Abstract
The enveloped alphavirus Semliki Forest virus (SFV) infects cells via a membrane fusion reaction mediated by the E1 membrane protein. Efficient SFV-membrane fusion requires the presence of cholesterol and sphingolipid in the target membrane. Here we report on two mutants, srf-4 and srf-5, selected for growth in cholesterol-depleted cells. Like the previously isolated srf-3 mutant (E1 proline 226 to serine), the phenotypes of the srf-4 and srf-5 mutants were conferred by single-amino-acid changes in the E1 protein: leucine 44 to phenylalanine and valine 178 to alanine, respectively. Like srf-3, srf-4 and srf-5 show striking increases in the cholesterol independence of growth, infection, membrane fusion, and exit. Unexpectedly, and unlike srf-3, srf-4 and srf-5 showed highly efficient fusion with sphingolipid-free membranes in both lipid- and content-mixing assays. Both srf-4 and srf-5 formed E1 homotrimers of decreased stability compared to the homotrimers of the wild type and the srf-3 mutant. All three srf mutations lie in the same domain of E1, but the srf-4 and srf-5 mutations are spatially separated from srf-3. When expressed together, the three mutations could interact to produce increased sterol independence and to cause temperature-sensitive E1 transport. Thus, the srf-4 and srf-5 mutations identify novel regions of E1 that are distinct from the fusion peptide and srf-3 region and modulate the requirements for both sphingolipid and cholesterol in virus-membrane fusion.
Membrane fusion is essential to the growth and function of eukaryotic cells during the processes of endocytosis, exocytosis, and cell-cell fusion, and it is also a critical step in the infectious entry of enveloped viruses into host cells (10, 11, 34). Research has begun to identify the cellular and viral proteins that carry out membrane fusion reactions and to characterize the protein domains and conformational changes involved in fusion. Our knowledge of the role of membrane lipids in fusion is less developed. Interestingly, recent work suggests that viral membrane fusion proteins may have specific interactions with lipids (13). One striking example is the Semliki Forest virus (SFV) fusion protein, which has specific lipid requirements during the fusion of the virus membrane with the target membrane.
SFV is a member of a group of small enveloped positive-strand RNA viruses termed “alphaviruses” (reviewed in reference 31). SFV infects cells by endocytic uptake and low-pH-triggered fusion of the virus and endosome membranes and exits the cell by budding from the plasma membrane (reviewed in references 12, 13, and 36). In vitro studies of SFV and another alphavirus, Sindbis virus (SIN), demonstrate that efficient virus fusion requires the presence of both cholesterol and sphingolipid in the target liposome membrane. Fusion is optimal at levels of about 1 molecule of cholesterol per 2 molecules of phospholipid (∼33 mol%), and is specific for sterols containing a 3β-hydroxyl group (16, 30, 35). Sphingolipids support fusion at lower concentrations (∼2 to 5 mol %), and minimal sphingolipids such as ceramide are active (25, 30). In vivo studies with cholesterol-depleted insect cells demonstrate that cholesterol is required for infection of cells by SFV and SIN (20, 26). Sterol-depleted cells were reduced by 3 to 4 logs in primary infection and fusion compared to control cells. Cholesterol is also required for the efficient exit of progeny virus from cells during budding from the plasma membrane (20, 22, 24).
Alphaviruses have a highly ordered structure composed of an icosahedral nucleocapsid surrounded by a lipid bilayer containing the viral envelope proteins (18, 27, 31). The envelope protein shell is also icosahedrally symmetrical. In the case of SFV, it contains 80 trimers (E1/E2/E3)3 of the transmembrane glycoproteins E1 and E2 (each about 50 kDa) and a peripheral glycoprotein, E3 (∼10 kDa). E1 and E2 are tightly associated as a noncovalent dimer during their biosynthesis and in the virus particle. E2 is the primary receptor-binding subunit and also acts to regulate the pH-dependent fusion activity of the E1 subunit. E1 is the viral fusion protein and contains the internal fusion peptide, a highly conserved apolar region between approximately residues 83 and 100 (12, 15). Structural studies of SFV E1 demonstrate that it is an elongated molecule that contains three domains (18) and is strikingly similar to the structure of the fusion protein E of the flavivirus tick-borne encephalitis (TBE) virus (28). The fusion peptide is at one end of E1 in domain II and is tightly associated with E2, while the transmembrane domain is at the other end of the molecule. E1 forms an icosahedral lattice on the virus surface (18, 27). Following exposure to low pH, a defined series of envelope protein conformational changes leads to the fusion of the viral and target membranes (reviewed in references 7, 12, and 13). The stable E1/E2 dimer dissociates, releasing the E1 protein. E1 then interacts hydrophobically with the target membrane and exposes previously hidden sites for monoclonal antibody (MAb) binding. Three E1 monomers associate to form a highly stable E1 homotrimer that is believed to be critical for fusion. The structural and functional similarities between the alphavirus E1 and flavivirus E proteins has led to their being grouped as “class II” viral fusion proteins, in contrast to the class I proteins exemplified by influenza virus hemagglutinin (18).
The molecular mechanism by which cholesterol and sphingolipid affect the alphavirus fusion reaction is not clear. Cholesterol is required for the hydrophobic interaction of virus with the target membrane (2, 16, 25). Both cholesterol and sphingolipid promote the association of E1*, the E1 ectodomain, with target membranes (17), leading to the strong interaction of the fusion peptide with cholesterol and sphingolipid-enriched membrane domains (1). Both cholesterol and sphingolipid increase the kinetics and efficiency of E1 acid epitope exposure and homotrimerization (3, 4).
The role of cholesterol in the virus life cycle was previously explored by selection for virus mutants that were able to infect cholesterol-depleted insect cells. A cholesterol-independent SFV mutant, srf-3 (sterol requirement in function), was isolated and shown to grow more efficiently in the absence of cholesterol due to increases in the cholesterol independence of both fusion and budding (24, 32). Although the overall fusion properties of srf-3 are very similar to those of wild-type SFV, the mutant shows increased fusion activity with sterol-deficient liposomes and decreased cholesterol dependence for E1 acid epitope exposure and homotrimerization (3). A single-amino-acid change in E1, proline 226→serine (P226S), is responsible for the srf-3 phenotype (32). P226S lies in domain II of E1 within a loop that is closely associated with the fusion peptide in the native E1 structure (Félix Rey, personal communication). In vitro mutagenesis studies of SIN show that mutations in the E1 226 loop can similarly increase SIN fusion and exit from sterol-depleted cells (20). Importantly, srf-3 retains the wild-type level of sphingolipid dependence for both fusion and E1 conformational changes (3), supporting the idea that sphingolipids and cholesterol play different roles in fusion.
In this report, we exploited cholesterol-depleted C6/36 cells to isolate two new cholesterol-independent SFV mutants, srf-4 and srf-5. Surprisingly, we found that, unlike the srf-3 mutant, the two new srf mutants were also dramatically increased in their ability to fuse with sphingolipid-deficient liposomes. Single-amino-acid changes in the E1 protein of each mutant conferred both the cholesterol and sphingolipid effects. While all three srf mutations were within domain II of the E1 protein, both of the sphingolipid-independent mutations were spatially separated from the srf-3 mutation and affected the stability of the E1 homotrimer.
MATERIALS AND METHODS
Cells and viruses.
C6/36, a clonal Aedes albopictus cell line, was cultured at 28°C in Dulbecco's modified Eagle's medium plus antibiotics (DME), containing 10% heat-inactivated fetal calf serum (HIFCS) (24). Cholesterol-depleted C6/36 cells were prepared by at least four passages in DME containing 10% HIFCS delipidated by colloidal silica treatment (Cab-O-Sil), as previously described (24, 26, 32). Such depleted cells had <2% of the control level of cholesterol and were used between passages 4 and 15 to avoid potential adaptation (23). BHK-21 cells were cultured at 37°C in DME containing 5% FCS and 10% tryptose phosphate broth (26). The srf-3 mutant was previously isolated from a well-characterized plaque-purified wild-type virus stock by selecting for growth on cholesterol-depleted C6/36 cells (32). For experimental purposes, we used either srf-3 virus stock or srf-3/ic, virus derived from the SFV infectious clone pSP6-SFV4 (19) containing the single point mutation E1 P226S (32). Either the wild-type virus stock or virus derived from the SFV wild-type infectious clone (wt/ic) was used as the starting virus for the selection of new cholesterol-independent SFV mutants.
