Abstract
Following infection with most reovirus strains, viral protein synthesis is robust, even when cellular translation is inhibited. To gain further insight into pathways that regulate translation in reovirus-infected cells, we performed a comparative microarray analysis of cellular gene expression following infection with two strains of reovirus that inhibit host translation (clone 8 and clone 87) and one strain that does not (Dearing). Infection with clone 8 and clone 87 significantly increased the expression of cellular genes characteristic of stress responses, including the integrated stress response. Infection with these same strains decreased transcript and protein levels of P58IPK, the cellular inhibitor of the eukaryotic initiation factor 2α (eIF2α) kinases PKR and PERK. Since infection with host shutoff-inducing strains of reovirus impacted cellular pathways that control eIF2α phosphorylation and unphosphorylated eIF2α is required for translation initiation, we examined reovirus replication in a variety of cell lines with mutations that impact eIF2α phosphorylation. Our results revealed that reovirus replication is more efficient in the presence of eIF2α kinases and phosphorylatable eIF2α. When eIF2α is phosphorylated, it promotes the synthesis of ATF4, a transcription factor that controls cellular recovery from stress. We found that the presence of this transcription factor increased reovirus yields 10- to 100-fold. eIF2α phosphorylation also led to the formation of stress granules in reovirus-infected cells. Based on these results, we hypothesize that eIF2α phosphorylation facilitates reovirus replication in two ways—first, by inducing ATF4 synthesis, and second, by creating an environment that places abundant reovirus transcripts at a competitive advantage for limited translational components.
As representative members of the Reoviridae family, mammalian orthoreoviruses (reoviruses) have genomes composed of 10 segments of double-stranded RNA (dsRNA) that are surrounded by two concentric protein capsids (reviewed in reference 54). In natural infections, reoviruses replicate in cells of the respiratory or enteric tract; however, in experimental infections of neonatal mice, their tropism is much broader. Pathogenesis studies with the mouse model have demonstrated that reoviruses replicate in cells of the brain, heart, liver, muscle, and pancreas (reviewed in reference 73). At the cellular level, the consequences of reovirus infection have been extensively analyzed and include the inhibition of DNA synthesis, cell cycle arrest at the G2/M stage, apoptosis induction, translational inhibition, and type I interferon (IFN) induction (54, 73). The extent of each of these effects varies in a strain-specific manner. Phenotypic analysis of reassortant viruses has provided insight into the viral determinants responsible for strain-specific differences. Less is known about the host cell determinants that impact reovirus replication.
Cellular protein synthesis decreases following infection with most strains of reovirus, a phenomenon referred to as host shutoff (81). However, the degree of reovirus-induced host shutoff varies depending on the infecting reovirus isolate (65, 67). The difference in cellular translational inhibition following infection with reovirus strains Dearing and Jones was genetically mapped to the viral S4 gene segment, which encodes the dsRNA-binding and outer capsid protein σ3 (67). Studies with knockout (KO) mouse embryo fibroblasts (MEFs) demonstrated that the dsRNA-activated, IFN-stimulated gene (ISG) product protein kinase R (PKR) contributes to reovirus-induced host shutoff (70). During infection with strain Jones, host shutoff is dependent on the ability of PKR to phosphorylate and inactivate the translation initiation factor eIF2α. The strong host shutoff induced as a consequence of infection with other strains of reovirus, specifically clone 87 (c87), clone 8 (c8), and clone 93 (c93), involves not only PKR but also additional cellular gene products, including the IFN-stimulated endoribonuclease RNase L (70). PKR and RNase L are believed to function globally within infected cells to inhibit both viral and cellular translation. Interestingly, viral protein synthesis is robust following infection with most reovirus strains, even when cellular translation is inhibited (65, 70). The molecular mechanisms that enable efficient viral translation under these conditions are completely unknown.
