Abstract
After identifying the interaction between the transcriptional coactivator lens epithelium-derived growth factor (LEDGF/p75) and the human immunodeficiency virus type 1 (HIV-1) integrase (IN), we have now investigated the role of LEDGF/p75 during HIV replication. Transient small interfering RNA-mediated knockdown of LEDGF/p75 in HeLaP4 cells resulted in a three- to fivefold inhibition of HIV-1 (strain NL4.3) replication. Quantitative PCR was used to pinpoint the replication block to the integration step. Next, polyclonal and monoclonal HeLaP4-derived cell lines were selected with a stable knockdown of LEDGF/p75 mediated by a lentiviral vector (lentivector) encoding a short hairpin RNA (shRNA) targeting this protein. Cell lines stably transduced with a lentivector encoding an unrelated hairpin or a double-mismatch hairpin served as controls. Again, a two- to fourfold reduction of HIV-1 replication was observed. The extent of LEDGF/p75 knockdown closely correlated with the reduction of HIV-1 replication. After the back-complementation of LEDGF/p75 in the poly- and monoclonal knockdown cell lines using an shRNA-resistant expression plasmid, viral replication was restored to nearly wild-type levels. The Q168A mutation in integrase has been shown to interfere with the interaction with LEDGF/p75 without reducing the enzymatic activity. Transduction by HIV-1-derived lentivectors carrying the Q168A IN mutant was severely hampered, pointing again to a requirement for LEDGF/p75. Altogether, our data validate LEDGF/p75 as an important cellular cofactor for HIV integration and as a potential target for antiviral drug development.
In the struggle against the AIDS pandemic, a continuous effort to validate new antiviral targets and to develop new drugs is mandatory. Our group targets the integration step of human immunodeficiency virus type 1 (HIV-1), an essential step in the retroviral life cycle. After integration, the provirus remains present throughout the life time of the infected cell and is passed on to its daughter cells (for reviews, see references 2 and 9). Integration of the viral cDNA into the host genome is carried out by the viral integrase (IN). However, insight has grown to show that HIV also relies on cellular proteins for completion of its replication cycle, including integration. The identification and characterization of these cellular cofactors will increase our understanding of the viral replication cycle and potentially lead to the development of new antiviral drugs. Various cofactors of the lentiviral integration process have been proposed (for a recent review, see reference 42). Integrase interactor 1 (Ini-1), a component of the SWI/SNF chromatin remodeling complex (43), interacts directly with HIV-1 IN and stimulates the integration reaction in vitro (19, 26). The minimal IN interaction domain of Ini-1 (amino acids 183 to 294), S6, acts as a specific trans-dominant inhibitor of the late steps of HIV-1 replication upon cellular overexpression (45), indicating a possible role of Ini-1 during the postintegration steps of HIV-1 replication.
The integration reaction is carried out by an ill-defined nucleoprotein complex derived from the core of the infecting virion, the preintegration complex (PIC). The high-mobility-group protein HMGA1, a nonhistone chromosomal protein involved in transcriptional control and nuclear architecture, was identified as a cellular host factor essential for PIC activity in vitro (14). A more recent report surprisingly showed that chicken cells lacking HMGA1 were not deficient in cell growth or in retroviral integration, suggesting that the HMGA1 protein is most probably not required for integration (4). To integrate successfully into the host DNA, the PIC must avoid self-destructive integration of the viral DNA into itself, a reaction termed autointegration. It was previously shown that PICs isolated from Moloney murine leukemia virus (MoMLV)infected cells contain a barrier-to-autointegration factor (BAF) (6, 21, 36). Relatively low concentrations of recombinant human BAF were shown to restore the integration activity of salt-disrupted HIV-1 PICs as well (6). Recently, BAF was found to be required for the segregation and enclosure of chromosomes within the nuclear envelope and for assembly of the nuclear lamina (25). In uninfected cells, BAF interacts with members of the LEM family of inner nuclear membrane and nucleoplasmic proteins. It was recently shown (37) that one of the LEM proteins, lamina-associated polypeptide 2α (LAP-2α), is a component of the MoMLV PIC. LAP-2α stabilizes the association of BAF with the PIC to stimulate intermolecular integration and suppress autointegration. Depletion of LAP-2α significantly inhibited MoMLV replication, as demonstrated with stable LAP-2α knockdown cell lines. However, the exact function of these potential cofactors during in vivo integration of MLV (and HIV) remains to be determined.
We previously reported that HIV-1 integrase associates with LEDGF/p75 in human cells and that recombinant LEDGF/p75 enhances the strand transfer activity of HIV-1 IN in vitro (7). This specific interaction was later confirmed by yeast two-hybrid (13) and coimmunoprecipitation (38) analyses. LEDGF/p75 is a member of the hepatoma-derived growth factor family (11). A high degree of homology exists between the N-terminal regions of the members of this protein family. The PWWP motif is located within the N-terminal homology regions of hepatoma-derived growth factors and relates them to an even larger and functionally diverse nuclear protein family that is characterized by a tight association with chromatin and that includes transcription factors, DNA repair enzymes, and DNA methylases. PWWP domains were originally believed to be implicated in protein-protein interactions (35); now they are assumed to be involved in chromatin association (28). The two splice variants of the protein, LEDGF/p75 and LEDGF/p52, are both karyophilic but differ in their nuclear distribution patterns (24). Whereas LEDGF/p75 tightly associates with HIV-1 IN, p52 neither interacts with IN in vitro nor colocalizes with fluorescently tagged IN inside cells (22).
The cellular function of LEDGF/p75 is not well understood. LEDGF/p75 was first described as the transcription cofactor PC4 interactor protein (17) and was therefore suggested to play a role in transcriptional regulation. Later, LEDGF/p75 was proposed to play a protective role during stress-induced apoptosis (15, 16, 33), and specific binding to stress response DNA elements has been reported (32).
We have previously shown that expression of LEDGF/p75 is required for the association of IN with nuclear DNA and mitotic chromosomes (24) and that recombinant LEDGF/p75, but not p52, strongly increases the affinity of HIV integrase for DNA (5). Apparently, the LEDGF/p75-mediated tethering of IN to the chromosomes is responsible for its nuclear retention (24). Although nuclear import of LEDGF/p75 itself is dependent on a nuclear localization signal (22, 23), and although LEDGF/p75 is proposed to be a component of the PIC (22), a clear role of LEDGF/p75 in nuclear import of the PIC remains to be demonstrated.
The integration of proviral HIV-1 genomes occurs preferentially in actively transcribed regions (31), in contrast to MoMLV, which preferentially integrates in the proximity of the promoter region (44). Our finding that LEDGF/p75 interacts with lentiviral but not with other retroviral integrases suggests a possible role of LEDGF/p75 in targeting lentiviral genomes to actively transcribed regions (5).
In this report, we examine the role of LEDGF/p75 in HIV-1 replication. A transient knockdown of LEDGF/p75 was obtained in HeLaP4 cells by using a specific small interfering RNA (siRNA), whereas stable knockdown in polyclonal or monoclonal HeLaP4 cells was achieved by using a lentiviral vector encoding a short hairpin RNA (shRNA) targeting LEDGF/p75. Inhibition of HIV-1 replication was observed in the cell lines exhibiting reduced levels of LEDGF/p75 but not in control cells, validating the importance of LEDGF/p75 for HIV replication.
MATERIALS AND METHODS
Cell culture.
HeLaP4 cells, obtained from the NIH Reagent Program, were grown in Dulbecco's modified Eagle's medium (DMEM) (Gibco-BRL) supplemented with 10% fetal calf serum (FCS) (International Medical, Belgium) and 20 μg/ml gentamicin (Gibco-BRL) (referred to herein as DMEM-complete), and Geneticin was added to a final concentration of 0.5 mg/ml, at 37°C and 5% CO2 in a humidified atmosphere. 293T cells were provided by O. Danos (Evry, France) and were grown in DMEM-complete. MT-4 cells were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, from Douglas Richman. The cells were grown in RPMI 1640 supplemented with 10% FCS and 20 μg/ml gentamicin (RPMI-complete).
Virus strains.
The molecular clone pNL4.3 (1) was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH (contributed by Malcolm Martin).
Transient knockdown of LEDGF/p75 in HeLaP4 cells.
