Abstract
Mammalian alphaherpesviruses normally establish latent infections in ganglia of the peripheral nervous system in their natural hosts. Occasionally, however, these viruses spread to the central nervous system (CNS), where they cause damaging, often fatal, infections. Attenuated alphaherpesvirus derivatives have been used extensively as neuronal circuit tracers in a variety of animal models. Their circuit-specific spread provides a unique paradigm to study the local and global CNS response to infection. Thus, we systematically analyzed the host gene expression profile after acute pseudorabies virus (PRV) infection of the CNS using Affymetrix GeneChip technology. Rats were injected intraocularly with one of three selected virulent and attenuated PRV strains. Relative levels of cellular transcripts were quantified from hypothalamic and cerebellar tissues at various times postinfection. The number of cellular genes responding to infection correlated with the extent of virus dissemination and relative virulence of the PRV strains. A total of 245 out of 8,799 probe sets, corresponding to 182 unique cellular genes, displayed increased expression ranging from 2- to more than 100-fold higher than in uninfected tissue. Over 60% thereof were categorized as immune, proinflammatory, and other cellular defense genes. Additionally, a large fraction of infection-induced transcripts represented cellular stress responses, including glucocorticoid- and redox-related pathways. This is the first comprehensive in vivo analysis of the global transcriptional response of the mammalian CNS to acute alphaherpesvirus infection. The differentially regulated genes reported here are likely to include potential diagnostic and therapeutic targets for viral encephalitides and other neurodegenerative or neuroinflammatory diseases.
All members of the human alphaherpesvirus subfamily, including herpes simplex virus type 1 (HSV-1), invade the peripheral nervous system to establish a reactivatable latent infection (55, 56). While such viral latency is the most common outcome after primary infection of healthy individuals, occasionally in neonates, and more rarely in adults, infection does not stop in the peripheral nervous system. Rather, productive replication continues unabated and virus spreads to the central nervous system (CNS) to cause life-threatening disease (71, 72).
Pseudorabies virus (PRV) is an alphaherpesvirus of swine, but it has an exceptionally broad host range to include lethal infection of a variety of mammals and some birds (19, 40, 48). PRV infections of baby pigs and non-natural hosts invariably involve invasion of the CNS. The CNS response to PRV infection has been studied in some detail (10, 11, 34, 49, 53). The interplay of neurons, support cells, glial cells, and infiltrating leukocytes with respect to immune and inflammatory reactions must play a critical role in limiting or potentiating virus-related CNS pathology in vivo (61). The capacity of PRV mutants with reduced virulence to spread through chains of connected neurons has been exploited to study neuronal circuitry and virus-induced neuropathogenesis (20). The attenuated live vaccine strain PRV-Bartha and its derivatives are widely used in neural circuit tracing (3). PRV-Bartha DNA carries a 3.4-kb deletion in the unique short region of the genome, removing sequences coding for gI, gE, US9, and US2 (36, 41, 46) as well point mutations within the glycoprotein C (gC), gM, and UL21 genes (16, 32, 37, 54). PRV-Bartha shows robust growth on most cell lines but is remarkably reduced for virulence in every permissive species tested (1, 2, 8, 12, 27-30, 42). Even at high inoculating doses, PRV-Bartha-infected animals survive longer with markedly reduced symptoms compared to similar infection by wild-type virus (8). Much of the attenuation of PRV-Bartha can be ascribed to the absence of the gE and gI proteins; however, the mechanisms that link gE/gI to virulence are essentially unknown (27, 51, 77).
Few studies have addressed the question of host molecular determinants of alphaherpesvirus-induced neuropathogenesis. We recently employed DNA array technology to study host gene expression on a genome-wide scale after PRV and HSV-1 infection of rat embryo fibroblasts (51). In this study, we used Affymetrix RG-U34A microarrays to identify cellular genes whose mRNA levels change during experimental PRV infection of rat brains in vivo. The resulting data define key pathways of host gene expression that characterize the host response to an acute CNS infection.
MATERIALS AND METHODS
Recombinant virus strains.
PRV-Becker is a virulent PRV isolate (11), and PRV99 is an attenuated isogenic Becker mutant carrying a deletion of the genes coding for gE and gI (70). PRV-Bartha is a live vaccine strain with reduced virulence (3). Construction of enhanced green fluorescent protein (EGFP)-expressing recombinant derivatives of the Becker (PRV151) and Bartha (PRV152) viruses was described elsewhere (14, 64, 65). PRV99G was constructed in a similar manner by homologous recombination between PRV99 and a plasmid containing an EGFP expression cassette inserted into the PRV gG gene. The EGFP-expressing viruses were kindly provided by Bruce W. Banfield (University of Colorado). All PRV strains were grown and titered on PK15 cells maintained in Dulbecco's modified Eagle's medium with 2% heat-inactivated fetal bovine serum.
Inoculation of animals and tissue preparation.
Male Sprague-Dawley rats (500 to 600 g; Charles River) were used in these experiments. Animals were maintained in a 12:12 light/dark cycle (lights on at 0700 h) in a vivarium with food and water freely available. At least three days prior to infection, animals were transferred to a biosafety level 2 laboratory maintained on a similar 12:12 light/dark cycle. Animals under deep anesthesia (80 mg/kg sodium pentobarbital) were injected with purified PRV virions (1 × 106 PFU in 2 μl) into the anterior chamber of the eye using a Hamilton syringe fitted with a 26-gauge needle. Control animals were mock infected with culture medium. At 48, 60, and 96 h postinfection, animals were anesthetized with CO2 prior to decapitation. The cerebellum, as well as a block of hypothalamic tissue containing the suprachiasmatic and paraventricular nuclei, was rapidly dissected (2 to 4 min), frozen in liquid nitrogen, and stored at −80°C for subsequent gene expression analysis. To control for potential diurnal changes in gene expression, all animals were killed at approximately 1100 h. The 60-h-postinfection animals were anesthetized and inoculated under dim red light at 2300 h during the dark phase of the light/dark cycle.
RNA purification and quantification by real-time RT-PCR.
Total RNA was isolated from frozen rat brain tissue using TRIzol reagent (Invitrogen) and Phase Lock Gel Heavy (Eppendorf) according to the manufacturers' instructions. A second purification step was performed on the isolated RNA using the RNeasy Mini kit from QIAGEN. During this step, RNA preparations for real-time reverse transcriptase (RT) PCR were subjected to on-column DNase digestion. Total RNA was converted into cDNA using reagents and protocols specified in the Affymetrix GeneChip expression manual (2000). Primers and hybridization probes used for cDNA quantification by real-time PCR were designed according to the guidelines in Roche Applied Science Technical Note no. LC 6/99. Their sequences are listed in Table 1. Quantitative PCR was performed in a Roche LightCycler instrument using either LightCycler FastStart DNA Master Hybridization Probes (GAPDH and EGFP) or the LightCycler FastStart DNA Master SYBR Green I kit (all templates except GAPDH and EGFP), according to the manufacturer's specifications. Amplification was carried out in quadruplicate on diluted first-strand cDNA in the presence of 0.5 μM (each) primer and 3 mM MgCl2. A touchdown protocol was used in which the annealing temperature was decreased during the first cycles from 64°C to 56°C with a ramping rate of 0.5°C per cycle. Relative changes in gene expression were calculated using the efficiency-corrected, calibrator-normalized relative quantification strategy described in Roche Applied Science Technical Note no. LC 13/2001. Expression of GAPDH was shown by microarray analysis to be unaffected by PRV infection, and its gene was used as the reference gene.
TABLE 1.
Oligonucleotides used in this studya
| Name | DNA sequence (5′ to 3′) |
|---|---|
| C3 primer fw | ACAAAGCCTTCTCCAACAAG |
| C3 primer rv | GAAGGACAGGCAGTCTTCTT |
| Cxcl10 primer fw | ACTACAGCGTGATGGACAAG |
| Cxcl10 primer rv | CTGCCTGAGGGAAGATTC |
| Dusp1 primer fw | CACCATCTGCCTTGCTTAC |
| Dusp1 primer rv | GCCTCTGCTTCACGAACT |
| EGFP primer fw | GACGACGGCAACTACAAGA |
| EGFP primer rv | GATGCCGTTCTTCTGCTT |
| EGFP probe 1 | GCACAAGCTGGAGTACAACTACAA CAG-FITC |
| EGFP probe 2 | LC705-CACAACGTCTATATCATGGCC GAC-Ph |
| GAPDH primer fw | GGAGAAACCTGCCAAGTATG |
| GAPDH primer rv | CATCAAAGGTGGAGGAATG |
| GAPDH probe 1 | GGCTACACTGAGGACCAGGTTGT-FITC |
| GAPDH probe 2 | LC640-TCCTGTGACTTCAACAGCAAC TCC-Ph |
| Gbp2 primer fw | GACGACAATCACCTGGGAAATG |
| Gbp2 primer rv | ATGCTGCCCAGTGGTCAGA |
| Irf7 primer fw | TCTGGACAGCAGCAGTCT |
| Irf7 primer rv | GCAGCAGTGGTTCTGAAC |
| Mt2 primer fw | TGCTCCTGTGCCACAGAT |
| Mt2 primer rv | CACTTGTCCGAAGCCTCTT |
| Mx1 primer fw | AGCAAACTTCCTCAGCAAC |
| Mx1 primer rv | GGCTCAACTTGTGGTAACAC |
| Nrgn primer fw | GTTCTGTGGGCAACGAAA |
| Nrgn primer rv | ACAGGCAGGTGTGGGATAG |
| Psmb9 primer fw | TCACCACAGACGCCATCA |
| Psmb9 primer rv | CAAAGAAGGGACTTCACTCATCG |
| Sgk primer fw | GTGACGAGCATCCAGATG |
| Sgk primer rv | ACGGCTCTGACTGACAACT |
fw, forward; rv, reverse; FITC, fluorescein isothiocyanate; LC705, LightCycler fluorochrome 705; Ph, phosphate; LC640, LightCycler fluorochrome 640.