To gain further insight into the cellular pathways that regulate translation in reovirus-infected cells, we performed a three-way comparative transcriptional profiling study with murine L929 cells. We compared cellular RNA profiles in cells infected with the host shutoff-inducing reovirus isolates c8 and c87 to those in cells infected with Dearing, a strain that does not induce host shutoff. This analysis revealed that c8 and c87 infections activate cellular pathways that regulate translation in response to stress. Specifically, a subset of genes characteristic of the integrated stress response (ISR) (31) and the endoplasmic reticulum (ER)-mediated unfolded protein response (UPR) (reviewed in references 36 and 68) were selectively induced following infection with c8 and c87. In addition, P58IPK, an inhibitor of the eIF2α kinases PKR (44) and PERK (77), was down-regulated at both the mRNA and protein levels in cells infected with the host-shutoff-inducing strains of reovirus. These findings led us to examine the capacity of reovirus to replicate in a variety of cell lines with mutations that affect eIF2α phosphorylation. Although eIF2α phosphorylation is a cellular response that typically restricts viral growth, we found that reovirus replication is more efficient in the presence of eIF2α kinases and when eIF2α can be phosphorylated at serine 51. eIF2α phosphorylation promotes the translation of activating transcription factor 4 (ATF4), a transcription factor that regulates cellular recovery from stress (61). Reovirus replication was compromised in cells deleted for ATF4, suggesting that one or more ATF4 target gene products facilitate reovirus infection. eIF2α phosphorylation also promotes the formation of stress granules (SGs), cytoplasmic aggregates of stalled translational preinitiation complexes (reviewed in references 2 and 3). Based on these results, we hypothesize that eIF2α phosphorylation facilitates reovirus replication in two ways—first, by inducing an ATF4-dependent gene expression program, and second, by creating an environment that places abundant reovirus transcripts at a competitive advantage for limited components of the translational machinery.
MATERIALS AND METHODS
Cells and viruses.
Murine L929 cells were maintained as suspension cultures as described previously (39). ATF4 KO and wild-type (wt) MEFs (33) were prepared by mating heterozygous parents and harvesting embryos at day 14.5. The animals were genotyped using PCR and the following primers: 5′-TTTGAGTTGTGCGCTCGGGTGTC-3′, 5′-GCTACCGGTGGATGTGGAATGTG-3′, 5′-GCAGCAGTGTTGCTGTAACGGAC-3′, and 5′-CTGGCCAAGCCATCATCCATAGC-3′. Using this primer mixture, the ATF4 mutant allele gives rise to an ∼400-bp product, and the wt allele gives rise to a 277-bp product (R. Kaufman, personal communication). Primary wt, PERK KO (31), and ATF4 KO MEFs, as well as immortalized PERK wt and KO MEFs, were maintained as monolayer cultures in high-glucose Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal calf serum (FCS) (HyClone Laboratories, Logan, UT), 2 mM glutamine, 0.1 mM nonessential amino acids, 55 μM 2-mercaptoethanol, 50 units/ml penicillin G, and 50 μg/ml streptomycin sulfate. Immortalized MEFs with wt or mutant (S51A) eIF2α (64) were maintained as monolayer cultures in high-glucose Dulbecco's modified Eagle's medium supplemented with 10% FCS, 2 mM glutamine, 1× essential amino acids, 0.1 mM nonessential amino acids, 50 units/ml penicillin G, and 50 μg/ml streptomycin sulfate. DU145 cells were maintained as monolayer cultures in minimum essential medium (MEM) supplemented with 10% heat-inactivated FCS, 2 mM glutamine, 0.1 mM nonessential amino acids, 1 mM sodium pyruvate, 50 units/ml penicillin G, and 50 μg/ml streptomycin sulfate. Experiments were performed with the maintenance medium for each cell type, except that 2-mercaptoethanol was omitted from the media for PERK and ATF4 MEFs. Unless otherwise noted, all tissue culture reagents were purchased from Invitrogen (Carlsbad, CA).
Reovirus strains Dearing, Jones, and c87/Abney are prototypic laboratory strains. Reovirus strain c8 was originally isolated by Rosen and colleagues and has been described previously (39, 60). Purified virions were prepared by CsCl density gradient centrifugation of extracts from cells infected with third-passage L929 cell lysate stocks (25). Intermediate subvirion particles (ISVPs) were prepared by treating purified virions with chymotrypsin (53). Because some MEFs restrict reovirus uncoating (27), MEF infections were performed with ISVPs.
Analysis of viral growth.
Cells were infected with ISVPs at a multiplicity of infection (MOI) of 2 PFU/cell, and adsorption was allowed to proceed for 1 h on ice at 4°C. After adsorption, cells were concentrated by low-speed centrifugation and resuspended in fresh medium. Virions and cells were then added to dram vials containing cold medium at cell densities to result in near-confluent monolayers. Triplicate samples were prepared for each time point. One set of samples was immediately frozen at −20°C. The remaining samples were incubated at 37°C until the desired time point was reached. Harvested samples were subjected to three cycles of freezing and thawing and then titrated by a plaque assay on L929 cells (76). In all cases, we found that infection of a pair of cells (wt and mutant) with a given strain of reovirus resulted in identical titers at time zero, indicating that adsorption to the different cell types was equivalent. Viral yields were calculated according to the following formula: log10yieldt = x = log10(PFU/ml)t = x − log10(PFU/ml)t=0, where t is time and x is the time postinfection.
