Abstract
Influenza virus hemagglutinin (HA) is a determinant of virus infectivity. Therefore, it is important to determine whether HA of a new influenza virus, which can potentially cause pandemics, is functional against human cells. The novel imaging technique reported here allows rapid analysis of HA function by visualizing viral fusion inside cells. This imaging was designed to detect fusion changing the spectrum of the fluorescence-labeled virus. Using this imaging, we detected the fusion between a virus and a very small endosome that could not be detected previously, indicating that the imaging allows highly sensitive detection of viral fusion.
The emergence of new subtypes of influenza A virus has been responsible for worldwide pandemics, such as the Spanish influenza pandemic in 1918-1919 (1). New virus subtypes are immunologically distinct from viruses circulating previously, because these viruses contain novel subtypes of hemagglutinin (HA), a viral membrane glycoprotein and major antigenic determinant of the virus. The antigenic shift is caused by reassortment of genomic RNA between human and avian viruses and replacement of human virus HA with avian virus HA (1). Although the antigenic shift is required for pandemics, not all reassortant viruses cause severe disease (3). When the HA of reassortant viruses functions efficiently in human cells, binding to sugar chains on the cell surface and fusing the viral and cellular membranes (1), the viruses presumably cause severe disease. In order to elucidate a correlation between HA function and viral infectivity to human cells, there is a need for methodologies to determine whether the HA of viruses is functional against the human cells. Here, we report a simple and rapid imaging technique that visualizes viral membrane fusion.
Membrane fusion of influenza virus is a good marker of HA functions. Influenza virus encloses its genomic RNA with a membrane that consists of a lipid bilayer and viral membrane proteins (1). When infecting a host cell, the virus attached to the cell surface is taken up into an endosome by cellular endocytosis. Viral HA is activated by endosome acidification, and activated HA fuses the viral and endosomal membranes, allowing viral RNA to be released into the cell cytoplasm (1, 13). Because virus-endosome fusion is the last step of the infectious process mediated by HA, efficiency of the fusion in human cells reflects HA function for human cells. The most widely used assay to detect virus-endosome fusion is based on dequenching of lipophilic fluorescence probes (9). In this assay, a fluorescence probe is incorporated into viral membranes at high concentrations to cause self-quenching. Virus-endosome fusion decreases the density of the probe and consequently increases the probe's fluorescence, i.e., dequenching. Commonly the dequenching is measured with a fluorometer. Spectroscopic measurements, however, do not allow proper quantitative analysis of virus-endosome fusion. Dequenching is also induced by other cellular membrane fusion activity, such as endosome-endosome or endosome-lysosome fusion, that can occur after virus-endosome fusion. Spectroscopic measurements cannot distinguish virus-endosome fusion from cellular membrane fusion events.
Recently, individual virus particles labeled with a fluorescent probe were monitored continuously in living cells under a fluorescence microscope, and dequenching induced by a single virus-endosome fusion event was detected (11, 16). This single-virus tracking technique potentially isolates virus-endosome fusion from cellular fusion events. This kind of imaging technique, however, follows only a few virus particles in a cell simultaneously and consequently requires considerable time and effort to obtain statistical data on virus-endosome fusion. In order to quantitatively analyze virus fusion activities in human cells, novel techniques that analyze numerous fusion events simultaneously are required. To improve the fluorescence dequenching assay, we have developed a dual wavelength imaging technique that detects individual virus-endosome fusion events as a shift in the fluorescence color of virus particles, which can be seen under a fluorescence microscope. We applied this technique to fusion of influenza A virus (PR/8/34 strain) in HeLa cells, in which virus-endosome fusion could not be observed by the previous imaging technique (11). We detected numerous virus-endosome fusion events in HeLa cells simultaneously and analyzed the fusion temporally and spatially. Moreover, we characterized virus fusion sites and found that in HeLa cells, the virus initiates fusion with endosomes at a stage earlier than the late endosome stage, which has previously been suggested to be the fusion site of the influenza A virus for other cell lines. This finding indicates that the dual wavelength imaging technique is useful for virological research on infectious processes, in addition to evaluating HA fusion activity in specific host cells.
