Abstract
Regulation of orf73 (LANA) gene expression is critical to the establishment and maintenance of latency following infection by members of the gamma-2 herpesvirus (rhadinovirus) family. Previous studies of murine gammaherpesvirus 68 (γHV68) have demonstrated that loss of LANA function results in a complete failure to establish virus latency in the spleens of laboratory mice. Here we report the characterization of alternatively spliced LANA and v-cyclin (orf72) transcripts encoded by γHV68. Similar to other rhadinoviruses, alternative splicing, coupled with alternative 3′ processing, of a ca. 16-kb transcriptional unit can lead to expression of either LANA or v-cyclin during γHV68 infection. Spliced LANA and v-cyclin transcripts were initially identified from an analysis of the γHV68 latently infected B-cell lymphoma cell line S11E, but were also detected during lytic infection of NIH 3T12 fibroblasts. 5′ Random amplification of cDNA ends (RACE) analyses identified two distinct promoters, p1 and p2, that drive expression of spliced LANA transcripts. Analysis of p1 and p2, using transiently transfected reporter constructs, mapped the minimal sequences required for promoter activity and demonstrated that both promoters are active in the absence of any viral antigens. Analysis of spliced LANA and v-cyclin transcripts in spleens recovered from latently infected mice at days 16 and 42 postinfection revealed that spliced v-cyclin transcripts can only be detected sporadically, suggesting that these may be associated with cells reactivating from latency. In contrast, spliced LANA transcripts were detected in ca. 1 in 4,000 splenocytes harvested at day 16 postinfection. Notably, based on the frequency of viral genome-positive splenocytes at day 16 postinfection (ca. 1 in 200), only 5 to 10% of viral genome-positive splenocytes express LANA. The failure of the majority of infected splenocytes at day 16 postinfection to express LANA may contribute to the contraction in the frequency of latently infected splenocytes as chronic infection is established, due to failure to maintain the viral episome in proliferating B cells.
Orf73 of gammaherpesvirus 68 (γHV68) encodes a protein that is essential for the establishment of splenic latency following intranasal infection (14, 23). Transcript analysis of infected cells has demonstrated that orf73 is an immediate-early transcript during lytic infection in vitro, as well as a latency-associated transcript in vivo (12, 21, 37, 38). The γHV68 orf73-encoded protein is a homolog of the LANA protein encoded by Kaposi's sarcoma-associated herpesvirus (KSHV), as well as herpesvirus saimiri orf73 and rhesus rhadinovirus orf73 (1, 26, 31, 37). Although the function of the γHV68 orf73-encoded protein has not been characterized, it is thought to function during viral latency to ensure faithful maintenance of viral genomes in proliferating latently infected cells in a manner analogous to the EBNA-1 protein encoded by Epstein-Barr virus (EBV) and KSHV LANA (4, 7, 9, 10, 17, 41).
The KSHV LANA transcript identified in KSHV-infected tumor PEL cell lines is a spliced polycistronic transcript generated from a promoter that also directs transcription of alternatively spliced bicistronic or monocistronic transcripts encoding either orf72 (v-cyclin) and orf71 (v-FLIP) or orf71 alone (11, 29, 34). Translation of v-FLIP from the bicistronic transcript proceeds from an internal ribosome entry site (IRES) located within the v-cyclin coding region (5, 15, 20). In the case of herpesvirus saimiri, orf73 is encoded as the first open reading frame of an unspliced polycistronic transcript that also contains sequences for v-cyclin (orf72) and v-FLIP (orf71) (16). A second, 5′-coterminal spliced transcript encodes v-cyclin and also contains sequences for v-FLIP (16). The γHV68 genome encodes homologues of KSHV v-cyclin and KSHV LANA, but does not encode a homolog of v-FLIP.
Using the γHV68 latently infected B-lymphoma cell line S11E as well as infected murine splenocytes, we have identified spliced transcripts encoding γHV68 LANA. In addition, we describe an alternatively spliced transcript encoding γHV68 v-cyclin that initiates from the same promoter regions that direct transcription of LANA. These transcripts can be found both in lytically infected cells in vitro as well as in latently infected cells in vivo.
Identification of spliced LANA transcripts using RNA ligase-mediated RACE.
To identify the 5′ termini of γHV68 LANA transcripts, polyadenylated RNA was prepared from the S11E lymphoma cell line, followed by treatment with alkaline phosphatase to remove the 5′ phosphate from degraded messages lacking a 5′ cap (Fig. 1A). The resulting RNA was further treated with tobacco acid phosphatase to remove 5′ caps, and an RNA adaptor was ligated to the 5′ end of the decapped mRNAs (Fig. 1A). This method of random amplification of cDNA ends (RACE) is highly selective since the alkaline phosphatase treatment of noncapped messages prevents ligation of the RNA adaptor to these transcripts (Fig. 1A), and thus ligation of the RNA adaptor is targeted only to the 5′ end of full-length transcripts. Using an oligo(dT) primer, cDNA was generated from the S11 5′ adapted mRNA. Reverse transcription (RT)-PCR was then performed using a primer specific for the 5′ adaptor oligonucleotide and a primer specific to the predicted orf73 coding sequence. RT-PCR products were subcloned and subsequently characterized by DNA sequencing.
FIG. 1.