Analysis of total protein synthesis.
Cells were plated in triplicate to result in near-confluent monolayers in 15-mm wells. After 4 h of incubation at 37°C, the medium was removed and cells were infected with purified virions or ISVPs at a multiplicity of 80 PFU/cell. This MOI maximizes strain differences in host shutoff (67, 70). Samples were incubated for 2 h at 37°C to allow particles to adsorb; medium was then added, and samples were incubated at 37°C. At 19 h postinfection (p.i.), cells were preincubated in methionine-free and l-glutamine-free MEM (ICN Biomedicals Inc., Aurora, OH) for 30 min at 37°C. The medium was removed, and cells were incubated with methionine- and l-glutamine-free MEM supplemented with 2 mM glutamine and 50 μCi/ml [35S]methionine-cysteine (EasyTag; NEN Life Science Products Inc., Boston, MA). After 30 min or 2 h at 37°C, lysates were prepared by resuspending cells in lysis buffer (0.1 M NaCl, 1 mM EDTA, 10 mM Tris, pH 7.4, 0.5% NP-40), and samples were adjusted with concentrated protein sample buffer to achieve final concentrations of 0.3 M sucrose, 0.125 M Tris (pH 8.0), 1% sodium dodecyl sulfate (SDS), 0.01% bromophenol blue, and 50 μl/ml 2-mercaptoethanol. Labeled proteins from equivalent numbers of cells were resolved by electrophoresis on SDS-polyacrylamide gels and visualized using a phosphorimager and ImageQuant software (Molecular Dynamics, Sunnyvale, CA).
Quantitative analysis of IFN-β mRNA induction and XBP1 mRNA splicing.
L929 cells were plated to result in near-confluent monolayers in 35-mm wells. After 3 to 4 h of incubation at 37°C, the medium was removed, and cells were infected with purified virions at an MOI of 80 PFU/cell. After 1 h of adsorption at 37°C, medium was added and samples were incubated at 37°C. In addition, at 13.5 h p.i., some samples were treated with 2 μg/ml tunicamycin (Sigma-Aldrich, St. Louis, MO) to inhibit protein glycosylation and activate the UPR. At the indicated times p.i., total RNA was extracted from cells by using Trizol reagent (Invitrogen) and the protocol supplied by Invitrogen. Contaminating DNAs were removed by treating RNAs with RNase-free DNase (Roche Diagnostics Corporation, Indianapolis, IN). RNAs were extracted with phenol-chloroform-isoamyl alcohol and precipitated with ethanol. For each IFN-β mRNA sample, 40-μl PCR mixtures were comprised of 1× AmpliTaq Gold buffer (Applied Biosystems, Foster City, CA), 4 mM MgCl2, 0.025 U/μl AmpliTaq Gold (Applied Biosystems), 0.25 U/μl Moloney murine leukemia virus reverse transcriptase (Invitrogen), 0.4 U/μl RNase inhibitor (Invitrogen), 0.5 μg/μl bovine serum albumin (Ambion, Austin, TX), 0.33× SYBR green I (Molecular Probes, Eugene, OR), 0.8 μM passive reference DNA oligohexamer, 5′-(6-carboxyrhodamine)-GATTAG-PO4-3′ (Rox standard I; Synthegen, Houston, TX), a 200 μM concentration of each deoxynucleoside triphosphate (Amersham Biosciences, Piscataway, NJ), a 50 nM concentration of each gene-specific primer (Integrated DNA Technologies, Inc., Coralville, IA), and 5 ng total RNA. Gene-specific primers were designed with the computer program Primer Express 1.5 (Applied Biosystems), using the default settings. The primers were 5′-CTGGAGCAGCTGAATGGAAAG-3′ and 5′-TGGATGGCAAAGGCAGTGT-3′. Using an ABI 7700 sequence detector (Applied Biosystems), the reactions were subjected to the following conditions: 30 min at 48°C, 10 min at 95°C, and 40 cycles comprised of 15 s at 95°C and 1 min at 60°C. Spliced XBP1 mRNA was quantified by preparing cDNA and performing real-time PCR as previously described (4).
Determination of cell viability and apoptosis induction.
L929 cells were plated in triplicate and infected as described above for quantitative analysis of XBP1 mRNA splicing. At 19.5 h p.i., all cells in the culture (adherent and nonadherent) were harvested and concentrated by low-speed centrifugation. Cells were stained with propidium iodide and annexin V-biotin according to the manufacturer's protocol (R&D Systems, Inc., Minneapolis, MN). Cells were subsequently labeled with fluorescein-conjugated streptavidin (eBioscience, San Diego, CA), sorted using a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA), and analyzed with FLOWJO software (TreeStar, San Carlos, CA).