MATERIALS AND METHODS
Fluorescent labeling.
Influenza virus (A/PR/8/34) was labeled with 3,3′-dioctadecyloxacarbocyanine (DiOC18) and octadecyl rhodamine B (R18). Purified virus (viral protein, 100 μg) was suspended in 1 ml of phosphate-buffered saline. DiOC18 and R18 (Molecular Probes, Inc.) were dissolved in ethanol and mixed at concentrations of 33 μM and 67 μM, respectively. The probe mixture (6 μl) was added to the virus suspension and mixed vigorously. The reaction mixture was gently shaken for 1 h at room temperature and then passed through a filter (0.22 μm pore size; Millipore). The mole fractions of DiOC18 and R18 for 1 mg of virus protein were 2 and 4 nmol, respectively. These values are several times smaller than the mole fraction (10 to 20 nmol/mg protein) of R18 in the traditional fusion assays (9). In the traditional fusion assays, unincorporated R18 was removed by gel filtration (9). However, we omitted this process because of the following two reasons. First, when we applied the reaction mixture to the gel filtration, we did not detect any unincorporated probes. Second, by testing virus fusion to red blood cells, we confirmed that there was no difference in fusion activity between gel-filtrated and unfiltrated viruses.
Imaging virus fusion.
HeLa cells, cultured in a glass dish (Iwaki), were incubated with labeled virus at 4°C for 10 min, washed with ice-cold phosphate-buffered saline, and then warmed to 37°C. After 10 to 60 min at 37°C, the dish was set on the stage of a confocal microscope (LSM410; Zeiss). The cells were scanned using an Ar-Kr laser light (488 nm). Two ranges of fluorescence wavelength, 510 to 525 nm and 575 to 640 nm, were detected simultaneously using two detectors. For quantitative analysis, the fluorescence intensities of individual fluorescent spots on both digital images were read with ImageJ software (http://rsb.info.nih.gov/ij/). To examine the colocalization of epidermal growth factor (EGF) in the virus fusion site, Cy5-labeled EGF and labeled virus were bound to cells simultaneously. To obtain images of Cy5-EGF, immediately after scanning with the Ar-Kr laser, the cells were scanned with a He-Ne laser light (633 nm) and fluorescence over 665 nm was detected.
RESULTS AND DISCUSSION
Labeling design.
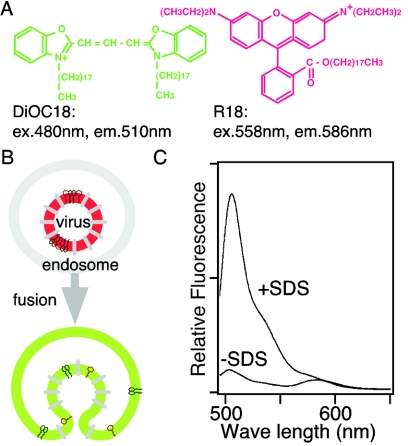
To visualize virus-endosome fusion under a fluorescence microscope, we designed a fluorescent labeling system that would have a change in fluorescence spectrum after fusion. Two lipophilic probes, DiOC18 and R18, were incorporated into the membrane of influenza A virus strain PR/8/34 (Fig. 1A and B). As seen in Fig. 1C, two peaks were observed in the fluorescence spectrum of the labeled virus (Fig. 1C, −SDS [sodium dodecyl sulfate]). The first peak represents the green (510 nm) fluorescence emitted from DiOC18, while the other peak represents the red (586 nm) fluorescence from R18. In the labeled virus, the green fluorescence is suppressed to a level similar to that of the red fluorescence by both self-quenching of DiOC18 and fluorescent resonance energy transfer (FRET) from DiOC18 to R18, whereas the red fluorescence from R18 is partly self-quenched. Removal of the self-quenching and FRET by solubilization with a detergent dramatically increased the green fluorescence of DiOC18 (Fig. 1C, +SDS). In spite of the dissolution of FRET from DiOC18 to R18 by solubilization with the detergent, total red fluorescence above 575 nm remained unchanged because DiOC18 fluorescence caused an increase in the red fluorescence intensity. The fluorescence spectra of DiOC18 and R18 and FRET between the probes were not altered between pH 4.5 and 7.5. When green and red fluorescence were monitored simultaneously using two detectors, the red channel detector was tuned with greater sensitivity than the green detector in order to clearly visualize the fluorescence color shift from red to green caused by membrane fusion, as illustrated in Fig. 1B.