Isolation and sequence analysis of cholesterol-independent SFV mutants.
New srf mutants were isolated by two different methods based on selection for growth on cholesterol-depleted C6/36 cells. The first procedure was similar to that used for the isolation of srf-3 (32). The wild-type SFV stock was mutagenized with nitrosoguanidine and passaged four times on individual plates of cholesterol-depleted cells in Opti-MEM medium (Gibco/BRL, Gaithersburg, Md.) containing 0.2% bovine serum albumin (BSA [O/B]). Mutants were then isolated by using three rounds of limiting dilution on cholesterol-depleted cells in 96-well tissue culture trays and amplification in sterol-depleted cells (32).
Alternatively, selection was performed without chemical mutagenesis, using wt/ic, which contains a silent codon change at E1 cysteine 96 compared to the sequences of wild-type and srf-3 viruses. Individual 35-mm-diameter plates of cholesterol-depleted C6/36 cells were infected with wt/ic virus and cultured for 64 h in either MEM containing 1% Cab-O-Sil-treated HIFCS and 10 mM HEPES (pH 7.0) or O/B, and the progeny virus was passaged four more times under the same conditions. Both srf-4 and srf-5 resulted from selections in medium containing Cab-O-Sil-treated serum. Since the srf-3 mutation is stable to passage in cholesterol-containing cells (32), mutants were isolated by plaque formation in BHK cells and expanded by growth in depleted cells at low multiplicity.
The virus mutants were sequenced by reverse transcription-PCR (RT-PCR) and automated sequence analysis as previously described (15, 32).
In vitro mutagenesis and generation of virus stocks.
Point mutations were introduced into the pSP6-SFV4 wt/ic construct (19) by overlap-extension PCR mutagenesis with Pfu DNA polymerase (32). Unique NsiI and SpeI sites were used to subclone the mutations into the pSP6-SFV4 infectious clone. Two isolates of each mutant construct were sequenced to confirm the presence of the desired mutations and the absence of other mutations in the subcloned fragment. In vitro transcription was used to prepare infectious viral RNA from wt/ic and mutant constructs, and the RNA was electroporated or transfected into cells for subsequent analysis or for preparation of virus stocks (32).
Assays of virus infection, virus-cell fusion, and growth properties.
The details of the methods for assaying virus infection, virus-cell fusion, and growth properties can be found in references 23 and 32. Primary virus infection of C6/36 cells was measured by an infectious center assay based on detection of infected cells by immunofluorescence. Virus-cell fusion was measured by prebinding virus to cells in the cold and treating the cells for 1 min at pH 5.5 at 28°C to trigger virus-plasma membrane fusion. The cells were then incubated for ∼16 h in O/B plus 20 mM NH4Cl to prevent secondary infection, and infected cells were quantitated by immunofluorescence. Virus growth kinetics were determined as in reference 23. To evaluate envelope protein localization, BHK cells were electroporated with wild-type or mutant RNA, cultured on coverslips as indicated, and fixed with 3% paraformaldehyde for surface staining followed by 0.1% Triton X-100 for internal staining. Immunofluorescence was performed by using MAb E1-1 against the E1 subunit or E2-1 against the E2 subunit (14).
Pulse-chase analysis of virus protein expression and virus exit.
BHK cells were electroporated with the appropriate viral RNA and analyzed by pulse-chase at either 37 or 28°C. The envelope proteins in the cells and the medium were immunoprecipitated with a polyclonal rabbit antibody to the SFV membrane proteins and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by fluorography, as described previously (6, 14). Exit of the wild-type and mutant viruses from control and cholesterol-depleted C6/36 cells was monitored by pulse-chase analysis and immunoprecipitation as described previously (32) and in the legend to Fig. 3. Quantitation of radiolabeled proteins was performed with a PhosphorImager and ImageQuant software (Molecular Dynamics, Sunnyvale, Calif.).
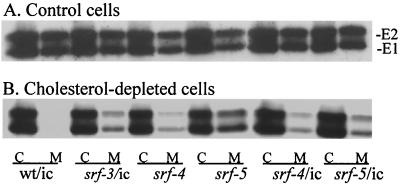
FIG. 3.
Exit of wild-type and mutant SFV from control (A) and cholesterol-depleted (B) C6/36 cells. Control (A) or cholesterol-depleted (B) cells were infected with the indicated viruses (labeled as in Fig. 2A) at a multiplicity of 100 PFU per cell, except for wt/ic in depleted cells, for which the multiplicity was 1,000 PFU/cell. After incubation at 28°C for 6 h for control cells and 22 h for depleted cells, the release of progeny virus was determined by pulse-labeling for 15 min with [35S]methionine/cysteine, chasing for 3 h in excess cold methionine and cysteine, collecting the chase medium, and lysing the cells. The amounts of radiolabeled virus envelope proteins in the cell lysate (C) and medium (M) were determined by immunoprecipitation and SDS-PAGE followed by fluorography. The positions of the E1 and E2 proteins are marked.
Virus-liposome fusion assays.
The methods to monitor pyrene-labeled SFV fusion with liposomes were essentially as previously described (2, 3). In brief, pyrene-labeled SFV was prepared by propagation of virus in BHK cells that were prelabeled by growth in the presence of C16-pyrene (Molecular Probes, Eugene, Oreg.). Liposomes were prepared by extrusion through two 0.2-μm-pore-diameter polycarbonate filters by using phospholipids from Avanti Polar Lipids (Alabaster, Ala.). Complete liposomes contained a 1:1:1:1.5 molar ratio of phosphatidylcholine (PC; from egg yolk), phosphatidylethanolamine (PE; prepared from egg PC by transphosphatidylation), sphingomyelin (Sph; from bovine brain), and cholesterol. Cholesterol-deficient liposomes had a PC/PE/Sph molar ratio of 1:1:1, sphingolipid-deficient liposomes had a PC/PE/cholesterol molar ratio of 1:1:1, and liposomes without sphingolipid and sterol had a 1:1 PC/PE ratio. Each fusion assay contained 0.6 μM virus phospholipid (calculated from a virus phospholipid/protein ratio of 0.45 μmol/mg) in a 2-ml volume. Fusion was triggered by adjusting the liposome-virus mix to pH 5.5, and the decrease in pyrene eximer fluorescence was monitored at an excitation wavelength of 343 nm and an emission wavelength of 480 nm with an Aminco-Bowman AB-2 fluorometer (Spectronic Unicam, Rochester, N.Y.) with a thermojacketed cuvette holder and 470-nm cutoff filter in the emission beam. The 0% fusion level was set to the initial virus eximer fluorescence, and 100% fusion was defined as the background fluorescence of target liposomes.
Virus-liposome fusion was also assayed by using purified [35S]methionine-labeled virus prepared in BHK cells, and liposomes containing entrapped trypsin (15). Nontrapped trypsin was removed from the liposome preparation by gel filtration as previously described. Liposomes were mixed with virus at a final concentration of 0.2 mM lipid in the presence of 125 μg of soybean trypsin inhibitor per ml. The samples were incubated at the indicated pH for 5 min at 37°C to trigger fusion, adjusted to pH 8.0, and then incubated for 1 h at 37°C to permit capsid digestion. Samples were then mixed with buffer containing detergent and protease inhibitors and immunoprecipitated with a mixture of polyclonal sera to capsid and envelope proteins. The amount of capsid protein in the samples was quantitated by SDS-PAGE and phosphorimaging, as previously described (15).
Assay of E1 homotrimer stability.