The day prior to transient transfection, 5 × 104 HeLaP4 cells were seeded per well in a 24-well plate for growth curve, Western blot, and quantitative PCR (Q-PCR) analyses. For HIV-1 infection, 2 × 105 cells were seeded in a six-well plate. After incubation overnight, cells were transfected with 20 nM siRNA following the guidelines of the siFECTamin protocol (ICVEC, United Kingdom). The target sequences of the synthetic siRNA duplexes are shown in Fig. 1. Synthetic siRNAs were designed with the following targets and sequences: L3, targeting nucleotides 1342 to 1361 (5′ AGA CAG CAU GAG GAA GCG dTdT 3′) (Dharmacon); L3MM, targeting the same nucleotides (1342 to 1361) harboring four mutations (underlined) (5′ AGA CAG CAU CUC CAA GCG dTdT 3′) (QIAGEN, The Netherlands); L4, targeting nucleotides 1301 to 1325 (5′ CCG UGA UCA CCC AAG UGC UGA AUA A 3′) (Invitrogen, Belgium); and siRNA specific for green fluorescent protein (siGFP) (siRNA libraries; Eurogentec, Belgium). In parallel, transfection without a siRNA duplex was performed as a control, and these cells are referred to as mock transfected.
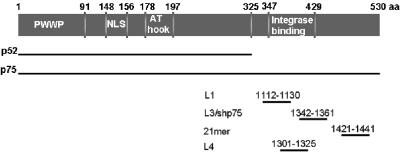
FIG. 1.
Overview of siRNAs targeting LEDGF/p75. LEDGF/p75 is a protein of 530 amino acids, and the splice variant p52 consists of the first 325 amino acids with eight additional amino acids. The different siRNAs used target the 3′ part of the mRNA of LEDGF/p75. L1 targets nucleotides 1112 to 1130 and has been described by Emiliani et al. (13), L3 targets nucleotides 1342 to 1361 and has been described previously (24), L3MM targets the same sequence as L3 but contains four mutations, the 21-mer targets nucleotides 1421 to 1441 and has been described by Devroe and Silver (10), and L4 is a stealth siRNA targeting nucleotides 1301 to 1325. The integrase binding domain is located between amino acids 347 and 429, as described previously (8, 13, 40).
HIV infection and analysis of transiently transfected HeLaP4 cells.
Three days after transfection, cells were detached from the six-well plates. Subsequently, 1.5 × 104 cells were seeded in a 24-well plate for 4 h at 37°C. After attachment, cells were infected with HIV-1. Infections were performed with the following amounts of virus, as determined by p24 measurement (Alliance HIV-1 p24 enzyme-linked immunosorbent assay [ELISA] kit; PerkinElmer): 0.43 × 104, 1.1 × 104, and 2.2 × 104 pg p24 in a total volume of 250 μl. Each experiment was run in triplicate. After 3 h of infection, the supernatant was removed, and cells were washed three times with phosphate-buffered saline (PBS) prior to the addition of 0.5 ml DMEM-complete. Twenty-four and 72 h after infection, a single well was analyzed for β-galactosidase activity (chemiluminescent β-Gal reporter gene assay; Roche Applied Science, Belgium). p24 analysis of the supernatant was performed after 72 h. For measurements of β-galactosidase activity, care was taken to measure reporter activities in adherent and detached cells. β-Galactosidase activity was measured following the protocol supplied by the manufacturer. Chemiluminescence was measured with a LumiCount instrument (Packard, Belgium). The protein concentration of each sample was determined (BCA protein assay kit; Perbio), and readouts were normalized to 1 mg/ml total protein. At each time point analyzed, the persistence of knockdown was demonstrated either by Western blotting or by Q-PCR for LEDGF mRNA.
Determination of knockdown by Q-PCR.
After 4 days of transfection, total RNA was isolated from wells of a 24-well plate, using RNAqueous-4PCR (Ambion). The RNA concentration was determined photometrically at a wavelength of 260 nm using GeneQuant Pro. After adjustment of the sample RNA concentration to 200 ng/μl, 50 μl of each sample was used for reverse transcription following the procedure of a High-Capacity cDNA archive kit (Applied Biosystems). Samples corresponding to 400 ng of RNA were subsequently used for Q-PCR analysis with the ABI Prism 7000 sequence detection system (Applied Biosystems). Positions of the primers and probe are given relative to the +1 position of LEDGF mRNA (ATG). The reaction mixtures contained TaqMan universal PCR master mix (Roche Applied Science) and 5′ AGC CAG AAG TTA AGA AAG TGG AGA AG 3′ (positions 1000 to 1025) as the forward primer, 5′ TCT TGA TGT GAA CAG ATG CAT TGA 3′ (positions 1100 to 1125) as the reverse primer, and 5′ (6-carboxyfluorescein)-AGC GAG AAA CAT CAA TGG ATT CTC GAC TTC A-(6-carboxytetramethylrhodamine) 3′ (positions 1027 to 1157) as the probe.
Growth kinetics.
For growth curve analysis, 50,000 cells of the different stable cell lines and HeLaP4 were seeded per well in a 24-well plate. Cell growth was followed on consecutive days by cytometry (Kovaslide; Hycor). For analysis of cell viability after transient knockdown, 5 × 104 HeLaP4 cells were seeded per well 1 day prior to transient knockdown. Two wells for each knockdown were trypsinized 3 days after transfection. Cells from both wells were combined, and the cell density was determined in a cytometer. Next, the cells were reseeded into two individual wells of a 24-well plate. This procedure was repeated on two subsequent days in order to analyze cell growth. Growth kinetics of stable cell lines were determined by seeding cells in a 24-well plate at a density of 5 × 104 cells/well. Cells were trypsinized and counted by cytometry every 72 h (data not shown). No significant difference in growth kinetics was observed between the different selected stable cell lines.
Construction of lentiviral vector transfer plasmids encoding shRNA targeting LEDGF/p75.
A fragment containing the mouse U6 promoter flanked by PpuMI restriction sites was generated by PCR with the forward primer 5′ AAA GGG ACC CAT CCG ACG CCG CCA TCT CTA GG 3′ and the reverse primer 5′ TTT GGG TCC CGT ACG TTT TCT AGA AAC AAG GCT TTT CTC CAA GGG 3′. The reverse primer contains an XbaI and a BsiWI site enabling cloning of short hairpin sequences. The PCR fragment was cloned into the PpuMI restriction site of pCHMWS (3) upstream of the central polypurine tract. The zeocin resistance gene was amplified from pBudCE4.1 (Invitrogen) by PCR using the forward primer 5′ AAA GGA TCC ATG GCC AAG TTG ACC AGT GC 3′ and the reverse primer 5′ AAA CTC GAG TCA GTC CTG CTC CTC GGC CAC G 3′. The PCR fragment was cloned into the multiple cloning site of pCHMWS using BamHI and XhoI, allowing zeocin selection. Two oligonucleotides, 5′ CTA GGA CAG CAT GAG GAA GCG AAC TCG AGT TCG CTT CCT CAT GCT GTC TTT TT 3′ and 5′ GTA CAA AAA GAC AGC ATG AGG AAG CGA ACT CGA GTT CGC TTC CTC ATG CTG TC 3′ (the loop region is shown in italics), encoding the hairpin targeting LEDGF/p75, were annealed and cloned at the XbaI and BsiWI restriction sites, creating the transfer plasmid shp75. To generate the control plasmid shp75mut, an shRNA encoding two point mutations (indicated in bold and underlined in the primer sequences) was constructed similarly. Two oligonucleotides, 5′ CTA GGA CAG CAT CTG GAA GCG AAC TCG AGT TCG CTT CCA GAT GCT GTC TTT TT 3′ and 5′ GTA CAA AAA GAC AGC ATC TGG AAG CGA ACT CGA GTT CGC TTC CAG ATG CTG TC 3′, were annealed and cloned at the XbaI and BsiWI sites.
Construction of lentiviral vector packaging plasmid containing Q168A integrase mutation.
p8.91Q168A was constructed by site-directed mutagenesis, using the method of Kirsch and Joly (20). Briefly, a sequence comprising the integrase coding region was first amplified by PCR, using pCMVΔR8.91 (a kind gift of D. Trono, University of Geneva, Switzerland) as a template, and cloned into pCR4-Topo (Invitrogen). In addition to the IN coding region, the PCR fragment harbored two restriction sites (SwaI and EcoRI) that are unique to pCMVΔR8.91. Secondly, the latter construct was used as a template to replace the Q168 amino acid with Ala (CAG to GCG), using the primers 5′ AGA AAT AGT AGC CAG CTG TG 3′ (sense) and 5′ AAG ATG TTC GGC CGC ATC TCT TAC CTG TCC 3′ (antisense). In addition, a silent EagI site was introduced (underlined), allowing selection of positive clones. Subsequently, the SwaI-EcoRI fragment in pCMVΔR8.91 was replaced by the fragment with the Q168A mutation. All PCRs were performed with Pwo proofreading polymerase (Roche) and confirmed by sequence analysis.