FIG. 1.
Influenza virus-endosome fusion visualization method. (A) Structures of DiOC18 and R18, which can be incorporated into the virus membrane. ex., excitation; em., emission. (B) Schema outlining the fluorescence color shift induced by membrane fusion between a labeled virus and an endosome. (C) Fluorescence spectra of labeled virus (−SDS) and solubilized virus (+SDS). Extreme dilution of the probes increases green (510 nm) fluorescence and faintly alters red (586 nm) fluorescence. From the spectra, virus fusion could be detected by monitoring green and red fluorescence simultaneously using two detectors with a confocal microscope. The labeled viruses were colored red before fusion, as shown in panel B, by tuning the red channel detector, which has greater sensitivity than the green channel detector.
DiOC18 and R18 do not affect virus infectivity. By use of a plaque-forming test with Madin-Darby canine kidney cells, plaque-forming activity of the labeled virus was 96% ± 11% of the activity of nonlabeled virus. These probes do not perturb the virus membrane. In fusion experiments using red blood cells, influenza virus induces leakage of hemoglobin (i.e., hemolysis), which is closely related to virus membrane stability (17). For testing hemolysis between pH 4.5 and 7.5, pH dependency of hemolysis induced by the labeled virus was the same as by nonlabeled virus (data not shown).
Imaging of virus-endosome fusion in living cells.
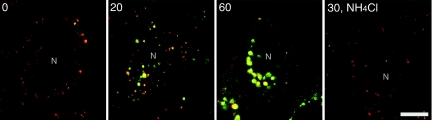
To experimentally detect the fluorescence color shift caused by the fusion of the labeled virus with an endosome, we incubated HeLa cells with the labeled virus for 10 min at 4°C and then at 37°C for various times. The green (510 to 525 nm) and red (575 to 640 nm) fluorescence images of cells were obtained simultaneously using two detectors on a confocal microscope, and both images were merged (Fig. 2). After virus binding (Fig. 2, 0 min), virus particles on cell surfaces were observed as red spots. After a 20-min incubation, orange, yellow, and green spots were seen all over the cells. These orange, yellow, and green spots were not observed in the presence of NH4Cl, which neutralizes the inside of endosomes and consequently blocks virus-endosome fusion, indicating that the color shifts in these spots were caused by fusion between the labeled viruses and endosomes. After 60 min, large spots with bright-green fluorescence were observed close to the cell nucleus. These spots are most likely lysosomes, since the viral membrane components, such as HA, are known to be transferred to lysosomes after 60 min (19).
FIG. 2.
Time-dependent color shift of viruses and virus-fused endosomes. Labeled virus was bound to HeLa cells for 10 min at 4°C and then incubated at 37°C for various time periods (times in min are indicated at upper left of each panel). Green and red fluorescence images were detected with two detectors by using a confocal microscope (LSM410; Zeiss). To detect the labeled viruses as red spots, the red channel detector was tuned with greater sensitivity than the green detector in all experiments. The two images have been merged. The color of fluorescent spots shifted from red to green with time. The shift was not observed in the presence of NH4Cl. The cell nucleus is indicated by N. Bar = 10 μm.
