Abstract
Epstein-Barr virus (EBV) nuclear antigen 1 (EBNA-1) is essential for replication of episomal EBV DNAs and maintenance of latency. Multifunctional EBNA-1 is phosphorylated, but the significance of EBNA-1 phosphorylation is not known. Here, we examined the effects on nuclear translocation of Ser phosphorylation of the EBNA-1 nuclear localization signal (NLS) sequence, 379Lys-Arg-Pro-Arg-Ser-Pro-Ser-Ser386. We found that Lys379Ala and Arg380Ala substitutions greatly reduced nuclear transport and steady-state levels of green fluorescent protein (GFP)-EBNA1, whereas Pro381Ala, Arg382Ala, Pro384Ala, and Glu378Ala substitutions did not. Microinjection of modified EBNA-1 NLS peptide-inserted proteins and NLS peptides cross-linked to bovine serum albumin (BSA) showed that Ala substitution for three NLS Ser residues reduced the efficiency of nuclear import. Similar microinjection analyses demonstrated that phosphorylation of Ser385 accelerated the rate of nuclear import, but phosphorylation of Ser383 and Ser386 reduced it. However, transfection analyses of GFP-EBNA1 mutants with the Ser-to-Ala substitution causing reduced nuclear import efficiency did not result in a decrease in the nuclear accumulation level of EBNA-1. The results suggest dynamic nuclear transport control of phosphorylated EBNA-1 proteins, although the nuclear localization level of EBNA-1 that binds to cellular chromosomes and chromatin seems unchanged. The karyopherin α NPI-1 (importin α5), a nuclear import adaptor, bound more strongly to Ser385-phosphorylated NLS than to any other phosphorylated or nonphosphorylated forms. Rch1 (importin α1) bound only weakly and Qip1 (importin α3) did not bind to the Ser385-phosphorylated NLS. These findings suggest that the amino-terminal 379Lys-Arg380 is essential for the EBNA-1 NLS and that Ser385 phosphorylation up-regulates nuclear transport efficiency of EBNA-1 by increasing its binding affinity to NPI-1, while phosphorylation of Ser386 and Ser383 down-regulates it.
Epstein-Barr virus (EBV) nuclear antigen 1 (EBNA-1) is essential for the replication of EBV plasmid DNAs in latently infected cells and is also a transcriptional transactivator (4, 41, 45, 46, 54). It has recently been suggested that EBNA-1 enhances B-cell growth and transformation and is critical for the continued survival of EBV-associated Burkitt's lymphoma cells (20, 24, 40). EBNA-1 is the only EBV-encoded protein expressed in all types of latently infected cells (54) and represents one of the most important viral antigens for seroepidemiologic studies and serodiagnostic tests (21, 23, 54, 64). EBNA-1 proteins in the form of homodimers (3, 13, 17) are highly localized in the nucleus (19, 28, 36, 51) and bind in a sequence-specific manner to the EBV DNA oriP region (1, 2, 17, 27, 44, 45), cellular chromosomes/chromatin (29, 48), and cellular replication foci (31). EBNA-1 binding to mitotic cellular chromosomes is widely considered to enable the partition of de novo-synthesized EBV episomal DNAs to daughter cells (29, 48). EBNA-1 proteins are phosphorylated at Ser residues (22) and two phosphorylation domains have been mapped (52). Nonetheless, the effect of phosphorylation on EBNA-1 function is not known.
We have previously reported that EBNA-1 proteins interact with NPI-1 (importin α5), which is a nuclear import adaptor karyopherin α (also known as importin α), in addition to Rchl (importin α1), using yeast two-hybrid screening and coimmunoprecipitation analyses of Raji cell lysates (30). Karyopherin β (importin β) family proteins recognize various nuclear localization signal (NLS) sequences with or without the aid of adaptor molecules such as karyopherin α (7, 11, 63). The EBNA-1 sequence 379Lys-Arg-Pro-Arg-Ser-Pro-Ser-Ser386 targets cross-linked carrier proteins to the nucleus (1), but it is not known whether all eight residues are necessary for NLS function. This 379KRPRSPSS386 sequence is referred to as the EBNA-1 NLS in this report.
Reported NLS amino acid sequences in different nuclear proteins are very diverse (12, 49, 57) and lack a strict consensus sequence, but generally are short and characterized by a high proportion of positively charged residues (7, 12). Monopartite NLSs have a single cluster of basic residues: the prototype sequences are the very basic simian virus 40 large T NLS (PKKKRKV) and the more hydrophobic c-Myc NLS (PAAKRVKLD) (10, 39). Bipartite NLSs have a motif with two interdependent basic domains and a mutation-tolerant spacer: the prototype sequences are the nucleoplasmin NLS (VKRPAATKKAGQAKKKKLD) and N1/N2 NLS (RKKRKTEEESPLKDKDAKKSKQEP) (12, 33, 56).
Many monopartite NLSs are 7 to 9 residues long and contain a helix-disrupting amino-terminal amino acid (proline or glycine) followed by at least three basic amino acids, while other NLSs contain basic amino acids flanking a proline residue (10). The EBNA-1 NLS belongs to the latter type. It has been proposed previously that Lys-x-x-Lys/Arg (where x can be any residue) (57), Lys-Arg/Lys-x-Arg/Lys (5), and several positively charged amino acids (Arg or Lys) associated with a Pro (18) represent consensus sequences of different groups of known monopartite NLSs. The EBNA-1 NLS fits this Lys-Arg/Lys-x-Arg/Lys sequence pattern. The KRPR tetrapeptide is also present in the NLSs of two other viral proteins, 281KRPRP of adenovirus E1A (47) and 189VSRKRPRP196 of polyomavirus large T, although the precise limits of the polyomavirus T NLS have not been defined (53).
The transport of proteins into the nucleus is a key regulatory step in functions of nuclear proteins (63). In the present study we examined whether and how phosphorylation of the Ser residues in the EBNA-1 NLS affects nuclear import and interaction with karyopherin α proteins. The EBNA-1 position 380 to 397 phosphorylation region includes almost the entire NLS and adjacent Ser-rich QSSSSGSP (amino acids 387 to 394) sequence (1, 30, 52) (Fig. 1C). We also analyzed which amino acids in the eight-residue EBNA-1 NLS are crucial for nuclear localization.
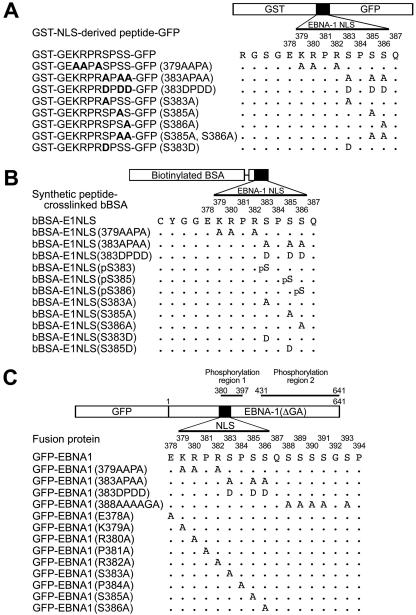
FIG. 1.
Schematic presentation of structures of EBNA-1 NLS substitution mutant-inserted chimeric proteins, biotinylated bovine serum albumin tagged with the substitution mutant synthetic peptides and Ser-phosphorylated forms of the EBNA-1 NLS, and green fluorescence protein fusion proteins of the EBNA-1 NLS mutants. (A) Chimeric proteins into which EBNA-1 NLS-derived sequences have been inserted. (B) Biotinylated bovine serum albumin cross-linked to synthetic peptides of substitution mutants or Ser-phosphorylated forms of the EBNA-1 NLS. (C) GFP fusion proteins of the NLS amino acid substitution mutants of EBNA-1.
MATERIALS AND METHODS
Construction of chimeric GST-NLS-GFP proteins.
EBNA-1 NLS substitution mutant-inserted GST-NLS-GFP proteins glutathione S-transferase (GST)-GEKRPRSPSS-GFP, GST-GEAAPASPSS-GFP(379AAPA), GST-GEKRPRAPAA-GFP(383APAA), GST-GEKRPRDPDD-GFP(383DPDD), GST-GEKRPRAPSS-GFP (S383A), GST-GEKRPRSPAS-GFP(S385A), GST-GEKRPRSPSA-GFP(S386A), GST-GEKRPRSPAA-GFP(S385A, S386A) and GST-GEKRPRDPSS-GFP(S383D) (Fig. 1A) were constructed by inserting NLS oligonucleotides GATCCGGAGAAAAGAGGCCCAGGAGTCCCAGTAGTCAGC for GST-GEKRPRSPSS-GFP, GATCCGGAGAAGCGGCGCCCGCGAGTCCCAGTAGTCAGC for GST-GEAAPASPSS-GFP(379AAPA), GATCCGGAGAAAAGAGGCCCAGGGCACCCGCAGCACAGC for GST-GEKRPRAPAA-GFP(383APAA), GATCCGGAGAAAAGAGGCCCAGGGACCCCGACGACCAGC for GST-GEKRPRDPDD-GFP(383DPDD), GATCCGGAGAAAAGAGGCCCAGGGCACCCAGTAGTCAGC for GST-GEKRPRAPSS-GFP (S383A), GATCCGGAGAAAAGAGGCCCAGGAGTCCCGCAAGTCAGC for GST-GEKRPRSPAS-GFP(S385A), GATCCGGAGAAAAGAGGCCCAGGAGTCCCAGTGCACAGC for GST-GEKRPRSPSA-GFP(S386A), GATCCGGAGAAAAGAGGCCCAGGAGTCCCGCAGCACAGC for GST-GEKRPRSPAA-GFP(S385A,S386A), and GATCCGGAGAAAAGAGGCCCAGGGACCCCAGTAGTCAGC for GST-GEKRPRDPSS-GFP(S383D), into the BamHI-SmaI site of pGEX2T (Pharmacia)-derived vectors for the previously described GST-NES-GFP (60) (substitutions are indicated in boldface).