Structure of orf73 and orf72 spliced transcripts. (A) Schematic illustration of the steps involved in the RNA ligase-mediated 5′ RACE protocol. Using the GeneRacer reagents (Invitrogen), polyadenylated RNA from S11E lymphoma cells was treated as described to generate adapted RNA templates for reverse transcription into cDNA. Following nested RT-PCRs using primers specific to the RNA adaptor region and orf73 (73RTo, ATCGTCTGTCTCTCCTACATCTAAA, and 73RTi, TCAACATCAACATCTGGTGATGGTG) products were subcloned and sequenced. (B) The region of the genome encoding γHV68 orf72 and orf73 is shown. Two major spliced orf73 transcripts were identified. The larger orf73 spliced transcript contained at least one copy of a 91-bp exon (E1) located within the viral terminal repeat, a 106-bp exon (E2) and the orf73 coding exon (E3). The second, smaller orf73 transcript identified contained only E2 and E3. RT-PCR performed with a primer specific to orf72 (72RTi, TCAACATCAACATCTGGTGATGGTG) and a primer specific to exon 2 of the orf73 transcript (73E2i, TCCCGACTCGTGAGTAGCGCCGACTAG) amplified a spliced product that contains the sequences encoding orf72 as well as an additional exon from within the orf73 coding region. Products were subsequently subcloned and sequenced. The positions of the 73p1 and 73p2 promoters are also indicated.
Based on results from 5′ RACE, a number of orf73 (LANA)-containing transcripts were identified from S11E tumor cells (Fig. 1B). In S11E cells, the largest orf73 transcript identified contained one full copy of a 91-bp exon (E1) extending from bp 118695 to bp 118605. This full copy of E1 was preceded by a partial copy of this exon extending from 118632 to 118605. The repeated 91-bp, E1 exon is within the boundaries of the terminal repeat of the virus. Thus, the maximum number of copies of the E1 exon present in a given orf73 transcript is equal to the number of terminal repeats present in a given viral genome.
The promoter that directs transcription of orf73 transcripts containing E1 was termed 73p1. Transcripts in S11 cells that contained E1 also contained a second 106-bp exon (E2) located in the unique region of the viral genome immediately adjacent to the terminal repeats (extending from bp 118160 to 118055). The E2 exon, in turn, was spliced to the (LANA) coding exon (E3) that began at bp 104871 just upstream of the translation initiation codon for the predicted orf73 (Fig. 1B). 3′ RACE from S11E polyadenylated RNA identified a 3′ end of the orf73 transcript at bp 103787 (Fig. 1B). Thus, the entire predicted orf73, beginning at bp 104868 and ending at bp 103927, was contained within this spliced transcript. Notably, the splicing to upstream exons did not extend the opening reading frame encoding LANA.
A second initiation site at bp 118163 for orf73-containing transcripts was also identified by 5′ RACE using the same S11E mRNA template. This initiation site was located at the 5′ end of the E2 exon and DNA sequence analysis of these RACE products revealed the presence of the E2/E3 splice junction observed in the E1 containing orf73 transcripts (Fig. 1B). Identification of spliced LANA transcripts that did not include E1 suggested the presence of an alternative orf73 promoter located upstream of E2 (73p2). The existence of multiple orf73 promoters is reminiscent of the differential regulation of EBNA-1 gene expression during latent EBV infection, where three distinct promoters driving EBNA1 gene transcription have been identified (6, 27, 28, 30, 32, 33). By analogy to EBV (19, 25), we are currently investigating whether the initiation of orf73 transcription upstream of E2 or E1 is specific to particular stages of virus infection in vivo. Because all of the orf73 3′ RACE products identified from S11E mRNA terminated at position bp 103787, we think it is likely that both p2- and p1-initiated orf73 transcripts are 3′ coterminal.
To confirm that the 5′ termini of spliced orf73 transcripts in latently infected splenocytes were similar to those identified in S11 lymphoma cells, polyadenylated RNA was prepared from mouse splenocytes harvested 16 days post-intranasal infection with γHV68. This RNA was prepared as previously described for 5′ RACE with S11 RNA, and cDNA reactions were carried out using a primer specific for the E2/E3 orf73 splice junction. RT-PCR products were subcloned and sequenced, and both 73p1- and 73p2-initiated transcripts were isolated. The 5′ termini identified in vivo for spliced orf73 transcripts containing the E1 exon were bp 118677 and 118683. Notably, each of the E1-containing orf73 transcripts characterized from in vivo 5′ RACE contained only a single copy of E1. This may reflect a bias toward amplification of shorter templates in the RT-PCR. The 5′ termini identified in vivo for spliced orf73 transcripts containing only the E2 and E3 exons (73p2-initiated) were bp 118097, 118125, 118161, and 118163.
Identification of spliced v-cyclin transcripts.
Based on the observation that the KSHV v-cyclin and LANA transcripts are generated from a common promoter, we used RT-PCR to determine whether γHV68 v-cyclin-encoding transcripts were generated from the γHV68 LANA gene promoter p1 or p2. RT-PCR was carried out using a primer specific for E2, in combination with a primer specific for orf72 (v-cyclin), employing as the template cDNA generated from S11E tumor cells. This analysis readily identified an alternatively spliced transcript that contains orf72 (Fig. 1B). RT-PCR on cDNA from lytically infected NIH 3T12 fibroblasts also resulted in the amplification of this transcript, raising the possibility that this transcript functions during lytic infection, as well as in transformed cells. Based on DNA sequencing of RT-PCR products from S11E cDNA, the structure of the splice orf72 transcript includes the same splice junction previously described between E2 and orf73. However, the transcript includes an additional splice within orf73 that defines an intron between bp 104715 and bp 103199 (Fig. 1B).
The 3′ terminus of the v-cyclin message was mapped by 3′RACE using RNA prepared from infected NIH 3T12 fibroblasts. Based on the DNA sequence of the subcloned 3′ RACE products, the polyadenylation signal used in transcription of the v-cyclin message is located within the final two codons of the v-cyclin orf (between nucleotide positions 102430 and 102435) and the 3′ end of the message extends to bp 102412. The full-length spliced 72 transcript we identified potentially encodes a truncated LANA protein and the full-length v-cyclin. Notably, the splice junction identified does not generate a fusion between the orf73 coding sequences and v-cyclin coding sequence. In addition, the v-cyclin translation initiation codon is the fifth AUG codon encoded in the 72 spliced transcript. Thus, it remains undetermined whether this transcript functions to encode v-cyclin and, if so, how translation of v-cyclin is initiated from this message. Alternatively, the truncated orf73 may be translated and serve an as yet undetermined role in viral infection.