The dual wavelength imaging technique allows highly sensitive detection of virus fusion. Although the previous imaging technique can track single fluorescently labeled viruses in a living cell, it was not capable of detecting the dequenching induced by the virus-endosome fusion in HeLa cells (11), which we could see as a fluorescence color shift. In the single-virus tracking experiments, influenza A virus strain X-31 was used (11). The X-31 virus has different subtypes of HA and neuramidase (H3N2) than PR/8/34 (H1N1), which we used in this study. However, the difference between virus strains is not crucial. Using Aichi/2/68, for which HA and neuramidase are identical to those of X-31, we obtained the same results as with PR/8/34 (data not shown). For the purpose of detection of fusion events, changes in fluorescence color may be more sensitive than changes in fluorescence intensity, because fluorescence intensity is easily affected by many factors other than dequenching, such as fluctuation of the illuminating light, the virus moving out of the focal plane, and the accumulation of viral particles in endosome vesicles.
Previous studies have observed that the virus-endosome fusion in CHO and BS-C-1 cells occurs after the viruses are transported close to the cell nuclei, whereas in HeLa cells the viruses stayed largely in peripheral regions and did not exhibit much motion (11, 16). We obtained consistent data for the distribution of viruses in HeLa cells. As seen in Fig. 2, endosomes containing the labeled virus were distributed peripherally even at 20 min. Furthermore, our data demonstrated that the virus-endosome fusion in HeLa cells occurred in peripheral regions before transport to the perinuclear regions (Fig. 2). This suggests that the virus fusion sites in HeLa cells are different than those in CHO and BS-C-1 cells. Therefore, in order to characterize the virus fusion site in HeLa cells, we analyzed the fusion quantitatively.
Quantitative analysis of virus-endosome fusion.
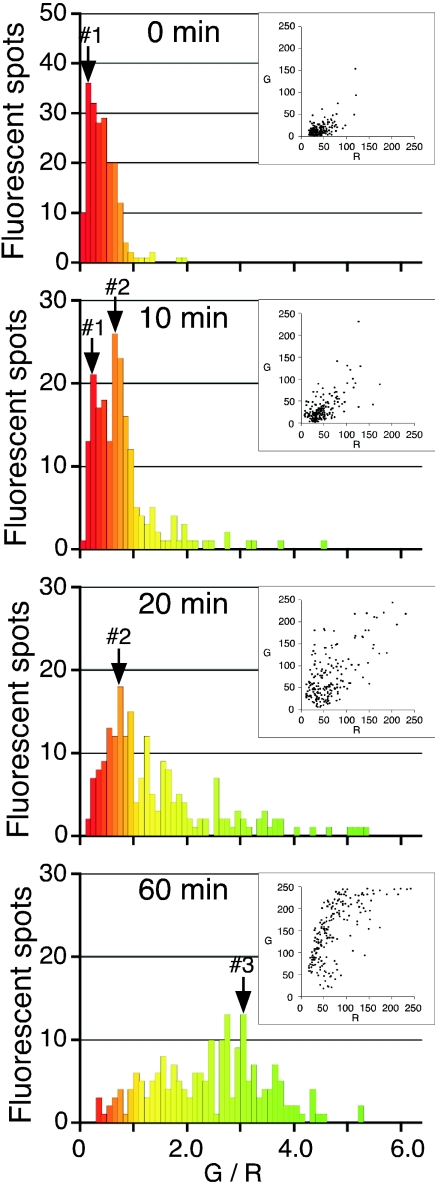
The dual wavelength imaging technique is advantageous in obtaining statistical data on virus fusion because it allows visualization of numerous virus particles (about 40 to 50 viruses per cell) as fluorescent spots, as seen in Fig. 2. To quantitatively analyze the virus-endosome fusion events, we measured the green and red fluorescence intensities of individual fluorescent spots from the digital images at various times (Fig. 3, insets). Each spot was characterized by the ratio of green to red (G/R). As seen in Fig. 3, the G/R values of unfused viruses were distributed close to peak no. 1 (G/R = 0.2 to 0.3) before the 37°C incubation (0 min). After 10 min, half of peak no. 1 shifted to peak no. 2 (G/R = 0.7 to 0.8), and after 20 min, the shift was complete. These results indicate that the subpopulation around peak no. 2 is probably the endosome fused with labeled virus and the half time of virus-endosome fusion is approximately 10 min. After 60 min, both peak no. 1 and peak no. 2 disappeared, and the G/R values increased and became widely spread. The increase in G/R values observed from 20 to 60 min is likely caused by endosome-endosome and endosome-lysosome fusion occurring after virus-endosome fusion. We next focused on the subpopulation where peak no. 2 corresponded with the orange spots in Fig. 2 to identify the endosomes with which the virus fused.