Synthetic peptides derived from the EBNA-1 NLS and their conjugation to biotinylated bovine serum albumin.
EBNA-1 NLS-derived peptides CYGGEKRPRSPSSQ, CYGGEAAPASPSSQ, CYGGEKRPRAPAAQ, CYGGEKRPRAPSSQ, CYGGEKRPRSPASQ, CYGGEKRPRSPSAQ, CYGGEKRPRDPDDQ, Ser383-phophorylated CYGGEKRPR-pS-PSSQ (pS stands for phosphoserine), Ser385-phosphorylated CYGGEKRPRSP-pS-SQ, Ser386-phosphorylated CYGGEKRPRSPS-pS-Q, CYGGEKRPRDPSSQ, and CYGGEKRPRSPDSQ (Fig. 1B) were synthesized by GenScript (New Jersey). The amino-terminal CYGG sequence was added to facilitate cross-linking of the synthetic peptides to biotinylated bovine serum albumin (bBSA) and the synthetic peptides were chemically conjugated to bBSA as previously described (26). The designations and structures of the peptide-tagged bBSAa are listed in Fig. 1A. Peptide-conjugated bBSA proteins contained 7 to 10 peptides per bBSA molecule, as judged by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). Rhodamine isothiocyanate-labeled BSA was prepared essentially as described previously (60).
Construction of GFP fusion proteins of the EBNA-1 NLS and flanking sequence mutants.
A GFP fusion polypeptide of EBNA-1 that lacks the Gly-Gly-Ala copolymer (65, 66), designated GFP-EBNA1, has been previously described (29, 31). GFP-EBNA1 mutants were constructed by the method of PCR-based site-directed mutagenesis. The GFP fusion protein of the EBNA-1 mutant with 379AAPA382 was designated GFP-EBNA1(379AAPA). The other GFP-EBNA-1 mutants were designated in the same fashion. The structures and names of a series of GFP fusion proteins of EBNA-1 mutants with an amino acid substitution mutation in the NLS, amino acids 379 to 394, and flanking sequences are as follows: GFP-EBNA1(383APAA), GFP-EBNA1(379AAPA), GFP-EBNA1(388AAAAGA), GFP-EBNA1(K379A), GFP-EBNA1(R380A), GFP-EBNA1(P381A), GFP-EBNA1(R382A), GFP-EBNA1(S383A), GFP-EBNA1(P384A), GFP-EBNA1(S385A), GFP-EBNA1(S386A), and GFP-EBNA1(E378A) (Fig. 1C).
The PCR primers used to construct the GFP-EBNA1 mutants are as follows: GACGTGGAGAAGCGGCGCCCGCGAGTCCCAGTAG for GFP-EBNA1(379AAPA), GTGGACGTGGAGAAGCGAGGCCCAGGAGTCCCAG for GFP-EBNA1NL1-1(K379A), GACGTGGAGAAAAGGCGCCCAGGAGTCCCAGTAG for GFP-EBNA1(R380A), GAGAAAAGAGGCCCGCGAGTCCCAGTAGTCAGTC for GFP-EBNA1(R382A), GAGGCCCAGGGCTCCCGCTGCTCAGTCATCATC for GFP-EBNA1(383APAA), GAGGCCCAGGGATCCCGATGATCAGTCATCATC for GFP-EBNA1(383DPDD), AGTAGTCAGGCAGCAGCAGCCGGGGCTCCACCGCGC for GFP-EBNA1(388AAAAGA), GGACGTGGAGCAAAGAGGCCCAGGAGTCCC for GFP-EBNA1(E378A), GGACGTGGAGAAAAGAGGGCCAGGAGTCCC for GFP-EBNA1(P381A), AAGAGGCCCAGGGCTCCCAGTAGTCAG for GFP-EBNA1(S383A), GAGGCCCAGGAGTGCCAGTAGTCAGTCATCATC forGFP-EBNA1(P384A), CCCAGGAGTCCCGCTAGTCAGTCATCA for GFP-EBNA1(S385A), and AGGAGTCCCAGTGCTCAGTCATCATCA for GFP-EBNA1(S386A). They were synthesized by Texas Genomics Japan (Tokyo, Japan). PCR was done using DNA polymerase KOD-Plus-201 (Toyobo, Osaka, Japan). Base sequences of PCR products were determined with an ABI PRISM 3100-Avant genetic analyzer (Applied Biosystems, Foster City, California).
Cell line, transfection, and laser scanning microscopy.
The cell line HeLa was obtained from the American Type Culture Collection through the Japanese Cancer Research Resources Bank (Japan) and was cultured in F-12 Ham's medium containing 10% fetal bovine serum. Transfection with plasmids expressing GFP fusion proteins of the EBNA-1 NLS and flanking sequence mutants was done with Lipofectin reagent (Invitrogen, California). Confocal laser scanning microscopy of transfected cells stained with Hoechst dye 33258 was described previously (29, 31). The mean and median values of percentages of nuclear GFP in 20 transfected cells per GFP-EBNA-1 mutant were calculated using Photoshop CS (Adobe Systems, California).
Microinjection assays.
Microinjection was carried out as previously described (58, 59). Briefly, GST-NLS-derived 10-amino-acid polypeptides tagged with GFP (3 mg/ml) were microinjected into the cytoplasm of HeLa cells grown on coverslips and incubated at 37°C. HeLa cells were fixed by 3.7% formaldehyde at the indicated times, and fluorescence of GFP was detected. bBSA conjugated with different peptides (3 mg/ml) was injected together with rhodamine isothiocyanate-BSA (50)into the HeLa cytoplasm and the cells were incubated at 37°C. HeLa cells were fixed and permeabilized at the indicated times, and the subcellular localization of conjugated proteins was detected by Alexa 488-labeled streptavidin (Molecular Probes) as described previously (58). The average fraction of GFP intensity present in the nuclei in three or four cells in each microscopic image was calculated using Photoshop CS (Adobe Systems, California).
Solution binding analyses of EBNA-1 NLS-derived peptides with karyopherin α proteins.
Recombinant karyopherin α proteins, Flag-NPI-1, Flag-Rch1, and Flag-Qip1, were described previously (58). bBSA proteins conjugated to EBNA-1 NLS-derived peptides were incubated with karyopherin α in the presence of avidin-agarose at 4°C for 2 h as described previously (58). After extensive washing, bound proteins were eluted, analyzed by SDS-PAGE, and stained with Coomassie brilliant blue (58).
Luciferase assays.
The effector plasmids pME18-EBNA1, pME18-EBNA1(379AAPA), pME18-EBNA1(383APAA), and pME18-EBNA1(383DPDD) were constructed on expression vector pME18S carrying the SRα promoter (constructed by K. Maruyama et al., Tokyo Medical and Dental University) so that they have the same EBNA-1 mutant sequences as those in the above-described GFP-EBNA1(379AAPA), GFP-EBNA1(383APAA), and GFP-EBNA1(383DPDD) plasmids, respectively. Activation of transcription from the oriP-BamHI C-Luc reporter (43), a kind gift from B. Sugden, was measured in HeLa cells; 1.0 μg of oriP-BamHI C-Luc and 1.0 μg of an effector were cotransfected into 1.0 × 105 or 2.0 × 105 cells per plate (60-mm diameter) using Lipofectamine (Invitrogen, Carlsbad, California). Luciferase assays were carried out using Promega's luciferase assay system (Promega, Madison, Wisconsin) according to the manufacturer's instruction.
RESULTS
EBNA-1 Lys379 and Arg380 are both essential for nuclear localization.
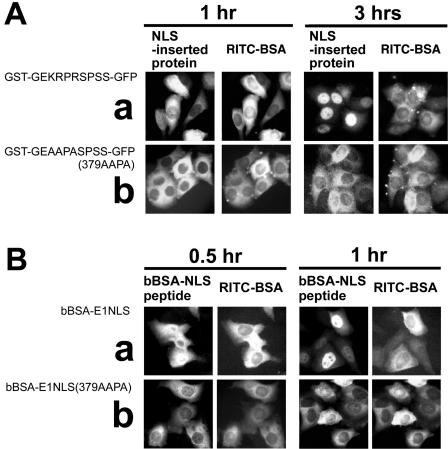
In order to identify amino acid residues in the EBNA-1 NLS that are essential for its activity, we performed point-mutation analyses. Microinjection experiments showed that a chimeric protein containing a wild-type EBNA-1 NLS, GST-GEKRPRSPSS-GFP, was transported into the nucleus with maximal accumulation within 3 h postinjection (Fig. 2A). In contrast, the nuclear import of a chimeric protein containing an NLS mutant with substitutions of Ala for the basic amino acids GST-GEAAPASPSS-GFP, was greatly reduced (Fig. 2A and Table 1). In addition, bBSA tagged with a synthetic EBNA-1 NLS peptide, bBSA-E1NLS, was transported into the nucleus 1 h postinjection (Fig. 2B). However, the nuclear import of bBSA tagged with the NLS mutant peptide with Ala substituted for the three basic amino acids, bBSA-E1NLS(379AAPA), was also greatly reduced (Fig. 2B; Table 2). In these assays, rhodamine isothiocyanate-BSA was added to the NLS sequence-inserted chimeric proteins or NLS peptide-tagged bBSA immediately before microinjection to serve as an internal injection marker in the same injected cell. The intracellular localization profiles of rhodamine isothiocyanate-BSA are also shown in Fig. 2.
