Biochem. J. 378:281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simons, K., and E. Ikonen. 1997. Functional rafts in cell membranes. Nature. 387:569–571. [DOI] [PubMed] [Google Scholar]
- 7.Lai, E. C. 2003. Lipid rafts make for slippery platforms. J. Cell Biol. 162:365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Munro, S. 2003. Lipid rafts: elusive or illusive? Cell. 115:377–388. [DOI] [PubMed] [Google Scholar]
- 9.Brown, R. E. 1998. Sphingolipid organization in biomembranes: what physical studies of model membranes reveal. J. Cell Sci. 111:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holopainen, J. M., A. J. Metso, J.-P. Mattila, A. Jutila, and P. K. J. Kinnunen. 2004. Evidence for the lack of a specific interaction between cholesterol and sphingomyelin. Biophys. J. 86:1510–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramstedt, B., and J. P. Slotte. 2002. Membrane properties of sphingomyelins. FEBS Lett. 531:33–37. [DOI] [PubMed] [Google Scholar]
- 12.Schmidt, C. F., Y. Barenholz, C. Huang, and T. E. Thompson. 1978. Monolayer coupling in sphingomyelin systems. Nature. 271:775–777. [DOI] [PubMed] [Google Scholar]
- 13.Levin, I. W. 1985. Two types of hydrocarbon chain interdigitation in sphingomyelin bilayers. Biochemistry. 24:6282–6286. [DOI] [PubMed] [Google Scholar]
- 14.Huang, C., and J. T. Mason. 1986. Structure and properties of mixed-chain phospholipid assemblies. Biochim. Biophys. Acta. 864:423–470. [DOI] [PubMed] [Google Scholar]
- 15.Lu, D., D. Singh, M. R. Morrow, and C. W. M. Grant. 1993. Effect of glycosphingolipid fatty acid chain length on behavior in unsaturated phosphatidylcholine bilayers. Biochemistry. 32:290–297. [DOI] [PubMed] [Google Scholar]
- 16.Mehlhorn, I. E., E. Florio, K. R. Barber, C. Lordo, and C. W. M. Grant. 1988. Evidence that trans-bilayer interdigitation of glycosphingolipid long chain fatty acids may be a general phenomenon. Biochim. Biophys. Acta. 939:151–159. [DOI] [PubMed] [Google Scholar]
- 17.Morrow, M. R., D. Singh, D. Lu, and C. W. M. Grant. 1993. Glycosphingolipid acyl chain orientational order in unsaturated phosphatidylcholine bilayers. Biophys. J. 64:654–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boggs, J. M., and K. M. Koshy. 1994. Do the long fatty acid chains of sphingolipids interdigitate across the center of a bilayer of shorter chain symmetric phospholipids? Biochim. Biophys. Acta. 1189:233–241. [DOI] [PubMed] [Google Scholar]
- 19.Ash, W. L., M. R. Zlomislic, E. O. Oloo, and D. P. Tieleman. 2004. Computer simulations of membrane properties. Biochim. Biophys. Acta. 1666:158–189. [DOI] [PubMed] [Google Scholar]
- 20.Feller, S. E. 2000. Molecular dynamics simulations of lipid bilayers. Curr. Opin. Colloid Interface Sci. 5:217–223. [Google Scholar]
- 21.Saiz, L., and M. L. Klein. 2002. Computer simulation studies of model biological membranes. Acc. Chem. Res. 35:482–489. [DOI] [PubMed] [Google Scholar]
- 22.Scott, H. L. 2002. Modeling the lipid component of membranes. Curr. Opin. Struct. Biol. 12:495–502. [DOI] [PubMed] [Google Scholar]
- 23.Tieleman, D. P., S. J. Marrink, and H. J. C. Berendsen. 1997. A computer perspective of membranes: Molecular dynamics studies of lipid bilayer systems. Biochim. Biophys. Acta. 1331:235–270. [DOI] [PubMed] [Google Scholar]
- 24.Chiu, S. W., S. Vasudevan, E. Jakobsson, R. J. Mashl, and H. L. Scott. 2003. Structure of sphingomyelin bilayers: A simulation study. Biophys. J. 85:3624–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hyvönen, M. T., and P. T. Kovanen. 2003. Molecular dynamics simulation of sphingomyelin bilayer. J. Phys. Chem. B. 107:9102–9108. [Google Scholar]
- 26.Mombelli, E., R. Morris, W. Taylor, and F. Fraternali. 2003. Hydrogen-bonding propensities of sphingomyelin in solution and in a bilayer assembly: A molecular dynamics study. Biophys. J. 84:1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niemelä, P., M. T. Hyvönen, and I. Vattulainen. 2004. Structure and dynamics of sphingomyelin bilayer: insight gained through systematic comparison to phosphatidylcholine. Biophys. J. 87:2976–2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khelashvili, G. A., and H. L. Scott. 2004. Combined Monte Carlo and molecular dynamics simulation of hydrated 18:0 sphingomyelin-cholesterol lipid bilayers. J. Chem. Phys. 120:9841–9847. [DOI] [PubMed] [Google Scholar]