Production of lentiviral vectors.
HIV-1-based lentiviral vector particles were produced in 8.5-cm dishes and in cell factories using the triple transient transfection system described by Geraerts et al. (18). A second-generation lentiviral vector was used. This HIV-1 vector is devoid of accessory genes. Transcription from the transfer plasmid is driven by the HIV long terminal repeat (LTR), and Rev and Tat are encoded by the packaging plasmid. For the production of lentiviral vectors expressing enhanced green fluorescent protein (EGFP) from the LTR promoter, the central polypurine tract/central termination sequence of plasmid p8.91 (27) was PCR amplified and ligated into the unique ClaI restriction site of the pRRLGFP-W (12) plasmid (both plasmids were a kind gift from D. Trono). Lentiviral vectors were produced in 8.5-cm dishes as described by Geraerts et al. (18).
Selection of LEDGF/p75 knockdown cell lines.
To establish cell lines stably expressing a specific shRNA, 2 × 105 293T or HeLaP4 cells were seeded in a 24-well plate and transduced with 104 RNA equivalents of lentiviral vector the following day. After 4 h of incubation, the supernatant was removed, cells were washed with PBS, and 1 ml of DMEM-complete was added. In parallel, 293T and HeLaP4 cells were transduced with a lentiviral vector encoding the double-mismatch shRNA (shp75mut). After 48 h, the transduced cells were trypsinized, and selection was initiated by diluting the cells 40-fold and adding 200 μg/ml zeocin (Invitrogen). Selection resulted in the polyclonal cell lines 293T/shp75PC, 293T/shp75mutPC, HeLaP4/shp75PC, and HeLaP4/shp75mutPC. Monoclonal HeLaP4 knockdown cell lines were selected as well. As an additional control, a polyclonal cell line, HeLaP4/shEGFP, expressing an irrelevant short hairpin targeting EGFP mRNA (39) was generated as described above. As additional controls, the HeLaP4/shp75PC and HeLaP4/shp75Cl15 cell lines were back-complemented. The back-complementation plasmid (a kind gift from E. Poeschla, Mayo Clinic College of Medicine) contains seven silent mutations in the DNA encoding the LEDGF/p75 mRNA region targeted by the short hairpin L3. To establish stable back-complementation cell lines expressing LEDGF/p75 from a short hairpin-resistant plasmid, 2 × 105 HeLaP4 cells were seeded in a 24-well plate and transfected with 0.8 μg DNA from the LEDGF/p75 back-complementation plasmid by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. After 10 days of puromycin selection (0.25 μg/ml), single colonies were picked to generate monoclonal cell lines, and the HeLaP4/shp75PC/BCCl3 and HeLaP4/shp75Cl15/BCCla cell lines were used for further experiments.
Virus production.
MT-4 cells (5 × 106) were spun down and resuspended in 800 μl RPMI-complete. Ten micrograms of the pNL4.3 plasmid was electroporated at 250 V and 1,500 μF. After electroporation, 5 ml RPMI-complete was added, and after 4 days of infection, supernatants were harvested and aliquots of 1.5 ml were stored at −80°C. The p24 antigen was measured and used to quantify the infectivity of the virus stock.
Western blotting.
Whole-cell extracts of different cell lines (293T and HeLaP4) were separated by 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electroblotted onto polyvinylidene difluoride membranes (Bio-Rad). Membranes were blocked with milk powder in PBS-0.1% Tween 20, and detection was carried out using specific monoclonal antibodies against LEDGF/p75 (BD Biosciences). Detection was performed using chemiluminescence (ECL+; Amersham) and a horseradish peroxidase (HRP)-conjugated anti-mouse antibody (Dako). After washing of the membrane overnight with PBS-0.1% Tween 20, equal loading was controlled using α-tubulin antibody (Sigma) and anti-mouse-HRP antibody.
Laser scanning microscopy.
One day before transfection, 2 × 104 HeLaP4, HeLaP4/shp75Cl15, HeLaP4/shp75mutCl1, or HeLaP4shp75Cl15/BCCla cells were seeded per chamber of a Lab-Tec 8 chamber cover glass (VWR International, Haasrode, Belgium). The following day, the cells were transfected with 0.8 μg of pEGFP-INs (24), using polyethyleneimine as described earlier (24). After 5 h, the medium was replaced with DMEM-complete. Twenty-four hours later, cells were washed with PBS and analyzed using an LSM510 laser scanning microscope (Zeiss, Zaventem, Belgium). Cells were visualized by phase-contrast microscopy, and EGFP was excited at a wavelength of 488 nm using an argon laser with a 530- to 550-nm-band-pass filter.
Transduction with HIV-1-based lentiviral vectors.
One day prior to transduction, 105 HeLaP4 cells were seeded in a 24-well plate. Transductions with the lentiviral vectors were carried out at a multiplicity of infection (MOI) between 0.5 and 5. Vector transduction was carried out in the presence of 4 μg/ml of Polybrene in DMEM-1% FCS. After 4 h of incubation, the medium was replaced with DMEM-complete. In each 24-well plate, mock-transduced HeLaP4 cells were grown in parallel. Cells were split 1/10 at 24 h posttransduction and reseeded. At 6 days posttransduction, the cells were harvested for fluorescence-activated cell sorting (FACS) analysis of EGFP expression and for Q-PCR analysis to determine the proviral copy number, using primers and a probe directed against the egfp gene.
HIV infection of stable knockdown cell lines.
HeLaP4, HeLaP4/shEGFP, HeLaP4/shp75PC, HeLaP4/shp75mutCl1, HeLaP4/shp75Cl15, HeLaP4/shp75PC/BCCl3 and HeLaP4/shp75Cl15/BCCla cells (105) were seeded in a 24-well plate 1 day prior to infection. Next, cells were incubated with different dilutions of HIV-1 for 3 h. Cells were washed twice with PBS, and 1 ml of DMEM-complete was added after washing. Cell-free supernatant was harvested and analyzed for p24 60 h after infection. For long-term infection experiments, HeLaP4 cells were reseeded at 72 h postinfection (i.e., at confluence) into six-well dishes. p24 values were obtained at 3, 6, and 9 days postinfection. The role of LEDGF/p75 during the late steps of HIV-1 replication was examined by transfecting 105 293T, 293T/shp75, or 293T/shp75mut cells in parallel with 0.8 μg of pNL4.3 DNA in a 24-well plate. The amount of virus released into the supernatant was measured 2 and 3 days after transfection by quantifying the p24 antigen via ELISA.
Quantification of different HIV-1 DNA species using real-time PCR.
Quantification of late reverse transcripts and of 2-LTR circles was done according to a previously described protocol (41). Quantification of integrated proviruses was done using a nested Alu-PCR, modified from the method described by O'Doherty et al. (29). The first-round PCR was performed with 400 ng cellular DNA, using the forward primer 5′ GCT AAC TAG GGA ACC CAC TGC TTA 3′ and the reverse primer 5′ TGC TGG GAT TAC AGG CGT GAG 3′. The reaction mixture contained 2 mM MgCl2, a 250 μM concentration of each deoxynucleoside triphosphate, a 400 nM concentration of each primer, 1.25 U AmpliTaq DNA polymerase (Applied Biosystems), 10 mM Tris-HCl (pH 8.3), and 50 mM KCl in a volume of 50 μl. Each sample was subjected to initial denaturation of 5 min at 95°C, followed by 15 amplification cycles of denaturation at 95°C for 30 s, annealing at 60°C for 40 s, and extension at 72°C for 1 min 30 s. The second-round, real-time TaqMan PCR was performed with 10 μl of first-round PCR product. The sequences of the primers and probe were as follows: forward primer, 5′ AGC TTG CCT TGA GTG CTT CAA 3′; reverse primer, 5′ TGA CTA AAA GGG TCT GAG GGA TCT 3′; and probe, 5′ (6-carboxyfluorescein)-TTA CCA GAG TCA CAC AAC AGA CGG GCA-(6-carboxytetramethylrhodamine) 3′. No-template controls (no DNA added to the PCR mixture) and no-amplification controls (with DNA extracted from cells that were not transduced) were run with each experiment. For each sample, RNaseP DNA was quantified as an endogenous control.
FACS analysis of reporter gene expression.
EGFP expression was analyzed at time points between 24 and 144 h posttransduction. Cell death was analyzed using 7-amino-actinomycin D (7-AAD) (BD Biosciences) as a marker. Cells were fixed in 2% paraformaldehyde prior to analysis with a FACSCalibur flow cytometer (Becton Dickinson). The data were analyzed using the CellQuest software package provided with the instrument.