FIG. 2.
Microinjection analyses of nuclear import directed by the EBNA-1 NLS and the mutant with basic amino acid residues changed to Ala (AAPASPSS). (A) EBNA-1 NLS-derived sequence-inserted chimeric proteins: (a) GST-GEKRPRSPSS-GFP, (b) GST-GEAAPASPSS-GFP(379AAPA). (B) bBSA molecules tagged with synthetic peptides of Ser-substituted EBNA-1 NLS: (a) bBSA-E1NLS, (b) bBSA-E1NLS(379AAPA).
TABLE 1.
Characteristics of nuclear transport directed by amino acid-substituted and Ser-phosphorylated forms of peptides derived from the EBNA-1 NLS assessed by microinjection and transfection analyses: nuclear transport of NLS-derived sequence-inserted proteinsa
| GST-NLS-derived peptide-GFP (amino acid sequence of NLS-derived peptide) | Nuclear GFP intensity in cells (%)
|
Nuclear transportb | |
|---|---|---|---|
| 1 h postmicroinjection | 2.5 or 3 h postmicroinjection | ||
| GST-GEKRPRSPSS-GFP | 28 (1.00) | 64 (1.00) | Wild type |
| GST-GEAAPASPSS-GFP(379AAPA) | 15 (0.54) | 18 (0.28) | Greatly reduced |
| GST-GEKRPRDPDD-GFP(383DPDD) | 22 (0.79) | 53 (0.83) | Wild type |
| GST-GEKRPRAPAA-GFP(383APAA) | 15 (0.54) | 14 (0.22) | Greatly reduced |
| GST-GEKRPRAPSS-GFP(S383A) | 22 (0.79) | 20 (0.31) | Greatly reduced |
| GST-GEKRPRSPAS-GFP(S385A) | 18 (0.64) | 36 (0.56) | Slightly reduced |
| GST-GEKRPRSPSA-GFP(S386A) | 23 (0.82) | 42 (0.66) | Slightly reduced |
| GST-GEKRPRSPAA-GFP(S385A, S386A) | 18 (0.64) | 25 (0.39) | Reduced |
| GST-GEKRPRDPSS-GFP(S383D) | 21 (0.75) | 16 (0.25) | Greatly reduced |
Nuclear transport was assayed at 1 h and 2.5 or 3 h postmicroinjection. The average fraction of GFP intensity in the nuclear areas of three or four cells in each photomicrograph in Fig. 2 and 4 is shown. The ratio of the fraction of GFP intensity in the nuclei of cells microinjected with each GST-NLS-derived peptide-GFP (NLS-derived peptide) to that of GST-GEKRPRSPSS-GFP is shown in parentheses.
The results are shown in terms of transport efficiency as follows. “Wild type” indicates that the nuclear transport rate is more than three-fourths that directed by the wild-type NLS GEKRPRSPSS; GST-wild-type NLS peptide-GFP proteins were transported fully into the nuclei 2.5 or 3 h postmicroinjection. “Greatly reduced” means that the nuclear transport rate of a GST-NLS-derived peptide-GFP protein was one-third or less than that directed by the wild-type NLS GEKRPRSPSS 2.5 or 3.0 h postmicroinjection. “Reduced” means that the nuclear transport rate of a GST-NLS-derived peptide-GFP protein was one-third to one-half of that directed by the wild-type NLS GEKRPRSPSS 2.5 or 3.0 h postmicroinjection. “Slightly reduced” means that the nuclear transport rate of a GST-NLS derived peptide-GFP protein was one-half to three-quarters of that directed by the wild-type NLS GEKRPRSPSS 2.5 or 3.0 h postmicroinjection.
TABLE 2.
Nuclear transport of bBSA cross-linked to synthetic peptides of phosphorylated forms of NLS and amino acid-substituted NLSa
| EBNA-1 NLS-derived peptide-conjugated bBSA | Amino acid sequence of NLS-derived peptides | Nuclear GFP intensity in cells (%)
|
Nuclear translocation rateb | Binding to NPI-1c | |
|---|---|---|---|---|---|
| 30 min postmicroinjection | 60 min postmicroinjection | ||||
| bBSA-E1NLS | GEKRPRSPSSQ | 33 (1.00) | 56 (1.00) | Wild type | + |
| bBSA-E1NLS1(379AAPA) | GEAAPASPSSQ | 19 (0.58) | 17 (0.30) | Reduced/greatly reduced | − |
| bBSA-E1NLS(383DPDD) | GEKRPRDPDDQ | 37 (1.12) | 50 (0.89) | Wild type | +++ |
| bBSA-E1NLS5(383APAA) | GEKRPRAPAAQ | 17 (0.52) | 19 (0.34) | Reduced | +/− |
| bBSA-E1NLS(pS383) | GEKRPRpSPSSQ | 26 (0.79) | 29 (0.52) | Slightly reduced | +/− |
| bBSA-E1NLS(S383A) | GEKRPRAPSSQ | 28 (0.85) | 35 (0.63) | Slightly reduced | +/− |
| bBSA-E1NLS(pS385) | GEKRPRSPpSSQ | 52 (1.58) | 50 (0.89) | Accelerated | +++ |
| bBSA-E1NLS(S385A) | GEKRPRSPASQ | 31 (0.94) | 31 (0.55) | Slightly reduced | +/− |
| bBSA-E1NLS(pS386) | GEKRPRSPSpSQ | 24 (0.73) | 42 (0.75) | Slightly reduced | +/− |
| bBSA-E1NLS(S386A) | GEKRPRSPSAQ | 43 (1.43) | 51 (0.91) | Accelerated | +/− |
| bBSA-E1NLS(S383D) | GEKRPRDPSSQ | 15 (0.46) | 16 (0.29) | Reduced/greatly reduced | +/− |
| bBSA-E1NLS(S385D) | GEKRPRSPDSQ | 44 (1.33) | 59 (1.05) | Accelerated | +/− |
Nuclear transport was assayed at 30 and 60 min postmicroinjection. The average fraction of GFP intensity in the nuclear areas of three or four cells in each photomicrograph in Fig. 2 and 5 is shown. The ratio of the fraction of GFP intensity in the nuclei of cells microinjected with each EBNA-1 NLS-derived peptide-conjugated bBSA to that of bBSA-E1NLS is shown in parentheses.
The results are shown in terms of transport efficiency as follows. “Wild type” indicates that the nuclear transport rate is more than three-quarters of that directed by the bBSA-E1NLS. “Accelerated” means that the nuclear transport rate of EBNA-1 NLS-derived peptide-conjugated bBSA proteins was ≥33% more 30 min postmicroinjection. “Greatly reduced” means that the nuclear transport rate of EBNA-1 NLS-derived peptide-conjugated bBSA proteins was one-third or less than that directed by the wild-type NLS GEKRPRSPSS 30 or 60 min postmicroinjection. “Reduced” means that the nuclear transport rate of EBNA-1 NLS-derived peptide-conjugated bBSA proteins was one-third to one-half of that directed by the wild-type NLS GEKRPRSPSS 30 or 60 min postmicroinjection. “Slightly reduced” means that the nuclear transport rate of EBNA-1 NLS-derived peptide-conjugated bBSA proteins was one-half to three-quarters of that directed by the wild-type NLS GEKRPRSPSS 30 or 60 min postmicroinjection.
Solution binding assays. +++, strong binding; +, weak binding; +/−, very weak binding; −, no binding.
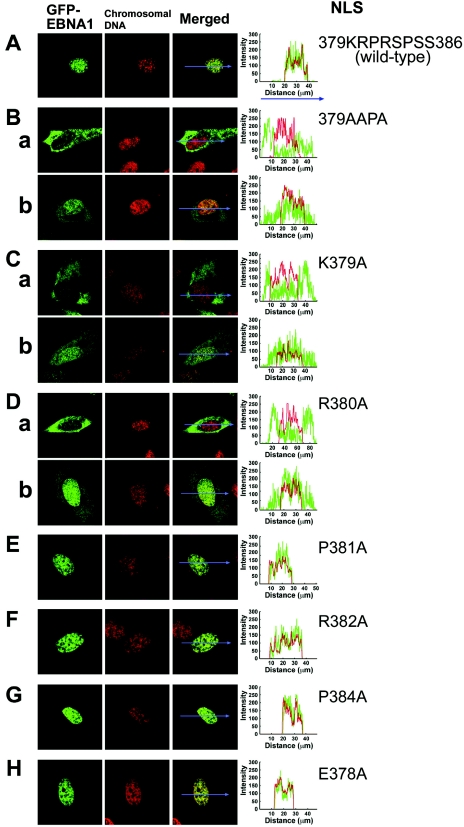
Transfection analyses of GFP-EBNA1 proteins with the basic amino acid-to-Ala substitutions in NLS were performed using confocal microscopy of living HeLa cells to analyze the effects of the mutation on the steady-state level of EBNA-1 in the nucleus. GFP-EBNA1 showed a highly nuclear localization profile (28-30), referred to as a “mostly nuclear localization profile” in this study (Fig. 3A). Nuclear GFP varied in the range of 55% to 100% among the 20 cells transfected with GFP-EBNA1 (mean, 90% ± 4% [standard deviation]; median, 98%) (Fig. 3A; Table 3). GFP-EBNA1(379AAPA) proteins, however, were highly localized in the cytoplasm (Fig. 3Ba) in the majority of cells (>75%) and in both the nucleus and cytoplasm in the remainder (<25%) (Fig. 3Bb) (mean, 21.8 ± 4%; median, 18%). It is to be noted that nuclear localization of GFP-EBNA1(379AAPA) is explained partially by its association with cellular chromosomes (25, 29, 48) during the nuclear envelope-disintegrated mitotic phase, as is the case with wild-type EBNA-1 (data not shown).