Characterization of the LANA p1 and p2 promoters.
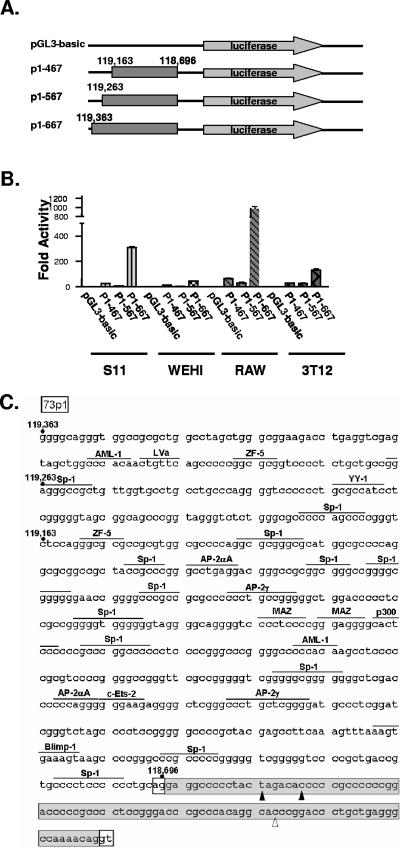
We tested the regions directly upstream of 73E2 and 73E1 (beginning at bp 118163 and 118696, respectively) for promoter activity in infected and uninfected lymphocytes, as well as uninfected fibroblasts and macrophages. Using luciferase reporter constructs, we identified a region extending from bp 118696 to 119363 that defined a promoter immediately upstream of the E1 exon (Fig. 2A and B). This promoter was active in all cell types tested, although activity in the uninfected WEHI B-cell line was quite modest (Fig. 2B). Taken together, the data demonstrated that 73p1 activity is not dependent on viral infection under these conditions (Fig. 2B). The promoter displayed the greatest activity in the macrophage-derived RAW cell line, generating luciferase levels 1,000-fold over that observed with the control reporter plasmid (Fig. 2B). Reporter constructs containing the region from bp 118696 to 118263 exhibited lower levels of luciferase activity in each of the cell types tested, suggesting that a major cis-regulatory sequence(s) for 73p1 lies within the region extending from bp 118263 to 118363 (Fig. 2B).
FIG. 2.
Orf73 promoter upstream of E1 functions independently of virus infection. (A) Characterization of the orf73 p1 promoter. Using wild-type γHV68 BAC DNA as template, PCR fragments encompassing the indicated genomic coordinates were generated with 5′ NheI restriction sites and 3′XhoI sites and subcloned into the pGL3-basic luciferase reporter vector (Promega). (B) 5 μg of each reporter construct was introduced into 106 S11E tumor cells, WEHI B-cell lymphoma cells, or RAW macrophage cells by nucleofection using optimized conditions according to the manufacturer's protocol (Amaxa), or transfected into NIH 3T12 fibroblasts using Superfect transfection reagent (QIAGEN); 24 h later, cell lysates were prepared using passive lysis buffer (Promega) and luciferase activity in each lysate was measured using a TD 20/20 luminometer (Turner Biosystems). (C) The genomic DNA fragments tested in panel B were analyzed for presence of consensus transcription factor binding sites (overlined) using Transcription Element Search Software (TESS; http://www.cbil.upenn.edu/tess/). The splice donor and splice acceptor sites for 73E1 are boxed in white and the sequence of 73E1 is boxed in gray. The 5′ terminus of E1/E2/E3 spliced LANA transcripts identified in S11E lymphoma cells is indicated with an empty arrowhead, and those termini identified in splenocytes harvested at day 16 postinfection are indicated with filled arrowheads.
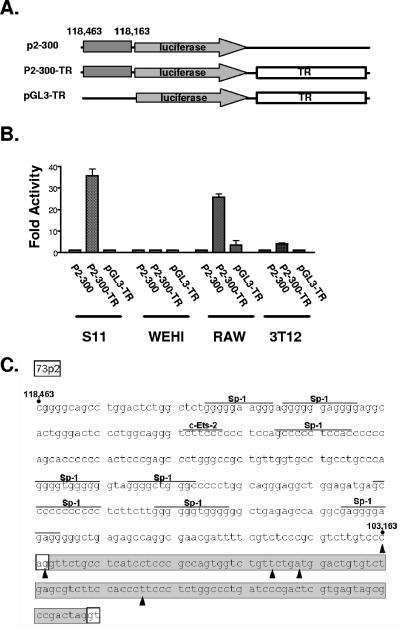
We also assessed the presence of promoter activity in the region immediately upstream of the E2 exon. Notably, little or no promoter activity of the 300-bp region directly upstream of position bp 118163 (73p2-300) was evident in the latently infected S11E B lymphoma cell line, the WEHI B-cell line, the macrophage-derived RAW cell line, or the murine fibroblast NIH 3T12 cell line (Fig. 2D). Based on the proximity of the putative p2 promoter to the terminal repeat, we assessed whether the terminal repeat might contain an enhancer that could augment p2-initiated transcription. An XcmI fragment, containing a single complete copy of the γHV68 terminal repeat, was cloned directly downstream of the luciferase reporter gene in the empty reporter vector (pGL3-TR), or in the context of the 73p2-300 reporter vector (73p2-300-TR) (Fig. 2C). As shown in Fig. 2D, the pGL3-TR construct exhibited luciferase activity levels comparable to the 73p2-300 construct. However, the levels of luciferase activity observed with the 73p2-300-TR construct, in either S11E tumor cells or RAW cells, resulted in a 25- to 30-fold increase activity compared to either 73p2-300 or pGL3-TR (Fig. 2D). Thus, the γHV68 terminal repeat acts to enhance transcription from 73p2. 73p2 exhibited little activity in the presence or absence of the terminal repeat in both WEHI and NIH 3T12 fibroblasts (Fig. 2D). As such, 73p2 may be dependent on other viral factors for activity in B cells and fibroblasts.