FIG. 3.
Temporal analysis of virus-endosome fusion. Green and red fluorescence intensities of individual fluorescent spots, as shown in Fig. 2, were measured. The fluorescent spots were characterized by a ratio of green to red fluorescence (G/R). Two hundred spots were analyzed at each time, as indicated in the histograms. The colors of the bars in the histograms correspond to the fluorescence colors of spots with the indicated G/R values. Insets show distribution of green and red fluorescence in each spot. Three subpopulations with peaks no. 1, no. 2, and no. 3 (labeled with arrows) represent labeled virus, virus-fused endosomes, and lysosomes, respectively.
Colocalization of endocytosed molecules in the virus fusion site.
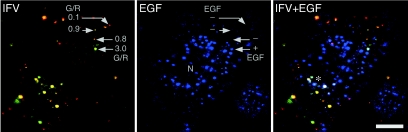
In order to characterize the influenza A virus fusion site, we examined colocalization of EGF in the fusion site. Both influenza virus and EGF are endocytosed through clathrin-coated pits, transferred through an early and a late endosome, and delivered to a lysosome (6, 13). Before delivery to a lysosome, EGF molecules accumulate in a late endosome (4). It has been hypothesized that fusion of influenza A virus occurs in late endosomes because the pH of late endosomes is 5.5 and fusion of influenza A virus (PR/8/34) is activated below pH 5.8 (1, 8, 12, 13). Therefore, EGF should accumulate at the virus fusion site. To examine this hypothesis, we incubated HeLa cells with DiOC18/R18-labeled virus and Cy5-labeled EGF simultaneously at 4°C and then warmed the cells up to 37°C. After 20 min at 37°C, EGF accumulated in organelles close to the nucleus (Fig. 4). The distribution of EGF-positive organelles was consistent with that of late endosomes or lysosomes, as reported previously (4). As seen in Fig. 4, orange spots corresponding to the virus fusion sites were still distributed all over the cell and many of them did not contain any detectable EGF. In contrast, some yellow and green spots close to the nucleus contained detectable EGF (Fig. 4). Virus fusion seemed to precede transfer of the virus to EGF-accumulating organelles. This implies that the virus fusion occurs before reaching late endosomes. To examine this possibility further, we next analyzed the size of endosomes that fused with the viruses.
FIG. 4.
Colocalization of EGF in virus-fused endosomes. DiOC18/R18-labeled virus and Cy5-labeled EGF were bound to HeLa cells for 10 min at 4°C and then incubated at 37°C for 20 min. Virus and virus-fused endosomes (left), EGF (center), and the merged image (right) are shown. Cy5-EGF is colored blue, which is a pseudocolor. The cell nucleus is indicated by N. G/R values for four fluorescent spots are indicated. The red and orange spots (G/R = 0.1 to 0.9) did not contain EGF, although the green spot (3.0) contained EGF. Yellow and green spots (*) close to the nucleus contained EGF. IFV, influenza virus. Bar = 10 μm.
Estimation of size of the virus fusion sites.