FIG. 3.
Confocal laser scanning microscopy of GFP fusion polypeptides of amino acid substitution NLS mutants in transfected cells. (A) GFP-EBNA1, (B) GFP-EBNA1(379AAPA), (C) GFP-EBNA1(K379A), (D) GFP-EBNA1(R380A), (E) GFP-EBNA1(P381A), (F) GFP-EBNA1(R382A), (G) GFP-EBNA1(P384A), (H) GFP-EBNA1(E378A). Chromosomal DNA was stained using Hoechst dye 33258. Localization profiles of GFP-EBNA1 mutants along straight lines drawn through the nucleus and cytoplasm are shown as graphs to the right of the photographs of transfected cells.
TABLE 3.
Intracellular localization characteristics of GFP fusion proteins of EBNA-1 with mutations in the NLS and C-terminal flanking sequences in transfected cells
| EBNA-1 mutant-GFP | Amino acid sequence | Subcellular localizationa
|
||
|---|---|---|---|---|
| Nuclear | Nuclear and cytoplasmic | Cytoplasmic | ||
| GFP-EBNA1 | −378EKRPRSPSS-QSSSSGSP-394 | Mostly | Some | |
| GFP-EBNA1(379AAPA) | −379AAPASPSS-QSSSSGSP-394 | +/− | <25% | >75% |
| GFP-EBNA1(K379A) | −379ARPRSPSS-QSSSSGSP-394 | +/− | >50% | <50% |
| GFP-EBNA1(R380A) | −379KAPRSPSS-QSSSSGSP-394 | +/− | >50% | <50% |
| GFP-EBNA1(R382A) | −379KRPASPSS-QSSSSGSP-394 | Mostly | Some | |
| GFP-EBNA1(E378A) | −378AKRPRSPSS-QSSSSGSP-394 | Mostly | +/− | |
| GFP-EBNA1(P381A) | −379KRARSPSS-QSSSSGSP-394 | Mostly | +/− | |
| GFP-EBNA1(P384A) | −379KRPRSASS-QSSSSGSP-394 | Mostly | Some | |
| GFP-EBNA1(383DPDD) | −379KRPRDPDD-QSSSSGSP-394 | Mostly | Some | |
| GFP-EBNA1(383APAA) | −379KRPRAPAA-QSSSSGSP-394 | Mostly | Some | |
| GFP-EBNA1(S383A) | −379KRPRAPSS-QSSSSGSP-394 | Mostly | Some | |
| GFP-EBNA1(S385A) | −379KRPRSPAS-QSSSSGSP-394 | Mostly | Some | |
| GFP-EBNA1(S386A) | −379KRPRSPSA-QSSSSGSP-394 | Mostly | Some | |
| GFP-EBNA1(388AAAAGA) | −379KRPRSPSS-QAAAAGAP-394 | Mostly | Some | |
GFP-wild type EBNA-1 was localized very highly in the nucleus in more than 95% of transfected cells. “Mostly” indicates that EBNA-1 wild type and mutants were highly localized in the nucleus, the nuclear-and-cytoplasmic localization profile indicates that GFP-EBNA1 mutant proteins were in both the nuclei and cytoplasm of the same cell, and the cytoplasmic localization profile indicates that GFP-EBNA1 mutant proteins were highly localized in the cytoplasm. “Some” indicates that the indicated localization profile of GFP-EBNA1 proteins was observed in some (<10%) of the transfected cells. Numbers indicate the percentage of transfected cells that showed the indicated localization profile. “+/−” indicates that GFP-EBNA-1 mutant proteins were localized in the nucleus of merely 5% or less of transfected cells, which is explained by the known association of EBNA-1 with mitotic cell chromosomes.
The single basic amino acid-substituted mutants GFP-EBNA1(K379A) (Fig. 3C) and GFP-EBNA1(R380A) (Fig. 3D) were found to localize primarily to the cytoplasm of <50% of transfected cells (Fig. 3Ca and Da) and to cytoplasm and nuclei in the remainder (Fig. 3Cb and Db); the mean nuclear GFP intensity in cells transfected with GFP-EBNA1(K379A) was 45 ± 6% (median, 43%). The mean nuclear GFP intensity in cells transfected with GFP-EBNA1(R380A) was similar (40 ± 7%; median, 43%). The other basic amino acid-substituted mutant GFP-EBNA1(R382A) proteins localized to the nucleus, to almost the same degree as wild-type EBNA-1 (Fig. 3F); the mean percentage of nuclear GFP intensity in cells transfected with GFP-EBNA1(R382A) was 84 ± 5% (median, 90%). The Pro381-substituted EBNA-1 NLS mutant, GFP-EBNA1(P381A) (Fig. 3E), and the Pro384-substituted GFP-EBNA1(P384A) mutant (Fig. 3G) also localized to the nucleus, as did wild-type EBNA-1; the mean nuclear GFP intensity in cells transfected with GFP-EBNA1(P381A) was 88 ± 2% (median, 91%) and that with GFP-EBNA1(P384A) was 94 ± 2% (median, 99%). The single amino acid substitution EBNA-1 NLS mutant GFP-EBNA1(E378A) also localized to the nucleus (Fig. 3H), like the wild-type (mean, 93 ± 4%; median, 99%). Localization profiles of GFP-EBNA1 mutants along straight lines arbitrarily drawn in the photomicrographs of transfected cells are included in Fig. 3 to show the relative intensities of GFP and Hoechst dye 33258 in the nucleus and cytoplasm.
These results indicate that the two basic amino acids, Lys379 and Arg380, are essential for nuclear localization, but Arg382, Pro381, Pro384, and Glu378 are not critical.
Microinjection shows that Ser383 and Ser385 residues of the EBNA-1 NLS are crucial for nuclear import of GST-NLS-GFP protein and NLS-tagged BSA.
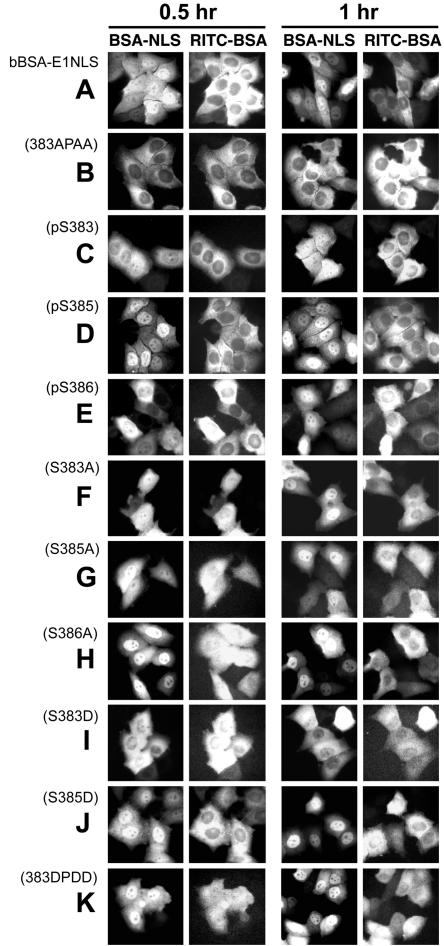
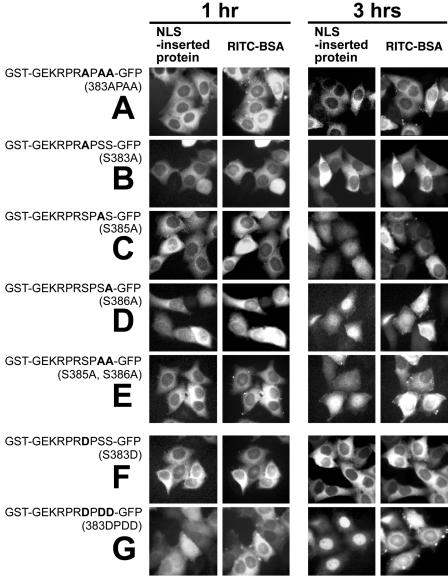
To test the effects of EBNA-1 NLS phosphorylation on nuclear localization, we analyzed nuclear import of NLS peptide-inserted GST-GFP chimeric proteins and bBSA cross-linked to the NLS peptide bBSA-E1NLS in HeLa cells by microinjection. GST-GEKRPRSPSS-GFP synthesized in Escherichia coli was not transported into the nucleus very quickly, but it had accumulated there 3 h after injection, as shown in Fig. 2Aa. bBSA-E1NLS was transported into the nucleus within 60 min (Fig. 5A). In contrast, the Ser-to-Ala substitution protein GST-GEKRPRAPAA-GFP(383APAA) was present in the nucleus only in small amounts 3.0 h postinjection (Fig. 4A; Table 1), and bBSA cross-linked to the Ser-to-Ala substitution NLS peptide, bBSA-E1NLS(383APAA), was also present only in small amounts 1.0 h postinjection (Fig. 5B; Table 2). The results indicate that Ser residues of the EBNA-1 NLS are crucial for efficient nuclear import of GST-NLS-GFP protein and NLS-tagged BSA.
FIG. 5.
Microinjection analyses of nuclear import of bBSA molecules tagged with synthetic peptides of Ser-substituted EBNA-1 NLS. (A) bBSA-E1NLS, (B) bBSA-E1NLS(383APAA), (C) bBSA-E1NLS(pS383), (D) bBSA-E1NLS(pS385), (E) bBSA-E1NLS(pS386), (F) bBSA-E1NLS(S383A), (G) bBSA-E1NLS(S385A), (H) bBSA-E1NLS(S386A), (I) bBSA-E1NLS(S383D), (J) bBSA-E1NLS(S385D), (K) bBSA-E1NLS(383DPDD).