GeneChip expression analysis.
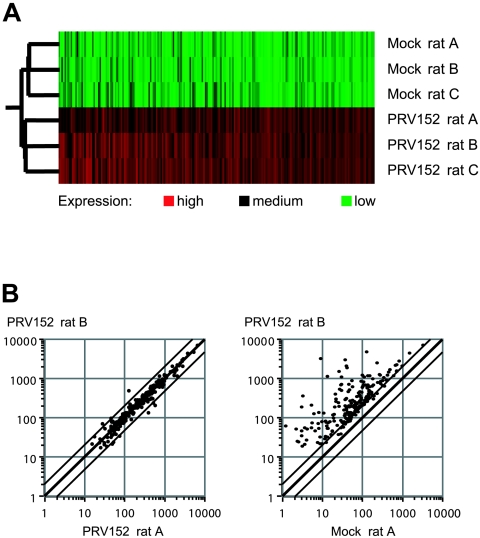
We used Affymetrix RG-U34A GeneChip rat genome arrays comprising 8,799 probe sets designed to detect more than 7,000 unique cellular transcripts. We probed a total of 60 microarrays, which allowed us to monitor the effects of three different PRV strains in two brain tissue types at two (PRV151, PRV99G) or three (PRV152) postinoculation time points compared to mock-infected samples. For each experimental condition, array data were obtained in triplicate (three rats per time point for each PRV strain and for mock-infected animals). Double-stranded cDNA derived from 16 μg of total RNA was used to produce biotinylated cRNA, and labeled cRNA targets were hybridized to GeneChips. After hybridization, probe arrays were washed and stained using the EukGE-WS2 v4 protocol and scanned in an Agilent GeneChip scanner. For more detailed descriptions, refer to the Affymetrix GeneChip expression manual and Microarray Suite 5.0 user's guide. Hybridization intensity data from every experiment were subjected to absolute expression analysis in Microarray Suite 5.0 to generate a present/marginal/absent call for each transcript. Affymetrix metrics files were loaded into GeneSpring 5.0 (Silicon Genetics) for normalization, filtering, and statistical analysis. The recorded fluorescent signal intensity values were subjected to default per-chip and per-gene normalizations to discriminate between real variations in gene expression levels and variations due to the measurement process and to center the data around a value of 1. Then, in a first filtering step, probe sets that were absent or only marginally present in PRV-infected and respective mock-infected samples were excluded from the analysis. Second, probe sets were filtered to retain only those whose expression varied twofold or more between infected and matched mock-infected samples. Third, only probe sets whose hybridization differed from the average mock value with a maximum analysis of variance P value of 0.05 were considered significant. Clustering analysis of the 245 probe sets identified as being significantly up-regulated illustrated that variations between triplicate samples were small (Fig. 1A), and very few mRNAs differed by a factor greater than 2 in replicate infections (Fig. 1B), showing that the hybridization signals observed in independent experiments were highly reproducible. Primary data can be accessed via the Princeton University MicroArray database (http://puma.princeton.edu/).
FIG. 1.
Graphic displays of microarray data. (A) Experiment tree showing reproducibility of gene expression data between triplicates (rats A, B, and C), as exemplified by complete sets of up-regulated genes from six GeneChips corresponding to mock- and PRV152-infected hypothalamic samples at 96 h postinfection. Data sets whose expression patterns are similar are clustered in nearby places in the tree; thus, the three mock-infected and the three virus-infected sets cluster together. Individual genes are shown as columns of rectangles, and expression levels are displayed colorimetrically in different shades of red or green or in black; expression of differentially regulated genes is low (green) or medium (black) in three out of three mock-infected samples and medium or high (red) in virus-infected samples. (B) Scatter plots showing variations in fluorescent signal intensities between two samples (rats A and B). Each dot corresponds to one differentially regulated gene, and parallel lines flanking the center line indicate twofold changes in intensities. Two samples from parallel infections with PRV152 (left) and an infected to a noninfected sample (right) are compared.
RESULTS
Experimental design: CNS infection after intraocular injection of PRV.
Intraocular (i.e., intravitreal or anterior chamber) injection of PRV-Bartha results in CNS infection via retrograde transneuronal transport of PRV through the autonomic circuitry that serves the eye, primarily via the innervation of the iris and ciliary muscle (47, 63). Thus, anterior chamber injection of PRV-Bartha produces infection of autonomic nuclei of the spinal cord, midbrain, and hypothalamus. It should be noted that wild-type PRV (e.g., PRV-Becker) is also capable of infecting the CNS after intraocular inoculation by anterograde transport via the optic nerve after infection of retinal ganglion cells. This anterograde transneuronal spread to retinorecipient targets occurs in addition to the infection via retrograde transneuronal transport through the autonomic circuitry (47).
We used EGFP-expressing variants of three different PRV strains that varied in virulence for our analyses (Table 2): (i) PRV-Becker-EGFP (PRV151), a highly virulent wild-type laboratory strain (11); (ii) an isogenic mutant (PRV99G) that encodes a deletion removing the coding sequences for the viral gE and gI (14, 70); and (iii) the live vaccine strain PRV-Bartha-EGFP (PRV152), which not only lacks the coding sequences for gE/gI but also carries other genetic modifications (3, 14). PRV152 has been used extensively as a tracer of neural circuitry (21, 47, 63, 64). The three PRV strains replicate with similar kinetics and to comparable titers in various cultured cells but display striking differences concerning their neurovirulence in animals (Table 2 and references 8 and 27). Thus, comparison among these strains could provide us with important clues about the cellular determinants of virus-induced neuropathogenesis and the effects of circuit-specific spread in the CNS.
TABLE 2.
Virus strains used in this study
| Name | Parental virus | Phenotype | Neurovirulencea |
|---|---|---|---|
| PRV151 | PRV-Becker | Wild-type, EGFP+ | +++ |
| PRV99G | PRV-Becker | gE−, gI−, EGFP+ | +(+) |
| PRV152 | PRV-Bartha | gE−, gI−,b EGFP+ | + |
+++, wild-type neurovirulence; ++, moderately attenuated neurovirulence; +, strongly attenuated neurovirulence.
Additional mutations in other viral genes (see introduction).
The effects of circuit-specific infection by the three PRV strains on cellular gene expression were analyzed in two different regions of the rat brain. Previous work has shown that intraocular injection of PRV results in retrograde, transneuronal infection of a restricted set of hypothalamic nuclei, including the suprachiasmatic and paraventricular nuclei, in hamsters and rats (47, 63). Therefore, our primary tissue for analysis was hypothalamic tissue. In parallel, samples from the cerebellum were analyzed, since this part of the rat brain is physically separated from the hypothalamus and has no direct connections with the eye. The comparison between the temporal gene expression patterns in hypothalamic and cerebellar tissues after infection enabled us to discriminate between local, direct effects of viral infection and more indirect, global consequences.
Based on previous work studying PRV infection of the rodent eye, we chose three postinoculation intervals to harvest infected tissue for gene expression studies: (i) 48 h, the time when PRV-Becker proteins are first detected in first- and second-order neurons in the brain; (ii) 60 h, the time point with maximum infection by the wild-type Becker strain and just before death; and (iii) 96 h, a time when all Becker-infected animals have succumbed but when PRV-Bartha infection is near its maximum. Rats infected with the modestly attenuated PRV99 die around 72 h postinfection. The EGFP-expressing strains, PRV151, PRV99G, and PRV152, gave mean times to death and spread of infection similar to those of PRV-Becker, PRV99, and PRV-Bartha, respectively. Accordingly, PRV151- and PRV99G-infected tissues were not available for the 96-h time point analysis.
Differential spread of PRV strains in the rat brain.
To correlate virus-regulated cellular gene expression with the extent of viral infection in the analyzed tissues, we monitored neuroinvasion by the three PRV strains in hypothalamic and cerebellar samples over time by quantitating EGFP mRNA expressed from the viral genomes (Table 3). At the 48-h time point, comparable amounts of EGFP mRNA were detected in tissue infected by PRV151 and PRV99G in the hypothalamus and there were hardly detectable EGFP transcripts in the cerebellum. After PRV152 infection, EGFP transcripts were present in the hypothalamus but were at least 20-fold lower than in the respective PRV151- and PRV99G-infected tissues, and no EGFP transcripts were detected in samples from cerebellum. At 60 h postinfection, all three viral genomes were present in both tissue types. However, while PRV151-infected hypothalamic tissue exhibited more than a 60-fold increase in EGFP mRNA accumulation compared to 48 h postinfection, transcription from PRV99G and PRV152 genomes increased by factors of only ∼10 and ∼20, respectively. As expected, cerebellar tissue infected with each of the three virus strains displayed a substantially lower extent of viral transcription than samples from the hypothalamus. At 96 h, PRV152 infection had increased exponentially compared to 60 h after infection (Table 3).
TABLE 3.