To form E1 homotrimers, radiolabeled virus preparations were mixed with 1 mM complete liposomes prepared as described above in morpholineethanesulfonic acid (MES)-saline buffer. The mixture was adjusted to pH 5.5 and incubated for 5 min at 20°C (3, 9). Samples were then adjusted to neutral pH and treated at 30°C for 4 min with SDS concentrations ranging from 0 to 4%. As indicated, the samples were then adjusted to final concentrations of 1% Triton-X-100 and 0.1% SDS and digested with 200 μg of trypsin per ml for 20 min at 37°C. Trypsin treatment was stopped by addition of phenylmethylsulfonyl fluoride to a final concentration of 2 mM. Trypsin-resistant homotrimers were immunoprecipitated with a rabbit polyclonal serum to E1 and E2, analyzed by SDS-PAGE, and quantitated as described above. The data were compared to the maximal trypsin-resistant homotrimer obtained without any SDS pretreatment.
RESULTS
Isolation of novel srf mutants.
Our original selection for SFV mutants that could grow in cholesterol-depleted C6/36 cells resulted in three isolates, srf-1, -2, and -3, all containing the same E1 P226S mutation and probably representing separate isolates of the same original mutant (32). Although selected on sterol-depleted cells repleted with 3β-chlorocholestene, a nonfusogenic analogue of cholesterol, these mutants do not require chlorocholestene for growth. To optimize for the isolation of new mutants with cholesterol-independent phenotypes, we selected for growth on independent plates of cholesterol-depleted cells in the absence of any sterol. The first selection, using a mutagenized wild-type virus stock, yielded two mutants, both of which carried the P226S mutation with the same codon change (CCC→UCC) as in srf-1, -2, and -3. Further isolations were then performed with a nonmutagenized wild-type virus stock derived from the infectious clone (termed wt/ic) and carrying a silent “marker” mutation not found in srf-3. Eight mutants having ∼100-fold greater infectivity on sterol-depleted cells were isolated. Sequence analysis showed that six mutants carried the E1 P226S mutation plus the parental silent mutation, indicating a marked selection preference for the independent isolation of the srf-3 mutant. The remaining two isolates were both new mutants, srf-4 and srf-5. Each contained the silent mutation plus one additional amino acid change in the E1 subunit, srf-4 L44F (UUG→UUU), and srf-5 V178A (GUC→GCC) (Fig. 1). To prove that these single-amino-acid changes in E1 were responsible for the cholesterol-independent phenotype, we introduced each into the SFV infectious clone (srf-4/ic, srf-5/ic). We then characterized in parallel the phenotypes of the mutant isolates and those of the virus stocks derived from the infectious clones. As shown below, the phenotypes of the original isolates and the point mutants were comparable in each case, confirming the identities of the mutations.
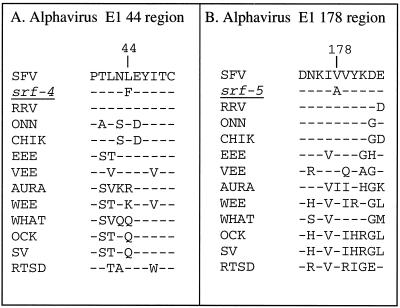
FIG. 1.
Sequence comparison of the alphavirus E1 44 and 178 regions. The alphavirus E1 sequences are listed for the region containing E1 residue 44 (A) and E1 residue 178 (B), as numbered in the SFV sequence. Dashed lines indicate sequence identity with wild-type SFV. Amino acid changes in the srf-4 and srf-5 mutants are indicated. Abbreviations are as follows, according to sequences listed in references 12 and 31 and in the National Center for Biotechnology Information database: RRV, Ross River virus; ONN, O'Nyong-nyong virus; CHIK, Chikungunya virus; EEE, Eastern equine encephalitis virus; VEE, Venezuelan equine encephalitis virus; AURA, Aura virus; WEE, Western equine encephalitis virus; WHAT, Whataroa virus; OCK, Ockelbo virus; SV, Sindbis virus; RTSD, Rainbow trout sleeping disease virus.
Cholesterol dependence of the srf mutants.
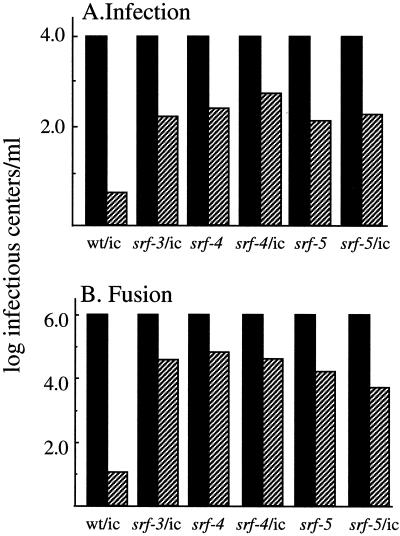
We first compared the ability of wild-type SFV, srf-3, and the new mutants to infect control and cholesterol-depleted C6/36 cells (Fig. 2A). In agreement with previous results (20, 32), infection by wt/ic virus was highly cholesterol dependent, showing a difference of ∼2,000-fold between the two cell types. In comparison, infection of cholesterol-depleted cells by the P226S mutant was increased by about 2 logs, in agreement with previous studies of this mutant. Infection of cholesterol-depleted cells with srf-4 and srf-5 was also markedly enhanced, showing increases similar to those observed with srf-3. Previous studies demonstrated that the decreased ability of wild-type SFV to infect sterol-depleted cells was due to a block in virus fusion, rather than to effects on virus receptor binding, endocytic uptake, and endosomal acidification (26, 32). To assay if the increased infectivity of srf-4 and srf-5 in cholesterol-depleted cells represented an increase in fusion activity, we compared the abilities of wild-type and mutant viruses to fuse with the plasma membrane of control and depleted cells upon exposure to acid pH (Fig. 2B). As previously observed, wild-type virus fusion with control cells was very efficient and was decreased by about 5 logs when the cells were cholesterol depleted (32). The srf-3 mutant, although still showing a preference for cholesterol-containing target membranes, demonstrated an increase of about 3 logs in its ability to fuse with sterol-depleted cells. The fusion activities of both the srf-4 and srf-5 mutants with sterol-depleted cells were markedly increased, to about the same level as that of srf-3. Both mutants still showed the highest levels of fusion with control cell plasma membranes.
FIG. 2.
Infection and fusion of wild-type and mutant SFV in control and cholesterol-depleted cells. (A) Infection. Control (solid bars) and cholesterol-depleted (hatched bars) C6/36 cells grown on duplicate coverslips were infected with serial dilutions of the srf-4 and srf-5 isolates, virus derived from the wild-type infectious clone (wt/ic), or virus derived from the infectious clone containing either the srf-3, srf-4, or srf-5 point mutations (srf-3/ic, srf-4/ic, and srf-5/ic, respectively). Primary infection was quantitated by immunofluorescence. Infection was normalized to 104 infectious centers per ml on control cells. The data shown are the average of three independent experiments, except for the srf-4/ic and srf-5/ic mutants, for which n = 2. (B) Fusion. Serial dilutions of the indicated virus stocks were bound to control (solid bars) and depleted (hatched bars) C6/36 cells on duplicate coverslips in the cold for 90 min and then warmed to 28°C at pH 5.5 for 1 min to trigger virus fusion with plasma membrane. The cells infected due to low-pH-mediated virus fusion were quantitated by immunofluorescence, and the titers were normalized to 106 infectious centers per ml on control cells. The data shown are the average of two independent experiments.
Studies of the srf-3 mutant revealed that the P226S mutation increases the cholesterol independence of both virus-membrane fusion and virus budding from the plasma membrane (20, 22, 24, 32). The effect of the srf-4 and srf-5 mutations on virus exit from sterol-depleted cells was evaluated by pulse-chase analysis (Fig. 3). High-multiplicity infection was used to overcome the wild-type SFV fusion block in depleted cells. Efficient and comparable exit of wild-type SFV and the srf mutants were observed in control cells, with ∼23 to 45% of the total radiolabeled virus envelope proteins released into the medium after the 3-h chase (Fig. 3A). The patterns of viral protein expression and processing were equivalent for the wild type and the mutants. In contrast, wild-type exit from cholesterol-depleted cells was very inefficient (2%), while release of srf-3 was considerably more efficient (23%) (Fig. 3B). Exit of both the srf-4 and srf-5 mutants from sterol-depleted cells was also more efficient than that of wild-type virus, with levels ranging from ∼13 to 35%. Thus, similar to srf-3, single point mutations in srf-4 and srf-5 confer increased cholesterol independence for both virus-membrane fusion and virus exit.