Infection, target preparation, and oligonucleotide microarray analysis.
Independent sets of infections were performed on three different days. For each set of samples, L929 cells were plated to result in near-confluent monolayers in 150-mm plates. After 4 h of incubation at 37°C, the medium was removed and cells were infected with purified virions (Dearing, c8, or c87) at an MOI of 80 PFU/cell. The reproducibility of gene expression studies is reported to improve with high-multiplicity infections (21), and this MOI maximizes the distinct reovirus translational phenotypes (67, 70). After a 1.5-h adsorption period at 37°C, medium was added, and the samples were incubated at 37°C. At 19.5 h p.i., total RNA was extracted from cells by using Trizol reagent (Invitrogen) according to the manufacturer's instructions. Extracted RNAs were further purified using an RNeasy column (QIAGEN, Valencia, CA). Total cellular RNA was converted to cDNAs using a Superscript custom kit (Invitrogen) with an oligo(dT)7 primer (Geneset, Boulder, CO). Biotin-labeled cRNAs were synthesized from cDNAs using an in vitro transcription kit (Enzo Bioarray Diagnostics, Farmingdale, NY), and purified with an RNeasy mini kit (QIAGEN). Fifteen micrograms of cRNA was hybridized to murine U74Av2 oligonucleotide microarrays (Affymetrix Inc., Santa Clara, CA) according to the manufacturer's protocol. Quantitative scanning of arrays was performed on a Hewlett Packard Agilent 2200 confocal scanner (Bio-Rad Laboratories, Hercules, CA). Affymetrix MAS 5.0 software was used to obtain signal values for each transcript. Signal values were then normalized so that the median expression level across the array was the same for all arrays. Normalized signals were logarithmically transformed (base 2), and the mean and standard deviation (SD) across all replicates were determined for each infection condition. The degree of change in gene expression between two different infection conditions was determined by using the ratio of the two means. In order to determine statistical significances of the differences in the means, P values were calculated using a two-sample t test assuming equal variances (59).
Analysis of P58IPK, eIF2α, and ATF4 protein expression in reovirus-infected cells.
L929 cells were plated and infected as described above for quantitative analysis of XBP1 mRNA splicing. At 3.5 h p.i., some samples were supplemented with 2 μg/ml tunicamycin. This treatment inhibits protein glycosylation, activates the UPR, and increases P58IPK expression (77). For analyses of P58IPK and ATF4 expression, cells were harvested at 19.5 h p.i. in phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, 1.47 mM KH2PO4), collected by centrifugation, and disrupted in lysis buffer (10 mM Tris [pH 7.5], 100 mM NaCl, 2.5 mM MgCl2, 0.5% Triton X-100, 1× Complete protease inhibitor cocktail [Roche], 1× protein phosphatase inhibitor cocktails 1 and 2 [Sigma]). After 30 min on ice, samples were clarified by centrifugation at 10,000 × g for 10 min at 4°C. For analyses of total and phosphorylated eIF2α, extracts were harvested as described above, except that cells were disrupted in a buffer containing 50 mM Tris [pH 7.4], 150 mM NaCl, 50 mM NaF, 10 mM β-glycerophosphate, 0.1 mM EDTA, 10% glycerol, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate, 2 μg/ml pepstatin, 2 μg/ml aprotinin, and 2 μg/ml leupeptin. Clarified cell lysates were normalized for protein content using a protein assay kit (DC protein assay; Bio-Rad Laboratories), solubilized in protein sample buffer, resolved by electrophoresis on SDS-polyacrylamide gels, and transferred to nitrocellulose membranes (Bio-Rad) by electroblotting for 1.75 h at 100 V in transfer buffer (25 mM Tris, 192 mM glycine, 20% methanol). Membranes for the detection of P58IPK, total eIF2α, and ATF4 were blocked with TBST (10 mM Tris [pH 8.0], 150 mM NaCl, 0.4% Tween 20) supplemented with 5% (P58IPK) or 10% (total eIF2α and ATF4) nonfat dry milk. P58IPK was detected using a P58IPK-specific monoclonal antibody (6) diluted in TBST and 1% milk. Total eIF2α and ATF4 were detected with commercially available rabbit polyclonal antibodies (sc-11386 and sc-200; Santa Cruz Biotechnology, Inc., Santa Cruz, CA) diluted in TBST. Membranes were washed with TBST and incubated for 1 h with horseradish peroxidase-conjugated anti-mouse (P58IPK) or anti-rabbit (total eIF2α and ATF4) immunoglobulin G (IgG; Pierce Chemical Company, Rockford, IL) diluted in TBST. Phosphorylated eIF2α was detected using a phospho-eIF2α (S51)-specific rabbit polyclonal antibody (Cell Signaling Technology, Beverly, MA) and horseradish peroxidase-conjugated anti-rabbit IgG (Pierce Chemical Company) according to the instructions provided by Cell Signaling Technology. Bound antibodies were detected using chemiluminescence (ECL detection reagents; Amersham, Arlington Heights, IL) and X-ray film (Full Speed Blue; Henry Schein, Melville, NY). Bands corresponding to the proteins of interest were quantitated with NIH Image.