Relative EGFP mRNA levels in PRV-infected rat brain tissue samples
| Strain | Relative EGFP mRNA level ata:
|
|||||
|---|---|---|---|---|---|---|
| 48 h pi
|
60 h pi
|
96 h pi
|
||||
| Cer | Hyp | Cer | Hyp | Cer | Hyp | |
| PRV151 | 0.01 | 0.95 | 0.21 | 58.21 | — | — |
| PRV99G | 0.01 | 0.86 | 0.16 | 9.56 | — | — |
| PRV152 | Neg | 0.04 | 0.02 | 0.78 | 41.65 | 368.05 |
mRNA preparations isolated at the indicated times (pi, postinfection) from cerebellum (Cer) or hypothalamus (Hyp) of three infected rats per time point and per virus strain were combined and quantified by real-time RT-PCR. Results for PRV151 in cerebellum at 48 h postinfection were set to 0.01. —, no sample available; Neg, below RT-PCR detection limit.
Temporal spread of PRV in the analyzed rat CNS tissues correlates strongly with the reported neurovirulence phenotypes of the respective virus strains. Nevertheless, rat brains infected with PRV151 showed modest viral gene expression shortly before death (60 h postinfection) compared to PRV152-infected animals, whose brains displayed extensive circuit-specific virus spread at a late predeath stage of infection (96 h postinfection). These findings resemble our previous observations showing that mice infected at skin surfaces with virulent PRV die before significant viral antigen is found in the brain, while PRV-Bartha-infected animals die much later with extensive viral antigen in the brain (8).
Increased transcription of host genes after PRV infection of the brain.
Most expressed genes remained unchanged after infection, and few showed a reduction in transcription by array analysis. For example, at the 60-h-postinfection time point, expression of only four or two genes was decreased more than twofold in the hypothalamus or cerebellum of infected animals, respectively (data not shown). Attempts to confirm decreased accumulation of several of these transcripts by RT-PCR analysis failed (data not shown). In contrast, a large number of host transcripts accumulated in response to PRV infection. Combining all time points, virus strains, and tissue types, we found a total of 245 probe sets corresponding to 182 different cellular mRNAs whose levels increased twofold or more.
To validate the results obtained by microarray analyses, we selected 10 genes with increased transcription for confirmation by quantitative real-time RT-PCR. These genes were representative of a broad range of expression changes and different mRNA accumulation profiles. PCR values were normalized to GAPDH mRNA levels, and the results are shown in Table 4. We confirmed the microarray data for all tested genes by RT-PCR. However, for those genes whose transcripts were induced less than fivefold in our microarrays, the RT-PCR data reported smaller increases. In contrast, the RT-PCR approach revealed much larger increases for those genes whose transcription was increased more than 50-fold according to array analysis (Table 4). Differential sensitivities, as well as the potential for chip saturation at high mRNA induction levels, may account for the differences in induction magnitudes by the two detection methods. The fact that increased transcription of the 10 tested genes was confirmed by an alternative approach strongly indicates that the background in our gene expression analysis is low and that essentially all of the 182 genes identified by GeneChip analysis are truly differentially expressed upon PRV infection compared to uninfected tissue.
TABLE 4.
Validation of microarray data by LightCycler kinetic RT-PCR
| Gene product (designation) | Samplea | Fold increaseb
|
|
|---|---|---|---|
| Microarray | RT-PCR | ||
| Chemokine, C-X-C motif, ligand 10 (Cxcl10) | PRV152, 96 h pi, Hyp | 194.7 | 636.4 |
| Complement component 3 (C3) | PRV152, 96 h pi, Cer | 2.9 | 2.1 |
| Dual-specificity phosphatase 1 (Dusp1) | PRV99G, 60 h pi, Cer | 3.3 | 1.9 |
| Guanylate nucleotide binding protein 2 (Gbp2) | PRV152, 96 h pi, Cer | 13.5 | 4.1 |
| Interferon regulatory factor 7 (Irf7) | PRV151, 60 h pi, Cer | 2.1 | 1.7 |
| Metallothionein 2 (Mt2) | PRV151, 60 h pi, Hyp | 14.8 | 6.8 |
| Myxovirus resistance 1 (Mx1) | PRV152, 96 h pi, Hyp | 50.7 | 298.2 |
| Neurogranin (Nrgn) | PRV152, 60 h pi, Cer | 9.4 | 13.5 |
| Proteosome subunit, beta type 9 (Psmb9) | PRV152, 96 h pi, Hyp | 10.8 | 33.4 |
| Serum/glucocorticoid-regulated kinase (Sgk) | PRV151, 48 h pi, Hyp | 5.4 | 3.7 |
| Serum/glucocorticoid-regulated kinase (Sgk) | PRV99G, 48 h pi, Cer | 3.1 | 2.1 |
pi, postinfection; Hyp, hypothalamus; Cer, cerebellum.
Comparison of relative increases in mRNA accumulation of 10 selected gene products between respective PRV- and mock-infected samples (set to 1.0) as determined by microarray and RT-PCR analysis.
Strain- and tissue-specific and temporal distribution of infection-stimulated genes.
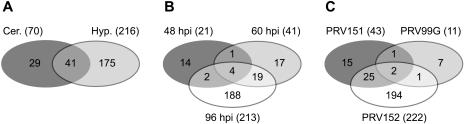
One hundred seventy-five of the 245 probe sets hybridized exclusively to RNA samples from the infected hypothalamus, while 29 mRNAs were specifically increased in the cerebellum. An overlapping set of 41 gene transcripts was regulated in common in both brain tissue types (Fig. 2A). The differences correlate with the extent of infection in these two regions of the brain (Table 3). This correlation is valid at 96 h postinfection, when the majority of differentially regulated genes was detected, but it does not hold for the early time points (Fig. 2B; Table 5). At the earlier time points of 48 or 60 h postinfection, the numbers of induced transcripts in the cerebellum, which shows barely detectable viral infection (Table 3), were similar to or even higher than those in hypothalamic tissue in response to all three virus strains (Table 5). The induction of gene expression in this tissue must reflect more indirect effects of CNS infection. Pooling of data from all three virus infections shows that the levels of 21 gene transcripts were increased at 48 h after infection, 41 transcripts at 60 h, and 213 transcripts at 96 h (Fig. 2B). The activation of most genes followed a specific pattern depending on time after infection, and a significant subset of transcripts was induced in at least two consecutive time points. For example, the transcription of 23 genes was increased at 60 and at 96 h postinfection, and transcription of 4 genes was increased at all time points tested (Fig. 2B).
FIG. 2.
Venn diagrams showing the distribution of differentially increased probe sets according to type of tissue (Cer., cerebellar; Hyp., hypothalamic) (A), time postinfection (hpi) (B), and virus strain (C).
TABLE 5.
Number of up-regulated probe sets for each experimental condition
| Strain | No. of up-regulated probe sets at time postinfectiona
|
|||||
|---|---|---|---|---|---|---|
| 48 h
|
60 h
|
96 h
|
||||
| Cer | Hyp | Cer | Hyp | Cer | Hyp | |
| PRV151 | 6 | 4 | 24 | 23 | — | — |
| PRV99G | 6 | 1 | 4 | 0 | — | — |
| PRV152 | 5 | 4 | 1 | 1 | 38 | 207 |
Cer, cerebellum; Hyp, hypothalamus; —, no sample available.
The numbers of genes with increased expression also differed depending on the virulence of the infecting virus (Fig. 2C; Table 5). The transcription of 43 genes increased after the most virulent virus infection (PRV151), while infection by the attenuated mutant PRV99G induced only 11 transcripts. Three mRNAs were regulated in common by both Becker strains. Infection by the most-attenuated strain, PRV152, induced transcription of more than 222 cellular genes, most of them at the 96-h time point. At the 60-h time point, we found significant overlap of cellular genes activated by PRV152 and the two more-virulent mutants.
Early host response to PRV (48 h postinfection): stimulation of a diverse group of functional classes.
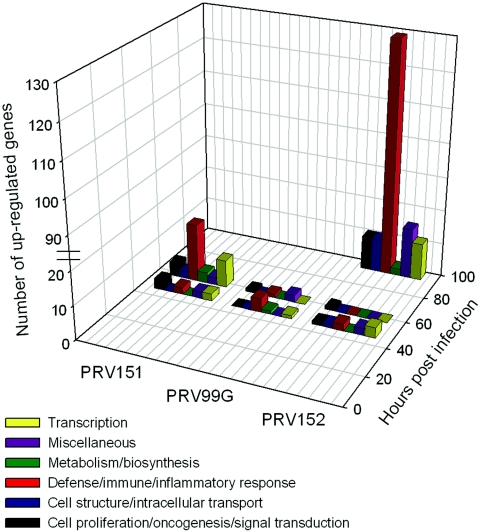
Infection by all three PRV strains at this earliest time point revealed increased expression of 21 different probe sets corresponding to 18 unique cellular genes (Fig. 2B and 3; Table 6). Of these, six (∼33%) were characteristic of the defense/immune/inflammatory response, four (∼22%) were transcription factors, three (∼17%) were involved with cell proliferation/oncogenesis/signal transduction, and five (∼28%) were linked to metabolism/biosynthesis, cell structure/intracellular transport, or miscellaneous functions. Thus, the earliest detectable host transcriptional response to PRV infection was not biased with respect to specific cellular processes (Fig. 3; Table 6). Most of these genes encoded enzymes, receptors, and/or transcription factors. The group of up-regulated enzymes comprised two transferases (sulfotransferase Sult1a1 and UDP glycosyltransferase Ugt2b), isopentenyl-diphosphate delta isomerase (Idi1), a DNA primase subunit (Prim1), serum/glucocorticoid-regulated kinase (Sgk, induced more than fivefold in PRV151 and more than threefold in PRV99G infections), and the tyrosine kinase encoded by proto-oncogene Ret, which serves as a membrane-bound receptor for glial cell-derived neurotrophic factor. Besides Ret, other induced receptor gene products included the neuropeptide-specific tachykinin receptor (Tacr3), an ionotropic glutamate receptor (Gria1), and a neuron-derived orphan nuclear receptor (Nr4a3). The latter also functions as a transcription factor, and other transcriptional regulators induced at 48 h are represented by the suppressor of Ty5 homolog (Supt5h, induced more than sixfold by PRV151), the Jun D proto-oncoprotein (Jund), and Krüppel-like factor 4 (Klf4). Interestingly, together with the Fos proto-oncogene (up-regulated at 60 h postinfection by PRV151 [Table 6]), Jund, Klf4, and Nr4a3 belong to the group of immediate-early response genes that, via intracellular signaling cascades, become rapidly activated following diverse stresses to alter patterns of gene expression (58, 66).