RESULTS
Characterization of transient knockdown of LEDGF/p75 in HeLaP4 cells.
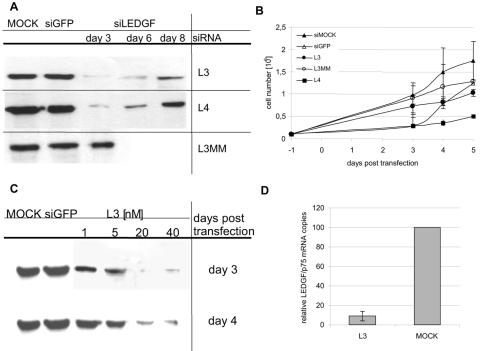
We first investigated the effect on HIV-1 replication of transient, siRNA-mediated knockdown of LEDGF/p75 in HeLaP4 cells. We used two siRNAs targeting the C terminus of LEDGF/p75 mRNA. L3 has been described before (24) and targets nucleotides 1342 to 1361, whereas L4 targets nucleotides 1301 to 1352 (Fig. 1). The efficiency of knockdown was verified by Western blotting of cell extracts obtained at different time intervals after transfection (Fig. 2A). Transfection with L3 and L4 clearly resulted in a pronounced and prolonged reduction in LEDGF/p75 levels for at least 6 days. Transfection with a mismatch siRNA (L3MM) did not reduce LEDGF/p75 levels. To further exclude off-target effects and nonspecific cellular toxicity, the growth kinetics of the transfected cells were determined (Fig. 2B). Transfection with L3 did not influence the growth of HeLaP4 cells significantly, whereas transient transfection with L4 resulted in impaired cell growth. Cells transfected with L3 or L3MM replicated similarly to control cells transfected with a siRNA targeting EGFP. These data were corroborated upon staining with 7-AAD, a marker for cell death, followed by FACS analysis (data not shown). Despite generating a strong knockdown, transfection with L4 resulted in cellular toxicity, with >50% of cells staining positive for 7-AAD. The number of 7-AAD-positive cells was similar or only slightly increased in cells transfected with L3 compared to mock-transfected cells. Hence, L4 was not included in further HIV-1 infection studies. Next, knockdown experiments were carried out with different concentrations of L3 (Fig. 2C). A concentration of 20 nM or higher during transfection was necessary to obtain a knockdown that persisted throughout the 7 days of the HIV-1 infection experiment. With lower concentrations of siRNA, no maximum level of knockdown was achieved after 3 days. This is important, as cells are infected with HIV-1 in our experimental setup 3 days after transfection. At 1 to 5 nM of L3, LEDGF/p75 was no longer suppressed 4 days after transfection (Fig. 2C). In addition, we confirmed LEDGF/p75 knockdown by using Q-PCR, showing a 90% reduction of LEDGF/p75 mRNA levels on day 4 (Fig. 2D). In all subsequent experiments, transient knockdown was performed with 20 nM L3 to ensure a prolonged suppression of LEDGF/p75 during HIV-1 replication.
FIG. 2.
Analysis of transient knockdowns with different siRNAs targeting LEDGF/p75 at different time points. (A) Western blot analysis of LEDGF/p75 transient knockdown. LEDGF/p75 expression levels were verified by Western blotting of HeLaP4 cells transiently transfected with 20 nM of L3 or L4 at different time points after transfection. Mock-, siGFP-, and L3MM-transfected cells were analyzed as well. (B) Cell viability of HeLaP4 cells transiently transfected with L3, L4, siGFP, or L3MM or of mock-transfected cells. Cell counts shown are average values ± standard deviations (SD) for four independent experiments. (C) Western blot analysis of knockdowns obtained after transient transfection of HeLaP4 cells with different concentrations of L3, determined at different time points after transfection. siGFP and mock transfections were included as controls. (D) Q-PCR analysis of LEDGF/p75 mRNA levels 4 days after transfection of HeLaP4 cells with L3 or mock siRNA. Average levels for three independent experiments ± SD using 20 nM of L3 are shown.
Transient knockdown of LEDGF/p75 impairs HIV replication and integration.
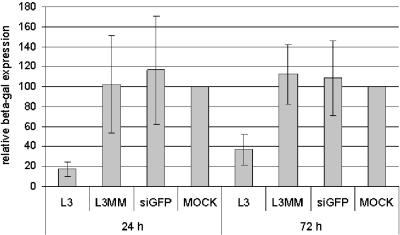
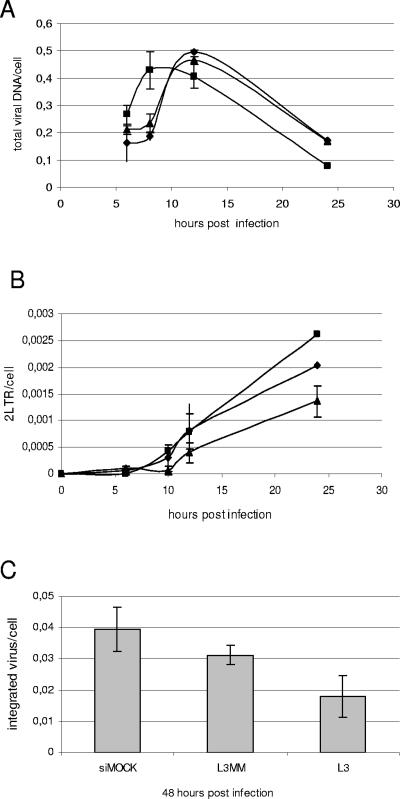
Next, the effect of L3-mediated depletion of LEDGF/p75 on the replication of HIV-1 strain NL4.3 in HeLaP4 cells was investigated. Three days after transfection with 20 nM siRNA, cells were reseeded at equal cell numbers and were analyzed by Western blotting on day 3 (the day of infection) and day 6 (72 h after infection) (Fig. 2A). HIV-1 replication was monitored by measuring the β-galactosidase reporter gene activities 24 and 72 h after infection (Fig. 3). Transfection with L3, but not with L3MM or siGFP, resulted in a three- to fivefold reduction in reporter gene activity. A fivefold reduction in p24 antigen levels 72 h after infection was observed in cells transfected with L3 (data not shown). The reduction in β-galactosidase activity 24 or 72 h after infection (Fig. 3) correlated with the extent of knockdown of LEDGF/p75 at each time point (Fig. 2A). In parallel, cells were harvested at different time points after infection, and DNAs were extracted to quantify late reverse transcripts, 2-LTR circles, and integrated proviruses using Q-PCR (41). Mock-transfected cells and cells treated with L3MM were used as controls. Reverse transcription was hardly affected by LEDGF/p75 knockdown (Fig. 4A). Likewise, the transient knockdown of LEDGF/p75 did not significantly reduce the number of 2-LTR circles, which reflects both prior DNA synthesis and nuclear import of the PIC (Fig. 4B). However, a clear reduction in the number of integrants was detected, pinpointing the replication block to the integration step (Fig. 4C).
FIG. 3.
Analysis of effect of transient knockdown of LEDGF/p75 on HIV-1 infection. (A) Inhibition of HIV-1 replication in HeLaP4 cells by knockdown of LEDGF/p75 using L3, as measured by reporter gene activity. HeLaP4 cells were transiently transfected with 20 nM of L3, L3MM, or siGFP prior to infection with HIV-1 at an MOI of 0.01. Intracellular β-galactosidase activities were measured 24 and 72 h after infection with HIV-1. The data shown are average values ± SD for 15 experiments. Mock-transfected cells were used as a control.
FIG. 4.
LEDGF/p75 knockdown impairs HIV-1 integration. HeLaP4 cells were transiently transfected with L3 (▪) or L3MM (▴) or mock transfected (⧫) before infection with NL4.3 at an MOI of 0.01. Total viral DNAs (A) and 2-LTR circles (B) were quantified by Q-PCR at different time points postinfection; integrated proviral DNAs were quantified by Alu-PCR at 48 h postinfection (C).
Generation of LEDGF/p75 stable knockdown and appropriate control cell lines.
Although RNA interference provides a powerful tool for target validation, caveats have been raised regarding the possible occurrence of artifacts. Indeed, siRNA may induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells (30). Furthermore, several research groups reported siRNA-mediated interferon induction affecting cell survival and, potentially, virus replication (34). Moreover, the transient nature of siRNA-based knockdown (illustrated in Fig. 2A) in dividing cells might complicate the interpretation of phenotypic effects in multiple-day infection experiments. Although we have carefully controlled potential artifacts related to the use of siRNA in this study, we decided to also select stable knockdown cell lines using lentiviral vector-encoded shRNA (39).