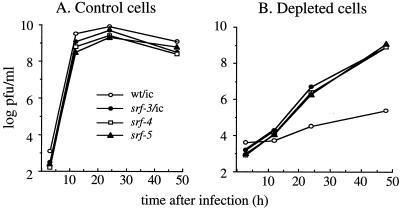
The growth properties of SFV in control and depleted cells reflect the dual requirements for cholesterol in virus entry and exit (20, 32). We compared the growth of wild-type and mutant SFV in control and cholesterol-depleted cells. Cells were infected at low multiplicity, and progeny virus production was quantitated by virus titration on BHK cells. All four viruses grew with comparable rates and efficiencies on control C6/36 cells, showing maximal titers of ∼109 PFU/ml within 12 h (Fig. 4A). As in previous studies, wild-type virus grew very inefficiently in sterol-depleted cells, with titers of only ∼105 PFU/ml after 48 h (Fig. 4B). All three srf mutants grew more slowly in depleted cells, but reached final titers of ∼109 PFU/ml after 48 h. The growth properties of virus from the srf-4/ic and srf-5/ic constructs were similar to those of the parental isolates (unpublished data). The overall increased cholesterol independence of the srf mutants was not due to effects on host range, since the mutants and the wild type showed similar growth kinetics in control C6/36 cells (Fig. 4A) and similar levels of infection and fusion in cholesterol-containing BHK cells (unpublished data). Taken together, these studies demonstrate that the mutations in srf-4 (E1 L44F) and srf-5 (E1 V178A), although in distinct positions in E1 compared to that in srf-3 (E1 P226S), confer similar increases in the cholesterol independence of virus infection, fusion, exit, and growth.
FIG. 4.
Growth of wild-type and mutant SFV in control and sterol-depleted C6/36 cells. Wild-type and mutant viruses (labeled as in Fig. 2A) were prebound to either control (A) or cholesterol-depleted (B) C6/36 cells on ice for 1 h at a multiplicity of 1 PFU/cell. To initiate infection, the cells were incubated for 2 h at 28°C. The cells were then washed to remove input virus and further incubated at 28°C, and the virus released in the medium at each time point was quantitated by plaque assay on BHK cells.
Lipid requirements of srf-4 and srf-5 fusion.
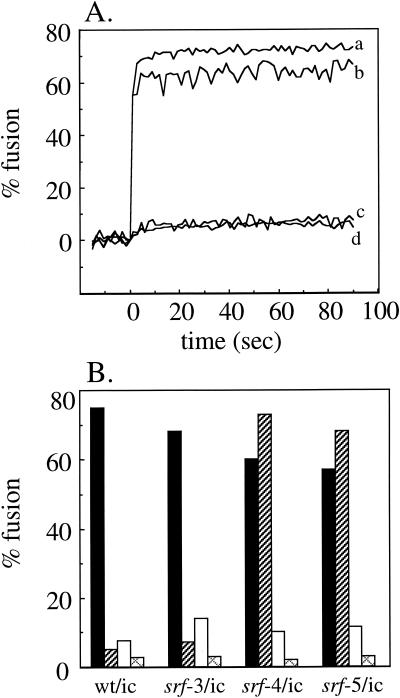
The cholesterol dependence of SFV fusion can be conveniently studied in insect cells, since these cells can be effectively sterol depleted. No such in vivo system for the efficient depletion of cellular sphingolipid exists. We have previously used a well-characterized in vitro system that monitors the fusion of pyrene-labeled SFV with unlabeled liposomes of defined composition (2, 3, 8, 33). Fusion studies of pyrene-labeled wild-type and srf-3 showed a small (approximately twofold) but reproducible increase in the extent of srf-3 fusion with cholesterol-deficient liposomes compared to that of wild-type SFV (3). No changes were observed in the rate, extent, pH, or temperature dependence of srf-3 fusion with either complete or sphingolipid-deficient liposomes. To characterize the lipid dependence of srf-4 and srf-5, the pyrene-labeled wild type and srf mutants were prepared, and their low-pH-triggered fusion with a variety of liposome target membranes was analyzed. Unexpectedly, both srf-4 and srf-5 showed a dramatic change in their ability to fuse in the absence of sphingolipid. Both mutants fused very efficiently with sphingolipid-deficient liposomes at either 37 or 20°C, with final extents of fusion ranging from 60 to 80% after 5 to 10 s (Fig. 5A). Wild-type SFV and srf-3 showed no significant fusion in the absence of sphingolipid, as previously observed. Fusion of srf-4 and srf-5 with sphingolipid-deficient liposomes was low pH dependent, similar to fusion with complete target membranes. The final extents of fusion after 90s at 37°C were determined for the wild type and the three srf mutants by using a variety of target membranes. Efficient fusion was observed for all four viruses with complete liposomes, as expected (Fig. 5B, black bars). At 37°C, the fusion of srf-4 and srf-5 with sphingolipid-deficient liposomes was reproducibly more efficient than with complete liposomes, while little fusion was observed in the absence of sphingolipid for wild-type SFV or srf-3 (Fig. 5B, hatched bars). When cholesterol-deficient liposomes were tested, srf-3 showed an approximately twofold increase over wild-type fusion (Fig. 5B, white bars). Fusion of srf-4 and srf-5 in the absence of cholesterol was reproducibly higher than that of wild-type SFV (results of two of nine experiments performed at various temperatures and liposome concentrations are presented), but the increase was smaller than that of srf-3. The small increases observed in srf mutant fusion with cholesterol-deficient liposomes are in keeping with the limited linear range of the pyrene fusion assay and the fact that fusion of all three srf mutants, although less cholesterol dependent, was still significantly more efficient in the presence than in the absence of cholesterol (as shown in Fig. 2B). Interestingly, the fusion observed with srf-4 and srf-5 in the absence of either cholesterol or sphingolipid alone was abrogated when both lipids were removed simultaneously, indicating that fusion was still dependent on the presence of one of the two critical lipids (Fig. 5B, cross-hatched bars).
FIG. 5.
Lipid dependence of virus-liposome fusion. (A) Real-time fluorescence recordings of the fusion of pyrene-labeled infectious clone-derived viruses with unlabeled sphingolipid-free liposomes (200 μM lipid) at 37°C. Samples were preequilibrated for 3 min at 37°C, and the mixture was adjusted to pH 5.5 at time 0. Lines: a, srf-4/ic; b, srf-5/ic; c, srf-3/ic; and d, wt/ic. Results from a representative example of six experiments are shown. (B) Pyrene-labeled infectious clone-derived viruses, as indicated, were mixed with different types of unlabeled liposomes (200 μM): complete (black bars), sphingolipid deficient (hatched bars), sterol deficient (white bars), and both sphingolipid and sterol deficient (cross-hatched bars). Fusion experiments were performed as described for panel A, and final extents of fusion at 90 s were plotted. The data shown are the average of two independent experiments, each with duplicate determinations.