FIG. 3.
Distribution of all virally up-regulated host genes among different PRV strains, time points, and functional classes.
TABLE 6.
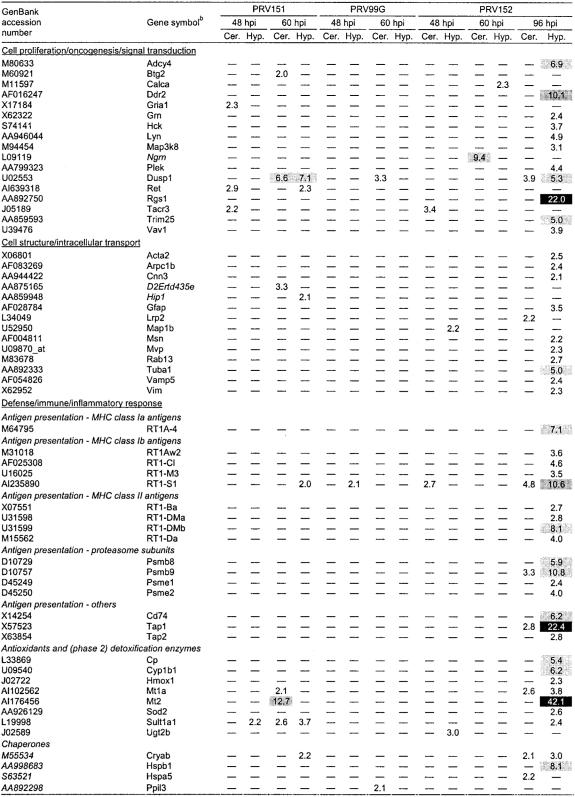
Host genes whose mRNA levels were increased by PRV infectiona
Continued on facing page
Table 6a.
Continued on facing page
Table 6b.
aAverage fold increases compared to mock-infected tissues are indicated for each experimental condition. The greatest increases (5.0-fold and higher) are indicated by different shades of grey and black, with the darker shading corresponding to higher magnitudes of induction. hpi, hours postinfection; Cer., cerebellum; Hyp., hypothalamus; —, absent or not significantly different from levels in mock-infected tissue. Italicized gene names indicate unofficial designations based mostly on homologous genes from related species.
Roughly one-third of all early-activated gene products have a role in innate defense responses (including the peroxisomal Mpv17 homolog; Sgk; testican-2 [Spock2]; Sult1a1; and Ugt2b), but most of them are dedicated to various different aspects of neuronal and glial cell activities, including synaptic function (e.g., Gria1, microtubule-associated protein 1b [Map1b]) and neurogenesis (e.g., Ret, Supt5h, Spock2, Map1b). Moreover, many of the early-response genes have been previously shown to facilitate (Gria1, Map1b, Nr4a3, Prim1) or inhibit (Jund, Ret, Sgk) (neuronal) cell death, thus being either neuroprotective (Ret, Ugt2b [69, 73]) or neurotoxic (Gria1). It is believed that imbalances in excitatory amino acids (e.g., glutamic acid) can kill neurons via binding to their receptors (excitotoxicity), leading to an uncontrollable rise in intracellular Ca2+ concentrations and hence cell lysis and death (18). A significant subset of transcripts induced at 48 h postinfection corresponded to genes that are responsive to activation by steroids such as glucocorticoids, which are considered to be the principal mammalian stress hormones (Sgk, Sult1a1, Ugt2b; see also 60-h-postinfection data, Table 7). Others have a role in signaling by steroid hormones (Nr4a3) and/or in steroid metabolism (Idi1, Sult1a1, Ugt2b). Furthermore, some of these gene products are known to be involved in the induction or inhibition of reactive oxygen species (ROS) production (Gria1, Mpv17, Ret), may be activated by ROS (Jund, Sgk; see also 60-h-postinfection data, Table 7), and/or have a known activity as antioxidative detoxification enzymes (Sult1a1, Ugt2b). Five of the gene products that we found up-regulated at 48 h postinfection were also induced at the 60-h time point (Idi1, Ret, the major histocompatibility complex class I [MHC-I] molecule RT1-S1, Sult1a1, and Sgk) (Fig. 2B; Table 6).
TABLE 7.
Redox-sensitive and glucocorticoid-induced genes up-regulated at late stage (60 h) of PRV151 infectiona
| Gene product | Designation | Tissue (fold increase) | Redox sensitive | GR inducible | Reference(s) |
|---|---|---|---|---|---|
| B-cell translocation gene 2 | Btg2 | Cer (2.0) | + | − | 59 |
| FBJ murine osteosarcoma viral oncogene homolog | Fos | Cer (3.4) | + | − | ROS-TRRDb |
| Crystallin, alpha B | Cryab | Hyp (2.2) | + | + | 26, 57 |
| Dual-specificity phosphatase 1 | Dusp1 | Cer (6.6) | + | + | ROS-TRRD, 74 |
| Hyp (7.1) | + | + | |||
| Glycerol-3-phosphate dehydrogenase 1 | Gpd1 | Hyp (14.7) | − | + | 38 |
| Metallothionein 1a | Mt1a | Cer (2.1) | + | + | ROS-TRRD, GR-TRRDc |
| Metallothionein 2 | Mt2 | Cer (12.7) | + | + | ROS-TRRD, GR-TRRD |
| Mitogen-inducible gene 6 protein homolog (gene 33) | Mig-6 | Cer (2.2) | − | + | 76 |
| Hyp (3.1) | − | + | |||
| Nuclear factor of kappa light-chain gene enhancer in B-cells inhibitor, alpha | Nfkbia | Hyp (2.1) | − | + | 15 |
| Serum/glucocorticoid-regulated kinase | Sgk | Cer (2.2) | + | + | 33 |
| Hyp (6.4) | + | + | |||
| Sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 | Sult1a1 | Cer (2.6) | − | + | 22 |
| Hyp (3.7) | − | + |
GR, glucocorticoid receptor; Cer, cerebellum; Hyp, hypothalamus.
ROS-TRRD, Redox-sensitive gene transcription regulatory regions database (http://wwwmgs.bionet.nsc.ru/mgs/papers/stepanenko/ros-trrd/).
GR-TRRD, Glucocorticoid-regulated transcription regulatory regions database (http://wwwmgs.bionet.nsc.ru/mgs/papers/merkulova/gluc/).
Late host response to virulent strain PRV151 (60 h postinfection): stimulation of innate immune and generalized stress responses.
At the 60-h-postinfection interval, a total of 41 probe sets corresponding to 34 unique cellular genes were induced by virus infection (Fig. 2B and 3; Table 6). The vast majority (29 [>85%]) were identified in PRV151-infected animals which, at this time point, were in a state just before death. A large fraction of virus-induced gene products at 60 h was associated with steroid hormone- and/or redox-related cellular processes. Roughly one-third of all up-regulated gene products (11 out of 34) were explicitly identified as glucocorticoid or redox sensitive or both, i.e., susceptible to induction by activated glucocorticoid receptors and/or ROS, respectively (Table 7). The top four most strongly up-regulated gene products at this postinfection time point (dual-specificity phosphatase 1 [Dusp1], glycerol-3-phosphate dehydrogenase 1 [Gpd1], metallothionein 2 [Mt2], and Sgk, each induced more than sixfold) could all be assigned to one or both of these two categories (Table 7).
Thirteen of the 34 stimulated genes at 60 h (∼38%) were components of the host immune/defense/inflammatory response (Table 6). With one exception (the nonclassical MHC-I protein RT1-S1), all were assigned to the innate defense system. Among these, the metallothioneins Mt1a/Mt2 and Sult1a1 have important roles in the immune and inflammatory response as scavengers of free radicals or detoxification enzymes, respectively. Interestingly, Mt2, which exhibits increased expression in astrocytes upon brain injury (13, 17), was induced by >12-fold, representing one of the most strongly activated genes at this postinfection time point (Table 6). Moreover, expression of critical components of the classical complement system (C1qb, C1s, and C4a) and interferon regulatory factor 7 (Irf7), along with interferon-stimulated GTPase2 (Iigp2), were increased. Notably, Irf7 has very recently been identified as the master regulator of type I interferon-dependent immune responses (25). Aside from Irf7, expression of several other transcription factors was also increased. Some of them belong to the class of stress-induced cellular immediate-early response genes, including members of the nuclear orphan receptor family (Nr4a1 and Nr4a2) as well as the cellular Fos gene (see results at 48 h postinfection, above).