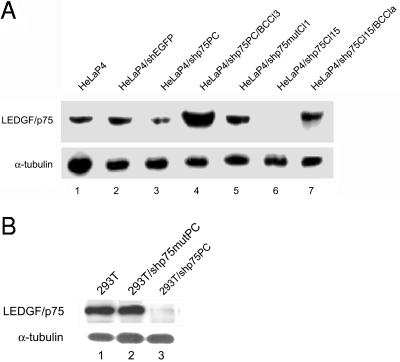
Polyclonal and monoclonal LEDGF/p75 knockdown cell lines HeLaP4/shp75PC and HeLaP4/shp75Cl15, respectively, were selected after transduction. Several precautions were taken in order to control off-target effects and specificity, as follows: (i) polyclonal and monoclonal stable cell lines were selected that encoded a mutant LEDGF/p75 hairpin (for the experiments shown, HeLaP4/shp75mutCl1 cells were used); (ii) the polyclonal cell line HeLaP4/shp75PC was back-complemented with an expression plasmid for LEDGF/p75 rendered resistant to the L3 shRNA, and subsequently, the monoclonal cell line HeLaP4/shp75PC/BCCl3 was selected; (iii) in addition, the monoclonal cell line HeLaP4/shp75Cl15 was back-complemented as well, and further experiments were done with the selected clone, HeLaP4/shp75Cl15/BCCla; and (iv) a polyclonal cell line encoding an irrelevant specific short hairpin against EGFP was included in the experiments. The LEDGF/p75 expression levels of selected HeLaP4 cell lines, as determined by Western blotting, are shown in Fig. 5A. All knockdown cell lines clearly demonstrated reduced levels of LEDGF/p75 protein. The suppression of LEDGF/p75 in knockdown cell lines was found to be stable for >100 passages after transduction. The most prominent knockdown of LEDGF/p75 was achieved in the monoclonal HeLaP4/shp75Cl15 cell line. Back-complementation of this cell line yielded wild-type expression levels, whereas HeLaP4/shp75PC/BCCl3 cells revealed LEDGF/p75 overexpression. Finally, wild-type levels of LEDGF/p75 were observed in cells carrying shRNA against EGFP or shp75mut.
FIG. 5.
Western blot analysis of LEDGF/p75 expression levels in stable knockdown cell lines. (A) Western blot analysis of LEDGF/p75 was performed with total cell extracts of HeLaP4 cells (lane 1), HeLaP4 cells stably transduced and selected with an irrelevant shRNA against EGFP (HeLaP4/shEGFP, lane 2), an shRNA targeting LEDGF/p75 (polyclonal cell line) (HeLaP4/shp75PC, lane 3), back-complementation of polyclonal LEDGF/p75 knockdown cells with a siRNA-resistant LEDGF/p75 expression plasmid (HeLaP4/shp75PC/BCCl3, lane 4), and an shRNA against LEDGF/p75 containing a double mismatch (HeLaP4/shp75mutCl1, lane 5). LEDGF/p75 expression is also shown for a monoclonal cell line (clone 15) expressing an shRNA against LEDGF/p75 (HeLaP4/shp75Cl15, lane 6) and for clone 15 after back-complementation (HeLaP4/shp75Cl15/BCCla, lane 7). (B) Western blot analysis of LEDGF/p75 expression in 293T cells (lane 1) and 293T cells stably transduced and selected with an shRNA against LEDGF/p75 containing a double mismatch (293T/shp75mutPC, lane 2) and an shRNA targeting LEDGF/p75 (polyclonal cell line) (293T/shp75PC, lane 3). Equal loading of gels was controlled by α-tubulin detection.
Growth kinetics and (co)receptor expression of selected cell lines.
The growth kinetics of all selected cell lines were similar (data not shown). To exclude selection artifacts affecting the HIV-1 replication capacity in the cell lines, the different HeLaP4 cell lines were analyzed for CD4 receptor and CXCR4 coreceptor expression by FACS. No significant differences in expression levels were found (data not shown). Equal percentages of dead cells, ranging between 2 and 4%, were measured for the different cell lines by FACS analysis using 7-AAD (data not shown).
Localization of EGFP-IN in stable LEDGF/p75 knockdown cell lines.
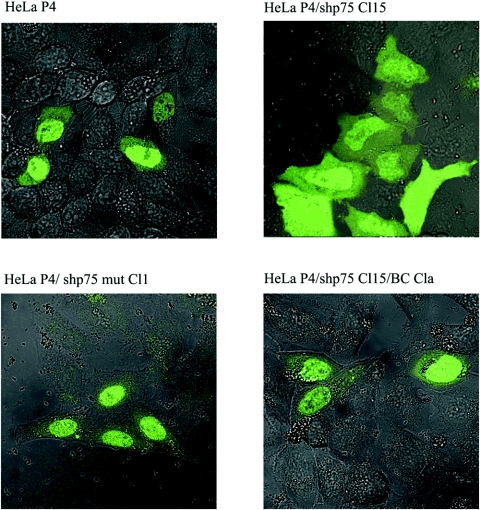
We and others have previously shown that LEDGF/p75 is required for the nuclear accumulation of HIV-1 integrase and its association with mitotic chromosomes (22, 24). To study the impact of LEDGF/p75 knockdown on the cellular localization of HIV-1 integrase in the established cell lines, HeLaP4/shp75Cl15, HeLaP4/shp75Cl15/BCCla, and HeLaP4/p75mutCl1 cells were transiently transfected with an expression plasmid for EGFP-IN (24) and analyzed by laser scanning microscopy (Fig. 6). In the parental HeLaP4 cell line and in HeLaP4/p75mutCl1 cells, HIV-1 EGFP-IN localized to the nucleus, whereas the stable knockdown of LEDGF/p75 drastically changed HIV-1 EGFP-IN localization. In HeLaP4/shp75Cl15 cells, EGFP-IN was distributed over the cytoplasm and the nucleus. Subsequent back-complementation of LEDGF/p75 (HeLaP4/shp75Cl15/BCCla) restored the nuclear accumulation of HIV-1 EGFP-IN.
FIG. 6.
Localization of EGFP-IN in different LEDGF/p75-expressing cell lines. HeLaP4, HeLaP4/shp75Cl15, HeLaP4/shp75mutCl1, and HeLaP4shp75Cl15/BCCla cells were transfected with pEGFP-INs prior to laser scanning microscopy. Cells were visualized by phase-contrast microscopy, and EGFP was excited at 488 nm using an argon laser with a 530- to 550-nm-band-pass filter.
HIV replication is impaired in stable LEDGF/p75 knockdown cell lines.
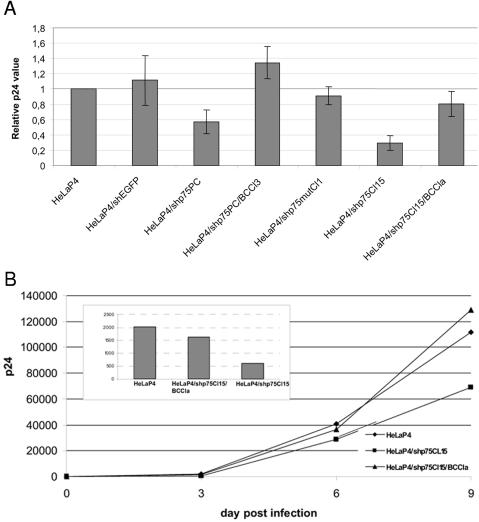
We infected the parental HeLaP4, HeLaP4/shEGFP, HeLaP4/shp75PC, HeLaP4/shp75PC/BCCl3, HeLaP4/shp75mutCl1, HeLaP4/shp75Cl15, and HeLaP4/shp75Cl15/ BCCla cell lines in parallel with HIV-1. Supernatants were harvested at 72 h postinfection, and the p24 antigen was measured by ELISA (Fig. 7A). We observed a 40% reduction in p24 antigen production in the polyclonal LEDGF/p75 knockdown cell line (HeLaP4/shp75PC) compared with the controls and a 70% reduction in the monoclonal LEDGF/p75 knockdown cell line HeLaP4/shp75Cl15. Viral replication could be restored to wild-type levels in HeLaP4/shp75PC/BCCl3 and HeLaP4/shp75Cl15/BCCla cells. In control cells encoding the mutant hairpin (HeLaP4/shp75mutCl1) or the unrelated hairpin (HeLaP4/shEGFP), no reduction of HIV-1 replication was observed. We also investigated the effect of LEDGF/p75 reduction on HIV-1 replication at later time points. For this purpose, infected cells were reseeded. Virus production, followed by the p24 antigen level in the supernatant, was persistently suppressed in HeLaP4/shp75Cl15 cells but not in back-complemented HeLaP4/shp75Cl15/BCCla cells (Fig. 7B).