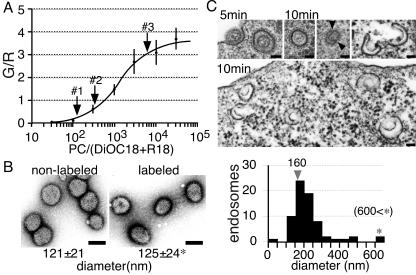
The size of endosomes that fused with the labeled viruses could be calculated from the G/R values in Fig. 3, since the G/R value depends on the density of the probes and the change of probe density induced by the fusion depends on the size of the endosome before the fusion event. We examined whether the influenza A virus fuses with the late endosome from the viewpoint of organelle size. To determine the relationship of the G/R value to probe concentration, we prepared egg phosphatidylcholine (PC) liposomes containing DiOC18 and R18 with various ratios of PC and measured the G/R value of these liposomes with the confocal microscope. The result is shown in Fig. 5A. On this calibration curve, the G/R values of spots no. 1, no. 2, and no. 3 in Fig. 3 were plotted and the corresponding concentrations were deduced (Fig. 5A). The concentration of spot no. 2 in Fig. 3 was 2.6 times less than that of spot no. 1. Since spot no. 1 represents the virus particle itself, this result suggests that the virus fused to an endosome with a membrane area 1.6 times larger than that of the virus particle (Table 1).
FIG. 5.
Size estimation of virus-fusing endosomes. (A) Determination of probe dilution. PC liposomes were made with various concentrations of DiOC18 and R18. Forty liposomes with various sizes (diameter ≤ 1 μm), which were attached to coverslips, were examined at each concentration, and G/R values of the liposomes were measured with the confocal microscope. Dilution rates corresponding to peaks no. 1 to 3 (labeled with arrows) in Fig. 3 were read and are summarized in Table 1. (B) Electron microscopic (EM) images of negative-stained influenza virus. The diameters of virus particles were measured and are summarized below the photographs. One hundred virus particles were examined in each experiment. *, this value is used in Table 1. Bar = 100 nm. (C) EM images of endosome containing virus. HeLa cells were bound with labeled virus for 10 min at 4°C and then incubated at 37°C for 5 min (upper left) and 10 min (others). The endosomal membrane joins to the viral membrane at the point indicated by the arrowhead. The C-shaped structures appear to be endosomes immediately after fusion with the virus. Bar = 100 nm. The distribution of diameters of endosomes containing the virus is presented in a histogram. From EM images, the diameters of 70 endosomes, which obviously contained virus particle(s), were measured.
TABLE 1.
Calculation of the size of the virus-fusing endosome
| Fluorescent spot no. | Area (relative) | Diam
|
|
|---|---|---|---|
| Relative | Actual (nm) | ||
| 1 | 1 | 1 | 125a |
| 2 | 2.6 | 1.6 | 200 |
| 2-1b | 1.6 | 1.3 | 160 |
| 3 | 50 | 7.1 | 890 |
This value is given in Fig. 5B.
2-1 corresponds to the endosome immediately before fusion.
To calculate the diameter of the endosome fusing with the virus, we used the diameter of virus particles determined from electron microscopic images of negative-stained virus (Fig. 5B). The diameter of labeled virus particles was the same as that of nonlabeled virus, that is, 125 ± 24 nm. Using this value, we estimated the diameter of the endosome fusing with the virus to be 160 nm (1.3 × 125 nm) (summarized in Table 1).
The estimated diameter of the endosome fusing with the virus is midway between the size of a clathrin-coated vesicle and the size of an early endosome and is several times smaller than that of a late endosome (5, 7, 10, 18). If viruses were accumulated through endosome-endosome fusion and consequently an endosome contained more than one virus particle, we could underestimate the size of the endosome fusing with the virus. However, accumulation of virus particles might be limited, because the red fluorescence intensity of endosomes containing the labeled virus, which is proportional to the number of virus particles, was not increased by much, even at 60 min (Fig. 3, insets). To test whether our estimation was correct, we estimated the size of spot no. 3 after 60 min (Fig. 3). After 60 min, membrane components of endocytosed influenza virus are transferred to lysosomes (19). By using the G/R value of spot no. 3, the diameter of this organelle was estimated to be approximately 900 nm by the same calculation procedure used above (Fig. 5A and Table 1). The estimated diameter, 900 nm, is equal to the diameter previously determined for lysosomes (5, 10). Therefore, this method appears to be a reliable estimate of size.