Unlike with PRV151 infection, the expression of only two gene products was induced significantly at 60 h in the PRV152 infection: calcitonin-related polypeptide (Calca) and neurogranin (Ngrn). Both are neuropeptides implicated in increased resistance to Ca2+-mediated toxicity (44, 62). In summary, the late (60-h) CNS response to the virulent PRV151 infection is dominated by innate immune reactions based on the cellular complement, interferon, and antioxidant/detoxification systems, as well as acute stress responses that involve steroid hormone release, redox dysregulation, and potentially other processes.
Late host response to attenuated strain PRV152 (96 h postinfection): massive stimulation of innate/adaptive immune and proinflammatory genes.
Only PRV152-infected animals survived to the 96-h time point. At this late time, strong stress responses were still evident in the cellular gene expression profile (Fig. 3; Table 6). For example, all but one (Btg2) of the glucocorticoid- and ROS-induced gene products that were activated at 60 h postinfection (Table 7) displayed increased expression at 96 h as well. In accordance with previous observations, expression of inducible nitric oxide synthase (iNOS) (Nos2) was also markedly increased (60), and two heat shock proteins (Hsp27 [Hspb1] and BIP [Hspa5]) were induced. However, the host transcriptional response to attenuated PRV infection at this late stage of infection was dominated by massive induction of immune and proinflammatory genes. The majority (∼61%) of mRNAs found to be up-regulated by infection, taking into account all time points, tissue types, and virus strains, fell into the functional category of immune/defense/inflammatory genes (Fig. 3; Tables 6 and 8). Moreover, of the 28 genes induced over 10-fold by viral infection, 23 (∼82%) belonged to this category. More strikingly, of the 11 gene products whose expression increased more than 20-fold, all (including the regulator of G-protein signaling 1, Rgs1, a B-cell activation protein) have known functions in immune and/or inflammatory responses.
TABLE 8.
Defense/immune/inflammatory response genes
| Functional subgroup | No. | Fold increasea |
|---|---|---|
| Antigen presentation | 16 | 5.4 |
| Antioxidants/detoxification enzymes | 8 | 6.7 |
| Chaperones | 4 | 3.3 |
| Complement | 9 | 5.7 |
| Cytokines/chemokines/receptors | 12 | 25.0 |
| Extracellular matrix/cell adhesion | 13 | 4.7 |
| Hemostasis/prostaglandin-related | 7 | 4.2 |
| Interferon response | 15 | 18.6 |
| Leukocyte antigens | 9 | 4.4 |
| Proteolysis | 5 | 4.0 |
| ROS/RNS generation | 6 | 5.4 |
| Others | 7 | 5.0 |
Average fold induction (all time points, virus strains, and tissue types).
Among the top ten most strongly induced cellular transcripts, nine encoded proteins related to innate immune control (Table 6). The CNS innate defense against pathogens is accomplished in part by the induction of proinflammatory cytokines. Accordingly, PRV-infected brain tissues display significant accumulation of the transcripts encoding interleukin 6 (IL-6), which is also implicated in neurodegenerative processes (45), IL-1-β (Il1b), and IL-18 (Il18), as well as IL-stimulated genes such as the one specifying expression of the IL-6-responsive glial fibrillary acidic protein (Gfap). Mediators of IL-targeted signaling pathways were also stimulated after PRV infection, such as signal transducer and activator of transcription 3 (Stat3), involved in gene induction of IL-6. Significantly, anti-inflammatory cytokines such as IL-10 and transforming growth factor β were not induced in virus-infected samples, reinforcing the view that the principal response to extended infection by PRV152 was proinflammatory. In addition, antiviral cytokine and response gene expression was induced, including IL-15 as well as a substantial number of type I and type II interferon-responsive gene products and transducers of interferon signaling, many of which have been shown to exhibit antiviral activity (e.g., Best5 protein, C-X-C motif chemokine ligand 10 [Cxcl10], guanylate nucleotide binding protein 2 [Gbp2], interferon consensus sequence binding protein 1 [Icsbp1], Irf7, Irf8, myxovirus resistance protein 1 [Mx1], diverse MHC-I and -II antigens, 2′,5′-oligoadenylate synthetase 1 [Oas1], double-stranded RNA-dependent protein kinase [Prkr], Stat1, and Stat2). Transcription of several of the interferon system components was strongly increased after 96 h of PRV infection (Best5, >60-fold; Cxcl10, >190-fold; Gbp2, >160-fold; Irf7, >40-fold; Mx1, >50-fold; average up-regulation of 15 interferon response genes, >18-fold [Table 8]). These results are in accordance with previous observations showing that interferon-stimulated gene expression represents a principal response to various viruses, including PRV, and that interferons are key cytokines in the immune control of viral infection (7, 9, 51, 78, 79). Other constituents of the innate defense system that were induced as part of the late host response to PRV-152 included a number of genes whose products function as antioxidants/detoxification enzymes (see above), e.g., Mt1a and Mt2 (increased >40-fold). Additionally, transcripts for multiple components of the classical and alternative pathways that form the complement system were increased, with those for C2 being most strongly induced (>30-fold; Tables 6 and 8). Not surprisingly, expression of the gene encoding cyclooxygenase 2 (Ptgs2) was also stimulated (Table 6). Cyclooxygenase 2 is a key mediator of prostaglandin biosynthesis, and it has been previously shown that its transcription is markedly induced after infection of fibroblasts with various herpesviruses, including human cytomegalovirus, HSV and PRV. Moreover, this enzyme is required for productive cytomegalovirus and PRV replication in culture (50, 80). Consistent with these findings, we observed that transcription of the prostaglandin F receptor (Ptgfr) was also stimulated (Table 6).
Another function of the CNS innate immune response is to facilitate the recruitment of leukocytes to the site of infection by the coordinated up-regulation of chemokines, adhesion molecules, and matrix metalloproteinases (MMPs). These molecules also are critical in the transition from innate tissue responses to the proper adaptive lymphocyte responses. Substantial proinflammatory CNS chemokine activity upon infection was evident in our data exclusively at late times after infection with PRV152. A marked increase in neutrophil and/or monocyte/macrophage-targeted chemokines was observed. For example, virus-induced Cxcl1 levels were >45-fold higher than in noninfected tissue (Table 6). Interestingly, the gene which proved to be most strongly (>190-fold) induced among all 180 differentially regulated cellular transcripts in our microarray analyses corresponded to the interferon-stimulated chemokine Cxcl10 (Table 6), which has a critical role in recruitment of T-helper cells and other lymphocytes. Moreover, expression of the proinflammatory chemoattractant/chemotactic C-C motif chemokines Ccl2 (also known as monocyte chemotactic protein 1 [MCP-1]; induced >50-fold), Ccl3 (also known as MIP-1α), Ccl4 (also known as MIP-1β), Ccl20, and the Ccl3/Ccl4 receptor Ccr5 were all increased by 96 h postinfection. All induced cytokines, chemokines, and chemokine receptors taken together displayed an average 25-fold increase, representing the strongest induction among all functional subclasses of defense/immune/inflammatory response genes (Table 8). Additionally, enhanced expression of cell adhesion molecules such as intercellular adhesion molecule 1 (Icam1), Cd44, and integrin alpha (Itgam) was detected (Table 6), which, when proceeding along the blood-brain barrier, allows leukocytes to attach and begin infiltration into the CNS. This process is augmented by MMP digestion of the endothelial layer. Surprisingly, our analyses did not find up-regulation of MMPs. Instead, transcription of an MMP inhibitor (tissue inhibitor of metalloproteases [Timp1]) was induced. A number of galactose-specific lectins were induced as well, including Lgals3, Lgals5, Lgals9 (Galectins), and Mgl. In agreement with the scenario of leukocyte infiltration through the blood-brain barrier at late times after CNS infection, we found accumulation of a large number of leukocyte-specific transcripts (e.g., Cd14, Cd38, and Cd86, etc.; Tables 6 and 8).
A key step in the transition of CNS innate immune functions to adaptive immunity is interaction of resident glia with infiltrating T cells in the context of MHC molecules. We observed consistent increases in both classical (class Ia) and nonclassical (class Ib) MHC-I transcript expression in virus-infected samples (Table 6). Little is known about the role of nonclassical MHC presentation, but it has been suggested that expression on neurons might be protective (31, 35). Transcription of other components of the MHC-I antigen presentation system were increased as well, including genes for the Tap transporters (Tap1 and Tap2) and (gamma interferon-inducible) activators of the immunoproteasome (PA subunits Psmb8, Psmb9, Psme1, and Psme2). Together with the fact that MHC-II and Cd74 transcripts were also induced, our data clearly indicate that late stages of brain infection by PRV152 are characterized by increased expression of the MHC-I- and MHC-II-dependent antigen presentation system (Tables 6 and 8). Moreover, the overall massive induction of immune and inflammatory functions suggests the potential for immunopathology at the late stages of CNS infection by PRV152.
DISCUSSION
Cellular response to CNS infection by PRV.