FIG. 7.
HIV-1 replication is impaired in LEDGF/p75 knockdown cell lines. (A) HeLaP4, HeLaP4/shEGFP, HeLaP4/shp75PC, HeLaP4/shp75PC/BCCl3, HeLaP4/shp75mutCl1, HeLaP4/shp75Cl15, and HeLaP4/shp75Cl15/BCCla cells were infected with HIV-1(NL4.3) at an MOI of 0.01. p24 antigen production in the supernatants 60 h after infection was measured by ELISA. Average levels ± SD for four experiments are shown. (B) HeLaP4, HeLaP4/shp75Cl15, and HeLaP4/shp75Cl15/BCCla cells were infected with HIV-1(NL4.3) at an MOI of 0.002. The cells were subsequently trypsinized at day 3 postinfection and reseeded into a six-well plate. Supernatants were harvested on days 3, 6, and 9 postinfection, and p24 antigen production was measured. The inset highlights the p24 values after 3 days.
The impact of knockdown and HIV-1 infection on cell death was analyzed again, using 7-AAD staining and FACS (data not shown). Dead cells ranged between 2 and 4% of the cells prior to infection with HIV-1. After 48 h, cell death in the different cell lines increased to 5 to 10% of noninfected cells and 25% of cells infected with HIV-1. Again, no difference in cell death was observed between the selected cell lines studied and the parental cell lines, indicating that the increase in cell death was caused by the HIV-1 infection as such and not due to the depletion of LEDGF/p75.
Effect of LEDGF/p75 knockdown on late steps of replication.
To examine a potential role of LEDGF/p75 during the late steps of HIV-1 replication, polyclonal cell lines were selected after lentiviral vector transduction (293T/shp75PC and 293T/shp75mutPC). The expression levels of LEDGF/p75 are shown in Fig. 5B. Next, 293T, 293T/shp75PC, and 293T/shp75mutPC cells were transfected with pNL4.3, and virus release in the supernatant was measured by p24 ELISA 2 and 3 days after transfection. No difference in p24 production between the three cell lines was detected (data not shown). The infectivities of the produced viral particles were measured by titration in MT-4 cells. Again, no difference in infectivity was observed between the different virus productions (data not shown). These results are at odds with a role of LEDGF/p75 in late replication steps.
Effect of LEDGF/p75 knockdown on HIV-1-derived lentiviral vector transduction and production.
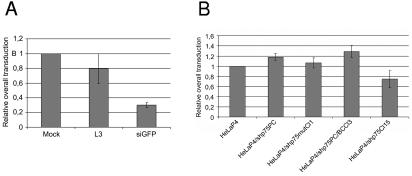
Since a block in viral integration was observed by Q-PCR during HIV replication in HeLaP4 cells after a transient knockdown of LEDGF/p75 (Fig. 4), we attempted to use HIV-1-derived lentiviral vectors to study the role of LEDGF/p75 during the early steps of viral replication. Therefore, we transduced transient and stable LEDGF/p75 knockdown cell lines with a second-generation vesicular stomatitis virus glycoprotein-pseudotyped lentiviral vector encoding EGFP as a reporter. Second-generation lentiviral vectors are devoid of accessory genes. Transcription from the transfer plasmid is driven by the HIV LTR, and Rev and Tat are encoded by the packaging plasmid. After L3-mediated transient knockdown of LEDGF/p75, HeLaP4 cells were transduced with lentiviral vectors at different MOIs. HeLaP4 cells transiently transfected with siGFP were used as a positive control. No significant reduction in overall transduction (i.e., the transduction efficiency multiplied by the mean fluorescence intensity [MFI]) could be detected (Fig. 8A). Next, stable polyclonal and monoclonal LEDGF/p75 knockdown cells were transduced with lentiviral vectors. In the polyclonal 293T/shp75PC (data not shown) and HeLaP4/shp75PC knockdown cells, no inhibition of lentiviral vector transduction was observed. In contrast, in the monoclonal knockdown cell line HeLaP4/shp75Cl15, overall transduction was reduced by 20% (Fig. 8B). A Student t test, with equal variances not assumed, applied to the results of transduction for HeLaP4/shp75Cl15 versus HeLaP4/shp75mutCl1 cells had a P value of 0.041 (P < 0.05). Transduction of the HeLaP4/shp75Cl15 cell line with an LTR-dependent lentiviral vector was not significantly reduced in comparison with control cells (wild-type HeLaP4, HeLaP4/shp75mutCl1, and HeLaP4/shp75PC/BCCl3 cells) (data not shown). We did not observe a difference in vector yield or in the transduction efficiency or MFI of lentiviral vectors obtained after triple transient transfections of 293T, 293T/shp75PC, and 293T/shp75mut cells (data not shown).
FIG. 8.
Impact of LEDGF/p75 knockdown on HIV-1 vector transduction. (A) Effect of transient LEDGF/p75 knockdown on lentiviral vector transduction. HeLaP4 cells were transfected with siRNA, as indicated, and subsequently transduced with a lentiviral vector encoding EGFP at an MOI of 3. Cells transfected with siRNA against EGFP were used as a positive control. Cells were analyzed by FACS at 48 h posttransduction. Average overall transduction values (transduction efficiencies × MFIs) ± SD for three experiments are shown. (B) Effect of stable LEDGF/p75 knockdown on lentiviral vector transduction. Stably selected cell lines were transduced with a lentiviral vector at an MOI of 3. Cells were analyzed by FACS at 48 h posttransduction. Average overall transduction values ± SD for three experiments are shown.
Transduction by Q168A IN mutant lentiviral vector is strongly impaired.
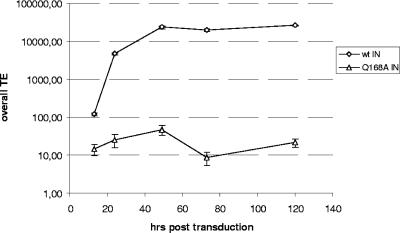
We recently identified and described a mutation (Q168A) in HIV-1 IN that critically reduces the interaction between LEDGF/p75 and HIV-1 IN without an apparent decrease of in vitro enzymatic activity (13). Viruses containing the Q168A IN mutant are replication defective due to a block at the integration step. The apparent lack of inhibition of vector transduction in LEDGF/p75-depleted cells could have multiple explanations. To exclude the possibility that lentiviral vector transduction is LEDGF/p75 independent, we constructed a packaging plasmid containing the Q168A IN mutation and produced the respective mutant lentiviral vector. The transduction efficiency of 293T cells transduced with the Q168A mutant lentiviral vector was severely reduced (Fig. 9).
FIG. 9.
Effect of Q168A IN mutation on lentiviral vector transduction efficiency. Transduction efficiencies (TE) in 293T cells are shown for the wild-type lentiviral vector (⋄) and a lentiviral vector containing the Q168A IN mutation (▵) at different time points after transduction. Cells were subcultured at 24, 48, and 120 h posttransduction. Results of a representative experiment are shown.