Analysis of P58IPK mRNA expression in reovirus-infected cells.
L929 cells were plated and infected as described for the analysis of XBP1 mRNA splicing. At 3.5 h p.i., the medium in some samples was replaced with medium supplemented with 2 μg/ml tunicamycin. At 19.5 h p.i., total RNA was prepared as described above. Five micrograms of total RNA from each sample was separated by electrophoresis through a denaturing formaldehyde-agarose gel, transferred to a GeneScreen Plus membrane (Perkin-Elmer Life Sciences, Inc., Boston, MA), and UV cross-linked in a Stratalinker 1800 cross-linker using the auto-cross-link setting (Stratagene, La Jolla, CA). The filters were prehybridized at 60°C with 10 ml ULTRAhyb solution (Ambion) and hybridized overnight at 42°C with a 32P-labeled DNA probe specific for P58IPK or glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The 1,070-bp PstI digestion fragment of mouse P58IPK cDNA (42) and the 1,300-bp EcoRI digestion fragment of mouse GAPDH cDNA were labeled with [α-32P]CTP using a random primer labeling kit (Invitrogen) for use as probes. Hybridized filters were washed sequentially in 2× SSC (0.3 M NaCl, 30 mM sodium citrate dihydrate, pH 7.0) with 0.1% SDS and 0.2× SSC with 0.1% SDS. Labeled RNAs were visualized using a phosphorimager and ImageQuant software.
Analysis of stress granule formation in reovirus-infected cells.
DU145 human prostate carcinoma cells were grown to near confluence on glass coverslips and infected with ISVPs at a multiplicity of 80 PFU/cell. Samples were incubated for 1 h at 37°C to allow particles to adsorb, medium was added, and samples were incubated at 37°C. To induce stress granule formation, some cells were treated with 1 mM sodium arsenite (Sigma) for 30 min at 16.5 h p.i. and then allowed to recover for 2 h (49). At 19 h p.i., cells were fixed in 2% paraformaldehyde, permeabilized in 100% methanol, and washed with PBS. Prior to primary antibody incubation, cells were blocked in PBS supplemented with 5% normal goat serum (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA). Cells were incubated with goat anti-T-cell internal antigen 1 (TIA-1)-related protein (TIAR) antibody (sc-1750; Santa Cruz Biotechnology, Inc.) and washed with PBS, and bound antibody was detected with a Cy3-conjugated donkey anti-goat secondary antibody (Jackson). Nuclei were visualized with Hoechst 33258 stain (Sigma). Samples were viewed on an Olympus BX60 fluorescence microscope linked to a video camera. Images were imported into Adobe Photoshop 7.0 (Adobe, San Jose, CA) and adjusted for contrast, and pairs of images were superimposed so that nuclei and TIAR could be visualized simultaneously.
RESULTS
Cellular responses to reovirus infection vary in a strain-dependent manner.
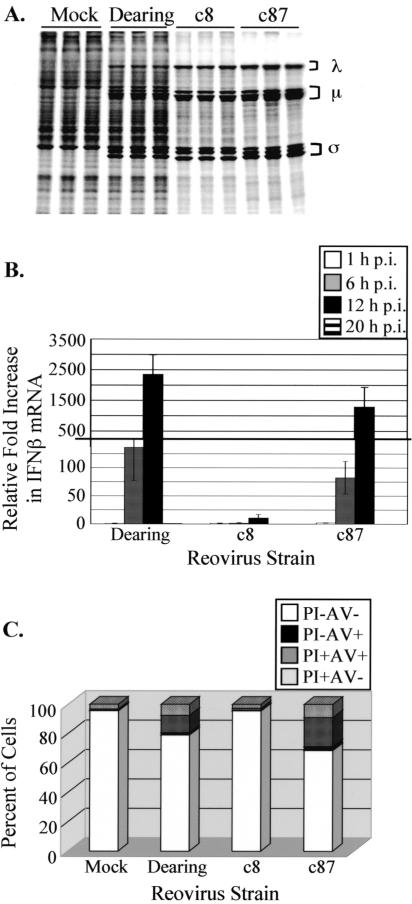
To gain insight into the mechanisms that regulate translation in reovirus-infected cells, we used a comparative transcriptional profiling approach to identify differences in cellular gene expression during infection with host shutoff (c8 and c87)- and non-host shutoff (Dearing)-inducing reovirus strains. In parallel with infections for transcriptional profiling, we used metabolic labeling to confirm that the three strains induced the anticipated host shutoff phenotypes in L929 cells. As expected (65, 70), infection with strains c8 and c87 resulted in the inhibition of cellular translation, whereas infection with strain Dearing did not (Fig. 1A). We also confirmed (data not shown) that the replication kinetics of these three reovirus isolates were similar when cells were infected at the high MOI used to maximize the distinct translational phenotypes (70).