FIG. 4.
Microinjection analyses of nuclear import of chimeric proteins containing a Ser-substituted NLS sequence. (A) GST-GEKRPRAPAA-GFP(383APAA), (B) GST-GEKRPRAPSS-GFP(S383A), (C) GST-GEKRPRSPAS-GFP(S385A), (D) GST-GEKRPRSPSA-GFP(S386A), (E) GST-GEKRPRSPAA-GFP(S385A, S386A), (F) GST-GEKRPRDPSS-GFP(S383D), (G) GST-GEKRPRDPDD-GFP(383DPDD).
Next, we separately analyzed the requirement for Ser383, Ser385, and Ser386 for nuclear import. GST-GEKRPRAPSS-GFP(S383A) was present in the nucleus only in small amounts 3.0 h postinjection (Fig. 4B; Table 1), suggesting that Ser 383 is crucial for efficient EBNA-1 nuclear import. bBSA cross-linked to the Ser383-to-Ala substitution NLS peptide, bBSA-E1NLS(S383A), was transported into the nucleus slightly more slowly than bBSA-E1NLS (Fig. 5F; Table 2), suggesting that multiples (7 to 10) of the Ser383-to-Ala substitution NLS peptide per bBSA molecule allowed bBSA-E1NLS(S383A) transport into the nucleus. GST-GEKRPRSPAS-GFP(S385A) and GST-GEKRPRSPSA-GFP(S386A) were transported into the nucleus at a slightly reduced rate (Fig. 4C and D; Table 1) and GST-GEKRPRSPAA-GFP(S385A, S386A) was transported at a more reduced rate (Fig. 4E; Table 1). bBSA-E1NLS(S385A) was also transported at a slightly reduced rate (Fig. 5G; Table 2), whereas bBSA-E1NLS(S386A) was transported into the nucleus even faster than bBSA-E1NLS (Fig. 5H; Table 2). Together, these results all suggest that Ser383 and Ser385 are important for nuclear translocation.
Phosphorylation of Ser residues in the EBNA-1 NLS regulates nuclear transport efficiency.
To analyze the impact of Ser phosphorylation on nuclear transport, synthetic peptides derived from the EBNA-1 NLS were cross-linked to bBSA, and nuclear import was assessed following microinjection. bBSA cross-linked to the NLS peptide, bBSA-E1NLS, was transported into the nucleus with almost maximal accumulation within 60 min (Fig. 5A).
bBSA tagged with the Ser385-phosphorylated peptide, bBSA-E1NLS(pS385), was more rapidly transported into the nucleus (within 30 min) than bBSA-E1NLS (Fig. 5D; Table 2). bBSA cross-linked to either the Ser383- or the Ser386-phosphorylated NLS peptide, bBSA-E1NLS(pS383) or bBSA-E1NLS(pS386), respectively, migrated more slowly into the nucleus than bBSA-E1NLS (Fig. 5C and E; Table 2). bBSA with the Ser385Asp substitution peptide, bBSA-E1NLS(S385D), was transported into the nucleus almost as rapidly as bBSA-E1NLS(pS385) (FIG. 5J; Table 2). GST-GEKRPRDPSS-GFP(S383D) was less efficiently transported than GST-GEKRPRSPSS-GFP (Fig. 4I), consistent with the slightly reduced nuclear transport rate of bBSA-E1NLS(pS383) (Fig. 5C; Table 2).
bBSA cross-linked to the NLS peptide in which all Ser at 383, 385, and 386 were replaced with Asp [bBSA-E1NLS(383DPDD)] was transported into the nucleus with an efficiency similar to bBSA-E1NLS (Fig. 5K; Table 2). The Ser-to-Asp substitution protein GST-GEKRPRDPDD-GFP, mimicking GST-KRPRpSPpSpS-GFP, was translocated to the nucleus within 3 h, similar to GST-GEKRPRSPSS-GFP (Fig. 4G; Table 1). These results possibly resulted from a balance between the effects of acceleration and deceleration of transport caused by phosphorylation of all three serine residues.
Ser-to-Ala substitution in the EBNA-1 NLS does not significantly affect nuclear accumulation levels of GFP-EBNA-1 in transfected cells.
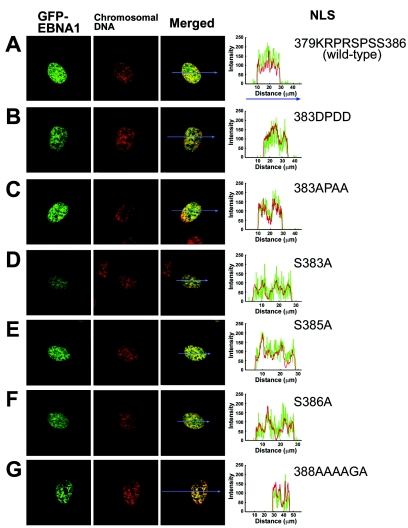
To analyze the effects on nuclear localization of NLS Ser phosphorylation, a set of GFP-EBNA-1 mutants with Ser-to-Ala or Ser-to-Asp substitutions in the 379KRPRSPSS386 sequence was constructed (Fig. 1C). Substitution of three Ser residues with Asp in the NLS [GFP-EBNA1(383DPDD)], mimicking simultaneous Ser phosphorylation, did not affect nuclear localization or accumulation levels (Fig. 6B), in agreement with the result of microinjection analysis of GST-GEKRPRDPDD-GFP(383DPDD) (Fig. 4G); the mean percentage of nuclear GFP in cells transfected with GFP-EBNA1(383DPDD) was 95 ± 3%, and the median was 99%.
FIG. 6.
Confocal laser scanning microscopy of cells transfected with GFP-EBNA1 mutants with Ser-to-Ala or Ser-to-Asp substitutions in the NLS or with Ser-to-Ala substitutions in the C-terminal flanking sequence. (A) GFP-EBNA1, (B) GFP-EBNA1(383DPDD), (C) GFP-EBNA1(383APAA), (D) GFP-EBNA1(S383A), (E) GFP-EBNA1(S385A), (F) GFP-EBNA1(S386A), (G) GFP-EBNA1(388AAAAGA). Chromosomal DNA was stained using Hoechst dye 33258. Localization profiles of GFP-EBNA1 mutants along straight lines drawn through the nucleus and cytoplasm are shown to the right of the photographs of transfected cells.
Surprisingly, GFP-EBNA1(383APAA), GFP-EBNA1(S383A), and GFP-EBNA1(S385A) also did not reduce nuclear accumulation levels (Fig. 6C to E). The mean percentages of nuclear GFP intensity in cells transfected with GFP-EBNA1(383APAA), GFP-EBNA1(S383A), and GFP-EBNA1(S385A) were 90 ± 5%, 89 ± 3%, and 90 ± 6% (median values, 96, 99, and 97%, respectively). GFP-EBNA1(S386A), which was included here for comparison, did not affect nuclear accumulation levels, as expected (Fig. 6F); mean nuclear GFP intensity in cells transfected with GFP-EBNA1(S386A) was 93 ± 3% (median, 99%). These results indicate that Ser at amino acid 383, 385, or 386 is not essential for nuclear accumulation of EBNA-1 to a high level.
We also asked whether the carboxyl-adjacent Ser-rich 387QSSSSGS393 sequence is involved in regulating the level of EBNA-1 in the nucleus. However, GFP-EBNA1(388AAAAGA) with Ser-to-Ala substitutions at all five Ser residues in the carboxyl-adjacent 387QSSSSGS393 (Fig. 6G) showed a localization profile similar to that of GFP-EBNA1; the mean proportion of nuclear GFP intensity in cells transfected with GFP-EBNA1(388AAAAGA) was 93 ± 9% (median, 96%). This indicates that phosphorylation of Ser residues in the QSSSSGSP sequence is not required for nuclear accumulation of EBNA-1.
Analyses of karyopherin α interactions with phosphorylated and nonphosphorylated forms of EBNA-1 NLS.
We previously reported that the karyopherin α NPI-1 (importin α5) binds to EBNA-1 (30) as well as the karyopherin α Rch1 (importin α1) (14, 30, 42). To examine the effects of phosphorylation of and amino acid substitutions in the EBNA-1 NLS, we analyzed interactions of NLS mutant peptides with recombinant NPI-1 (importin α5), Rch1 (importin α1), and Qip1 (importin α3) (61) using the biotin-avidin affinity binding assays.
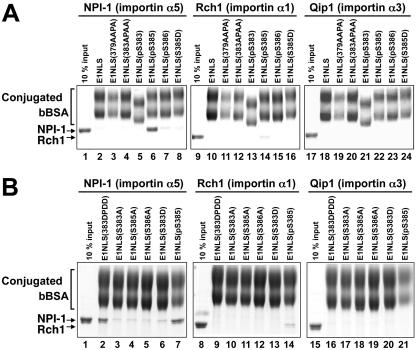
NPI-1 did not bind to bBSA-E1NLS(379AAPA) (Fig. 7A), consistent with failure of nuclear import of bBSA-E1NLS(379AAPA) (Fig. 2B). NPI-1 bound to bBSA-E1NLS(pS385) much more strongly than to bBSA-E1NLS (Fig. 7A), a finding in agreement with the accelerated nuclear import rate of bBSA-E1NLS(pS385) (Table 2). In contrast, NPI-1 bound to bBSA-E1NLS(pS383) and bBSA-E1NLS(pS386) only weakly (Fig. 7A; Table 2). NPI-1 bound to bBSA-E1NLS(383APAA) more weakly than to bBSA-E1NL (Fig. 7A), in agreement with its greatly reduced nuclear import rate (Table 4). These data agree with our previous findings that EBNA-1 protein synthesized in Escherichia coli interacted with NPI-1 much less efficiently than EBNA-1 synthesized in Raji cells (30) and indicate that phosphorylation of S385, but neither S383 nor S386, is required for the interaction of EBNA-1 with NPI-1. NPI-1 bound to bBSA-E1NLS(S385D) and bBSA-E1NLS(S383D) only weakly, but bound to bBSA-E1NLS(383DPDD) as strongly as bBSA-E1NLS(pS385) (Fig. 7B). NPI-1 bound to bBSA-E1NLS(S383A), bBSA-E1NLS(S385A) and bBSA-E1NLS(S386A) only weakly (Fig. 7B; Table 2).
