Abstract
The tRNAs used to prime reverse transcription in human immunodeficiency virus type 1 (HIV-1), Rous sarcoma virus (RSV), and Moloney murine leukemia virus (Mo-MuLV) are  , tRNATrp, and tRNAPro, respectively. Using antibodies to the three cognate human aminoacyl-tRNA synthetases, we found that only lysyl-tRNA synthetase (LysRS) is present in HIV-1, only tryptophanyl-tRNA synthetase (TrpRS) is present in RSV, and neither these two synthetases nor prolyl-tRNA synthetase (ProRS) is present in Mo-MuLV. LysRS and TrpRS are present in HIV-1 and RSV at approximately 25 and 12 molecules/virion, respectively. These results support the hypothesis that, in HIV-1 and RSV, the cognate aminoacyl-tRNA synthetase may be used as the signal for targeting the selective packaging of primer tRNAs into retroviruses. The absence of ProRS in Mo-MuLV is consistent with reports that selective packaging of tRNAPro in this virus is less important for achieving optimum annealing of the primer to Mo-MuLV genomic RNA.
, tRNATrp, and tRNAPro, respectively. Using antibodies to the three cognate human aminoacyl-tRNA synthetases, we found that only lysyl-tRNA synthetase (LysRS) is present in HIV-1, only tryptophanyl-tRNA synthetase (TrpRS) is present in RSV, and neither these two synthetases nor prolyl-tRNA synthetase (ProRS) is present in Mo-MuLV. LysRS and TrpRS are present in HIV-1 and RSV at approximately 25 and 12 molecules/virion, respectively. These results support the hypothesis that, in HIV-1 and RSV, the cognate aminoacyl-tRNA synthetase may be used as the signal for targeting the selective packaging of primer tRNAs into retroviruses. The absence of ProRS in Mo-MuLV is consistent with reports that selective packaging of tRNAPro in this virus is less important for achieving optimum annealing of the primer to Mo-MuLV genomic RNA.
Specific cellular tRNA isoacceptors are packaged into retroviruses during viral assembly, and some of these are annealed onto the primer-binding site of viral genomic RNA and used to initiate the synthesis of minus-strand cDNA by reverse transcriptase. Different primer tRNAs are used by different retroviral families. tRNATrp is the primer for all members of the avian sarcoma virus-avian leukosis virus group (19, 20, 24). The major tRNALys isoacceptors in mammalian cells are 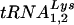 and
and 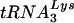 , the latter being the primer tRNA for reverse transcriptase in lentiviruses, including human immunodeficiency virus type 1 (HIV-1) (18). Primer tRNAs are selectively packaged into retroviruses during their assembly, i.e., their relative concentration in the low-molecular-weight RNA population increases in moving from the cytoplasm to the virus. For example, the relative concentration of tRNATrp has been reported to increase from 1.4% to 32% in avian myeloblastosis virus (24) while tRNALys (i.e.,
, the latter being the primer tRNA for reverse transcriptase in lentiviruses, including human immunodeficiency virus type 1 (HIV-1) (18). Primer tRNAs are selectively packaged into retroviruses during their assembly, i.e., their relative concentration in the low-molecular-weight RNA population increases in moving from the cytoplasm to the virus. For example, the relative concentration of tRNATrp has been reported to increase from 1.4% to 32% in avian myeloblastosis virus (24) while tRNALys (i.e.,  and
and  ) increases from 5 to 6% in the cytoplasm to 50 to 60% in HIV-1 (16). On the other hand, selective packaging of primer tRNAPro in Moloney murine leukemia virus (Mo-MuLV) has been reported to be much less, with the relative concentration of tRNAPro within low-molecular-weight RNA changing from 5 to 6% in the cytoplasm to 12 to 24% in the virion (24). Furthermore, while mutations in the reverse transcriptase eliminate selective packaging of primer tRNA in all three of these retroviruses, primer tRNA annealing to the viral genome is reduced in only HIV-1 and the avian retroviruses and not in Mo-MuLV (7, 14-16, 19). Increases in the concentration of primer
) increases from 5 to 6% in the cytoplasm to 50 to 60% in HIV-1 (16). On the other hand, selective packaging of primer tRNAPro in Moloney murine leukemia virus (Mo-MuLV) has been reported to be much less, with the relative concentration of tRNAPro within low-molecular-weight RNA changing from 5 to 6% in the cytoplasm to 12 to 24% in the virion (24). Furthermore, while mutations in the reverse transcriptase eliminate selective packaging of primer tRNA in all three of these retroviruses, primer tRNA annealing to the viral genome is reduced in only HIV-1 and the avian retroviruses and not in Mo-MuLV (7, 14-16, 19). Increases in the concentration of primer  in HIV-1 result in both greater
in HIV-1 result in both greater  annealing to the genome and greater viral infectivity (8).