FIG. 1.
Reovirus strains vary in their effects on host cells. (A) L929 cells were mock infected or infected in triplicate with the indicated reovirus strains at a multiplicity of 80 PFU/cell. At 19.5 h p.i., cells were pulse labeled with [35S]methionine-cysteine for 30 min. Extracts were prepared, and proteins were separated by SDS-polyacrylamide gel electrophoresis. Reovirus proteins are labeled with brackets to the right of the gel image. (B) L929 cells were mock infected or infected in triplicate with the indicated strains of reovirus at an MOI of 80 PFU/cell. At 1, 6, 12, and 20 h p.i., total RNAs were extracted, and the levels of IFN-β mRNA were measured by real-time reverse transcription-PCR. The relative increase in IFN-β mRNA compared to that in mock-infected cells at each time p.i. was calculated. Each bar represents the average (± SD) from three samples. (C) L929 cells were mock infected or infected in triplicate with the indicated reovirus strains at a multiplicity of 80 PFU/cell. At 19.5 h p.i., cells were harvested, stained with propidium iodide and annexin V-biotin, sorted by flow cytometry using a FACSCalibur machine, and analyzed with FLOWJO software. Each bar represents the average from three samples. PI-AV− indicates viable cells, PI−AV+ indicates early apoptotic cells, PI+AV+ indicates late apoptotic/necrotic cells, and PI+AV− indicates necrotic cells.
Because some reovirus strains induce type I IFN (IFN-α/β) (69), which would be expected to impact the expression of a large number of cellular genes, we assessed IFN-β mRNA induction under our infection conditions. We performed quantitative real-time reverse transcription-PCR on total RNA extracted from mock-infected or reovirus-infected L929 cells. The induction of IFN-β mRNA was apparent by 6 h p.i., with maximal levels observed at 12 h p.i. (Fig. 1B). By 20 h p.i., the amounts of IFN-β mRNA in reovirus-infected cells had returned to levels detected in mock-infected cells. At 6 and 12 h p.i., the amounts of IFN-β mRNA detected in Dearing- and c87-infected cells were substantially greater than those in c8-infected cells. At 12 h p.i., Dearing and c87 infections led to 2,400- and 1,300-fold increases, respectively, compared to mock-infected cells. This is in contrast to the 11-fold increase in IFN-β mRNA during c8 infection. These results reveal that the level of IFN-β mRNA induced as a consequence of reovirus infection does not correlate with the extent to which cellular translation is inhibited, even though the ISG products PKR and RNase L contribute to reovirus-induced host shutoff (70). Therefore, comparing these three reovirus strains in a transcriptional profiling study would enable us to distinguish changes in cellular gene expression that are a consequence of robust type I IFN induction (characteristic of Dearing and c87) from those that are associated with host shutoff (characteristic of c87 and c8).
Infection with the strong IFN-β mRNA-inducing strains of reovirus, Dearing and c87, leads to cell death via apoptosis (15). Since the apoptosis-inducing potential of c8 infection was unknown, and since some pathways that contribute to host shutoff also lead to apoptosis (17), we examined apoptosis in mock-infected L929 cells and cells infected with each of the three reovirus isolates. We performed this analysis at 19.5 h p.i., the time at which we isolated RNAs for transcriptional profiling and a time at which host shutoff is well established (24, 67, 70). Cells were treated with annexin V to label exposed phosphatidylserine and propidium iodide to stain DNA and then subjected to flow cytometry to determine the percentages of living, apoptotic, and necrotic cells. Consistent with previous reports (14, 22, 74), we found that significant fractions of L929 cells infected with strains Dearing and c87 undergo apoptosis (Fig. 1C), as shown by the number of cells that scored positively for annexin V (characteristic of early apoptosis) or both annexin V and propidium iodide (characteristic of late apoptosis) staining. c8 induced minimal apoptosis. These results reveal that there is no correlation between the extent to which a reovirus strain induces apoptosis and the degree to which it induces host translational shutoff. In contrast, apoptosis does appear to be associated with the induction of IFN-β mRNA; more apoptosis occurs following infection with reovirus strains that induce high levels of IFN-β mRNA. This is consistent with the fact that a number of ISG products mediate apoptosis (12).