FIG. 7.
Solution-binding assay of interactions of phosphorylated forms and basic amino acids to Ala-substituted forms of EBNA-1 NLS peptides with karyopherin α proteins. (A) Synthetic peptide-cross-linked bBSA proteins bBSA-E1NLS (wild type), bBSA-E1NLS(379AAPA), bBSA-E1NLS(383APAA), bBSA-E1NLS(pS383), bBSA-E1NLS(pS385), bBSA-E1NLS(pS386), and bBSA-E1NLS(S385D), indicated on top of the lanes, were incubated with NPI-1 (importin α5) (lanes 2 to 8), Rch1 (importin α1) (lanes 10 to 16), or Qip1 (importin α3) (lanes 18 to 24) in the presence of avidin-agarose. The bound proteins were eluted and analyzed by SDS-PAGE. Controls are a 10% input of NPI-1 (lane 1), Rch1 (lane 9) and Qip1 (lane 17). (B) Synthetic peptide-cross-linked bBSA proteins bBSA-E1NLS(383DPDD), bBSA-E1NLS(S383A), bBSA-E1NLS(S385A), bBSA-E1NLS(S386A), bBSA-E1NLS(S383D), and bBSA-E1NLS(pS385), indicated at the top of the lanes, were incubated with NPI-1 (importin α5) (lanes 2 to 7), Rch1 (importin α1) (lanes 8 to 14), or Qip1 (importin α3) (lanes 15 to 21) in the presence of avidin-agarose. The bound proteins were eluted and analyzed by SDS-PAGE. Controls are a 10% input of NPI-1 (lane 1), Rch1 (lane 8), and Qip1(lane 15).
TABLE 4.
Activation of transcription from the BamHI-C promoter by the EBNA-1 NLS mutants EBNA1(379AAPA), EBNA1(383APAA), and EBNA1(383DPDD) examined by cotransfection luciferase assaysc
| Effector | Relative fold transactivationa
|
||||||
|---|---|---|---|---|---|---|---|
| Expt 1
|
Expt 2
|
||||||
| 24 h | 48 h | 72 h | 24 h | 48 h | 72 h | 96 h | |
| Vector | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| EBNA-1 | 3.33 | 13.4 | 20.23 | 3.92 | 30.15 | 44.99 | 60.46 |
| EBNA1(379AAPA) | 2.32 | 6.55 | 6.51 | 3.48 | 7.42 | 23.69 | 27.02 |
| EBNA1(383APAA) | 3.29 | 13.9 | 19.6 | 4.14 | 28.63 | 43.50 | 60.28 |
| EBNA1(383DPDD) | 3.42 | 13.11 | 20.1 | 3.64 | 30.38 | 40.94 | 58.32 |
Relative fold transactivation is shown as the average for duplicate plates for each effector harvested at the indicated time. The cell numbers at the time of cotransfection were 105 cells and 2.0 × 105 cells per plate in experiments 1 and 2, respectively.
Interaction of Rch1 with bBSA-E1NLS was minimal (Fig. 7A). Rch1 also bound more strongly to bBSA-E1NLS(pS385) than to bBSA-E1NLS(pS383) and bBSA-E1NLS(pS386) (Fig. 7A). However, Rch1 binding to bBSA-E1NLS(pS385) was much weaker than NPI-1 binding (Fig. 7A). Also, the interaction of Rch1 with bBSA cross-linked to the other EBNA-1 NLS-derived peptides was very weak (Fig. 7A and B). Interactions of Qip1 with any of these EBNA-1 NLS-derived peptides were absent (Fig. 7A and B).
Effects of NLS mutations on the activation of transcription from the BamHI C promoter by EBNA-1 expressed transiently in cotransfection luciferase assays.
To test whether the above described regulation of nuclear import efficiency of EBNA-1 is involved in its multifunctional properties, we first performed DNA replication/retention assays by cotransfection of plasmid pME18-EBNA1 with oriP-BamHI C-Luc or oriP-minus (43), which were kind gifts from B. Sugden (University of Wisconsin), and by growing HeLa cells. However, the activity of transiently expressed EBNA-1(ΔGA), the expression of which was confirmed by Western blotting performed as previously described (31), to support oriP-plasmid once per cell cycle replication/retention was too low to detect a de novo-synthesized DpnI-resistant band of linearized oriP-BamHI C-Luc by Southern blotting, the detection sensitivity of which was 2.04 copies per cell (0.01 ng per lane of gel electrophoresis) using a 32P-labeled probe (1.9 × 109 cpm/μg) (data not shown). In parallel analyses, in HeLa S3#8 cells that constitutively express full-length-EBNA-1 (N. Yoshizawa-Sugata and H. Masai, unpublished) a de novo-synthesized oriP-BamHI C-Luc band was detected (data not shown).
To analyze whether the nuclear import regulation of EBNA-1 affects the activation of transcription by EBNA-1, we constructed plasmids pME18-EBNA1, pME18-EBNA1(379AAPA), pME18-EBNA1(383APAA), and pME18-EBNA1(383DDPD) to assess the effects of the NLS mutation in the absence of GFP. Luciferase assays using HeLa cells cotransfected with the oriP-BamHI C-Luc reporter plasmid (43) (kindly provided by B. Sugden) and pME18-EBNA1(379AAPA) showed that the Ala substitution mutation 379AAPA, which greatly reduces nuclear import and localization of EBNA-1, caused a significant reduction in the activation of transcription from the BamHI C-Luc reporter at 48, 72, and 96 h after transfection, compared to pME18-EBNA1 and pME18-EBNA1(383DPDD) (Table 4). However, pME18-EBNA-1(383APAA), which greatly reduces the efficiency of nuclear import of EBNA-1, did not reduce the activation of transcription from the BamHI C-Luc reporter (Table 4). These results are consistent with the finding that the 383APAA Ala substitution mutation did not reduce the nuclear intensity of GFP-EBNA1 (Fig. 6C to E). Further, they indicate that under the present experimental conditions of transient expression of EBNA-1 in cotransfection assays, the Ser-to-383APAA mutation that causes the great reduction in the nuclear transport efficiency of EBNA-1 proteins does not affect the transcriptional activator function of EBNA-1.
DISCUSSION
Regulation of nuclear transport by phosphorylation of sites that are adjacent to NLSs, but not within them, has been reported previously (16, 32-34, 37). The best studied example is that the nuclear import of simian virus 40 T antigen directed by the NLS (126PKKKRKV132) is enhanced by phosphorylation of S112 (34), but is inhibited by phosphorylation of T124 (35, 55, 62). The findings of the present study are summarized in Tables 1 to 3, and the major conclusions are as follows. First, our data suggest that only the amino-terminal 379Lys-Arg380 is essential for both nuclear transport and nuclear accumulation directed by the EBNA-1 NLS and for interaction with the karyopherin α NPI-1 protein. This remarkable variability of the N-terminal and C-terminal sequences flanking the EBNA-1 NLS 379KR380 is similar to the simian virus 40 T antigen NLS (amino acids 126 to 132) in which the Lys residue at position 128 is essential for NLS integrity (6, 38). Crystallographic studies of complexes of karyopherin α and NLS peptides have shown that the basic residues of the simian virus 40 large T (8, 9) and c-Myc NLSs (8, 15) interact with the binding pockets of the major binding site of the karyopherin α molecule. In contrast, no Lys in the downstream cluster of nucleoplasmin (167KKKK170) can be substituted (56). It is not yet clear whether any Lys or Arg residues in the smallest known NLS, KRPRP, can be mutated without loss of function.
Second, the Ser residues of 383SPSS386 in the EBNA-1 NLS are important for efficient nuclear translocation, as shown by microinjection experiments.
Third, Ser383 is crucial for efficient nuclear import, because replacement of Ser383 with either Ala, Asp, or phosphoserine reduced nuclear import. Ser386 is not essential for nuclear transport, but phosphorylation of Ser at position 386 reduces nuclear translocation efficiency, as demonstrated by microinjection analyses.
Fourth, Ser385 is important for nuclear transport and Ser385 phosphorylation accelerates the rate of EBNA-1 nuclear translocation.
Fifth, a reduction of nuclear transport rate does not result in a decrease in the intensity of nuclear GFP-EBNA-1 relative to that of cytoplasmic EBNA-1. The results described in this report suggest that nuclear levels of EBNA-1 are regulated not only by import rate but also by the retention of EBNA-1 in the nucleus due to the association of EBNA-1 with cellular chromosomes and chromatin (29, 48).