The transcripts we analyzed represent both direct and indirect cellular responses, because circuit-specific PRV infection of the CNS results in a direct infection of neurons and a local response of non-neuronal cells that leads to more global responses. The immediate or acute response to infection by 48 h by the most virulent virus (PRV151) is remarkably similar to that stimulated by the most attenuated strain, PRV152. However, animals infected with PRV152 live long enough (96 h) to allow extensive invasion of those CNS circuits that innervate the anterior chamber of the eye and, consequently, the appearance of delayed CNS-specific responses to the infection that were not detected early. This late transcriptional response to PRV152 infection accounts for the majority of PRV-induced genes identified by our microarray analysis. As infection progresses, cellular innate defense and stress responses, including glucocorticoid- and redox-related pathways, predominate, followed by adaptive immune reactions. Previously, we noted the spatio-temporal response of peripheral immune and resident glial cells to CNS infection with PRV (10, 34, 49, 53). CD45+ leukocytes traffic to the specific sites of CNS infection; cells of the monocyte/macrophage lineage then appear prior to the appearance of CD4+ T cells, which are in turn followed by CD8+ cytotoxic lymphocytes. Given that the CNS lacks significant numbers of antigen-presenting cells, it is likely that infiltrating T cells are activated in the periphery, probably via lymph nodes that drain the eye and then gain access to the CNS (49). Our present gene expression study clearly supports this view of a staged response to infection.
Although a correlation exists between the extent of infection, the neurovirulence of a given strain, and the number of genes with increased transcription at late times after infection, early time point data suggest that infection triggers activation of a subset of cellular genes in noninfected cells via more systemic or global mechanisms that must involve intercellular communication. While the gE/gI protein complex is known to be a strong PRV virulence factor, we could not identify a simple pattern of host gene response that might provide insight into the mechanism of virulence by comparison of PRV99G to PRV151 or PRV152. It is more likely that gE/gI affect virulence in peripheral tissues rather than in the CNS (8).
Hormonal stress response.
Glucocorticoids are immunomodulatory steroid hormones produced by the adrenal cortex under both physiological and pathological conditions (6, 39). Cortisol is the major endogenous glucocorticoid in humans, while corticosterone is its functional equivalent in mice and rats (67). Both of these hormones are similarly regulated within the framework of the hypothalamic-pituitary-adrenal (HPA) axis. The adrenal cortex releases cortisol/corticosterone in response to adrenocorticotropin (ACTH), a peptide hormone derived from the anterior lobe of the pituitary gland. The production of ACTH is under the regulatory control of the hypothalamus, by virtue of neurons in the paraventricular nucleus that release corticotrophin-releasing factor (CRF) into the hypophyseal portal circulation that leads to the anterior pituitary gland. CRF is the main neurohormone responsible for stimulating ACTH release.
Viral infection stimulates glucocorticoid production by indirect mechanisms or by direct infection of the various endocrine tissues comprising the HPA axis. The HPA axis is also vulnerable to functional perturbation by direct viral infection of the hypothalamus. For example, hypothalamic infection of rats with a neurovirulent strain of HSV results in loss of CRF content, despite increased plasma ACTH (4, 5). Glucocorticoids are regarded as being anti-inflammatory and important for neuronal function, but their release in conjunction with a brain injury is known to intensify neuronal death (43). Since many of the genes activated by PRV were glucocorticoid responsive (Tables 6 and 7), it is conceivable that overproduction of steroid hormones contributes to neuronal death and neuropathogenesis in the context of viral infection. In support of this view, Clase and Banfield (13a) recently reported that systemic delivery of the corticosteroid dexamethasone did not protect chicken embryo CNS from damage due to PRV-Becker infection but rather increased virulence of the virus.
Oxidative stress response.
Reactive nitrogen species (RNS), like nitric oxide (NO) and reactive oxygen species (ROS), exert multiple modulating effects on inflammation and play a key role in the regulation of the immune response (24). Low concentrations of NO produced by constitutive and neuronal iNOS inhibit cytokine and chemokine synthesis, as well as leukocyte adhesion and transmigration. In contrast, iNOS produces large amounts of NO in a sustained manner that can be proinflammatory and toxic. In brain tissue, iNOS is expressed both by glial cells and neurons. Our microarray analysis found significant increases in iNOS transcripts at late times after infection in accordance with previous observations (Nos2 [Table 6]) (60). Similarly, superoxide free radical O2− produced by all cell types participating in inflammation may lead to toxic effects when produced at high levels during oxidative burst. The effects of both RNS and ROS in immune regulation are exerted through multiple mechanisms, which include interaction with cell signaling molecules, such as cGMP, cAMP, G-proteins, STAT proteins, or various transcription factors, and in this way modulate the expression of multiple other mediators of inflammation. On the other hand, induction of specific protection functions, such as antioxidants and detoxifying enzymes (e.g., Mt1a, Mt2, and superoxide dismutase [Sod2]) observed in our study, can counteract oxidative stress responses, including protection from large iNOS-generated amounts of NO. NO has also been implicated in regulating the release of hormones that control the inflammatory process. For example, NO plays a marked inhibitory role in CRF-induced ACTH release and inhibits corticosterone secretion (23).
Different causes of death by virulent PRV151 or attenuated PRV152 infection.
Although innate immunity and its subsequent impact on adaptive responses have the potential to control virus infections, the immune response can also contribute to virally induced immunopathology that can manifest as inflammatory neurological disease. For example, cytokines and chemokines can contribute to both defense against and neuropathogenesis of CNS infections. In addition, free radicals such as ROS and RNS, produced by activated microglia and other cells, are regarded as important mediators of defense of the nervous system against intracellular microorganisms and, when these toxic molecules are released into the extracellular milieu, of potential damage to neurons (see above).
The eye infection model used in this study results in neuroinvasion with limited peripheral infection. Nevertheless, this mode of infection still results in the characteristic symptoms of virulent PRV infection and rapid death characteristic of a global peripheral response, as opposed to direct effects of CNS infection as described and reviewed by Brittle et al. (8). The rather limited induction of host gene transcription in PRV151-infected CNS tissue at the time of death provides no real insight into the mechanism of death. Instead, the response observed is more characteristic of an early innate immune response and protective reaction by CNS tissue. The stimulation of a toxic global stress response due to a peripheral local overreaction as proposed by Brittle et al. (8) seems more likely to be the cause of the animals' early demise. By contrast, the prolonged survival of PRV152-infected animals leads to increased CNS spread and accumulation of host gene transcripts that reflect both local and global CNS responses to the circuit-specific spread of these less virulent PRV strains. These animals show little or no signs of the toxic peripheral response so typical of virulent PRV infections despite an extensive infection of the CNS. Death of these animals is likely to be due to a combination of accumulated compromised central circuitry and overreactive local and global host defense reactions (immunopathogenesis).
Other considerations.
While the majority of gene transcripts detected at late times after PRV infection reflected innate defense and stress responses, several other genes, encoding various serine/threonine proteases, a variety of Ca2+-binding/regulating proteins, and transcription factors, were stimulated (Table 6). The role of these gene products in the CNS response to infection remains to be understood. A number of similarities exist in the host response to PRV infection in our study of infected CNS and those of primary cultures of embryonic rat fibroblasts (51). However, given the complexity of the infected tissue, it is virtually impossible to determine what responses reflect directly infected cells and indirect responses of uninfected tissue. The list of rat CNS genes that respond to PRV infection is remarkably similar to those genes responsive to rodent CNS infection by diverse agents such as mouse hepatitis virus (52), prions (75), and West Nile virus (68). It is likely that the rodent CNS response to infection is not tailored to a specific infectious agent but rather represents a conserved defensive reaction to neuronal damage characterized by a vigorous intrinsic and innate immune response, followed by the infiltration of adaptive immune cells from the periphery.
Acknowledgments
We thank Bruce W. Banfield (University of Colorado) for providing virus strains PRV151, PRV152, and PRV99G. C.P. thanks Michael Nevels (University of Regensburg) for help with the manuscript and Hans Wolf (University of Regensburg) for generous support.
This work was funded by an Emmy-Noether fellowship from the Deutsche Forschungsgemeinschaft (PA 815/1-1) to C.P. and by grants from the National Institutes of Health to L.W.E. (5P01 CA87661) and to G.E.P. (R21 NS46719).