annealing to the genome and greater viral infectivity (8).
It was recently reported that human lysyl-tRNA synthetase (LysRS), the enzyme that aminoacylates tRNALys, is also selectively packaged into HIV-1 (2). In contrast, two other aminoacyl-tRNA synthetases (aaRSs), isoleucyl-tRNA synthetase and prolyl-tRNA synthetase (ProRS), were not detected in HIV-1 (2). This result suggests that LysRS may represent the factor which signals HIV-1 proteins to interact with a tRNALys/LysRS complex for selective packaging of tRNALys into the virus. This hypothesis is further strengthened by studies with 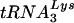 anticodon mutants showing that the extent of
anticodon mutants showing that the extent of  aminoacylation is directly correlated with its incorporation into the virion (11). This suggests that productive
aminoacylation is directly correlated with its incorporation into the virion (11). This suggests that productive 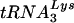 interaction with LysRS is a prerequisite for packaging. The aim of the present study was to determine whether the cognate aaRS for primer tRNAs in Rous sarcoma virus (RSV) and Mo-MuLV (i.e., tryptophanyl-tRNA synthetase [TrpRS] and ProRS, respectively) are also selectively packaged into the virion.
interaction with LysRS is a prerequisite for packaging. The aim of the present study was to determine whether the cognate aaRS for primer tRNAs in Rous sarcoma virus (RSV) and Mo-MuLV (i.e., tryptophanyl-tRNA synthetase [TrpRS] and ProRS, respectively) are also selectively packaged into the virion.
Purified HIV-1, RSV, and Mo-MuLV were obtained from HIV-1-transfected human 293T cells, RSV-infected turkey embryo fibroblasts (TEFs), and Mo-MuLV-infected murine NIH 3T3 cells as described in the legend to Fig. 1. Figure 1A shows Western blots of lysates from HIV-1, RSV, and Mo-MuLV, probed, respectively, with mouse antiserum to CAp24, rabbit antiserum to CAp27, and rabbit antiserum to CAp30. Panels B through D show Western blots of cell and viral lysates (human 293T cells and HIV-1, TEFs and RSV, and murine NIH 3T3 cells and Mo-MuLV, respectively) probed with anti-human LysRS (panel B), anti-human TrpRS (panel C), and anti-human ProRS (panel D). The major cytoplasmic LysRS species in all cell types migrated with an apparent molecular weight (Mr) of 68,000 to 70,000. In 293T cells, this was also accompanied by a minor cytoplasmic species with an Mr of 62,000 (lane 2), while in RSV, a minor species with an Mr of approximately 45,000 was observed (lane 4). As reported previously (2), panel B, lane 1, shows two species of LysRS in HIV-1, one with an Mr of approximately 68,000 to 70,000 and one with that of approximately 63,000. LysRS was not detected in RSV or Mo-MuLV (lanes 3 and 5).
FIG. 1.