DISCUSSION
With carefully controlled experiments, we have shown that transient or stable knockdown of LEDGF/p75 expression interferes with HIV-1 replication, presumably at the integration step. Moreover, a lentiviral vector engineered not to interact with LEDGF/p75 by the introduction of a single amino acid exchange in integrase (Q168A) was found to be transduction deficient. Together, our results provide strong evidence for an important role of this integrase cofactor in HIV-1 replication. Previously, some doubts have been raised concerning the role of LEDGF/p75 during HIV integration and replication. We had to rule out the possibility that the effect of the L3 siRNA on HIV-1 infectivity was due to nonspecific effects not directly related to LEDGF/p75. In agreement with our findings, the members of the Stevenson lab reported a significant reduction in HIV-1 infection of primary macrophages after transient transfection with L3 (M. Stevenson, personal communication). Our data indicate that the different potencies of siRNAs used might be based on differences in the time course and/or magnitude of the knockdown. Since HIV-1 infection experiments take place over several days, it is essential to demonstrate knockdown persistence during the full duration of the experiment. Our data suggest that not all siRNAs targeting LEDGF/p75 have this characteristic and therefore are suitable for elucidating the role of LEDGF/p75 in HIV-1 replication. Yet careful characterization of the influence of L3 on cellular function does not indicate the occurrence of effects, regardless of reductions in LEDGF/75 expression. We have shown here that a potent knockdown of LEDGF/p75 to 10% at the mRNA level and to hardly detectable at the protein level has a pronounced impact on HIV replication. This pinpoints LEDGF/p75 as an important player in productive viral replication. Benarous and coworkers obtained a similar knockdown resulting in a reduction in HIV-1 replication, using another siRNA (L1; Fig. 1) targeting LEDGF/p75 (personal communication). To address the controversy regarding the in vivo relevance of transient knockdown experiments, we took our study one step further by generating stable knockdown cell lines. Moreover, we could reverse the observed phenotype by back-complementation of our stable knockdown cell lines with a short-hairpin-insensitive LEDGF/p75 construct. Again, these experiments were carefully controlled and included all relevant control cell lines. Intriguingly, HIV replication was reduced up to fourfold after the depletion of LEDGF/p75. Interferon-mediated inhibition of HIV-1 replication or off-target effects are unlikely under these conditions. This strong correlation of transient and stable siRNA experiments strongly supports our conclusions on the importance of LEDGF/p75 for HIV replication. Poeschla and coworkers reported that a stable knockdown of LEDGF/p75 expression in a Jurkat T-cell line did not affect HIV-1 replication in comparison with a back-complemented cell line (22). We obtained both cell lines and were able to reproduce both the Western blot and HIV-1 replication data (data not shown). We have at present no explanation for why a 10-fold reduction in LEDGF/p75 in Jurkat cells apparently does not hamper HIV replication. Possibly, the levels of LEDGF/p75 required for HIV replication differ from cell line to cell line. The HeLaP4-derived cell lines presented here were carefully controlled for (co)receptor expression, cell growth, and cell viability prior to and after HIV-1 infection. In order to exclude cell type specificities, we have generated knockdown T-cell lines. Preliminary experiments corroborate the data obtained with HeLaP4 cells, demonstrating a pronounced decrease in HIV replication in cells that are nearly depleted of LEDGF/p75.
Recently, Llano and coworkers showed that transduction by HIV-1- and feline immunodeficiency virus-derived lentiviral vectors was not affected in 293T cells with stably suppressed LEDGF/p75 levels (22). We obtained similar results with our 293T and HeLaP4 LEDGF/p75 knockdown cells. The discrepancy between the inhibition of virus replication on the one hand and the lack of inhibition of lentiviral vector transduction in the same HeLaP4/shp75PC cells on the other hand required further investigation. Since the lentiviral vector is pseudotyped with the vesicular stomatitis virus glycoprotein envelope, it enters the cell through endocytosis, bypassing the normal HIV entry process based on membrane fusion and interaction with the CD4 receptor and CXCR4 coreceptor. We investigated whether the requirement for LEDGF/p75 during HIV integration depends on HIV entry, possibly through entry-induced signal transduction cascades and posttranslational modifications of LEDGF/p75. To examine this hypothesis, we infected the HeLaP4/shp75PC cell line with a high MOI of HIV-1 (NL4.3) before transduction with a high MOI of lentiviral vectors (data not shown). Yet no inhibition of lentiviral vector transduction in HIV-1-primed polyclonal HelaP4-p75 knockdown cell lines was observed compared with transduction in control cell lines.
It is conceivable that the extent of knockdown of the abundant LEDGF/p75 protein achieved by RNA interference is insufficient to result in a clear inhibition of vector transduction (or HIV infection in particular cell lines). We recently presented a mutant of integrase (Q168A) (13) that is incapable of interaction with LEDGF/p75 but exhibits enzymatic activities in vitro similar to those of wild-type IN. Moreover, a mutant virus bearing this amino acid substitution lost the capability of replication. For this study, we studied the effect of this mutation on vector transduction and clearly showed that the Q168A mutant is also suicidal for the lentiviral vector. We have hypothesized a possible role for LEDGF/p75 in targeting integration into transcriptionally active regions (5, 13). Lentiviral vectors that drive reporter gene expression from an internal cytomegalovirus promoter may therefore be less dependent on LEDGF/p75 activity. However, in our hands, the transduction efficiency of an LTR-dependent viral vector was not hampered after the knockdown of LEDGF/p75. In our opinion, two possibilities remain open to explain the vector/virus paradox, as follows: (i) the genomic sizes of vectors and virus differ significantly, and this may affect PIC nuclear import, targeting, and integration; and (ii) since LEDG/p75 is an abundant protein in cells, the knockdown obtained may be insufficient to demonstrate an inhibition of transduction by lentiviral vectors used at the rather high MOI required for detecting reporter gene expression. Most probably, the generation of LEDGF/p75 knockout cell lines will provide the answer. In conclusion, we employed different approaches to elucidate the function of LEDGF/p75 in HIV-1 replication and vector transduction. Despite the potential pitfalls of the individual techniques applied, all the data combined consistently demonstrate the key role of LEDGF/p75 in productive HIV replication, unveiling LEDGF/p75 as a new target for antiviral therapy.
Acknowledgments
We acknowledge M. Michiels, B. Poddesu, B. Van Remoortele, and A. Nijs for excellent technical assistance. We acknowledge Y. Engelborghs (KULeuven) for use of the confocal microscope. We thank the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, for providing different cell lines and the HIV pNL4.3 molecular clone. We are grateful to E. Poeschla (Mayo Clinic College of Medicine) for providing us the back-complementation plasmid and the different Jurkat cell lines.
Work at KULeuven was financially supported by the European Commission (LSHB-CT-2003-503480) and the SBO program of the Flemish IWT (530-030239). L.V., J.D.R., and B.V.M. are funded by grants from the Flemish Institute Supporting Scientific-Technological Research in Industry (IWT).
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asante-Appiah, E., and A. M. Skalka. 1997. Molecular mechanisms in retrovirus DNA integration. Antiviral Res. 36:139-156. [DOI] [PubMed] [Google Scholar]
- 3.Baekelandt, V., A. Claeys, K. Eggermont, E. Lauwers, B. De Strooper, B. Nuttin, and Z. Debyser. 2002. Characterization of lentiviral vector-mediated gene transfer in adult mouse brain. Hum. Gene Ther. 13:841-853. [DOI] [PubMed] [Google Scholar]
- 4.Beitzel, B., and F. Bushman. 2003. Construction and analysis of cells lacking the HMGA gene family. Nucleic Acids Res. 31:5025-5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Busschots, K., J. Vercammen, S. Emiliani, R. Benarous, Y. Engelborghs, F. Christ, and Z. Debyser. 2005. The interaction of LEDGF/p75 with integrase is lentivirus-specific and promotes DNA binding. J. Biol. Chem. 280:17841-17847. [DOI] [PubMed] [Google Scholar]
- 6.Chen, H., and A. Engelman. 1998. The barrier-to-autointegration protein is a host factor for HIV type 1 integration. Proc. Natl. Acad. Sci. USA 95:15270-15274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherepanov, P., G. Maertens, P. Proost, B. Devreese, J. Van Beeumen, Y. Engelborghs, E. De Clercq, and Z. Debyser. 2003. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 278:372-381. [DOI] [PubMed] [Google Scholar]