REFERENCES
- 1.Babic, N., B. Klupp, A. Brack, T. C. Mettenleiter, G. Ugolini, and A. Flamand. 1996. Deletion of glycoprotein gE reduces the propagation of pseudorabies virus in the nervous system of mice after intranasal inoculation. Virology 219:279-284. [DOI] [PubMed] [Google Scholar]
- 2.Banfield, B. W., G. S. Yap, A. C. Knapp, and L. W. Enquist. 1998. A chicken embryo eye model for the analysis of alphaherpesvirus neuronal spread and virulence. J. Virol. 72:4580-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartha, A. 1961. Experimental reduction of virulence of Aujeszky's disease virus. Magy. Állatorv. Lapja 16:42-45. [Google Scholar]
- 4.Ben-Hur, T., N. Conforti, A. Itzik, and J. Weidenfeld. 1995. Effects of HSV-1, a neurotropic virus, on the hypothalamic-pituitary-adrenocortical axis in rats. Brain Res. 702:17-22. [DOI] [PubMed] [Google Scholar]
- 5.Ben-Hur, T., J. Rosenthal, A. Itzik, and J. Weidenfeld. 1996. Adrenocortical activation by herpes virus: involvement of IL-1 beta and central noradrenergic system. Neuroreport 7:927-931. [PubMed] [Google Scholar]
- 6.Besedovsky, H. O., and A. del Rey. 1989. Mechanism of virus-induced stimulation of the hypothalamus-pituitary-adrenal axis. J. Steroid Biochem. 34:235-239. [DOI] [PubMed] [Google Scholar]
- 7.Bonjardim, C. A. 2005. Interferons (IFNs) are key cytokines in both innate and adaptive antiviral immune responses—and viruses counteract IFN action. Microbes Infect. 7:569-578. [DOI] [PubMed] [Google Scholar]
- 8.Brittle, E. E., A. E. Reynolds, and L. W. Enquist. 2004. Two modes of pseudorabies virus neuroinvasion and lethality in mice. J. Virol. 78:12951-12963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Browne, E. P., B. Wing, D. Coleman, and T. Shenk. 2001. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J. Virol. 75:12319-12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Card, J. P., L. Rinaman, R. B. Lynn, B. H. Lee, R. P. Meade, R. R. Miselis, and L. W. Enquist. 1993. Pseudorabies virus infection of the rat central nervous system: ultrastructural characterization of viral replication, transport, and pathogenesis. J. Neurosci. 13:2515-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Card, J. P., L. Rinaman, J. S. Schwaber, R. R. Miselis, M. E. Whealy, A. K. Robbins, and L. W. Enquist. 1990. Neurotropic properties of pseudorabies virus: uptake and transneuronal passage in the rat central nervous system. J. Neurosci. 10:1974-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Card, J. P., M. E. Whealy, A. K. Robbins, and L. W. Enquist. 1992. Pseudorabies virus envelope glycoprotein gI influences both neurotropism and virulence during infection of the rat visual system. J. Virol. 66:3032-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung, R. S., P. A. Adlard, J. Dittmann, J. C. Vickers, M. I. Chuah, and A. K. West. 2004. Neuron-glia communication: metallothionein expression is specifically up-regulated by astrocytes in response to neuronal injury. J. Neurochem. 88:454-461. [DOI] [PubMed] [Google Scholar]
- 13a.Clase, A. C., and B. W. Banfield. 2003. Corticosteroids are unable to protect against pseudorabies virus-induced tissue damage in the developing brain. J. Virol. 77:4979-4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demmin, G. L., A. C. Clase, J. A. Randall, L. W. Enquist, and B. W. Banfield. 2001. Insertions in the gG gene of pseudorabies virus reduce expression of the upstream Us3 protein and inhibit cell-to-cell spread of virus infection. J. Virol. 75:10856-10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deroo, B. J., and T. K. Archer. 2001. Glucocorticoid receptor activation of the I kappa B alpha promoter within chromatin. Mol. Biol. Cell 12:3365-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dijkstra, J. M., T. C. Mettenleiter, and B. G. Klupp. 1997. Intracellular processing of pseudorabies virus glycoprotein M (gM): gM of strain Bartha lacks N-glycosylation. Virology 237:113-122. [DOI] [PubMed] [Google Scholar]
- 17.Dittmann, J., S. J. Fung, J. C. Vickers, M. I. Chuah, R. S. Chung, and A. K. West. 2005. Metallothionein biology in the ageing and neurodegenerative brain. Neurotox. Res. 7:87-93. [DOI] [PubMed] [Google Scholar]
- 18.Doble, A. 1995. Excitatory amino acid receptors and neurodegeneration. Therapie 50:319-337. [PubMed] [Google Scholar]
- 19.Enquist, L. W. 1999. Life beyond eradication: veterinary viruses in basic science. Arch. Virol. 15(Suppl.):87-109. [DOI] [PubMed] [Google Scholar]
- 20.Enquist, L. W., and J. P. Card. 2003. Recent advances in the use of neurotropic viruses for circuit analysis. Curr. Opin. Neurobiol. 13:603-606. [DOI] [PubMed] [Google Scholar]
- 21.Enquist, L. W., M. J. Tomishima, S. Gross, and G. A. Smith. 2002. Directional spread of an alpha-herpesvirus in the nervous system. Vet. Microbiol. 86:5-16. [DOI] [PubMed] [Google Scholar]
- 22.Fang, H. L., S. Shenoy, Z. Duanmu, T. A. Kocarek, and M. Runge-Morris. 2003. Transactivation of glucocorticoid-inducible rat aryl sulfotransferase (SULT1A1) gene transcription. Drug Metab. Dispos. 31:1378-1381. [DOI] [PubMed] [Google Scholar]
- 23.Givalois, L., S. Li, and G. Pelletier. 2002. Central nitric oxide regulation of the hypothalamic-pituitary-adrenocortical axis in adult male rats. Brain Res. Mol. Brain Res. 102:1-8. [DOI] [PubMed] [Google Scholar]
- 24.Guzik, T. J., R. Korbut, and T. Adamek-Guzik. 2003. Nitric oxide and superoxide in inflammation and immune regulation. J. Physiol. Pharmacol. 54:469-487. [PubMed] [Google Scholar]
- 25.Honda, K., H. Yanai, H. Negishi, M. Asagiri, M. Sato, T. Mizutani, N. Shimada, Y. Ohba, A. Takaoka, N. Yoshida, and T. Taniguchi. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434:772-777. [DOI] [PubMed] [Google Scholar]
- 26.Iwaki, T., A. Iwaki, J. Tateishi, Y. Sakaki, and J. E. Goldman. 1993. Alpha B-crystallin and 27-kd heat shock protein are regulated by stress conditions in the central nervous system and accumulate in Rosenthal fibers. Am. J. Pathol. 143:487-495. [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobs, L. 1994. Glycoprotein E of pseudorabies virus and homologous proteins in other alphaherpesvirinae. Arch. Virol. 137:209-228. [DOI] [PubMed] [Google Scholar]
- 28.Jacobs, L., W. A. Mulder, J. T. Van Oirschot, A. L. Gielkens, and T. G. Kimman. 1993. Deleting two amino acids in glycoprotein gI of pseudorabies virus decreases virulence and neurotropism for pigs, but does not affect immunogenicity. J. Gen. Virol. 74:2201-2206. [DOI] [PubMed] [Google Scholar]
- 29.Jacobs, L., H. J. Rziha, T. G. Kimman, A. L. Gielkens, and J. T. Van Oirschot. 1993. Deleting valine-125 and cysteine-126 in glycoprotein gI of pseudorabies virus strain NIA-3 decreases plaque size and reduces virulence in mice. Arch. Virol. 131:251-264. [DOI] [PubMed] [Google Scholar]
- 30.Kimman, T. G., N. de Wind, N. Oei-Lie, J. M. Pol, A. J. Berns, and A. L. Gielkens. 1992. Contribution of single genes within the unique short region of Aujeszky's disease virus (suid herpesvirus type 1) to virulence, pathogenesis and immunogenicity. J. Gen. Virol. 73:243-251. [DOI] [PubMed] [Google Scholar]
- 31.Kimura, T., and D. E. Griffin. 2000. The role of CD8+ T cells and major histocompatibility complex class I expression in the central nervous system of mice infected with neurovirulent Sindbis virus. J. Virol. 74:6117-6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klupp, B. G., H. Kern, and T. C. Mettenleiter. 1992. The virulence-determining genomic BamHI fragment 4 of pseudorabies virus contains genes corresponding to the UL15 (partial), UL18, UL19, UL20, and UL21 genes of herpes simplex virus and a putative origin of replication. Virology 191:900-908. [DOI] [PubMed] [Google Scholar]
- 33.Leong, M. L., A. C. Maiyar, B. Kim, B. A. O'Keeffe, and G. L. Firestone. 2003. Expression of the serum- and glucocorticoid-inducible protein kinase, Sgk, is a cell survival response to multiple types of environmental stress stimuli in mammary epithelial cells. J. Biol. Chem. 278:5871-5882. [DOI] [PubMed] [Google Scholar]
- 34.Levine, J. M., L. W. Enquist, and J. P. Card. 1998. Reactions of oligodendrocyte precursor cells to alpha herpesvirus infection of the central nervous system. Glia 23:316-328. [PubMed] [Google Scholar]
- 35.Lidman, O., T. Olsson, and F. Piehl. 1999. Expression of nonclassical MHC class I (RT1-U) in certain neuronal populations of the central nervous system. Eur. J. Neurosci. 11:4468-4472. [DOI] [PubMed] [Google Scholar]
- 36.Lomniczi, B., M. L. Blankenship, and T. Ben-Porat. 1984. Deletions in the genomes of pseudorabies virus vaccine strains and existence of four isomers of the genomes. J. Virol. 49:970-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lomniczi, B., A. S. Kaplan, and T. Ben-Porat. 1987. Multiple defects in the genome of pseudorabies virus can affect virulence without detectably affecting replication in cell culture. Virology 161:181-189. [DOI] [PubMed] [Google Scholar]
- 38.Masters, J. N., C. E. Finch, and N. R. Nichols. 1994. Rapid increase in glycerol phosphate dehydrogenase mRNA in adult rat brain: a glucocorticoid-dependent stress response. Neuroendocrinology 60:23-35. [DOI] [PubMed] [Google Scholar]
- 39.McEwen, B. S., C. A. Biron, K. W. Brunson, K. Bulloch, W. H. Chambers, F. S. Dhabhar, R. H. Goldfarb, R. P. Kitson, A. H. Miller, R. L. Spencer, and J. M. Weiss. 1997. The role of adrenocorticoids as modulators of immune function in health and disease: neural, endocrine and immune interactions. Brain Res. Brain Res. Rev. 23:79-133. [DOI] [PubMed] [Google Scholar]