Reovirus-induced alterations in cellular gene expression vary dramatically depending on the infecting strain.
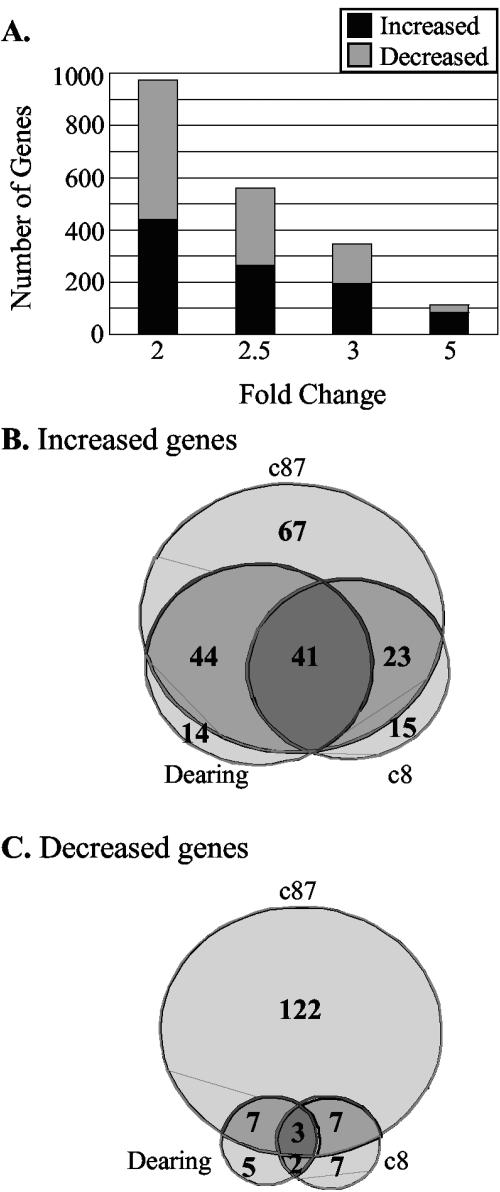
With the goal of identifying cellular pathways that regulate translation in reovirus-infected cells, we used Affymetrix U74Av2 murine-specific chips to examine L929 cellular gene expression profiles at 19.5 h p.i. with strains Dearing, c8, and c87. As mentioned above, at this time point c8- and c87-induced host shutoff is well established (24, 67, 70) and cell viability is high (90 to 100% relative to mock-infected L929 cells, as measured by trypan blue exclusion) (Fig. 1C). An independent set of infections was performed on each of three days. On each day, we also harvested a mock-infected sample. Using a cutoff of a twofold change with a P value of ≤0.05, we found that 974 genes represented by one or more probes on the Affymetrix chip had altered transcript levels after infection with at least one of the three strains of reovirus (Fig. 2A). The expression of 561 genes was altered ≥2.5-fold (P ≤ 0.05), while that of 344 genes was altered ≥3-fold (P ≤ 0.05) and that of 113 genes changed at least 5-fold (P ≤ 0.05). We chose to focus our initial analysis on the 344 cellular genes whose expression increased or decreased at least threefold (P ≤ 0.05) following infection with at least one of the reovirus strains in our analysis. The genes with increased expression are listed in Supplementary Table 1, and those with decreased expression are shown in Supplementary Table 2 (all Supplementary Tables may be found at http://www.ccgb.umn.edu/∼schiff/Array/).
FIG. 2.
Reovirus-induced changes in levels of L929 transcripts. (A) Number of genes on the Affymetrix U74Av2 chip that increased (black portion of each bar) or decreased (gray portion of each bar) 2-, 2.5-, 3-, or 5-fold, with all P values being ≤0.05, after infection with at least one of the three strains of reovirus examined. (B and C) Venn diagrams of cellular genes whose expression increased (B) and decreased (C) ≥3-fold, illustrating which infection conditions (light gray, one reovirus strain; gray, two viruses; and dark gray, all three reoviruses) altered the level of cellular transcripts. Ten genes of the 344 total were included in more than one category because they were represented by more than one probe and the intensity values for the different probes were not identical.
TABLE 1.