Analysis of spliced LANA and v-cyclin transcripts in vivo.
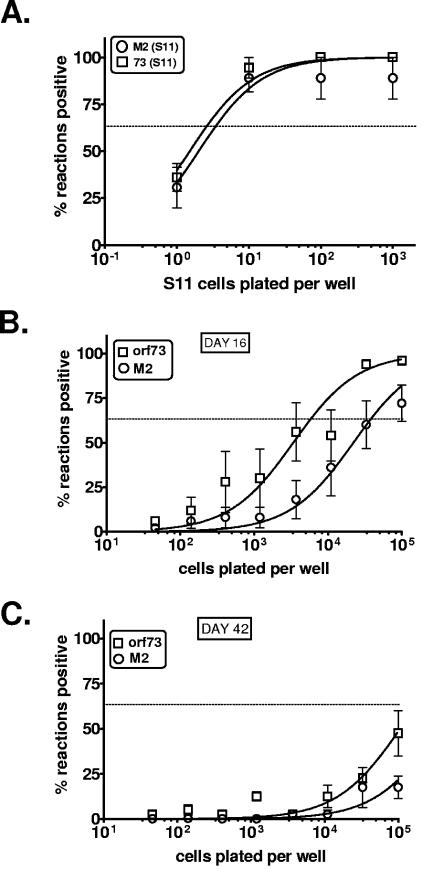
We next sought to establish the significance of the spliced LANA and v-cyclin transcripts during latent infection of splenocytes in vivo. Using RNA prepared from whole infected spleens 16 days post-intranasal infection with 1,000 PFU of wild-type γHV68, we were able to weakly detect the presence of both the spliced orf73 and orf72 transcripts by single-round RT-PCR (Fig. 3 and 4A). To better determine the relative frequency of infected splenocytes transcribing these two messages, we designed a nested RT-PCR assay that employed a pair of primers specific to 73E2 in combination with either a nested pair of primers specific to the orf73 (LANA) spliced transcript or the orf72 (v-cyclin) spliced transcript (Fig. 4B). Using S11 cells, diluted in a background of 105 uninfected splenocytes, we were able to demonstrate that these nested RT-PCR assays can detect the orf73 spliced transcripts in as few as two latently infected S11 cells (Fig. 4B and 5A). In addition, appropriate water controls were consistently negative for the presence of these spliced transcripts, demonstrating the absence of PCR product contamination.
FIG. 3.
Viral terminal repeat acts to enhance transcription from a promoter upstream of orf73E2. (A) Characterization of the orf73 p2 promoter. Reporter constructs carrying DNA fragments from within the indicated genomic coordinates were generated as described for p1 reporter constructs. For constructs containing the γHV68 terminal repeat, an XcmI fragment containing a single terminal repeat sequence was subcloned into the BamHI site of pGL3. (B) Transfections and luciferase assays were carried out in the same manner as described for p1 promoter constructs in Fig. 2B. Luciferase activity in cells transfected with p2-300-TR or pGL3-TR was plotted in comparison to activity in lysates from cells transfected with the p2-300 construct. (C) The genomic DNA fragments tested in panel B were analyzed for presence of consensus transcription factor binding sites (overlined) as described in the legend to Fig. 2. The splice donor and splice acceptor sites for 73E2 are boxed in white and the sequence of 73E2 is boxed in gray. The 5′ termini identified in S11E lymphoma cells and day 16-infected mouse splenocytes for 73 transcripts containing only 73E2 and 73E3 exons are indicated with solid arrowheads.
FIG. 4.
Spliced orf73 and orf72 transcripts are present in S11E tumor cells, day 16-infected splenocytes, and infected NIH 3T12 fibroblasts. (A) Total RNA was extracted from 107 cultured cells or mouse splenocytes and 5 μg of total RNA was used as template for oligo(dT)-primed cDNA synthesis. RT-PCR with primers specific to 73E2 (73E2o, TCTTCCACCCTTCCCTCTGGCCCTG) and orf73 (73RTo) or orf72 (72RTo, GGGGAAAGACGTTGTTATCCTGACG) was performed and products of the reaction were electrophoresed in a 1% agarose gel. Using cDNA from S11E tumor cells, infected day 16 splenocytes, or infected NIH 3T12 fibroblasts, RT-PCR with primers 73E2o and 73E3o generated a 466-bp product, and reactions using primers 73E2o and 72RTo generated a 433-bp product. Negative control reactions using cDNA generated from either naïve splenocytes (naïve SPL) or mock-infected NIH 3T12 fibroblasts did not generate any detectable RT-PCR products. (B) Mice were infected with 1,000 PFU of γHV68 intranasally and spleens were harvested from infected animals 16 days postinfection. Single cell splenocyte suspensions were prepared using a Dounce homogenizer, and subsequently treated with red blood cell lysis buffer. Splenocytes were then counted and resuspended at a concentration of 105 cells/ml and 10-fold serial dilutions of infected splenocytes (10 replicates at each dilution) were prepared in a background of 106 uninfected splenocytes. Cells were centrifuged and pellets were resuspended in 1 ml of guanidine isothiocyanate/phenol solution (2 M guanidine isothiocyanate, 0.05 M β-mercaptoethanol, 0.25% Sarcosyl, 0.1 M sodium acetate). Following extraction with phenol-chloroform-isoamyl alcohol (25:24:1, EM Science), RNA was precipitated and resuspended in 12 μl of RNase-free water. Each sample was then used as template in a 20-μl random hexamer-primed cDNA reaction using Superscipt II reverse transcriptase (Invitrogen); 4 μl of each cDNA reaction was used as the template in nested PCRs specific for detection of orf73 spliced transcripts or orf72 spliced transcripts. The orf73E2-specific primer set used in both reactions was 73E2o and 73E2i. The orf73 spliced transcript-specific primer set was 73RTo and 73RTi. The orf72 spliced transcript-specific primer set was 72RTo and 72RTi. S11E cells were diluted in a background of 106 uninfected splenocytes and used as a control template to determine the sensitivity of the primer sets.