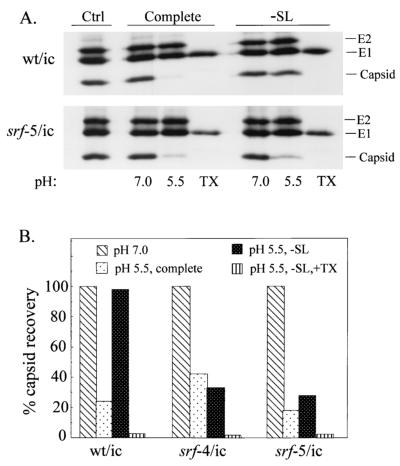
Fusion studies of pyrene-labeled virus monitor lipid mixing, and thus do not differentiate between full fusion (mixing of both leaflets of the viral and target membranes) versus hemifusion (mixing of only the outer leaflets of the two membranes). It was thus possible that the fusion of srf-4 and srf-5 with sphingolipid-deficient liposomes represented hemifusion rather than full fusion. We therefore tested for full fusion by following digestion of the viral capsid protein by trypsin encapsulated within target liposomes (Fig. 6A) (15, 25). The capsid protein of wild-type virus was efficiently digested by entrapped trypsin when the virus was mixed with complete liposomes and treated at pH 5.5 for 5 min at 37°C, but not when fusion was carried out at pH 7.0. No wild-type fusion was observed at either pH with sphingolipid-deficient liposomes, as expected (25), although when these liposomes were disrupted with the detergent Triton X-100, the entrapped trypsin was sufficient to completely digest the capsid protein. In contrast, the srf-5 mutant showed efficient low-pH-triggered fusion with both complete and sphingolipid-deficient liposomes. A series of similar experiments were performed for the wild type and mutants, and the capsid recovery was quantitated for each condition and plotted as the percentage of the capsid recovered at pH 7.0 (Fig. 6B). Clearly, both srf-4 and srf-5 show efficient low-pH-dependent content mixing with liposomes containing no sphingolipid. Thus, srf-4 and srf-5 have a unique phenotype in which a single point mutation increases their ability to fuse with membranes deficient in either cholesterol or sphingolipid.
FIG. 6.
Content-mixing assay of virus-liposome fusion. (A) Radiolabeled viruses were mixed with soybean trypsin inhibitor and trypsin-containing liposomes of the indicated lipid composition. (−SL indicates sphingolipid-deficient liposomes.) Samples were treated at the indicated pH for 5 min at 37°C. The samples were then adjusted to pH 8.0, incubated for 1 h at 37°C to permit capsid digestion, and analyzed by immunoprecipitation and SDS-PAGE. The TX samples were treated at pH 5.5 with liposomes as indicated, but in the absence of soybean trypsin inhibitor, and the 1-h incubation was performed in the presence of 1% Triton X-100 (TX). Control (Ctrl) samples were incubated on ice at neutral pH in the absence of liposomes and immunoprecipitated as described above. (B) The capsid protein was quantitated in experiments performed as described for panel A and expressed as a percentage of the capsid recovered at pH 7.0. The data shown are averages of two experiments each for the wild type and srf-4 mutant and one experiment for the srf-5 mutant. The liposome designations are as in panel A.
E1 homotrimer stability in the srf mutants.
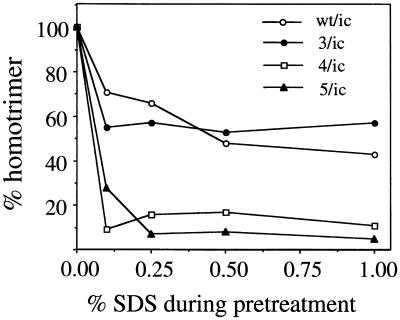
The role of sphingolipid in SFV fusion is unclear. Sphingolipids are not required for the initial low-pH-triggered attachment of virus to the target membrane, and it has been proposed that they act in the formation of the fusion-active E1 homotrimer (25, 36). However, when the E1* ectodomain is assayed, both membrane interaction and homotrimerization are strongly promoted by sphingolipid and cholesterol (1, 17). We examined homotrimer formation in srf-4 and srf-5 as a way of addressing the role of sphingolipid in this process. The wild-type homotrimer is remarkably stable and can be assayed by its resistance to dissociation in SDS-sample buffer at 30°C, its resistance to trypsin digestion in Triton-containing buffers, and its position in sucrose gradient sedimentation assays (8, 33). We treated wild-type and mutant viruses at low pH in the presence of complete liposomes and then directly solubilized the samples at 30°C in sample buffer containing 4% SDS. The presence of E1 in the homotrimer position was assessed by SDS-PAGE. While wild-type SFV and srf-3 showed efficient homotrimerization (33 to 40% trimeric E1) as previously observed (3), surprisingly srf-4 and srf-5 showed little or no E1 migrating in the position of the SDS-resistant homotrimer (0 to 7%, respectively) (data not shown). We then assayed E1 homotrimer formation by its resistance to trypsin digestion in 1% Triton X-100 (8, 15). Wild-type SFV and the three srf mutants all showed very efficient homotrimer formation by this assay, with levels ranging from 50 to 90% trimeric E1 (unpublished data). These results indicated that the homotrimer was formed efficiently in the srf-4 and srf-5 mutants, but that it appeared to have altered stability to SDS-containing buffers. We tested this directly by pretreating wild-type and mutant homotrimers with increasing concentrations of SDS at 30°C, diluting out the SDS in Triton-containing buffer, and then assaying any homotrimer remaining in the reaction mixture by its resistance to trypsin digestion. The data were compared to those for the maximal trypsin-resistant homotrimer obtained for each mutant without any SDS pretreatment. As expected, wild-type and srf-3 homotrimers were stable to pretreatment with SDS concentrations up to 4% at 30°C (Fig. 7) (unpublished data). In contrast, even SDS concentrations as low as 0.1% caused the homotrimers from srf-4 and srf-5 to dissociate, allowing the E1 monomer to be digested with trypsin. At SDS concentrations of 0.25% or higher, srf-4 and srf-5 homotrimer dissociation was essentially complete. When very low concentrations of SDS (0.05 to 0.1%) were tested, all of the srf-4 homotrimer was dissociated, while ∼40% of the srf-5 homotrimer was recovered, suggesting that the srf-4 homotrimer was even less stable than the srf-5 homotrimer (unpublished data). Although their decreased stability was clearly revealed by SDS dissociation, the srf-4 and srf-5 homotrimers were similar to wild-type and srf-3 homotrimers (8) in their resistance to dissociation by urea or by heat treatment in the absence of SDS (unpublished data). Taken together, these data demonstrate that the sphingolipid independence of the srf-4 and srf-5 mutants correlates with their formation of homotrimers that are fusion active, but less stable.
FIG. 7.
Stability of E1 homotrimers to SDS pretreatment. E1 homotrimers were formed by treating radiolabeled wild-type and mutant viruses at pH 5.5 in the presence of 1.0 mM complete liposomes. The samples were then treated at 30°C for 4 min with the indicated concentrations of SDS. All samples were then adjusted to a final concentration of 0.1% SDS and 1% Triton X-100 and digested with trypsin at 200 μg/ml for 20 min at 37°C. The samples were analyzed by immunoprecipitation and SDS-PAGE, and the trypsin-resistant homotrimers at each SDS concentration were compared to the trypsin-resistant homotrimer recovered without any SDS pretreatment. The data shown are a representative example of two experiments.
Interactions among the srf mutations.
From the known domain structure of the native SFV E1 protein (18), it is clear that all three of the srf mutations map to domain II of E1. While P226S is within a loop close to the fusion peptide, both the srf-4 and srf-5 mutations are located in more distal parts of domain II (Rey, personal communication). The three mutations have similar effects on cholesterol dependence, but differ markedly in their effects on sphingolipid dependence. In order to address whether the mutations act by affecting the same or different functions of E1 in fusion, we used the SFV infectious clone to prepare constructs bearing all possible combinations of the mutations. The combination mutants were termed srf-3+4, -3+5, -4+5, and -3+4+5.