Western blots of cell and viral lysates probed with antibodies to viral proteins or aaRSs. HIV-1 was produced and isolated from HIV-1-transfected human 293T cells as previously described for HIV-1-transfected COS7 cells (2). RSV was produced and isolated from infected TEFs as previously described (5). Mo-MuLV was produced and isolated from infected murine NIH 3T3 cells and purified by banding twice in sucrose (and concentrated 5,000 times) and was provided by the AIDS Vaccine Program, SAIC Frederick, National Cancer Institute, Frederick, Frederick, Md. Purified viruses and cells were lysed by suspension in 1× radioimmunoprecipitation assay buffer (10 mM Tris [pH 7.4], 100 mM NaCl, 1% deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1% Nonidet P-40, protease inhibitor cocktail tablets [Boehringer Mannheim]), and the protein concentration in the samples was determined by the Bradford assay (1). One hundred micrograms of cellular protein or 30 μg of viral protein was resolved by SDS-10% polyacrylamide gel electrophoresis followed by blotting onto nitrocellulose membranes (Amersham Life Sciences) and detection with antibody. (A) Western blots of viral lysates of HIV-1, RSV, and Mo-MuLV were probed, respectively, with mouse antiserum to HIV capsid (Intracel Corporation), rabbit antiserum to RSV (25), and rabbit antiserum to Mo-MuLV capsid (AIDS Vaccine Program). (B through D) Western blots of viral and cellular lysates were probed with rabbit antiserum to human LysRS (B), human TrpRS (C), and human ProRS (D). Rabbit antisera to these aaRSs were isolated following three subcutaneous injections of purified protein with 3- to 4-week intervals between injections (150 to 300 μg of total protein). The isolations of human LysRS (21) and TrpRS (23) were as previously described. Human ProRS is derived from the C-terminal domain (amino acid residues 926 to 1440) of GluProRS and was purified as described previously (9). Western blots were analyzed by enhanced chemiluminescence (ECL kit; Amersham Life Sciences) with goat anti-mouse or donkey anti-rabbit antibody (Amersham Life Sciences) as a secondary antibody. The sizes of the detected protein bands were estimated by using prestained high-molecular-mass protein markers (GIBCO/BRL), represented by the numbers to the left of each panel. V, viral lysate; C, cell lysate.
As shown in Fig. 1C, a similar blot was probed with rabbit anti-human TrpRS (23). The major species seen in 293T cells (lane 2) migrated with an Mr of approximately 55,000, somewhat larger than the Mr of the major TrpRS species seen in NIH 3TC cells (approximately 45,000 [lane 6]). The major cytoplasmic TrpRS species in TEFs appeared to migrate with an Mr equal to 62,000 (lane 4) and appeared larger than TrpRS in mouse or human cells. TrpRS was not detected in HIV-1 (lane 1) or Mo-MuLV (lane 5) but was detected in RSV as two bands with Mrs of approximately 62,000 and 55,000.
In Fig. 1D, a Western blot was probed with rabbit anti-human ProRS. The two major species seen in 293T cells migrated with Mrs of approximately 160,000 and 55,000. In higher eukaryotes, such as the rat (22) and Drosophila melanogaster (3), sheep (13), and humans (6, 12), ProRS is the C-terminal part of a fusion with glutamyl-tRNA synthetase, and the large species on this gel corresponds to the human glutamyl-prolyl-tRNA synthetase (GluProRS) fusion protein. The smaller species migrates according to purified human ProRS, which has an Mr of approximately 55,000 to 60,000. The lower-molecular-weight band was also seen in the cytoplasm of TEFs (lane 4) and murine NIH 3T3 cells (lane 6). In TEFs, the larger species was detected as a protein with an Mr of 80,000, while the larger species in murine NIH 3T3 cells (lane 6) migrated somewhat faster than the large species found in the human 293T cells. ProRS was not detected in HIV-1, RSV, or Mo-MuLV (lanes 1, 3, and 5).
We next determined the number of molecules of LysRS and TrpRS present in HIV-1 and RSV and in the cells producing these viruses. Human His6-LysRS was expressed in bacteria and purified as previously described (21). Human TrpRS was purified from Escherichia coli as previously described (23). Western blots of known amounts of purified LysRS (Fig. 2A, lanes 1 to 7) or TrpRS (Fig. 3A, lanes 1 to 7) were quantitated by phosphorimaging and used to generate the LysRS and TrpRS standard curves shown in Fig. 2C and 3C, respectively. These curves were used to quantitate the amount of LysRS and TrpRS present in viral and cell lysates. Total protein in the viral and cell lysates was determined by the Bradford assay (1), and the nanograms of aaRS/microgram of protein for both the cell and viral lysates are listed in Table 1. The standard curves in Fig. 2 and 3 indicate the lower limit of detection of either LysRS or TrpRS at approximately 0.15 ng, and this was used to calculate the limits of detection listed in Table 1 when no aaRS signal was detected. In both human 293T cells and TEFs, the relative concentrations of LysRS and TrpRS were very similar. However, whereas the relative amount of LysRS in HIV-1 was comparable to that found in the cell lysate, TrpRS was not detected in HIV-1. Similarly, the relative amount of TrpRS in RSV was similar to the amount found in TEFs whereas LysRS was not detected in RSV (Table 1). Viruses with large differences in the relative concentrations of LysRS and TrpRS were produced in cells in which there was little difference in the cytoplasmic concentrations of these two aaRSs. This indicates the existence of a selective packaging mechanism for the aaRS cognate to the primer tRNA found in either HIV-1 or RSV.