Sixth, Ser385 phosphorylation in the EBNA-1 NLS increases the interactions of EBNA-1 with the karyopherin α NPI-1 protein. Rch1, on the other hand, bound to EBNA-1 NLS peptides, either phosphorylated or nonphosphorylated, far less strongly than NPI-1, and Qip1 did not bind to any of the EBNA-1 NLS peptides. The very weak binding of NPI-1 to bBSA-E1NLS(S385D) indicates that an effect of phosphorylation of serine at position 385 on the interaction of E1NLS with NPI-1 is highly specific. The finding that NPI-1 bound to bBSA-E1NLS(383DPDD) as strongly as bBSA-E1NLS(pS385) but without resulting in accelerated nuclear transport rates of bBSA-E1NLS(383DPDD) indicates that the strong binding of NPI-1 to the EBNA-1 NLS is not sufficient for accelerated nuclear import. These data need emphasizing because it was previously reported that phosphorylation of Ser112 in the vicinity of the NLS (126PKKKRKV132) of simian virus 40 T antigen enhanced nuclear import but not the affinity of binding between the T-antigen NLS and importin α2 (16). These results indicate that NPI-1 is a major karyopherin α member that mediates the nuclear import of EBNA-1 and that phosphorylation of S385 accelerates the nuclear import rate through an increased affinity of NLS for NPI-1.
Lastly, the great decrease in the efficiency of nuclear import of EBNA-1(383APAA) did not reduce activation of transcription from the BamHI C-Luc promoter, in contrast to EBNA-1(379AAPA). However, the high transient expression level of the mutant EBNA-1 proteins may have hindered detection of any possible effects of the reduced nuclear transport efficiency of EBNA-1 on the activation of transcription. Functional effects of the reduced nuclear transport efficiency of EBNA-1 may be complex because of stable colocalization of EBNA-1 with cellular chromosomes and chromatin (29) and may act in concert with other cellular factors, such as cell cycle position or infection with EBV. Furthermore, the regulation of nuclear import efficiency by phosphorylation of the EBNA-1 NLS may be correlated with phosphorylation of other Ser residues of EBNA-1 outside the NLS (22, 52) and required for adequate functional association of EBNA-1 to cellular proteins in EBV-infected cells.
From the present study, we conclude that the amino-terminal 379Lys-Arg380 and Ser383 are the essential elements of the EBNA-1 NLS (amino acids 379 to 386) required for function. It has been found that phosphorylation of Ser385 increases nuclear transport efficiency of EBNA-1 by modulating its affinity to NPI-1, whereas phosphorylation of Ser383 or Ser386 decreases it. This is the first study to suggest any role for Ser phosphorylation in the context of EBNA-1. The results suggest dynamic transport control of phosphorylated EBNA-1 proteins, whereas the steady-state nuclear level of EBNA-1 seems unchanged.
Acknowledgments
We thank Bill Sugden, University of Wisconsin, for his kind gift of the oriP-BamHI C-Luc (1033) and oriP-minus (1381) plasmids (43).
Financial support for this research was provided by a grant-in-aid from the Ministry of Health, Welfare and Labor.
REFERENCES
- 1.Ambinder, R. F., M. A. Mullen, Y. N. Chang, G. S. Hayward, and S. D. Hayward. 1991. Functional domains of Epstein-Barr virus nuclear antigen EBNA-1. J. Virol. 65:1466-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bochkarev, A., J. A. Barwell, R. A. Pfuetzner, E. Bochkareva, L. Frappier, and A. M. Edwards. 1996. Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin-binding protein, EBNA1, bound to DNA. Cell 84:791-800. [DOI] [PubMed] [Google Scholar]
- 3.Bochkarev, A., J. A. Barwell, R. A. Pfuetzner, W. Furey, Jr., A. M. Edwards, and L. Frappier. 1995. Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin-binding protein EBNA 1. Cell 83:39-46. [DOI] [PubMed] [Google Scholar]
- 4.Ceccarelli, D. F., and L. Frappier. 2000. Functional analyses of the EBNA1 origin DNA binding protein of Epstein-Barr virus. J. Virol. 74:4939-4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chelsky, D., R. Ralph, and G. Jonak. 1989. Sequence requirements for synthetic peptide-mediated translocation to the nucleus. Mol. Cell. Biol. 9:2487-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colledge, W. H., W. D. Richardson, M. D. Edge, and A. E. Smith. 1986. Extensive mutagenesis of the nuclear location signal of simian virus 40 large-T antigen. Mol. Cell. Biol. 6:4136-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conti, E. 2002. Structures of importins. Results Probl. Cell Differ. 35:93-113. [DOI] [PubMed] [Google Scholar]
- 8.Conti, E., and J. Kuriyan. 2000. Crystallographic analysis of the specific yet versatile recognition of distinct nuclear localization signals by karyopherin alpha. Structure Fold Des. 8:329-338. [DOI] [PubMed] [Google Scholar]
- 9.Conti, E., M. Uy, L. Leighton, G. Blobel, and J. Kuriyan. 1998. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell 94:193-204. [DOI] [PubMed] [Google Scholar]
- 10.Dang, C. V., and W. M. Lee. 1989. Nuclear and nucleolar targeting sequences of c-erb-A, c-myb, N-myc, p53, HSP70, and HIV tat proteins. J. Biol. Chem. 264:18019-18023. [PubMed] [Google Scholar]
- 11.Dingwall, C., and R. A. Laskey. 1998. Nuclear import: a tale of two sites. Curr. Biol. 8:R922-924. [DOI] [PubMed] [Google Scholar]
- 12.Dingwall, C., and R. A. Laskey. 1991. Nuclear targeting sequences—a consensus? Trends Biochem. Sci. 16:478-481. [DOI] [PubMed] [Google Scholar]
- 13.Eda, H., Y. Ishii, M. Obayashi, S. Harada, S. Ito, T. Fujita, M. Ikeda, S. Kusano, R. Kitamura, C. Suzuki, T. Hara, M. Watanabe, H. Satoh, K. Sugihara, and K. Yanagi. 2005. Monoclonal antibodies against regions topologically surrounding the homodimeric beta-barrel interface of Epstein-Barr virus nuclear antigen-1. Virus Res. 109:87-94. [DOI] [PubMed] [Google Scholar]
- 14.Fischer, N., E. Kremmer, G. Lautscham, N. Mueller-Lantzsch, and F. A. Grasser. 1997. Epstein-Barr virus nuclear antigen 1 forms a complex with the nuclear transporter karyopherin alpha2. J. Biol. Chem. 272:3999-4005. [DOI] [PubMed] [Google Scholar]
- 15.Fontes, M. R., T. Teh, and B. Kobe. 2000. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-alpha. J. Mol. Biol. 297:1183-1194. [DOI] [PubMed] [Google Scholar]
- 16.Fontes, M. R., T. Teh, G. Toth, A. John, I. Pavo, D. A. Jans, and B. Kobe. 2003. Role of flanking sequences and phosphorylation in the recognition of the simian-virus-40 large T-antigen nuclear localization sequences by importin-alpha. Biochem. J. 375:339-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujita, T., M. Ikeda, S. Kusano, M. Yamazaki, S. Ito, M. Obayashi, and K. Yanagi. 2001. Amino acid substitution analyses of the DNA contact region, two amphipathic alpha-helices and a recognition-helix-like helix outside the dimeric beta-barrel of Epstein-Barr virus nuclear antigen 1. Intervirology 44:271-282. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Bustos, J., J. Heitman, and M. N. Hall. 1991. Nuclear protein localization. Biochim. Biophys. Acta 1071:83-101. [DOI] [PubMed] [Google Scholar]
- 19.Grogan, E. A., W. P. Summers, S. Dowling, D. Shedd, L. Gradoville, and G. Miller. 1983. Two Epstein-Barr viral nuclear neoantigens distinguished by gene transfer, serology, and chromosome binding. Proc. Natl. Acad. Sci. USA 80:7650-7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammerschmidt, W., and B. Sugden. 2004. Epstein-Barr virus sustains Burkitt's lymphomas and Hodgkin's disease. Trends Mol. Med. 10:331-336. [DOI] [PubMed] [Google Scholar]
- 21.Harada, S., Y. Kamata, Y. Ishii, H. Eda, R. Kitamura, M. Obayashi, S. Ito, F. Ban, J. Kuranari, H. Nakajima, T. Kuze, M. Hayashi, N. Okabe, H. Senpuku, N. Miyasaka, Y. Nakamura, H. Kanegane, and K. Yanagi. 2004. Maintenance of serum immunoglobulin G antibodies to Epstein-Barr virus (EBV) nuclear antigen 2 in healthy individuals from different age groups in a Japanese population with a high childhood incidence of asymptomatic primary EBV infection. Clin. Diagn. Lab Immunol. 11:123-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hearing, J. C., and A. J. Levine. 1985. The Epstein-Barr virus nuclear antigen (BamHI K antigen) is a single-stranded DNA binding phosphoprotein. Virology 145:105-116. [DOI] [PubMed] [Google Scholar]
- 23.Henle, W., G. Henle, J. Andersson, I. Ernberg, G. Klein, C. A. Horwitz, G. Marklund, L. Rymo, C. Wellinder, and S. E. Straus. 1987. Antibody responses to Epstein-Barr virus-determined nuclear antigen (EBNA)-1 and EBNA-2 in acute and chronic Epstein-Barr virus infection. Proc. Natl. Acad. Sci. USA 84:570-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Humme, S., G. Reisbach, R. Feederle, H. J. Delecluse, K. Bousset, W. Hammerschmidt, and A. Schepers. 2003. The EBV nuclear antigen 1 (EBNA1) enhances B cell immortalization several thousandfold. Proc. Natl. Acad. Sci. USA 100:10989-10994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hung, S. C., M. S. Kang, and E. Kieff. 2001. Maintenance of Epstein-Barr virus (EBV) oriP-based episomes requires EBV-encoded nuclear antigen-1 chromosome-binding domains, which can be replaced by high-mobility group-I or histone H1. Proc. Natl. Acad. Sci. USA 98:1865-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Imamoto, N., T. Tachibana, M. Matsubae, and Y. Yoneda. 1995. A karyophilic protein forms a stable complex with cytoplasmic components prior to nuclear pore binding. J. Biol. Chem. 270:8559-8565. [DOI] [PubMed] [Google Scholar]