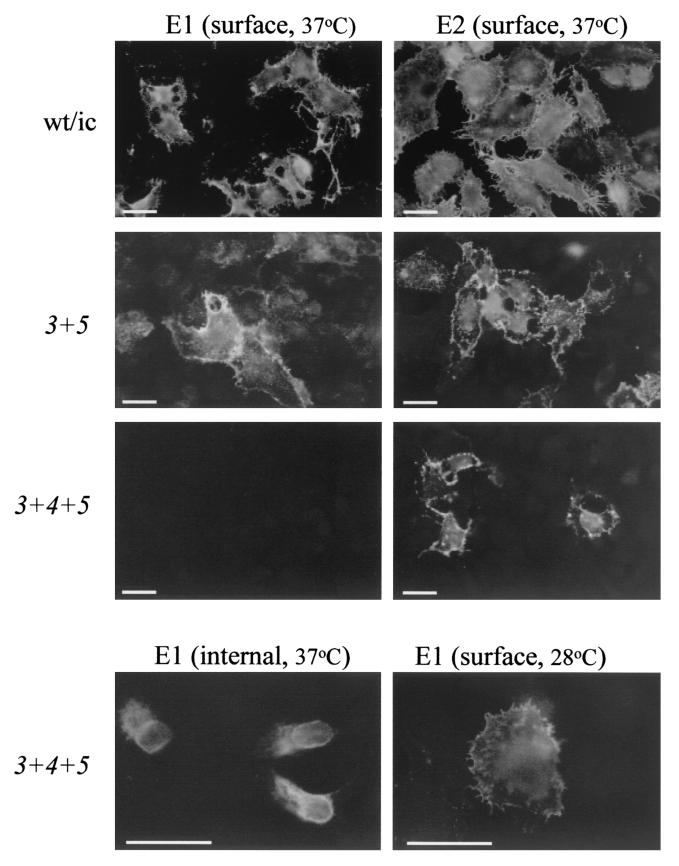
We first assayed the ability of the mutant RNAs to infect cells and produce virus particles. The RNAs were electroporated into BHK cells, the cells were incubated at 37°C, and the distribution of the E1 and E2 proteins was detected by staining nonpermeabilized cells with specific MAbs. Both E1 and E2 were efficiently expressed on the surface of cells infected with wild type or srf-3+5 or srf-3+4 (Fig. 8) (unpublished data). In contrast, cells infected with srf-3+4+5 or srf-4+5 showed efficient expression of E2 on the cell surface, but did not stain with the E1-specific MAb (Fig. 8) (unpublished data). Staining of permeabilized cells demonstrated that the E1 protein from these two mutants was localized to the endoplasmic reticulum and Golgi apparatus (Fig. 8, lower panel) (unpublished data). When cells infected with srf-3+4+5 or srf-4+5 were incubated at 28°C, a less-stringent temperature for protein folding (6), the E1 protein from both mutants was then detected on the cell surface (Fig. 8, lower panel) (unpublished data). The E1 and E2 proteins from all of the combination mutants were efficiently expressed on the cell surface of either control or sterol-depleted C6/36 cells, which are propagated at 28°C (unpublished data). Thus the combination mutants can be divided into two distinct groups: those resembling the wild-type virus (srf-3+4 and srf-3+5) and those with a temperature-sensitive defect in E1 transport (srf-4+5 and srf-3+4+5).
FIG. 8.
Localization of virus envelope proteins in wild-type- and mutant-infected cells. BHK cells were electroporated with RNA derived from the indicated infectious clones, cultured on coverslips for 12 h at 37°C or 18 h at 28°C, and fixed with 3% paraformaldehyde (surface staining) followed by 0.1% Triton X-100 (internal staining). The cells were then stained with MAbs against the E1 or E2 proteins, followed by fluorescein-conjugated secondary antibody. Representative fields were photographed by fluorescence microscopy. This is a representative example of five experiments. Bar, 20 μm.
We compared the growth properties of the wild-type virus with those of one mutant from each of the two groups. BHK cells were electroporated with the indicated RNAs and incubated at either 37 or 28°C. Progeny virus production was quantitated with an infectious center assay in BHK cells at 28°C. At 37°C, both the wild type and the srf-3+4 mutant grew to high titers (∼109 infectious centers per ml), while the srf-4+5 mutant produced 4 to 5 logs less infectious virus (∼105 infectious centers per ml) even after 25 h of incubation (Table 1). At 28°C, both the wild type and srf-3+4 grew more slowly, but produced titers of ∼108 infectious centers per ml by 40 h, about 10-fold less virus than was produced at 37°C. Under the permissive 28°C temperature conditions, srf-4+5 produced titers of ∼107 infectious centers per ml after either 25 or 40 h. Although the final titer of srf-4+5 was ∼10-fold less than those of the wild type and srf-3+4, it was increased about 100-fold compared to the titer of srf-4+5 at 37°C. Electron microscopy of cells infected with wild type or srf-4+5 showed that at 28°C, srf-4+5 produced virus particles similar to those of the wild type, while at 37°C, this mutant showed cytoplasmic nucleocapsids, but only rare examples of budding virus particles (unpublished data). Pulse-chase analysis of cells infected with the wild type and the various combination mutants showed efficient production of viral proteins and their release into the medium by budding at 28°C. At the nonpermissive temperature of 37°C, cells infected with either srf-4+5 or srf-3+4+5 released mainly E1s, a cleaved form of E1 that is produced in vivo under a variety of conditions in which budding is impaired (21). Taken together, these data indicate that the srf mutations could be simultaneously expressed to produce infectious virus. The combination of the srf-4 and srf-5 mutations led to a temperature-sensitive phenotype due to the lack of efficient expression of the E1 protein on the cell surface.
TABLE 1.
Temperature-dependent growth of combination mutants
| Virus | Virus production (infectious centers/ml)a
|
|||
|---|---|---|---|---|
| 37°C
|
28°C
|
|||
| 17 h | 25 h | 25 h | 40 h | |
| Wild type | 3.7 × 109 | 3.2 × 109 | 1.1 × 108 | 7.0 × 108 |
| srf-3 + 4 | 2.9 × 109 | 1.3 × 109 | 1.2 × 108 | 2.2 × 108 |
| srf-4 + 5 | 1.5 × 104 | 1.5 × 105 | 1.1 × 107 | 1.3 × 107 |
BHK cells were electroporated with the indicated viral RNA, plated for 2 h at 37°C, and cultured for the indicated times at 28 or 37°C. Progeny virus production was quantitated by using an infectious center assay in BHK cells at 28°C.
Cholesterol dependence of the combination mutants.
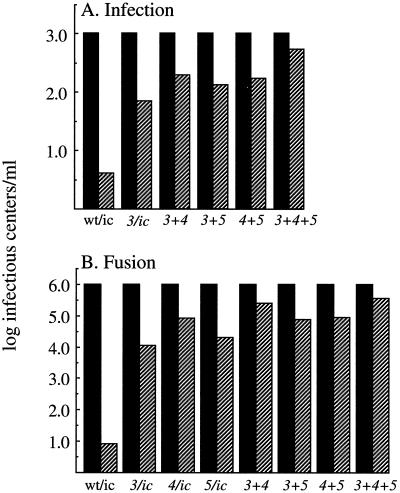
The effect of the combined srf mutations on E1's cholesterol requirement was then tested by using mutant virus stocks produced at 28°C. Infection by the wild-type SFV and the srf-3 mutant showed the expected patterns of strong cholesterol dependence and increased cholesterol independence, respectively (Fig. 9A). All of the combination mutants infected sterol-depleted cells at least as efficiently as the srf-3 mutant, with the highest efficiency observed in the srf-3+4+5 mutant. Fusion studies with control and sterol-depleted cells showed that the srf-4+5 combination mutant was more cholesterol independent than the srf-5 mutant and had levels similar to those of the srf-4 mutant (Fig. 9B). The combination of the srf-3 mutation with either the srf-4 or srf-5 mutation led to increased cholesterol independence beyond that of any single mutation by itself. The highest level of cholesterol independence was observed for the triple mutant, which was somewhat increased over the level of sterol independence of the srf-3+4 mutant. None of the combination mutants showed a return to the wild-type level of cholesterol dependence. Thus, multiple regions of the SFV E1 protein can regulate virus cholesterol dependence and can interact to produce increased sterol independence.
FIG. 9.