FIG. 2.
Quantitation of LysRS in HIV-1. (A) Western blots probed with anti-LysRS. Purified viruses and cells were lysed by suspension in 1× radioimmunoprecipitation assay buffer, and 100 μg of cell lysate protein or 30 μg of viral lysate protein was resolved by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis, followed by blotting onto nitrocellulose membranes and probing with anti-human LysRS. (B) Total viral RNA from 30 μg of viral protein of HIV-1 was extracted by the guanidinium isothiocyanate procedure (4). HIV-1 genomic RNA was measured by dot blot hybridization with a radioactive DNA probe specific for the HIV-1 genomic RNA sequence from nucleotides 338 to 354. (C) The first seven lanes in the Western blot shown in panel A contained decreasing amounts of His6-human LysRS, purified on Ni+ columns as previously described (21), and were used to generate the LysRS concentration curve shown in panel C. This standard curve was used to determine the amount of LysRS present in lysates of HIV-1 (lane 8), 293T cells (lane 9), RSV (lane 10), and TEFs (lane 11), shown in panel A. (D) Known amounts of in vitro synthetic HIV-1 genomic RNA (lanes 1 to 7) were used to establish a standard RNA concentration curve as previously described (17). This curve was used to establish the concentration of viral RNA in the HIV-1 RNA sample in lane 8 of panel B. The mock lane (lane 9) contained a total RNA preparation from the culture medium of nontransfected 293T cells.
FIG. 3.
Quantitation of TrpRS in RSV. (A) A Western blot was prepared similar to that shown in Fig. 2A, except purified LysRS was replaced with purified TrpRS, and the blot was probed with anti-human TrpRS. The expression of TrpRS in E. coli and its purification were done as previously described (23). (B) Total viral RNA from 30 μg of viral protein of RSV was extracted by the guanidinium isothiocyanate procedure (4). RSV genomic RNA was measured by dot blot hybridization with a radioactive oligomer probe (5′ ACA GCC AAT AGC TGT TCC GCA 3′) that hybridized specifically to the RSV genomic RNA sequence from nucleotides 848 to 868. (C) The first seven lanes in the Western blot shown in panel A contained decreasing amounts of purified human TrpRS and were used to generate the TrpRS concentration curve shown in panel C. This standard curve was used to determine the amount of TrpRS present in lysates of HIV-1 (lane 8), human 293T cells (lane 9), RSV (lane 10), and TEFs (lane 11), as shown in panel A. (D) To generate the RSV RNA concentration curve, a 193-bp fragment coding the RSV genome from nucleotides 763 to 956 was amplified by PCR from RSV proviral cDNA. For the PCR, a sense primer (5′ GGA TCC TAA TAC GAC TCA TAG GGA GAA ACA ACT GTG CAG CGA GAT 3′) was used to introduce a T7 promoter 5′ terminal to the amplified fragment and an antisense primer sequence (5′ CCT TGG CCC TGC TCT CCC A 3′) was used. Then the purified PCR product was used as a template to in vitro transcribe RSV genomic RNA, and a standard curve was established by using known amounts of this synthetic RNA (panel B, lanes 1 to 7). This curve was used to establish the concentration of viral RNA in the RSV RNA sample in lane 8 of panel B. The mock lane (lane 9) contained a total RNA preparation from the culture medium of nontransfected TEF cells.
TABLE 1.