- 8.Cherepanov, P., Z. Y. Sun, S. Rahman, G. Maertens, G. Wagner, and A. Engelman. 2005. Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75. Nat. Struct. Mol. Biol. 12:526-532. [DOI] [PubMed] [Google Scholar]
- 9.Craigie, R. 2001. HIV integrase, a brief overview from chemistry to therapeutics. J. Biol. Chem. 276:23213-23216. [DOI] [PubMed] [Google Scholar]
- 10.Devroe, E., and P. A. Silver. 2002. Retrovirus-delivered siRNA. BMC Biotechnol. 2:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dietz, F., S. Franken, K. Yoshida, H. Nakamura, J. Kappler, and V. Gieselmann. 2002. The family of hepatoma-derived growth factor proteins: characterization of a new member HRP-4 and classification of its subfamilies. Biochem. J. 366:491-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dull, T., R. Zufferey, M. Kelly, R. J. Mandel, M. Nguyen, D. Trono, and L. Naldini. 1998. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 72:8463-8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emiliani, S., A. Mousnier, K. Busschots, M. Maroun, B. Van Maele, D. Tempe, L. Vandekerckhove, F. Moisant, L. Ben-Slama, M. Witvrouw, F. Christ, J. C. Rain, C. Dargemont, Z. Debyser, and R. Benarous. 2005. Integrase mutants defective for interaction with LEDGF/p75 are impaired in chromosome tethering and HIV-1 replication. J. Biol. Chem. 280:25517-25523. [DOI] [PubMed] [Google Scholar]
- 14.Farnet, C. M., and F. D. Bushman. 1997. HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell 88:483-492. [DOI] [PubMed] [Google Scholar]
- 15.Fatma, N., D. P. Singh, T. Shinohara, and L. T. Chylack, Jr. 2001. Transcriptional regulation of the antioxidant protein 2 gene, a thiol-specific antioxidant, by lens epithelium-derived growth factor to protect cells from oxidative stress. J. Biol. Chem. 276:48899-48907. [DOI] [PubMed] [Google Scholar]
- 16.Ganapathy, V., T. Daniels, C. A. Casiano, D. P. Singh, A. Kimura, L. T. Chylack, Jr., T. Shinohara, N. Ohguro, T. Kikuchi, T. Sueno, V. N. Reddy, K. Yuge, and N. Fatma. 2003. LEDGF/p75: a novel nuclear autoantigen at the crossroads of cell survival and apoptosis. Autoimmun. Rev. 2:290-297. [DOI] [PubMed] [Google Scholar]
- 17.Ge, H., Y. Si, and A. P. Wolffe. 1998. A novel transcriptional coactivator, p52, functionally interacts with the essential splicing factor ASF/SF2. Mol. Cell 2:751-759. [DOI] [PubMed] [Google Scholar]
- 18.Geraerts, M., M. Michiels, V. Baekelandt, Z. Debyser, and R. Gijsbers. 2005. Upscaling of lentiviral vector production by tangential flow filtration. J. Gene Med. 7:1299-1310. [DOI] [PubMed] [Google Scholar]
- 19.Kalpana, G. V., S. Marmon, W. Wang, G. R. Crabtree, and S. P. Goff. 1994. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science 266:2002-2006. [DOI] [PubMed] [Google Scholar]
- 20.Kirsch, R. D., and E. Joly. 1998. An improved PCR-mutagenesis strategy for two-site mutagenesis or sequence swapping between related genes. Nucleic Acids Res. 26:1848-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, M. S., and R. Craigie. 1994. Protection of retroviral DNA from autointegration: involvement of a cellular factor. Proc. Natl. Acad. Sci. USA 91:9823-9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Llano, M., M. Vanegas, O. Fregoso, D. Saenz, S. Chung, M. Peretz, and E. M. Poeschla. 2004. LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral preintegration complexes. J. Virol. 78:9524-9537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maertens, G., P. Cherepanov, Z. Debyser, Y. Engelborghs, and A. Engelman. 2004. Identification and characterization of a functional nuclear localization signal in the HIV-1 integrase interactor LEDGF/p75. J. Biol. Chem. 279:33421-33429. [DOI] [PubMed] [Google Scholar]
- 24.Maertens, G., P. Cherepanov, W. Pluymers, K. Busschots, E. De Clercq, Z. Debyser, and Y. Engelborghs. 2003. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J. Biol. Chem. 278:33528-33539. [DOI] [PubMed] [Google Scholar]
- 25.Margalit, A., M. Segura-Totten, Y. Gruenbaum, and K. L. Wilson. 2005. Barrier-to-autointegration factor is required to segregate and enclose chromosomes within the nuclear envelope and assemble the nuclear lamina. Proc. Natl. Acad. Sci. USA 102:3290-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, M. D., and F. D. Bushman. 1995. HIV integration. Ini1 for integration? Curr. Biol. 5:368-370. [DOI] [PubMed] [Google Scholar]
- 27.Naldini, L., U. Blomer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263-267. [DOI] [PubMed] [Google Scholar]
- 28.Nameki, N., N. Tochio, S. Koshiba, M. Inoue, T. Yabuki, M. Aoki, E. Seki, T. Matsuda, Y. Fujikura, M. Saito, M. Ikari, M. Watanabe, T. Terada, M. Shirouzu, M. Yoshida, H. Hirota, A. Tanaka, Y. Hayashizaki, P. Guntert, T. Kigawa, and S. Yokoyama. 2005. Solution structure of the PWWP domain of the hepatoma-derived growth factor family. Protein Sci. 14:756-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Doherty, U., W. J. Swiggard, D. Jeyakumar, D. McGain, and M. H. Malim. 2002. A sensitive, quantitative assay for human immunodeficiency virus type 1 integration. J. Virol. 76:10942-10950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scacheri, P. C., O. Rozenblatt-Rosen, N. J. Caplen, T. G. Wolfsberg, L. Umayam, J. C. Lee, C. M. Hughes, K. S. Shanmugam, A. Bhattacharjee, M. Meyerson, and F. S. Collins. 2004. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc. Natl. Acad. Sci. USA 101:1892-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schroder, A. R., P. Shinn, H. Chen, C. Berry, J. R. Ecker, and F. Bushman. 2002. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110:521-529. [DOI] [PubMed] [Google Scholar]
- 32.Singh, D. P., N. Fatma, A. Kimura, L. T. Chylack, Jr., and T. Shinohara. 2001. LEDGF binds to heat shock and stress-related element to activate the expression of stress-related genes. Biochem. Biophys. Res. Commun. 283:943-955. [DOI] [PubMed] [Google Scholar]
- 33.Singh, D. P., N. Ohguro, T. Kikuchi, T. Sueno, V. N. Reddy, K. Yuge, L. T. Chylack, Jr., T. Shinohara, N. Fatma, and A. Kimura. 2000. Lens epithelium-derived growth factor: effects on growth and survival of lens epithelial cells, keratinocytes, and fibroblasts. Biochem. Biophys. Res. Commun. 267:373-381. [DOI] [PubMed] [Google Scholar]
- 34.Sledz, C. A., M. Holko, M. J. de Veer, R. H. Silverman, and B. R. Williams. 2003. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 5:834-839. [DOI] [PubMed] [Google Scholar]
- 35.Stec, I., S. B. Nagl, G. J. van Ommen, and J. T. den Dunnen. 2000. The PWWP domain: a potential protein-protein interaction domain in nuclear proteins influencing differentiation? FEBS Lett. 473:1-5. [DOI] [PubMed] [Google Scholar]
- 36.Suzuki, Y., and R. Craigie. 2002. Regulatory mechanisms by which barrier-to-autointegration factor blocks autointegration and stimulates intermolecular integration of Moloney murine leukemia virus preintegration complexes. J. Virol. 76:12376-12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki, Y., H. Yang, and R. Craigie. 2004. LAP2alpha and BAF collaborate to organize the Moloney murine leukemia virus preintegration complex. EMBO J. 23:4670-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turlure, F., E. Devroe, P. A. Silver, and A. Engelman. 2004. Human cell proteins and human immunodeficiency virus DNA integration. Front. Biosci. 9:3187-3208. [DOI] [PubMed] [Google Scholar]
- 39.Van den Haute, C., K. Eggermont, B. Nuttin, Z. Debyser, and V. Baekelandt. 2003. Lentiviral vector-mediated delivery of short hairpin RNA results in persistent knockdown of gene expression in mouse brain. Hum. Gene Ther. 14:1799-1807. [DOI] [PubMed] [Google Scholar]
- 40.Vanegas, M., M. Llano, S. Delgado, D. Thompson, M. Peretz, and E. Poeschla. 2005. Identification of the LEDGF/p75 HIV-1 integrase-interaction domain and NLS reveals NLS-independent chromatin tethering. J. Cell Sci. 118:1733-1743. [DOI] [PubMed] [Google Scholar]
- 41.Van Maele, B., J. De Rijck, E. De Clercq, and Z. Debyser. 2003. Impact of the central polypurine tract on the kinetics of human immunodeficiency virus type 1 vector transduction. J. Virol. 77:4685-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Maele, B., and Z. Debyser. 2005. HIV-1 integration: an interplay between HIV-1 integrase, cellular and viral proteins. AIDS Rev. 7:26-43. [PubMed] [Google Scholar]
- 43.Wang, W., J. Cote, Y. Xue, S. Zhou, P. A. Khavari, S. R. Biggar, C. Muchardt, G. V. Kalpana, S. P. Goff, M. Yaniv, J. L. Workman, and G. R. Crabtree. 1996. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 15:5370-5382. [PMC free article] [PubMed] [Google Scholar]
- 44.Wu, X., Y. Li, B. Crise, and S. M. Burgess. 2003. Transcription start regions in the human genome are favored targets for MLV integration. Science 300:1749-1751. [DOI] [PubMed] [Google Scholar]
- 45.Yung, E., M. Sorin, A. Pal, E. Craig, A. Morozov, O. Delattre, J. Kappes, D. Ott, and G. V. Kalpana. 2001. Inhibition of HIV-1 virion production by a transdominant mutant of integrase interactor 1. Nat. Med. 7:920-926. [DOI] [PubMed] [Google Scholar]