- 40.Mettenleiter, T. C. 2000. Aujeszky's disease (pseudorabies) virus: the virus and molecular pathogenesis—state of the art, June 1999. Vet. Res. 31:99-115. [DOI] [PubMed] [Google Scholar]
- 41.Mettenleiter, T. C., N. Lukacs, and H. J. Rziha. 1985. Pseudorabies virus avirulent strains fail to express a major glycoprotein. J. Virol. 56:307-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mettenleiter, T. C., L. Zsak, A. S. Kaplan, T. Ben-Porat, and B. Lomniczi. 1987. Role of a structural glycoprotein of pseudorabies in virus virulence. J. Virol. 61:4030-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mirescu, C., and E. Gould. 2004. From neurotoxin to neurotrophin. Nat. Neurosci. 7:899-900. [DOI] [PubMed] [Google Scholar]
- 44.Muff, R., W. Born, T. A. Lutz, and J. A. Fischer. 2004. Biological importance of the peptides of the calcitonin family as revealed by disruption and transfer of corresponding genes. Peptides 25:2027-2038. [DOI] [PubMed] [Google Scholar]
- 45.Papassotiropoulos, A., C. Hock, and R. M. Nitsch. 2001. Genetics of interleukin 6: implications for Alzheimer's disease. Neurobiol. Aging 22:863-871. [DOI] [PubMed] [Google Scholar]
- 46.Petrovskis, E. A., J. G. Timmins, T. M. Gierman, and L. E. Post. 1986. Deletions in vaccine strains of pseudorabies virus and their effect on synthesis of glycoprotein gp63. J. Virol. 60:1166-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pickard, G. E., C. A. Smeraski, C. C. Tomlinson, B. W. Banfield, J. Kaufman, C. L. Wilcox, L. W. Enquist, and P. J. Sollars. 2002. Intravitreal injection of the attenuated pseudorabies virus PRV Bartha results in infection of the hamster suprachiasmatic nucleus only by retrograde transsynaptic transport via autonomic circuits. J. Neurosci. 22:2701-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pomeranz, L. E., A. E. Reynolds, and C. J. Hengartner. 2005. Molecular biology of pseudorabies virus: impact on neurovirology and veterinary medicine. Microbiol. Mol. Biol. Rev. 69:462-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rassnick, S., L. W. Enquist, A. F. Sved, and J. P. Card. 1998. Pseudorabies virus-induced leukocyte trafficking into the rat central nervous system. J. Virol. 72:9181-9191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ray, N., M. E. Bisher, and L. W. Enquist. 2004. Cyclooxygenase-1 and -2 are required for production of infectious pseudorabies virus. J. Virol. 78:12964-12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ray, N., and L. W. Enquist. 2004. Transcriptional response of a common permissive cell type to infection by two diverse alphaherpesviruses. J. Virol. 78:3489-3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rempel, J. D., L. A. Quina, P. K. Blakely-Gonzales, M. J. Buchmeier, and D. L. Gruol. 2005. Viral induction of central nervous system innate immune responses. J. Virol. 79:4369-4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rinaman, L., J. P. Card, and L. W. Enquist. 1993. Spatiotemporal responses of astrocytes, ramified microglia, and brain macrophages to central neuronal infection with pseudorabies virus. J. Neurosci. 13:685-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robbins, A. K., J. P. Ryan, M. E. Whealy, and L. W. Enquist. 1989. The gene encoding the gIII envelope protein of pseudorabies virus vaccine strain Bartha contains a mutation affecting protein localization. J. Virol. 63:250-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 56.Roizman, B., and P. E. Pellett. 2001. The family herpesviridae: a brief introduction, p. 2381-2397. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 57.Scheier, B., A. Foletti, G. Stark, A. Aoyama, U. Dobbeling, S. Rusconi, and R. Klemenz. 1996. Glucocorticoids regulate the expression of the stress protein alpha B-crystallin. Mol. Cell. Endocrinol. 123:187-198. [DOI] [PubMed] [Google Scholar]
- 58.Senba, E., and T. Ueyama. 1997. Stress-induced expression of immediate early genes in the brain and peripheral organs of the rat. Neurosci. Res. 29:183-207. [DOI] [PubMed] [Google Scholar]
- 59.Seo, M. S., M. S. Lee, and I. K. Lim. 1999. Expression of rat BTG(3) gene, Rbtg3, is regulated by redox changes. Gene 240:165-173. [DOI] [PubMed] [Google Scholar]
- 60.Serrano, F., L. W. Enquist, and J. P. Card. 2002. Pseudorabies virus-induced expression of nitric oxide synthase isoforms. Physiol. Behav. 77:557-563. [DOI] [PubMed] [Google Scholar]
- 61.Shrikant, P., and E. N. Benveniste. 1996. The central nervous system as an immunocompetent organ: role of glial cells in antigen presentation. J. Immunol. 157:1819-1822. [PubMed] [Google Scholar]
- 62.Slemmon, J. R., B. Feng, and J. A. Erhardt. 2000. Small proteins that modulate calmodulin-dependent signal transduction: effects of PEP-19, neuromodulin, and neurogranin on enzyme activation and cellular homeostasis. Mol. Neurobiol. 22:99-113. [DOI] [PubMed] [Google Scholar]
- 63.Smeraski, C. A., P. J. Sollars, M. D. Ogilvie, L. W. Enquist, and G. E. Pickard. 2004. Suprachiasmatic nucleus input to autonomic circuits identified by retrograde transsynaptic transport of pseudorabies virus from the eye. J. Comp. Neurol. 471:298-313. [DOI] [PubMed] [Google Scholar]
- 64.Smith, B. N., B. W. Banfield, C. A. Smeraski, C. L. Wilcox, F. E. Dudek, L. W. Enquist, and G. E. Pickard. 2000. Pseudorabies virus expressing enhanced green fluorescent protein: a tool for in vitro electrophysiological analysis of transsynaptically labeled neurons in identified central nervous system circuits. Proc. Natl. Acad. Sci. USA 97:9264-9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith, G. A., and L. W. Enquist. 2000. A self-recombining bacterial artificial chromosome and its application for analysis of herpesvirus pathogenesis. Proc. Natl. Acad. Sci. USA 97:4873-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sng, J. C., H. Taniura, and Y. Yoneda. 2004. A tale of early response genes. Biol. Pharm. Bull. 27:606-612. [DOI] [PubMed] [Google Scholar]
- 67.Turnbull, A. V., and C. L. Rivier. 1999. Regulation of the hypothalamic-pituitary-adrenal axis by cytokines: actions and mechanisms of action. Physiol. Rev. 79:1-71. [DOI] [PubMed] [Google Scholar]
- 68.Venter, M., T. G. Myers, M. A. Wilson, T. J. Kindt, J. T. Paweska, F. J. Burt, P. A. Leman, and R. Swanepoel. 2005. Gene expression in mice infected with West Nile virus strains of different neurovirulence. Virology 342:119-140. [DOI] [PubMed] [Google Scholar]
- 69.Wang, Y., C. F. Chang, M. Morales, Y. H. Chiang, and J. Hoffer. 2002. Protective effects of glial cell line-derived neurotrophic factor in ischemic brain injury. Ann. N. Y. Acad. Sci. 962:423-437. [DOI] [PubMed] [Google Scholar]
- 70.Whealy, M. E., J. P. Card, A. K. Robbins, J. R. Dubin, H. J. Rziha, and L. W. Enquist. 1993. Specific pseudorabies virus infection of the rat visual system requires both gI and gp63 glycoproteins. J. Virol. 67:3786-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Whitley, R. J. 2001. Herpes simplex viruses, p. 2461-2509. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 72.Whitley, R. J., and J. W. Gnann. 2002. Viral encephalitis: familiar infections and emerging pathogens. Lancet 359:507-513. [DOI] [PubMed] [Google Scholar]
- 73.Wu, G., Z. H. Lu, X. Xie, L. Li, and R. W. Ledeen. 2001. Mutant NG108-15 cells (NG-CR72) deficient in GM1 synthase respond aberrantly to axonogenic stimuli and are vulnerable to calcium-induced apoptosis: they are rescued with LIGA-20. J. Neurochem. 76:690-702. [DOI] [PubMed] [Google Scholar]
- 74.Wu, W., T. Pew, M. Zou, D. Pang, and S. D. Conzen. 2005. Glucocorticoid receptor-induced MAPK phosphatase-1 (MPK-1) expression inhibits paclitaxel-associated MAPK activation and contributes to breast cancer cell survival. J. Biol. Chem. 280:4117-4124. [DOI] [PubMed] [Google Scholar]
- 75.Xiang, W., O. Windl, G. Wunsch, M. Dugas, A. Kohlmann, N. Dierkes, I. M. Westner, and H. A. Kretzschmar. 2004. Identification of differentially expressed genes in scrapie-infected mouse brains by using global gene expression technology. J. Virol. 78:11051-11060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu, D., A. Makkinje, and J. M. Kyriakis. 2005. Gene 33 is an endogenous inhibitor of epidermal growth factor (EGF) receptor signaling and mediates dexamethasone-induced suppression of EGF function. J. Biol. Chem. 280:2924-2933. [DOI] [PubMed] [Google Scholar]
- 77.Yang, M., J. P. Card, R. S. Tirabassi, R. R. Miselis, and L. W. Enquist. 1999. Retrograde, transneuronal spread of pseudorabies virus in defined neuronal circuitry of the rat brain is facilitated by gE mutations that reduce virulence. J. Virol. 73:4350-4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhu, H., J. P. Cong, G. Mamtora, T. Gingeras, and T. Shenk. 1998. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:14470-14475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu, H., J. P. Cong, and T. Shenk. 1997. Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs. Proc. Natl. Acad. Sci. USA 94:13985-13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhu, H., J. P. Cong, D. Yu, W. A. Bresnahan, and T. E. Shenk. 2002. Inhibition of cyclooxygenase 2 blocks human cytomegalovirus replication. Proc. Natl. Acad. Sci. USA 99:3932-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]