To further confirm the size of the virus-fusing endosome, we attempted to observe the endosome through an electron microscope. HeLa cells were infected with virus, incubated at 37°C for 5 to 20 min, and subjected to electron microscopy. A large number of small endosomes containing a virus particle were observed. After incubation for 10 min, which was the half time of the fusion, the diameter of these endosomes remained similar to or slightly larger than that of endosomes immediately after virus internalization (Fig. 5C). The diameter of endosomes after the 10-min incubation was around 160 nm (Fig. 5C). This value is consistent with the diameter estimated from the G/R values of spot no. 2. In addition, the endosomes with membranes continuous with viral membranes and a C-shaped membrane organelle that can be seen in Fig. 5C (10 min) imply that these small endosomes had just fused with the virus. These C-shaped structures were detected more often at 10 to 20 min after infection (data not shown). The electron microscopic observations confirmed that the influenza A viruses fused with the endosomes several times smaller than late endosomes in HeLa cells.
Endosomes with a diameter of 160 nm have a size midway between that of a coated vesicle and that of an early endosome. An endosome of such an early stage was reported to have a pH of 6.3, which is insufficient for fusion of the influenza virus strain (A/PR/8/34) (12, 14). To resolve this problem, further study is needed. However, a similar inconsistency between endosomal pH and optimal pH for biological activity has also been pointed out in the case of iron ion release from endocytosed transferrin (14). It is generally accepted that iron ion release occurs in early endosomes, since transferrin is not transferred to late endosomes (14). However, the pH of early endosomes is 6.0 to 6.5, which is higher than the optimal pH of iron release from transferrin (pH 5.5) (2, 8). Therefore, the pH of early endosomes, especially small early endosomes, must be considered carefully, since pH 6 corresponds to only two to three protons in a small endosome (a diameter of ∼200 nm).
Conclusion.
We developed a dual wavelength imaging technique for visualizing individual viral fusion events as a fluorescence color shift. Because the color shift is directly recognized from fluorescence microscopic images, this technique has advantages that allow rapid analysis of viral fusion activity. Therefore, this technique should be useful not only for surveillance of virulent viral strains but also for screening of antiviral drugs blocking HA and development of influenza virus vectors. Since labeling virus with DiOC18 and R18 is very simple and rapid, this technique could be widely applied to membrane viruses. Many viruses, including Epstein-Barr and vesicular stomatitis viruses, have been reported to have unknown fusion sites, which could not be deduced from pH profiles of viral fusion activity (15). This technique should be a convenient tool for identifying fusion sites of membrane viruses because of spatial and temporal resolutions and high detectability.
Acknowledgments
We thank Keiko Isoda and Kenzo Uehira for technical assistance in electron microscopy. We also thank Yasuko Tanaka for laboratory assistance and Naoko Akazawa for preparation of the manuscript.
This work was partially supported by the Japanese Ministry of Education, Science, and Culture (15570140) and by the Kawasaki Medical School (11504, 15-401A, and 17-401M).