FIG. 5.
Determination of the frequency of infected splenocytes transcribing orf73 or M2 in vivo. (A) Determination of detection sensitivity for M2 and 73 primer sets. S11E lymphoma B cells were serially diluted 10-fold in a background of 105 uninfected splenocytes and multiple, 1-ml replicates of each dilution were processed for RNA extraction and reverse transcription as described for Fig. 3B. For PCR detection of the spliced M2 mRNA, the primers used in the primary reaction were M2E1o (ACTTTCAGCTTTCGGGAAGGGTTTAGGCAC) and M2E2o (GGACTGTCAGTCGAGCCAGAGTCCAACATC), and the primers used in the nested reaction were M2E1i (CAGGACTTCCTGCAGGGTTAACTTCTTCAG) and M2E2i (TTCCCCTCTCAAGCTGCTTCCTTAGCCAGT). For the orf73 nested PCR, the primers used in the primary reaction were 73E2o and 73RTo, and the primers used in the nested reaction were 73E2i and 73RTi. Data were analyzed using nonlinear regression analysis and sensitivities were calculated as the point at which 63% of the PCRs were positive for each primer set. (B) At 16 days following intranasal infection of C57BL/6 mice with 1,000 PFU of wild-type γHV68, whole splenocytes were harvested from five mice, pooled, and treated as described for Fig. 3B. Single-cell suspensions of infected splenocytes were then diluted (threefold serial dilutions beginning with 105 splenocytes/ml and ending with 4.6 × 101 splenocytes/ml) in a background of uninfected splenocytes. Ten 1.0-ml replicates at each dilution point were processed for RNA extraction and reverse transcription as described for Fig. 3B. The PCR conditions used were those described for panel A. Results were compiled from five independent experiments. To obtain the frequency for each set of limiting-dilution experiments, data were subjected to nonlinear regression (using a sigmoidal dose curve with nonvariable slope to fit the data). Frequencies of transcript-positive cells were obtained by calculating the cell density at which 63% of the PCRs were positive for orf73 or M2 transcript based on a Poisson distribution. (C) Experiments were performed as described for panel B on splenocytes isolated from C57BL/6 mice 42 days postinfection.
Analysis of latently infected splenocytes harvested at day 16 postinfection revealed a clear disparity in the frequency of splenocytes expressing spliced LANA transcripts versus. the frequency of splenocytes expressing spliced v-cyclin transcripts (Fig. 4B). While spliced LANA transcripts were detected in the majority of replicates using 105 input splenocytes, we did not detect the spliced v-cyclin transcripts in any of these replicates (Fig. 4B). The spliced LANA transcripts, as expected, were detected less frequently upon serial dilution, while spliced v-cyclin transcripts were only sporadically detected over the range of input cell numbers assessed (Fig. 4B). Based on the known role of the γHV68 v-cyclin in virus reactivation from latency (35, 36), we speculate that the sporadic detection of spliced v-cyclin transcripts at day 16 postinfection likely reflects a small population of latently infected splenocytes which undergo spontaneous virus reactivation.
Because of the structures described for the orf73 and orf72 transcripts, transcription of the spliced orf72 message precludes transcription of the orf73 spliced message following a given transcriptional initiation event. The degree to which the presence of these two transcripts in the same cell favors latent or lytic infection has yet to be determined. Targeted disruption of orf73 results in the complete loss of latency establishment in the spleens of infected mice (14, 23). In contrast, viruses lacking a functional orf72 establish latency to wild-type levels, but are severely attenuated for virus reactivation (35, 36). With these mutant phenotypes in mind, it is worth considering that the orf73 transcript detected in splenocytes functions to establish and maintain a latent viral infection and that spliced orf72 transcripts may be associated with reactivation of virus from latency. The latter could explain the paucity of splenocytes expressing the spliced orf72 transcripts since there is little evidence of virus replication in the spleen post-clearance of acute virus replication (39).
To further characterize the frequency of infected splenocytes expressing spliced LANA transcripts, limiting-dilution nested RT-PCR analyses were carried out at both day 16 and day 42 postinfection (Fig. 5A and B). In addition, the frequency of cells expressing another γHV68 latency-associated gene, M2, was also assessed at these times postinfection (Fig. 5A and B). These analyses revealed that the frequency of splenocytes expressing LANA at day 16 is ca. 1 in 4,000, while at day 42 it is in the range of 1 in 10,000 to 20,000 cells. Notably, the frequency of splenocytes expressing the M2 gene is significantly lower than that of LANA-expressing cells at both days 16 and 42, suggesting that only a fraction of LANA-expressing splenocytes also express the M2 antigen. Importantly, based on the frequency of viral genome-positive splenocytes routinely observed at day 16 postinfection (ca. 1 in 200 to 400) (13, 40), we estimate that only ca. 5 to 10% of the viral genome-positive splenocytes have detectable spliced LANA transcripts. As discussed below, this may have profound implications with respect to the steady-state frequency of latently infected cells observed during chronic γHV68 infection.
Presence of both p1- and p2-initiated transcripts in vivo.