- 29.Pandit, S. A., E. Jakobsson, and H. L. Scott. 2004. a. Simulation of the early stages of nano-domain formation in mixed bilayers of sphingomyelin, cholesterol, and dioleylphosphatidylcholine. Biophys. J. 87:3312–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pandit, S. A., S. Vasudevan, S. W. Chiu, R. J. Mashl, E. Jakobsson, and H. L. Scott. 2004. b. Sphingomyelin-cholesterol domains in phospholipid membranes: atomistic simulation. Biophys. J. 87:1092–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berendsen, H. J. C., D. van der Spoel, and R. van Drunen. 1995. Gromacs: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 91:43–56. [Google Scholar]
- 32.Lindahl, E., B. Hess, and D. van der Spoel. 2001. Gromacs 3.0: A package for molecular simulation and trajectory analysis. J. Mol. Model. 7:306–317. [Google Scholar]
- 33.Koynova, R., and M. Caffrey. 1995. Phases and phase transitions of the sphingolipids. Biochim. Biophys. Acta. 1255:213–236. [DOI] [PubMed] [Google Scholar]
- 34.Ramstedt, B., P. Leppimäki, M. Axberg, and J. P. Slotte. 1999. Analysis of natural and synthetic sphingomyelins using high-performance thin-layer chromatography. Eur. J. Biochem. 266:997–1002. [DOI] [PubMed] [Google Scholar]
- 35.Tieleman, D. P., and H. J. C. Berendsen. 1998. A molecular dynamics study of the pores formed by Escherichia coli OmpF porin in a fully hydrated palmitoyloleoylphosphatidylcholine bilayer. Biophys. J. 74:2786–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bachar, M., P. Brunelle, D. P. Tieleman, and A. Rauk. 2004. Molecular dynamics simulation of a polyunsaturated lipid bilayer susceptible to lipid peroxidation. J. Phys. Chem. B. 108:7170–7179. [Google Scholar]
- 37.Berendsen, H. J. C., J. P. M. Postma, W. F. van Gunsteren, and J. Hermans. 1981. Interaction models for water in relation to protein hydration. In Intermolecular Forces. B. Pullman, editor. Reidel, Dordrecht, The Netherlands. 331–342.
- 38.Essmann, U., L. Perera, M. L. Berkowitz, T. Darden, H. Lee, and L. G. Pedersen. 1995. A smooth particle mesh Ewald potential. J. Chem. Phys. 103:8577–8592. [Google Scholar]
- 39.Patra, M., M. Karttunen, M. T. Hyvönen, E. Falck, P. Lindqvist, and I. Vattulainen. 2003. Molecular dynamics simulations of lipid bilayers: major artifacts due to truncating electrostatic interactions. Biophys. J. 84:3636–3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patra, M., M. Karttunen, M. T. Hyvönen, E. Falck, and I. Vattulainen. 2004. Lipid bilayers driven to a wrong lane in molecular dynamics simulations by subtle changes in long-range electrostatic interactions. J. Phys. Chem. B. 108:4485–4494. [Google Scholar]
- 41.Hess, B., H. Bekker, H. J. C. Berendsen, and J. G. E. M. Fraaije. 1997. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 18:1463–1472. [Google Scholar]
- 42.Miyamoto, S., and P. A. Kollman. 1992. SETTLE: An analytical version of the SHAKE and RATTLE algorithms for rigid water models. J. Comput. Chem. 13:952–962. [Google Scholar]
- 43.Maulik, P. R., P. K. Sripada, and G. G. Shipley. 1991. Structure and thermotropic properties of hydrated N-stearoyl sphingomyelin bilayer membranes. Biochim. Biophys. Acta. 1062:211–219. [DOI] [PubMed] [Google Scholar]
- 44.Maulik, P. R., and G. G. Shipley. 1995. X-ray diffraction and calorimetric study N-lignoceryl sphingomyelin membranes. Biophys. J. 69:1909–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berendsen, H. J. C., J. P. M. Postma, W. F. van Gunsteren, A. DiNola, and J. R. Haak. 1984. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81:3684–3690. [Google Scholar]
- 46.Hoover, W. G. 1985. Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. A. 31:1695–1697. [DOI] [PubMed] [Google Scholar]
- 47.Nosé, S. 1984. A molecular dynamics method for simulations in the canonical ensemble. Mol. Phys. 52:255–268. [Google Scholar]
- 48.Nosé, S., and M. L. Klein. 1983. Constant pressure molecular dynamics for molecular systems. Mol. Phys. 50:1055–1076. [Google Scholar]
- 49.Parrinello, M., and A. Rahman. 1981. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 52:7182–7190. [Google Scholar]
- 50.Bar, L. K., Y. Barenholz, and T. E. Thompson. 1997. Effect of sphingomyelin composition on the phase structure of phosphatidylcholine-sphingomyelin bilayers. Biochemistry. 36:2507–2516. [DOI] [PubMed] [Google Scholar]
- 51.Sripada, P. K., P. R. Maulik, J. A. Hamilton, and G. G. Shipley. 1987. Partial synthesis and properties of a series of N-acyl sphingomyelins. J. Lipid Res. 28:710–718. [PubMed] [Google Scholar]