REFERENCES
- 1.Cox, N. J., and Y. Kawaoka. 1998. Orthomyxoviruses: influenza, p. 385-433. In B. W. J. Mahy and L. H. Coller (ed.), Topley and Wilson's microbiology and microbial infections, 9th ed., vol. 1. Virology. Arnold, London, United Kingdom. [Google Scholar]
- 2.Dautry-Varsat, A., A. Ciechanover, and H. F. Lodish. 1983. pH and the recycling of transferrin during receptor-mediated endocytosis. Proc. Natl. Acad. Sci. USA 80:2258-2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dowdle, W. R., and J. D. Millar. 1978. Swine influenza: lessons learned. N. Y. State J. Med. 62:1047-1057. [DOI] [PubMed] [Google Scholar]
- 4.Futter, C. E., A. Pearse, L. J. Hewlett, and C. R. Hopkins. 1996. Multivesicular endosomes containing internalized EGF-EGF receptor complexes mature and then fuse directly with lysosomes. J. Cell Biol. 132:1011-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geuze, H. J., J. W. Slot, G. J. A. M. Strous, H. F. Lodish, and A. L. Schwartz. 1983. Intracellular site of asialoglycoprotein receptor-ligand uncoupling: double label immunoelectron microscopy during receptor-mediated endocytosis. Cell 32:277-287. [DOI] [PubMed] [Google Scholar]
- 6.Goldstein, J. L., M. S. Brown, R. G. W. Anderson, D. W. Russell, and W. J. Schneider. 1985. Receptor-mediated endocytosis: concepts emerging from the LDL receptor system. Annu. Rev. Cell Biol. 1:1-39. [DOI] [PubMed] [Google Scholar]
- 7.Gruenberg, J., G. Griffiths, and K. E. Howell. 1989. Characterization of the early endosome and putative endocytic carrier in vivo and with an assay of vesicle fusion in vitro. J. Cell Biol. 108:1301-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gruenberg, J., and F. R. Maxfield. 1995. Membrane transport in the endocytic pathway. Curr. Opin. Cell Biol. 7:552-563. [DOI] [PubMed] [Google Scholar]
- 9.Hoekstra, D., T. de Boer, K. Klappe, and J. Wilschut. 1984. Fluorescence method for measuring the kinetics of fusion between biological membranes. Biochemistry 23:5675-5681. [DOI] [PubMed] [Google Scholar]
- 10.Hopkins, C. R., A. Gibson, M. Shipman, D. K. Strickland, and I. S. Trowbridge. 1994. In migrating fibroblasts, recycling receptors are concentrated in narrow tubules in the pericentriolar area, and then routed to the plasma membrane of the leading lamella. J. Cell Biol. 125:1265-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lakadamyali, M., M. J. Rust, H. P. Babcock, and X. Zhuang. 2003. Visualizing infection of individual influenza viruses. Proc. Natl. Acad. Sci. USA 100:9280-9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maeda, T., K. Kawasaki, and S. Ohnishi. 1981. Interaction of influenza virus hemagglutinin with target membrane lipid is a key step in virus-induced hemolysis and fusion at pH 5.2. Proc. Natl. Acad. Sci. USA 78:4133-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marsh, M., and A. Helenius. 1989. Virus entry into animal cells. Adv. Virus Res. 36:107-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maxfield, F. R., and D. J. Yamashiro. 1991. Acidification of organelles and the intracellular sorting of proteins during endocytosis, p. 157-182. In C. J. Steer and J. A. Hanover (ed.), Intracellular trafficking of proteins. Cambridge University Press, Cambridge, United Kingdom.
- 15.Russell, D. G., and M. Marsh. 2001. Endocytosis in pathogen entry and replication, p. 247-280. In M. Marsh (ed.), Endocytosis. Oxford University Press, New York, N.Y.
- 16.Rust, M. J., M. Lakadamyli, F. Zang, and X. Zhuang. 2004. Assembly of endocytic machinery around individual influenza viruses during viral entry. Nat. Struct. Mol. Biol. 11:567-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watanabe, Y., M. Tashiro, F. Kitame, and M. Homma. 1985. Enhancement of influenza virus hemolysis by physical and serological treatments. Microbiol. Immunol. 29:825-837. [DOI] [PubMed] [Google Scholar]
- 18.Yamashiro, D. J., B. Tycko, S. R. Fluss, and F. R. Maxfield. 1984. Segregation of transferrin to a mildly acidic (pH 6.5) para-Golgi compartment in the recycling pathway. Cell 37:789-800. [DOI] [PubMed] [Google Scholar]
- 19.Yoshimura, A., and S. Ohnishi. 1984. Uncoating of influenza virus in endosomes. J. Virol. 51:497-504. [DOI] [PMC free article] [PubMed] [Google Scholar]