Quantitation of aaRSs in HIV-1 and RSVa
| Sample | Concn (ng/μg cell or viral protein)
|
No. of molecules/virion
|
||
|---|---|---|---|---|
| LysRS | TrpRS | LysRS | TrpRS | |
| HIV-1 | 0.069 ± 0.027 | <0.005 | 25 ± 9 | <1.7 |
| 293T cells | 0.082 ± 0.017 | 0.087 ± 0.022 | —b | — |
| RSV | <0.005 | 0.051 ± 0.014 | <1.1 | 12 ± 5 |
| TEFs | 0.095 ± 0.028 | 0.078 ± 0.019 | — | — |
Determinations of the number of aaRSs per cell or viral protein and the number of aaRSs per virion were done in triplicate. The average values are listed, including the range of values obtained from the different experiments.
—, not applicable.
To determine the number of aaRS molecules/virion, we first determined the number of genomic RNA molecules present in the viral lysates. Total viral RNA from HIV-1, RSV, or Mo-MuLV was extracted by the guanidinium isothiocyanate procedure (4). Viral genomic RNA was quantitated by hybridizing dot blots of viral RNA with the following DNA oligonucleotides: HIV-1 (5′ CTG ACG CTC TCG CAC CC3 ′), complementary to HIV-1 genomic RNA nucleotides 338 to 354 (Fig. 2B); RSV (5′ ACA GCC AAT AGC TGT TCC GCA3 ′), complementary to RSV genomic RNA nucleotides 848 to 868 (Fig. 3B); and Mo-MuLV (5′ ATC CCG GAC GAG CCC CCA 3′), complementary to Mo-MuLV primer-binding site nucleotides 595 to 612 (Fig. 4B). Standard curves were established by using known amounts of synthetic viral RNA, which were transcribed in vitro from PCR-amplified fragments encoding HIV-1 nucleotides 473 to 1420 (Fig. 2B and D), RSV nucleotides 763 to 956 (Fig. 3B and D), or Mo-MuLV nucleotides 456 to 797 (Fig. 4B and D). Based on the data shown in Fig. 2 and 3, and assuming there were two RNA molecules/virion, we estimated the amounts of LysRS in HIV and TrpRS in RSV to be approximately 25 and 12 molecules, respectively (Table 1). These estimates were determined by assuming that the genomic RNA probes hybridize with equal efficiency to both the full-length viral RNA in our samples and the much shorter synthetic RNA fragments used to generate the genomic RNA standard curves.
FIG. 4.
Quantitation of ProRS in Mo-MuLV. (A) A Western blot was prepared similar to that shown in Fig. 2A, except purified LysRS was replaced with purified ProRS, only Mo-MuLV and murine NIH 3T3 cells were examined, and the blot was probed with anti-human ProRS. The expression of ProRS in E. coli and its purification were done as previously described (9). (B) Total viral RNA from 30 μg of viral protein of Mo-MuLV was extracted by the guanidinium isothiocyanate procedure (4). Mo-MuLV genomic RNA was measured by dot blot hybridization with a radioactive oligomer probe (5′ ATC CCG GAC GAG CCC CCA 3′) complementary to the Mo-MuLV primer-binding site from nucleotides 595 to 612. (C) The first seven lanes in the Western blot shown in panel A contained decreasing amounts of purified human ProRS and were used to generate the ProRS concentration curve shown in panel C. This standard curve was used to determine the amount of ProRS present in lysates of Mo-MuLV (lane 8) and murine NIH 3T3 cells (lane 9), as shown in panel A. To generate the Mo-MuLV RNA concentration curve shown in panel D, a DNA fragment coding the Mo-MuLV genome from nucleotides 456 to 797 was amplified by PCR from Mo-MuLV proviral cDNA with the primer sequences 5′ AGTCCTCCGATTGACTGA 3′ and 5′ CCGAACTCGTCAGTTCCA 3′. The PCR fragment was ligated into the pCR-Blunt vector (Invitrogen), and this vector was linearized at the BamHI site. In vitro transcription was performed using the Megascript kit (Ambion). The size of the RNA is 341 bases. In vitro-transcribed Mo-MuLV genomic RNA was used to establish a standard curve (panel B, lanes 1 to 7). This curve was used to establish the concentration of viral RNA in the Mo-MuLV RNA sample (lane 8). The mock lane (lane 9) contained a total RNA preparation from the culture medium of nontransfected murine NIH 3T3 cells.