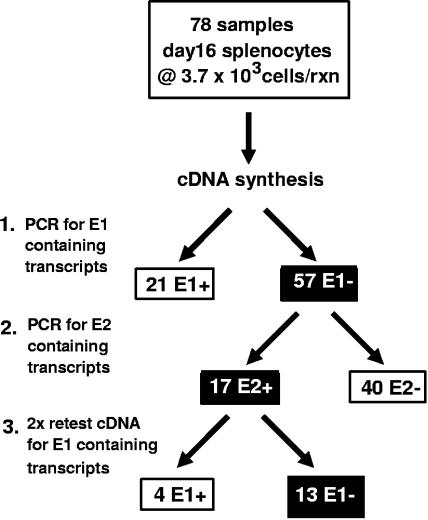
As described in Fig. 1, our studies have thus far identified two distinct types of orf73 encoding transcripts. Transcripts containing exon 1 (E1), exon 2 (E2) and the LANA coding exon are initiated from p1, while transcripts containing only the E2 exon and the LANA coding exon are initiated from p2. To determine the relative extent of p1 and p2 promoter usage during latent infection, we generated 78 replicate cDNA samples from 1.3 × 103 splenocytes harvested 16 days postinfection (Fig. 6). First, nested RT-PCR to detect E1-containing transcripts was performed on all 78 replicates. Those replicates that did not score positive for E1 were then tested for E2-containing transcripts. Replicates that scored positive for E2 were then tested twice more for the presence of E1-containing transcripts. Transcripts initiating from p1 were initially detected in 21 of the 78 samples. Of the remaining 57 samples that were E1 exon negative, 17 were positive following nested RT-PCR to detect E2-containing transcripts. Repeated RT-PCR for E1-containing transcripts on these E2-positive replicates identified E1-containing transcripts in 4 of the 17 E2-positives replicates. Thus, in total, 25 of the 78 samples contained p1-initiated transcripts, while 13 of the E1 exon-negative replicates contained p2-initiated transcripts. Based on these analyses, we conclude that both p1 and p2 are utilized during latent infection to direct transcription of orf73. The degree to which utilization of the either p1 or p2 defines distinct stages of latency is currently under investigation.
FIG. 6.
Detection of orf73p1 and orf73p2 initiation events in infected splenocytes. Sixteen days following intranasal infection of C57BL/6 mice with 1,000 PFU of wild-type γHV68 bulk splenocytes were harvested from five mice, pooled, and treated as described for Fig. 3B. A total of 78 replicates were prepared with 3.7 × 103 cells per sample in a background of 105 uninfected splenocytes. RNA was extracted and reverse transcribed as detailed for Fig. 3B. Initially, all 78 replicates were screened for the presence of E1-containing (E1+) orf73 transcripts using the first-round primer set 73E1o (GACCCCCGCCCCTCCGGGACCCGCC) and 73RTo, and the nested primer set 73E1i (GCACCCGGACCCTGCTGAGGGCCAA) and 73RTi. cDNA reactions that were negative following orf73-E1 RT-PCR were then screened for the presence of E2-containing (E2+) orf73 transcripts using the first-round primer set 73E2o and 73RTo, and the nested primers 73E2i and 73RTi. Any cDNA reactions that scored E1 negative (E1−) but were E2+ were then rescreened two further times for the presence of E1 exon-containing transcripts using the orf73-E1 nested primer pair.
Here we have described the initial characterization of spliced orf73 and orf72 transcripts, detected in the latently γHV68-infected S11 lymphoma cell line. Although there are clear differences in the organization of genes in this region of the KSHV and γHV68 genomes, the transcription pattern shown here bears significant similarity to transcription of KSHV LANA (orf73) and v-cyclin (orf72) (11, 29, 34). In addition, the identification of multiple promoters (73p2 and multiple copies of 73p1, which are encoded within the terminal repeats) driving orf73 transcription is reminiscent of EBNA gene transcription in latently EBV-infected B cells (6, 27, 28, 30, 32, 33). The utilization of distinct EBNA gene promoters in EBV correlates with distinct latency programs, which have been proposed to facilitate the orderly differentiation of latently infected naïve B cells through a germinal center reactivation and ultimately into resting memory B cells (2, 18, 22). It is worth noting that for γHV68 infection of mice, at early times postinfection latent infection of naïve, germinal center memory B cells can be detected in the spleen. However, as infection progresses, latency is predominantly established in isotype-switched memory B cells (13, 40). Thus, like EBV, γHV68 may encode distinct latency programs to facilitate access to the memory B-cell reservoir.
The p1 promoter described here corresponds closely with an orf73 promoter recently described by Coleman et al. (8). Furthermore, RT-PCR analyses carried out by Coleman et al. demonstrated transcription of the orf73 spliced transcript emanating from p1 in bulk infected splenocytes, as well as sorted infected CD19+ PNA+ B cells (8). Based on this result, they concluded that orf73 is transcribed from p1 in proliferating germinal center B cells (8). Coleman et al. (8), characterizing viral transcription in lytically infected NIH 3T3 fibroblasts, also defined a promoter that includes the region upstream of orf73 E2 which directs transcription of an unspliced message that appears to encode the γHV68 orf75a protein. Our RACE results in S11E tumor cells define the region upstream of E2, as well as the region upstream of E1, as regions involved in the regulation of orf73 and, potentially, orf72 transcription. To what degree transcription initiation from 73p1 or 73p2 defines specific stages of lytic or latent viral infection is currently under investigation.
Because of the structures described for the orf73 and orf72 transcripts, transcription of the spliced orf72 message precludes transcription of the orf73 spliced message following a given transcriptional initiation event. The degree to which the presence of these two transcripts in the same cell favors latent or lytic infection has yet to be determined. Targeted disruption of orf73 results in the complete loss of latency establishment in the spleens of infected mice (14, 23). In contrast, viruses lacking a functional orf72 establish latency to wild-type levels, but are severely attenuated for virus reactivation (35, 36).
With these mutant phenotypes in mind, it is worth considering that the orf73 transcript detected in splenocytes functions to establish and maintain a latent viral infection and that spliced orf72 transcripts may be associated with reactivation of virus from latency. This could explain the paucity of splenocytes expressing the spliced orf72 transcripts since there is little evidence of virus replication in the spleen post-clearance of acute virus replication (39).