Infection and fusion of wild-type and combination mutants in control and cholesterol-depleted cells. (A) Infection. Virus infection of control (solid bars) and cholesterol-depleted (hatched bars) C6/36 cells was assayed as described for Fig. 2A, using virus stocks derived from infectious clones containing the indicated mutations. Virus titers on control cells were normalized to 103 infectious centers per ml. (B) Fusion. Fusion of the indicated viruses with control (solid bars) and cholesterol-depleted (hatched bars) C6/36 cells was measured as described in the legend to Fig. 2B. Virus titers on control cells were normalized to 106 infectious centers per ml. The data in panels A and B are the averages of two independent experiments.
DISCUSSION
We have used cholesterol-depleted mosquito cells to select two new SFV mutants with increased cholesterol independence for growth, infection, fusion, and exit. Similar to the previously described srf-3 mutant, the srf-4 and srf-5 mutations coordinately affect the virus sterol requirements for entry and exit. Unlike srf-3, which is relatively cholesterol independent but retains the wild-type sphingolipid dependence, the srf-4 and srf-5 mutations not only decrease the sterol requirement for fusion, but essentially make the virus sphingolipid independent. The mutations also destabilize the structure of the E1 homotrimer. To our knowledge, this is the first observation of sphingolipid-independent alphavirus fusion and the first identification of E1 regions that are involved in the sphingolipid requirement. While a mutation in the virus fusion peptide was previously shown to block homotrimer formation and fusion (15), this is the first report of a functional but destabilized homotrimer. Studies with combination mutants showed that multiple regions of the SFV E1 protein could interact to further reduce the cholesterol requirement for fusion. Interestingly, whenever the srf-4 and srf-5 mutations were present in the same virus genome, the combination caused a transport defect in E1. Taken together, our data demonstrate a correlation between sphingolipid independence, a destabilized homotrimer structure, and mutations in the midregion of E1 domain II.
All three of the srf mutations map to domain II, an elongated domain that is composed primarily of β-strands and that contains the internal fusion peptide in a loop at the tip (18). The srf-3 mutation lies within a loop adjacent to and associated with the fusion peptide. In contrast, both the srf-4 and srf-5 mutations are located in a more central region of domain II (Rey, personal communication). Comparison of the E1 amino acid sequences between a number of alphaviruses shows that L44, the residue identified by srf-4, is not highly conserved and can be arginine, lysine, or glutamine in other alphaviruses (Fig. 1A). The flanking sequences vary, although proline 40 and residues 45 to 49 are highly conserved. V178, the residue identified by srf-5, is quite conserved, differing only in Aura virus, in which it is an isoleucine (Fig. 1B). The N-terminal flanking sequences are invariant at positions 174 and 176, while the C-terminal flanking sequences are highly variable. Although alanine is considered a fairly neutral substitution, the V178A mutant nonetheless has potent effects on both srf-5’s lipid dependence and on the transport properties of the srf-4+5 double mutant. In contrast, although there were additive effects on sterol independence for the srf-3+4 and srf-3+5 double mutants, no interference with protein transport was observed. Thus the negative interactions between the srf-4 and -5 mutations correlate with their positions in the same general region of domain II. Clearly, in the future, it will be important to analyze these mutations in the context of the refined E1 structure and to determine the amino acid interactions within E1 that may be affected.
It is striking that several independent selections repeatedly resulted in the isolation of the srf-3 P226S mutation. This is indicative of a strong selection preference and suggests that the srf-4 and srf-5 mutations, although equally sterol independent for growth, may be more detrimental to the virus life cycle in tissue culture cells. This could be due to the dual effects of these mutations on cholesterol and sphingolipid dependence or to their decreased homotrimer stability. One interesting question is the mechanism by which selection for cholesterol independence resulted in two mutations with such significant effects on the virus sphingolipid requirement. The original studies of cholesterol-depleted insect cells, including the C6/36 line, indicated that cholesterol depletion is not deleterious to insect cells (29). No replacement sterol is detected, the fatty acid and phospholipid compositions do not change, and the cells do not appear to compensate for the absence of sterol. However, the effect of sterol depletion on sphingolipid composition and distribution in C6/36 cells was not measured. It is thus possible that under depletion conditions, the sphingolipid properties of the cells are altered and that the virus is being coselected for both cholesterol and sphingolipid independence. Given the infection and fusion properties of srf-3 on these cells, though, such a coselection appears unlikely. The srf-3 mutant is highly sphingolipid dependent for fusion with liposomes, and yet shows efficient fusion and infection on cholesterol-depleted C6/36 cells, suggesting that the effect of cellular sterol depletion is primarily to decrease cholesterol levels. We favor the interpretation that the sphingolipid independence of srf-4 and srf-5 is a distinct and additional effect of the amino acid changes selected to confer cholesterol independence in these mutants.
Recent studies have demonstrated that following membrane insertion, the E1 fusion peptide efficiently associates with cholesterol and sphingolipid-enriched membrane microdomains, or “rafts,” that are resistant to Triton solubilization in the cold (1). Low-pH-triggered insertion of E1 into the target membrane is promoted by cholesterol and sphingolipid, but both insertion and fusion can occur with sterols that do not produce a detergent-resistant membrane, suggesting that the physical properties of the raft are not essential (1, 16, 17). A plausible model is that both cholesterol and sphingolipid act to promote E1 conformational changes, and that E1-raft association reflects the interaction of cholesterol with the membrane-inserted fusion peptide. The srf mutants are a source of E1 proteins with alterations in either the cholesterol requirement alone or the sterol and sphingolipid requirements. Analysis of the E1-membrane interactions of these mutants will be important in determining the relationship between the fusion requirements for cholesterol and sphingolipid and the roles of these two lipids in E1 conformational changes, membrane insertion, and raft association. All three srf mutants retain a preference for target membranes containing cholesterol, and it will be interesting to see if this preference reflects an interaction of cholesterol with the fusion peptides, which are identical in sequence between the wild type and srf mutants.
A number of studies have defined the sphingolipid dependence of SFV and SIN fusion and the important structural features of sphingolipids in this process (reviewed in references 13, 30, and 36). The data indicate that the initial hydrophobic interaction of viral E1 with the target membrane can occur in the absence of sphingolipid, but that subsequent fusion requires sphingolipid (25). This has led to the suggestion that sphingolipids are involved in the formation of the fusion-active homotrimer (36), and indeed both sphingolipids and cholesterol promote E1 homotrimer formation in vitro (3, 4, 17). The mechanism by which sphingolipid influences homotrimer formation is unknown. Since the fusion of srf-4 and srf-5 occurred in the absence of sphingolipid, their E1 proteins are presumably independent of any requirements for sphingolipid in the fusogenic conformational changes in E1. Intriguingly, both mutations also affect the structure of the final E1 homotrimer to make it less stable than that of the wild type. It is possible that the srf-4 and srf-5 mutations affect the function of the homotrimer in fusion by uncoupling its normal dependence on sphingolipid. This remains speculative until further information on the location of the mutations in the native E1 and homotrimer structures is available. It is worth noting that TBE fusion, although promoted by cholesterol, is independent of sphingolipid (5), and thus there is precedent for sphingolipid-independent activity of a class II fusion protein.
In conclusion, our data identify two E1 regions that are involved in both the cholesterol and sphingolipid requirements for SFV fusion. These regions differ from that previously identified by srf-3, which affects only sterol dependence, and from the fusion peptide, which is the only portion of the E1 ectodomain that has been directly demonstrated to interact with membranes (1). Future studies will examine the effects of these mutations on E1 conformational changes and elucidate the mechanism by which they act with each other and srf-3 to cause increased sterol independence. The effects of the mutations have here been defined in fusion assays with liposomes and cell membranes and in infection assays in tissue culture cells. In the future, the mutants may be useful in determining the role of specific lipids in infection of mammalian and invertebrate hosts.
Acknowledgments
We thank the members of our laboratory for helpful discussions and suggestions and Duncan Wilson, Félix Rey, and the members of our laboratory for critical reading of the manuscript. We also thank Matthew Scharff for helpful suggestions on mutant isolation.