- 52.Maulik, P. R., D. Atkinson, and G. G. Shipley. 1986. X-ray scattering of vesicles of N-acyl sphingomyelins: determination of bilayer thickness. Biophys. J. 50:1071–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maulik, P. R., and G. G. Shipley. 1996. N-palmitoyl sphingomyelin bilayers: Structure and interactions with cholesterol and dipalmitoylphosphatidylcholine. Biochemistry. 35:8025–8034. [DOI] [PubMed] [Google Scholar]
- 54.Li, X.-M., J. M. Smaby, M. M. Momsen, H. L. Brockman, and R. E. Brown. 2000. Sphingomyelin interfacial behavior: the impact of changing acyl chain composition. Biophys. J. 78:1921–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nagle, J. F., and S. Tristam-Nagle. 2000. Structure of lipid bilayers. Biochim. Biophys. Acta. 1469:159–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaznessis, Y. N., K. Sangtae, and R. G. Larson. 2002. Simulations of zwitterionic and anionic phospholipid monolayers. Biophys. J. 82:1731–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Petrache, H. I., S. W. Dodd, and M. F. Brown. 2000. Area per lipid and acyl length distributions in fluid phosphatidylcholines determined by 2H NMR spectroscopy. Biophys. J. 79:3172–3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chiu, S. W., E. Jakobsson, and H. L. Scott. 2001. Combined Monte Carlo and molecular dynamics simulation of hydrated lipid-cholesterol lipid bilayers at low cholesterol concentration. Biophys. J. 80:1104–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hyvönen, M. T., and P. T. Kovanen. 2005. Molecular dynamics simulations of unsaturated lipid bilayers: effects of varying the numbers of double bonds. Eur. Biophys. J. 34:294–305. [DOI] [PubMed] [Google Scholar]
- 60.McIntosh, T. J., S. A. Simon, D. Needham, and C.-H. Huang. 1992. Interbilayer interactions between sphingomyelin and sphingomyelin/cholesterol bilayers. Biochemistry. 31:2020–2024. [DOI] [PubMed] [Google Scholar]
- 61.Seelig, J., and N. Waespe-Šarčevic. 1978. Molecular order in cis and trans unsaturated phospholipid bilayers. Biochemistry. 17:3310–3315. [DOI] [PubMed] [Google Scholar]
- 62.Li, S., H. Lin, Z. Wang, and C. Huang. 1994. Identification and characterization of kink motifs in 1-palmitoyl-2-oleoyl-phosphatidylcholines: a molecular mechanics study. Biophys. J. 66:2005–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Falck, E., M. Patra, M. Karttunen, M. T. Hyvönen, and I. Vattulainen. 2004. Lessons of slicing membranes: interplay of packing, free area, and lateral diffusion in phospholipid/cholesterol bilayers. Biophys. J. 87:1076–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kupiainen, M., E. Falck, S. Ollila, P. Niemelä, A. A. Gurtovenko, M. T. Hyvönen, M. Patra, M. Karttunen, and I. Vattulainen. 2005. Free volume properties of sphingomyelin, DMPC, DPPC, and PLPC bilayers. J. Comput. Theor. Nanosci. 2:401–413. [Google Scholar]
- 65.Vaz, W. L. C., R. M. Clegg, and D. Hallmann. 1985. Translational diffusion of lipids in liquid crystalline phase phosphatidylcholine multibilayers: a comparison of experiment with theory. Biochemistry. 24:781–786. [DOI] [PubMed] [Google Scholar]
- 66.Dustman, J. M., R. S. Casas, H. A. Scheidt, N. V. Eldho, W. E. Teague, and K. Gawrisch. 2005. Lipid hydrocarbon chain dynamics and lateral diffusion in a polyunsaturated lipid matrix. Biophys. J. 88:27a. [Google Scholar]
- 67.Filippov, A., G. Orädd, and G. Lindblom. 2003. The effect of cholesterol on the lateral diffusion of phospholipids in oriented bilayers. Biophys. J. 84:3079–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Róg, T., K. Murzyn, R. Gurbiel, Y. Takaoka, A. Kusumi, and M. Pasenkiewicz-Gierula. 2004. Effects of phospholipid unsaturation on the bilayer nonpolar region: a molecular simulation study. J. Lipid Res. 45:326–336. [DOI] [PubMed] [Google Scholar]
- 69.Murzyn, K., T. Róg, G. Jezierski, Y. Takaoka, and M. Pasenkiewicz-Gierula. 2001. Effects of phospholipid unsaturation on the membrane/water interface: a molecular simulation study. Biophys. J. 81:170–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Feller, S. E., K. Gawrisch, and A. D. MacKerell, Jr. 2002. Polyunsaturated fatty acids in lipid bilayers: intrinsic environmental contributions to their unique physical properties. J. Am. Chem. Soc. 124:318–326. [DOI] [PubMed] [Google Scholar]
- 71.Gawrisch, K., N. V. Eldho, and L. L. Holte. 2003. The structure of DHA in phospholipid membranes. Lipids. 38:445–452. [DOI] [PubMed] [Google Scholar]