Based on our observation that a latency-associated gene (LANA) and a reactivation-associated gene (v-cyclin) can be generated from a single shared promoter, it is also interesting to consider the regulatory potential of polyadenylation site usage in the determination of specific outcomes during virus infection. In the case of EBV latency, generation of the long primary transcripts encoding the EBV EBNA gene products involves readthrough of numerous polyadenylation signals utilized during lytic virus replication (3). The mechanism(s) involved in the recognition and utilization of these signals during infection remains unresolved.
Of particular interest with respect to the analysis of spliced LANA gene transcripts in vivo during the establishment of chronic infection is the observation that there is a significant disparity between the frequency of splenocytes at day 16 harboring viral genomes versus the frequency of these cells containing LANA transcripts. Our previous analyses have repeatedly shown that there is a major contraction in the frequency of splenocytes harboring viral genome from day 16 to day 42 postinfection (40). This could result from immune system-mediated clearance of latently infected cells, migration of latently infected splenocytes to other organs, and/or failure of the virus to establish a long-lived infection in some population(s) of infected cells.
While there are likely multiple mechanisms that contribute to the contraction in the frequency of latently infected splenocytes, the data presented here provide evidence that failure to express LANA in a large percentage of viral genome-positive splenocytes at day 16 may contribute significantly to the attrition of latently infected splenocytes. It is important to point out, however, that our RACE analyses may have failed to detect the presence of alternatively spliced orf73 transcripts (e.g., transcripts lacking the E1 and E2 exons) and, if so, would underestimate the frequency of latently infected splenocytes expressing LANA at days 16 and 42 postinfection. We have recently shown that at both days 16 and 42 postinfection, the vast majority of viral genome-positive cells are actively proliferating (24). Thus, if a key function of the γHV68 LANA is to maintain the viral episome in proliferating cells, then failure to express LANA will likely result in loss of viral genomes in proliferating cell populations. The mechanism(s) underlying the failure to transcribe the LANA gene in a high percentage of infected splenocytes remains to be determined.
Acknowledgments
We thank the members of the Speck laboratory for helpful discussions and advice.
This research was supported by NIH grant CA87650. During a portion of this research. R.D.A. was supported by a Leukemia and Lymphoma Society Postdoctoral Fellowship. S.H.S. is supported by NIH grants R01 CA43143, CA52004, CA87650, CA95318, and AI59057 and the Yerkes National Primate Research Center Base Grant P51 RR00165.
REFERENCES
- 1.Albrecht, J. C., J. Nicholas, D. Biller, K. R. Cameron, B. Biesinger, C. Newman, S. Wittmann, M. A. Craxton, H. Coleman, B. Fleckenstein, et al. 1992. Primary structure of the herpesvirus saimiri genome. J. Virol. 66:5047-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babcock, G. J., L. L. Decker, R. B. Freeman, and D. A. Thorley-Lawson. 1999. Epstein-barr virus-infected resting memory B cells, not proliferating lymphoblasts, accumulate in the peripheral blood of immunosuppressed patients. J. Exp. Med. 190:567-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baer, R., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. Hatfull, G. S. Hudson, S. C. Satchwell, C. Seguin, et al. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310:207-211. [DOI] [PubMed] [Google Scholar]
- 4.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 5.Bieleski, L., and S. J. Talbot. 2001. Kaposi's sarcoma-associated herpesvirus vCyclin open reading frame contains an internal ribosome entry site. J. Virol. 75:1864-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bodescot, M., M. Perricaudet, and P. J. Farrell. 1987. A promoter for the highly spliced EBNA family of RNAs of Epstein-Barr virus. J. Virol. 61:3424-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaudhuri, B., H. Xu, I. Todorov, A. Dutta, and J. L. Yates. 2001. Hum. DNA replication initiation factors, ORC and MCM, associate with oriP of Epstein-Barr virus. Proc. Natl. Acad. Sci. USA 98:10085-10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coleman, H. M., S. Efstathiou, and P. G. Stevenson. 2005. Transcription of the murine gammaherpesvirus 68 ORF 73 from promoters in the viral terminal repeats. J. Gen. Virol. 86:561-574. [DOI] [PubMed] [Google Scholar]
- 9.Cotter, M. A., 2nd, C. Subramanian, and E. S. Robertson. 2001. The Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen binds to specific sequences at the left end of the viral genome through its carboxy-terminus. Virology 291:241-259. [DOI] [PubMed] [Google Scholar]
- 10.Dhar, S. K., K. Yoshida, Y. Machida, P. Khaira, B. Chaudhuri, J. A. Wohlschlegel, M. Leffak, J. Yates, and A. Dutta. 2001. Replication from oriP of Epstein-Barr virus requires human ORC and is inhibited by geminin. Cell 106:287-296. [DOI] [PubMed] [Google Scholar]
- 11.Dittmer, D., M. Lagunoff, R. Renne, K. Staskus, A. Haase, and D. Ganem. 1998. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J. Virol. 72:8309-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ebrahimi, B., B. M. Dutia, K. L. Roberts, J. J. Garcia-Ramirez, P. Dickinson, J. P. Stewart, P. Ghazal, D. J. Roy, and A. A. Nash. 2003. Transcriptome profile of murine gammaherpesvirus-68 lytic infection. J. Gen. Virol. 84:99-109. [DOI] [PubMed] [Google Scholar]
- 13.Flano, E., I. J. Kim, D. L. Woodland, and M. A. Blackman. 2002. Gamma-herpesvirus latency is preferentially maintained in splenic germinal center and memory B cells. J. Exp. Med. 196:1363-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fowler, P., S. Marques, J. P. Simas, and S. Efstathiou. 2003. ORF 73 of murine herpesvirus-68 is critical for the establishment and maintenance of latency. J. Gen. Virol. 84:3405-3416. [DOI] [PubMed] [Google Scholar]