This work was supported by a grant to M.K. from the Public Health Service (R01 GM57454), by the Jack K. and Helen B. Lazar Fellowship in Cell Biology, and by Cancer Center Core Support grant NIH/NCI P30-CA13330.
REFERENCES
- 1.Ahn, A., D. L. Gibbons, and M. Kielian. 2002. The fusion peptide of Semliki Forest virus associates with sterol-rich membrane domains. J. Virol. 76:3267-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bron, R., J. M. Wahlberg, H. Garoff, and J. Wilschut. 1993. Membrane fusion of Semliki Forest virus in a model system: correlation between fusion kinetics and structural changes in the envelope glycoprotein. EMBO J. 12:693-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chatterjee, P. K., M. Vashishtha, and M. Kielian. 2000. Biochemical consequences of a mutation that controls the cholesterol dependence of Semliki Forest virus fusion. J. Virol. 74:1623-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corver, J. 1998. Membrane fusion activity of Semliki Forest virus. Ph.D. thesis. Groningen University, Groningen, The Netherlands.
- 5.Corver, J., A. Ortiz, S. L. Allison, J. Schalich, F. X. Heinz, and J. Wilschut. 2000. Membrane fusion activity of tick-borne encephalitis virus and recombinant subviral particles in a liposomal model system. Virology 269:37-46. [DOI] [PubMed] [Google Scholar]
- 6.Duffus, W. A., P. Levy-Mintz, M. R. Klimjack, and M. Kielian. 1995. Mutations in the putative fusion peptide of Semliki Forest virus affect spike protein oligomerization and virus assembly. J. Virol. 69:2471-2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garoff, H., J. Wilschut, P. Liljeström, J. M. Wahlberg, R. Bron, M. Suomalainen, J. Smyth, A. Salminen, B. U. Barth, and H. Zhao. 1994. Assembly and entry mechanisms of Semliki Forest virus. Arch. Virol. 9:329-338. [DOI] [PubMed] [Google Scholar]
- 8.Gibbons, D. L., A. Ahn, P. K. Chatterjee, and M. Kielian. 2000. Formation and characterization of the trimeric form of the fusion protein of Semliki Forest virus. J. Virol. 74:7772-7780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibbons, D. L., and M. Kielian. 2002. Molecular dissection of the Semliki Forest virus homotrimer reveals two functionally distinct regions of the fusion protein. J. Virol. 76:1194-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernandez, L. D., L. R. Hoffman, T. G. Wolfsberg, and J. M. White. 1996. Virus-cell and cell-cell fusion. Annu. Rev. Cell Dev. Biol. 12:627-661. [DOI] [PubMed] [Google Scholar]
- 11.Jahn, R., and T. C. Sudhof. 1999. Membrane fusion and exocytosis. Annu. Rev. Biochem. 68:863-911. [DOI] [PubMed] [Google Scholar]
- 12.Kielian, M. 1995. Membrane fusion and the alphavirus life cycle. Adv. Virus Res. 45:113-151. [DOI] [PubMed] [Google Scholar]
- 13.Kielian, M., P. K. Chatterjee, D. L. Gibbons, and Y. E. Lu. 2000. Specific roles for lipids in virus fusion and exit: examples from the alphaviruses, p. 409-455. In H. Hilderson and S. Fuller (ed.), Subcellular biochemistry, vol. 34. Fusion of biological membranes and related problems. Plenum Publishers, New York, N.Y. [DOI] [PubMed]
- 14.Kielian, M., S. Jungerwirth, K. U. Sayad, and S. DeCandido. 1990. Biosynthesis, maturation, and acid-activation of the Semliki Forest virus fusion protein. J. Virol. 64:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kielian, M., M. R. Klimjack, S. Ghosh, and W. A. Duffus. 1996. Mechanisms of mutations inhibiting fusion and infection by Semliki Forest virus. J. Cell Biol. 134:863-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kielian, M. C., and A. Helenius. 1984. The role of cholesterol in the fusion of Semliki Forest virus with membranes. J. Virol. 52:281-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klimjack, M. R., S. Jeffrey, and M. Kielian. 1994. Membrane and protein interactions of a soluble form of the Semliki Forest virus fusion protein. J. Virol. 68:6940-6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lescar, J., A. Roussel, M. W. Wien, J. Navaza, S. D. Fuller, G. Wengler, and F. A. Rey. 2001. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105:137-148. [DOI] [PubMed] [Google Scholar]
- 19.Liljeström, P., S. Lusa, D. Huylebroeck, and H. Garoff. 1991. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J. Virol. 65:4107-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu, Y. E., T. Cassese, and M. Kielian. 1999. The cholesterol requirement for Sindbis virus entry and exit and characterization of a spike protein region involved in cholesterol dependence. J. Virol. 73:4272-4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu, Y. E., C. H. Eng, S. G. Shome, and M. Kielian. 2001. In vivo generation and characterization of a soluble form of the Semliki Forest virus fusion protein. J. Virol. 75:8329-8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu, Y. E., and M. Kielian. 2000. Semliki Forest virus budding: assay, mechanisms, and cholesterol requirement. J. Virol. 74:7708-7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marquardt, M. T., and M. Kielian. 1996. Cholesterol-depleted cells that are relatively permissive for Semliki Forest virus infection. Virology 224:198-205. [DOI] [PubMed] [Google Scholar]
- 24.Marquardt, M. T., T. Phalen, and M. Kielian. 1993. Cholesterol is required in the exit pathway of Semliki Forest virus. J. Cell Biol. 123:57-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nieva, J. L., R. Bron, J. Corver, and J. Wilschut. 1994. Membrane fusion of Semliki Forest virus requires sphingolipids in the target membrane. EMBO J. 13:2797-2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phalen, T., and M. Kielian. 1991. Cholesterol is required for infection by Semliki Forest virus. J. Cell Biol. 112:615-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pletnev, S. V., W. Zhang, S. Mukhopadhyay, B. R. Fisher, R. Hernandez, D. T. Brown, T. S. Baker, M. G. Rossmann, and R. J. Kuhn. 2001. Locations of carbohydrate sites on alphavirus glycoproteins show that E1 forms an icosahedral scaffold. Cell 105:127-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2A resolution. Nature 375:291-298. [DOI] [PubMed] [Google Scholar]
- 29.Silberkang, M., C. M. Havel, D. S. Friend, B. J. McCarthy, and J. A. Watson. 1983. Isoprene synthesis in isolated embryonic Drosophila cells. I. Sterol-deficient eukaryotic cells. J. Biol. Chem. 258:8303-8311. [PubMed] [Google Scholar]
- 30.Smit, J. M., R. Bittman, and J. Wilschut. 1999. Low-pH-dependent fusion of Sindbis virus with receptor-free cholesterol- and sphingolipid-containing liposomes. J. Virol. 73:8476-8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vashishtha, M., T. Phalen, M. T. Marquardt, J. S. Ryu, A. C. Ng, and M. Kielian. 1998. A single point mutation controls the cholesterol dependence of Semliki Forest virus entry and exit. J. Cell Biol. 140:91-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wahlberg, J. M., R. Bron, J. Wilschut, and H. Garoff. 1992. Membrane fusion of Semliki Forest virus involves homotrimers of the fusion protein. J. Virol. 66:7309-7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weissenhorn, W., A. Dessen, L. J. Calder, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1999. Structural basis for membrane fusion by enveloped viruses. Mol. Membr. Biol. 16:3-9. [DOI] [PubMed] [Google Scholar]
- 35.White, J., and A. Helenius. 1980. pH-dependent fusion between the Semliki Forest virus membrane and liposomes. Proc. Natl. Acad. Sci. USA 77:3273-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilschut, J., J. Corver, J. L. Nieva, R. Bron, L. Moesby, K. C. Reddy, and R. Bittman. 1995. Fusion of Semliki Forest virus with cholesterol-containing liposomes at low pH: a specific requirement for sphingolipids. Mol. Membr. Biol. 12:143-149. [DOI] [PubMed] [Google Scholar]