- 27.Inoue, N., S. Harada, T. Honma, T. Kitamura, and K. Yanagi. 1991. The domain of Epstein-Barr virus nuclear antigen 1 essential for binding to oriP region has a sequence fitted for the hypothetical basic-helix-loop-helix structure. Virology 182:84-93. [DOI] [PubMed] [Google Scholar]
- 28.Ito, S., H. Eda, F. Ban, and K. Yanagi. 2003. Epstein-Barr virus nuclear antigen-1 colocalizes with lamin B1 in the nucleoplasm and along the nuclear rim. Arch. Virol. 148:1633-1642. [DOI] [PubMed] [Google Scholar]
- 29.Ito, S., E. Gotoh, S. Ozawa, and K. Yanagi. 2002. Epstein-Barr virus nuclear antigen-1 is highly colocalized with interphase chromatin and its newly replicated regions in particular. J. Gen. Virol. 83:2377-2383. [DOI] [PubMed] [Google Scholar]
- 30.Ito, S., M. Ikeda, N. Kato, A. Matsumoto, Y. Ishikawa, S. Kumakubo, and K. Yanagi. 2000. Epstein-Barr virus nuclear antigen-1 binds to nuclear transporter karyopherin alpha1/NPI-1 in addition to karyopherin alpha2/Rch1. Virology 266:110-119. [DOI] [PubMed] [Google Scholar]
- 31.Ito, S., and K. Yanagi. 2003. Epstein-Barr virus (EBV) nuclear antigen 1 colocalizes with cellular replication foci in the absence of EBV plasmids. J. Virol. 77:3824-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jans, D. A. 1995. The regulation of protein transport to the nucleus by phosphorylation. Biochem. J. 311:705-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jans, D. A., and S. Hubner. 1996. Regulation of protein transport to the nucleus: central role of phosphorylation. Physiol. Rev. 76:651-685. [DOI] [PubMed] [Google Scholar]
- 34.Jans, D. A., and P. Jans. 1994. Negative charge at the casein kinase II site flanking the nuclear localization signal of the SV40 large T-antigen is mechanistically important for enhanced nuclear import. Oncogene 9:2961-2968. [PubMed] [Google Scholar]
- 35.Jans, D. A., C. Y. Xiao, and M. H. Lam. 2000. Nuclear targeting signal recognition: a key control point in nuclear transport? Bioessays 22:532-544. [DOI] [PubMed] [Google Scholar]
- 36.Jiang, W. Q., W.-H. V. Lundkvist, A. Ringertz, N. Klein, and G. Rosen. 1991. Intranuclear distribution of Epstein-Barr virus-encoded nuclear antigens EBNA-1, -2, -3 and -5. J. Cell Sci. 99:497-502. [DOI] [PubMed] [Google Scholar]
- 37.Kaffman, A., and E. K. O'Shea. 1999. Regulation of nuclear localization: a key to a door. Annu. Rev. Cell Dev. Biol. 15:291-339. [DOI] [PubMed] [Google Scholar]
- 38.Kalderon, D., W. D. Richardson, A. F. Markham, and A. E. Smith. 1984. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature 311:33-38. [DOI] [PubMed] [Google Scholar]
- 39.Kalderon, D., B. L. Roberts, W. D. Richardson, and A. E. Smith. 1984. A short amino acid sequence able to specify nuclear location. Cell 39:499-509. [DOI] [PubMed] [Google Scholar]
- 40.Kennedy, G., J. Komano, and B. Sugden. 2003. Epstein-Barr virus provides a survival factor to Burkitt's lymphomas. Proc. Natl. Acad. Sci. USA 100:14269-14274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kieff, E., and A. B. Rickinson. 2001. Epstein-Barr virus and its replication, p. 2511-2573. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 42.Kim, A. L., M. Maher, J. B. Hayman, J. Ozer, D. Zerby, J. L. Yates, and P. M. Lieberman. 1997. An imperfect correlation between DNA replication activity of Epstein-Barr virus nuclear antigen 1 (EBNA1) and binding to the nuclear import receptor, Rch1/importin alpha. Virology 239:340-351. [DOI] [PubMed] [Google Scholar]
- 43.Kirchmaier, A. L., and B. Sugden. 1997. Dominant-negative inhibitors of EBNA-1 of Epstein-Barr virus. J. Virol. 71:1766-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kusano, S., K. Tamada, H. Senpuku, S. Harada, S. Ito, and K. Yanagi. 2001. Epstein-Barr virus nuclear antigen-1-dependent and -independent oriP-binding cellular proteins. Intervirology 44:283-290. [DOI] [PubMed] [Google Scholar]
- 45.Leight, E. R., and B. Sugden. 2000. EBNA-1: a protein pivotal to latent infection by Epstein-Barr virus. Rev. Med. Virol. 10:83-100. [DOI] [PubMed] [Google Scholar]
- 46.Leight, E. R., and B. Sugden. 2001. Establishment of an oriP replicon is dependent upon an infrequent, epigenetic event. Mol. Cell. Biol. 21:4149-4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lyons, R. H., B. Q. Ferguson, and M. Rosenberg. 1987. Pentapeptide nuclear localization signal in adenovirus E1a. Mol. Cell. Biol. 7:2451-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marechal, V., A. Dehee, R. Chikhi-Brachet, T. Piolot, M. Coppey-Moisan, and J. C. Nicolas. 1999. Mapping EBNA-1 domains involved in binding to metaphase chromosomes. J. Virol. 73:4385-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mattaj, I. W., and L. Englmeier. 1998. Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 67:265-306. [DOI] [PubMed] [Google Scholar]
- 50.McDonagh, P. F., and S. K. Williams. 1984. The preparation and use of fluorescent-protein conjugates for microvascular research. Microvasc. Res. 27:14-27. [DOI] [PubMed] [Google Scholar]
- 51.Petti, L., C. Sample, and E. Kieff. 1990. Subnuclear localization and phosphorylation of Epstein-Barr virus latent infection nuclear proteins. Virology 176:563-574. [DOI] [PubMed] [Google Scholar]
- 52.Polvino-Bodnar, M., J. Kiso, and P. A. Schaffer. 1988. Mutational analysis of Epstein-Barr virus nuclear antigen 1 (EBNA 1). Nucleic Acids Res. 16:3415-3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Richardson, W. D., B. L. Roberts, and A. E. Smith. 1986. Nuclear location signals in polyoma virus large-T. Cell 44:77-85. [DOI] [PubMed] [Google Scholar]
- 54.Rickinson, A. B., and E. Kieff. 2001. Epstein-Barr virus, p. 2575-2627. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 55.Rihs, H. P., D. A. Jans, H. Fan, and R. Peters. 1991. The rate of nuclear cytoplasmic protein transport is determined by the casein kinase II site flanking the nuclear localization sequence of the SV40 T-antigen. EMBO J. 10:633-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robbins, J., S. M. Dilworth, R. A. Laskey, and C. Dingwall. 1991. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell 64:615-623. [DOI] [PubMed] [Google Scholar]
- 57.Roberts, B. 1989. Nuclear location signal-mediated protein transport. Biochim. Biophys. Acta 1008:263-280. [DOI] [PubMed] [Google Scholar]
- 58.Sekimoto, T., M. Fukumoto, and Y. Yoneda. 2004. 14-3-3 suppresses the nuclear localization of threonine 157-phosphorylated p27(Kip1). EMBO J. 23:1934-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sekimoto, T., N. Imamoto, K. Nakajima, T. Hirano, and Y. Yoneda. 1997. Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1. EMBO J. 16:7067-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tachibana, T., M. Hieda, T. Sekimoto, and Y. Yoneda. 1996. Exogenously injected nuclear import factor p10/NTF2 inhibits signal-mediated nuclear import and export of proteins in living cells. FEBS Lett. 397:177-182. [DOI] [PubMed] [Google Scholar]
- 61.Tsuji, L., T. Takumi, N. Imamoto, and Y. Yoneda. 1997. Identification of novel homologues of mouse importin alpha, the alpha subunit of the nuclear pore-targeting complex, and their tissue-specific expression. FEBS Lett. 416:30-34. [DOI] [PubMed] [Google Scholar]
- 62.Vancurova, I., J. Jochova-Rupes, W. Lou, and P. L. Paine. 1995. Distinct phosphorylation sites differentially influence facilitated transport of an NLS-protein and its subsequent intranuclear binding. Biochem. Biophys. Res. Commun. 217:419-427. [DOI] [PubMed] [Google Scholar]
- 63.Weis, K. 2002. Nucleocytoplasmic transport: cargo trafficking across the border. Curr. Opin. Cell Biol. 14:328-335. [DOI] [PubMed] [Google Scholar]
- 64.Yamazaki, M., R. Kitamura, S. Kusano, H. Eda, S. Sato, M. Okawa-Takatsuji, S. Aotsuka, and K. Yanagi. 2005. Elevated immunoglobulin G antibodies to the proline-rich amino-terminal region of Epstein-Barr virus nuclear antigen-2 in sera from patients with systemic connective tissue diseases and from a subgroup of Sjogren's syndrome patients with pulmonary involvements. Clin. Exp. Immunol. 139:558-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yates, J. L., and S. M. Camiolo. 1988. Dissection of DNA replication and enhancer activation function of Epstein-Barr virus nuclear antigen 1. Cancer Cells 6:197-205. [Google Scholar]
- 66.Yates, J. L., N. Warren, and B. Sugden. 1985. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature 313:812-815. [DOI] [PubMed] [Google Scholar]