- 15.Grundhoff, A., and D. Ganem. 2001. Mechanisms governing expression of the v-FLIP gene of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:1857-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall, K. T., M. S. Giles, D. J. Goodwin, M. A. Calderwood, I. M. Carr, A. J. Stevenson, A. F. Markham, and A. Whitehouse. 2000. Analysis of gene expression in a human cell line stably transduced with herpesvirus saimiri. J. Virol. 74:7331-7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu, J., A. C. Garber, and R. Renne. 2002. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus supports latent DNA replication in dividing cells. J. Virol. 76:11677-11687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joseph, A. M., G. J. Babcock, and D. A. Thorley-Lawson. 2000. EBV persistence involves strict selection of latently infected B cells. J. Immunol. 165:2975-2981. [DOI] [PubMed] [Google Scholar]
- 19.Kerr, B. M., A. L. Lear, M. Rowe, D. Croom-Carter, L. S. Young, S. M. Rookes, P. H. Gallimore, and A. B. Rickinson. 1992. Three transcriptionally distinct forms of Epstein-Barr virus latency in somatic cell hybrids: cell phenotype dependence of virus promoter usage. Virology 187:189-201. [DOI] [PubMed] [Google Scholar]
- 20.Low, W., M. Harries, H. Ye, M. Q. Du, C. Boshoff, and M. Collins. 2001. Internal ribosome entry site regulates translation of Kaposi's sarcoma-associated herpesvirus FLICE inhibitory protein. J. Virol. 75:2938-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez-Guzman, D., T. Rickabaugh, T. T. Wu, H. Brown, S. Cole, M. J. Song, L. Tong, and R. Sun. 2003. Transcription program of murine gammaherpesvirus 68. J. Virol. 77:10488-10503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyashita, E. M., B. Yang, G. J. Babcock, and D. A. Thorley-Lawson. 1997. Identification of the site of Epstein-Barr virus persistence in vivo as a resting B cell. J. Virol. 71:4882-4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moorman, N. J., D. O. Willer, and S. H. Speck. 2003. The gammaherpesvirus 68 latency-associated nuclear antigen homolog is critical for the establishment of splenic latency. J. Virol. 77:10295-10303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moser, J. M., J. W. Upton, R. D. Allen, C. B. Wilson, and S. H. Speck. 2005. Role of B-cell proliferation in the establishment of gammaherpesvirus latency. J. Virol. 79:9480-9491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rowe, M., A. L. Lear, D. Croom-Carter, A. H. Davies, and A. B. Rickinson. 1992. Three pathways of Epstein-Barr virus gene activation from EBNA1-positive latency in B lymphocytes. J. Virol. 66:122-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sample, J., L. Brooks, C. Sample, L. Young, M. Rowe, C. Gregory, A. Rickinson, and E. Kieff. 1991. Restricted Epstein-Barr virus protein expression in Burkitt lymphoma is due to a different Epstein-Barr nuclear antigen 1 transcriptional initiation site. Proc. Natl. Acad. Sci. USA 88:6343-6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sample, J., E. B. Henson, and C. Sample. 1992. The Epstein-Barr virus nuclear protein 1 promoter active in type I latency is autoregulated. J. Virol. 66:4654-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sarid, R., J. S. Wiezorek, P. S. Moore, and Y. Chang. 1999. Characterization and cell cycle regulation of the major Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter. J. Virol. 73:1438-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schaefer, B. C., M. Woisetschlaeger, J. L. Strominger, and S. H. Speck. 1991. Exclusive expression of Epstein-Barr virus nuclear antigen 1 in Burkitt lymphoma arises from a third promoter, distinct from the promoters used in latently infected lymphocytes. Proc. Natl. Acad. Sci. USA 88:6550-6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Searles, R. P., E. P. Bergquam, M. K. Axthelm, and S. W. Wong. 1999. Sequence and genomic analysis of a rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 73:3040-3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith, P. R., and B. E. Griffin. 1992. Transcription of the Epstein-Barr virus gene EBNA-1 from different promoters in nasopharyngeal carcinoma and B-lymphoblastoid cells. J. Virol. 66:706-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Speck, S. H., A. Pfitzner, and J. L. Strominger. 1986. An Epstein-Barr virus transcript from a latently infected, growth-transformed B-cell line encodes a highly repetitive polypeptide. Proc. Natl. Acad. Sci. USA 83:9298-9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Talbot, S. J., R. A. Weiss, P. Kellam, and C. Boshoff. 1999. Transcriptional analysis of human herpesvirus-8 open reading frames 71, 72, 73, K14, and 74 in a primary effusion lymphoma cell line. Virology 257:84-94. [DOI] [PubMed] [Google Scholar]
- 35.van Dyk, L. F., H. W. T. Virgin, and S. H. Speck. 2003. Maintenance of gammaherpesvirus latency requires viral cyclin in the absence of B lymphocytes. J. Virol. 77:5118-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Dyk, L. F., H. W. T. Virgin, and S. H. Speck. 2000. The murine gammaherpesvirus 68 v-cyclin is a critical regulator of reactivation from latency. J. Virol. 74:7451-7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Virgin, H. W. T., P. Latreille, P. Wamsley, K. Hallsworth, K. E. Weck, A. J. Dal Canto, and S. H. Speck. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 71:5894-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Virgin, H. W. T., R. M. Presti, X. Y. Li, C. Liu, and S. H. Speck. 1999. Three distinct regions of the murine gammaherpesvirus 68 genome are transcriptionally active in latently infected mice. J. Virol. 73:2321-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weck, K. E., S. S. Kim, H. I. Virgin, and S. H. Speck. 1999. B cells regulate murine gammaherpesvirus 68 latency. J. Virol. 73:4651-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Willer, D. O., and S. H. Speck. 2003. Long-term latent murine Gammaherpesvirus 68 infection is preferentially found within the surface immunoglobulin D-negative subset of splenic B cells in vivo. J. Virol. 77:8310-8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wysokenski, D. A., and J. L. Yates. 1989. Multiple EBNA1-binding sites are required to form an EBNA1-dependent enhancer and to activate a minimal replicative origin within oriP of Epstein-Barr virus. J. Virol. 63:2657-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]