As shown in Fig. 1 and 4, we were unable to detect ProRS in Mo-MuLV. The standard ProRS curve in Fig. 4 indicates a lower limit of detection of ProRS at approximately 0.15 ng, and this was used to calculate the numbers listed in Table 1 when no aaRS signal was detected. Based upon this, we were unable to detect the presence of ProRS in fewer than 1.6 molecules/virion. These data also indicate that the concentration of ProRS was 0.061 ± 0.018 ng/μg of NIH 3T3 cell protein, i.e., very similar to the cellular concentrations of LysRS and TrpRS shown in Table 1.
The similarity between the number of molecules of LysRS/virion and the previously determined number of tRNALys molecules/virion (20 to 25 [10]) supports the hypothesis that LysRS is a limiting factor in tRNALys incorporation into HIV-1. (Indeed, overexpression of LysRS in HIV-1-transfected cells doubles the number of tRNALys molecules/virion [8].) The tRNATrp content in RSV has not been reported, but the determined number of avian TrpRS molecules in RSV may be an underestimate since the standard on which it was based (Fig. 3C) used antibodies specific for human TrpRS and differences which may exist between human and avian TrpRS may reduce the antibody's affinity for avian TrpRS.
Our data indicate that the aaRS cognate to the primer tRNA in both HIV-1 and RSV is selectively packaged into the virion and supports the hypothesis that aaRS may act as a signal for targeting the primer tRNA for incorporation into these viruses. On the other hand, our inability to detect ProRS in Mo-MuLV is further evidence (7, 14, 15) that the mechanism for ensuring optimal primer tRNA annealing to the viral genome in HIV-1 and avian retroviruses, i.e., selective packaging, is of less importance in Mo-MuLV.
Acknowledgments
This work was supported by grants from the Canadian Institutes for Health Research and the Canadian Foundation for AIDS Research.
We thank Sandy Fraiberg for assistance in preparation of the manuscript.
REFERENCES
- 1.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 2.Cen, S., A. Khorchid, H. Javanbakht, J. Gabor, T. Stello, K. Shiba, K. Musier-Forsyth, and L. Kleiman. 2001. Incorporation of lysyl-tRNA synthetase into human immunodeficiency virus type 1. J. Virol. 75:5043-5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cerini, C., P. Kerjan, M. Astier, D. Gratecos, M. Mirande, and M. Semeriva. 1991. A component of the multisynthetase complex is a multifunctional aminoacyl-tRNA synthetase. EMBO J. 10:4267-4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chomczynski, P., and N. Sacchi. 1987. RNA isolation from cultured cells. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 5.Craven, R. C., A. E. Leure-DuPree, R. A. Weldon, and J. W. Wills. 1995. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. J. Virol. 69:4213-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fett, R., and R. Knippers. 1991. The primary structure of human glutaminyl-tRNA synthetase. A highly conserved core, amino acid repeat regions, and homologies with translation elongation factors. J. Biol. Chem. 266:1448-1455. [PubMed] [Google Scholar]
- 7.Fu, W., B. A. Ortiz-Conde, R. J. Gorelick, S. H. Hughes, and A. Rein. 1997. Placement of tRNA primer on the primer-binding site requires pol gene expression in avian but not murine retroviruses. J. Virol. 71:6940-6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gabor, J., S. Cen, H. Javanbakht, M. Niu, and L. Kleiman. 2002. Effect of altering the tRNA3Lys3 concentration in human immunodeficiency virus type 1 upon its annealing to viral RNA, GagPol incorporation, and viral infectivity. J. Virol. 76:9096-9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heacock, D., C. J. Forsyth, K. Shiba, and K. Musier-Forsyth. 1996. Synthesis and aminoacyl-tRNA synthetase inhibitory activity of prolyl adenylate analogs. Bioorg. Chem. 24:273-289. [Google Scholar]
- 10.Huang, Y., J. Mak, Q. Cao, Z. Li, M. A. Wainberg, and L. Kleiman. 1994. Incorporation of excess wild-type and mutant tRNA3Lys into human immunodeficiency virus type 1. J. Virol. 68:7676-7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Javanbakht, H., S. Cen, K. Musier-Forsyth, and L. Kleiman. 2002. Correlation between tRNALys3 aminoacylation and incorporation into HIV-1. J. Biol. Chem. 277:17389-17396. [DOI] [PubMed] [Google Scholar]
- 12.Kaiser, E., B. Hu, S. Becher, D. Eberhard, B. Schray, M. Baack, H. Hameister, and R. Knippers. 1994. The human EPRS locus (formerly the QARS locus): a gene encoding a class I and a class II aminoacyl-tRNA synthetase. Genomics 19:280-290. [DOI] [PubMed] [Google Scholar]
- 13.Kerjan, P., M. Triconnet, and J.-P. Waller. 1992. Mammalian prolyl-tRNA synthetase corresponds to the approximately 150 kDa subunit of the high-M(r) aminoacyl-tRNA synthetase complex. Biochimie 74:195-205. [DOI] [PubMed] [Google Scholar]
- 14.Levin, J. G., S. C. Hu, A. Rein, L. I. Messer, and B. I. Gerwin. 1984. Murine leukemia virus mutant with a frameshift in the reverse transcriptase coding region: implications for pol gene structure. J. Virol. 51:470-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levin, J. G., and J. G. Seidman. 1981. Effect of polymerase mutations on packaging of primer tRNAPro during murine leukemia virus assembly. J. Virol. 38:403-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mak, J., M. Jiang, M. A. Wainberg, M.-L. Hammarskjold, D. Rekosh, and L. Kleiman. 1994. Role of Pr160gag-pol in mediating the selective incorporation of tRNALys into human immunodeficiency virus type 1 particles. J. Virol. 68:2065-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mak, J., A. Khorchid, Q. Cao, Y. Huang, I. Lowy, M. A. Parniak, V. R. Prasad, M. A. Wainberg, and L. Kleiman. 1997. Effects of mutations in Pr160gag-pol upon tRNALys3 and Pr160gag-pol incorporation into HIV-1. J. Mol. Biol. 265:419-431. [DOI] [PubMed] [Google Scholar]
- 18.Mak, J., and L. Kleiman. 1997. Primer tRNAs for reverse transcription. J. Virol. 71:8087-8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peters, G. G., and J. Hu. 1980. Reverse transcriptase as the major determinant for selective packaging of tRNA's [sic] into avian sarcoma virus particles. J. Virol. 36:692-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sawyer, R. C., and J. E. Dahlberg. 1973. Small RNAs of Rous sarcoma virus: characterization by two-dimensional polyacrylamide gel electrophoresis and fingerprint analysis. J. Virol. 12:1226-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shiba, K., T. Stello, H. Motegi, T. Noda, K. Musier-Forsyth, and P. Schimmel. 1997. Human lysyl-tRNA synthetase accepts nucleotide 73 variants and rescues E. coli double-defective mutant. J. Biol. Chem. 272:22809-22816. [DOI] [PubMed] [Google Scholar]
- 22.Ting, S. M., P. Bogner, and J. D. Dignam. 1992. Isolation of prolyl-tRNA synthetase as a free form and as a form associated with glutamyl-tRNA synthetase. J. Biol. Chem. 267:17701-17709. [PubMed] [Google Scholar]
- 23.Wakasugi, K., B. M. Slike, J. Hood, A. Otani, K. L. Ewalt, M. Friedlander, D. A. Cheresh, and P. Schimmel. 2000. A human aminoacyl-tRNA synthetase as a regulator of angiogenesis. Proc. Natl. Acad. Sci. USA 99:173-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waters, L. C., and B. C. Mullin. 1977. Transfer RNA in RNA tumor viruses. Prog. Nucleic Acid Res. Mol. Biol. 20:131-160. [DOI] [PubMed] [Google Scholar]
- 25.Weldon, R. A. J., C. R. Erdie, M. G. Oliver, and J. W. Wills. 1990. Incorporation of chimeric Gag protein into retroviral particles. J. Virol. 64:4169-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]