Cellular genes that had significantly higher expression following Dearing and c87 infections than following c8 infectiona
| Function and gene nameb | Accession no.c | FC, P value for c87/c8 | FC, P value for D/c8 | FC, P value for c87/D |
|---|---|---|---|---|
| Cell cycle | ||||
| Schlafen 2 (Slfn2) | AF099973 | 2, 0.034 | 2.5, 0.011 | 0.8, 0.474 |
| Gene/protein expression—DNA binding | ||||
| Histone 1, H1c (H1c) | J03482 | 4.4, 0.005 | 2.1, 0.021 | 2.1, 0.058 |
| H2B histone family, member S (H2bfs) | X05862 | 2.8, 0.012 | 2.2, 0.006 | 1.3, 0.313 |
| Gene/protein expression—protein degradation | ||||
| Serine protease inhibitor 6 (Spi6) | U96700 | 2.4, 0.037 | 2.1, 0.004 | 1.1, 0.769 |
| Ubiquitin-conjugating enzyme 8 (Ubce8) | AW048912 | 3.1, 0.046 | 2.6, 0.046 | 1.2, 0.669 |
| Gene/protein expression—transcription | ||||
| Glutamine repeat protein 1 (Glrp1) | U46463 | 2.8, 0.041 | 2.5, 0.009 | 1.1, 0.862 |
| IFN response and other defenses | ||||
| Chemokine (C-X-C motif) ligand 10 (Cxcl10) | M33266 | 5.1, 0.015 | 9.3, 0.004 | 0.6, 0.016 |
| Guanylate nucleotide binding protein 1 (GBP1) | M55544 | 14.2, 0.005 | 10.3, 0.001 | 1.4, 0.651 |
| Guanylate nucleotide binding protein 2 (GBP2) | AJ007970 | 13, 0.004 | 10.1, 0.001 | 1.3, 0.689 |
| Histocompatibility 2, D region locus 1 (H2-D1) | X00246 | 3.2, 0.013 | 2.6, 0.000 | 1.2, 0.616 |
| Histocompatibility 2, T region locus 10 (H2-T10) | M35244 | 3.1, 0.002 | 3.3, 0.000 | 0.9, 0.629 |
| Histocompatibility 2, T region locus 17 (H2-T17) | M35247 | 2.8, 0.002 | 3.1, 0.000 | 0.9, 0.393 |
| IFN-β | K00020 | 3.4, 0.005 | 2.3, 0.015 | 1.4, 0.016 |
| IFN-γ-induced GTPase (GTPI) | AJ007972 | 2.4, 0.004 | 2.8, 0.005 | 0.9, 0.486 |
| IFN-γ-induced GTPase (Igtp) | U53219 | 2.6, 0.004 | 3.3, 0.001 | 0.8, 0.221 |
| IFN-γ-inducible protein, 47 kDa (ISG47) | M63630 | 11.2, 0.001 | 17.3, 0.000 | 0.6, 0.132 |
| IFN-induced protein 35 (ISG35) | AW121732 | 2.9, 0.000 | 2, 0.001 | 1.5, 0.012 |
| IFN-induced protein with tetratricopeptide repeats 2 (ISG54) | U43085 | 3.9, 0.026 | 4.4, 0.005 | 0.9, 0.576 |
| IFN-induced protein with tetratricopeptide repeats 3 (ISG49) | U43086 | 4.5, 0.002 | 5.6, 0.001 | 0.8, 0.099 |
| IFN-inducible GTPase (Iigp) | AA914345 | 7.4, 0.028 | 10.9, 0.002 | 0.7, 0.352 |
| IFN regulatory factor 7 (IRF7) | U73037 | 2, 0.039 | 2.4, 0.018 | 0.8, 0.158 |
| Myeloid differentiation primary response gene 88 (Myd88) | X51397 | 2.9, 0.000 | 2, 0.001 | 1.4, 0.030 |
| Myxovirus resistance 2 (Mx2) | J03368 | 2.8, 0.029 | 2.3, 0.022 | 1.2, 0.581 |
| Nuclear antigen Sp100 (Sp100) | AF040242 | 2.8, 0.027 | 2.7, 0.022 | 1, 0.907 |
| AF040242 | 3.9, 0.012 | 3.7, 0.004 | 1.1, 0.907 | |
| Proteasome subunit, beta type 9 (Psmβ9) | D44456 | 4, 0.015 | 3.8, 0.000 | 1, 0.865 |
| Proteasome subunit, beta type 10 (Psmβ10) | Y10875 | 6.2, 0.000 | 4.9, 0.000 | 1.3, 0.010 |
| Transporter 1, ATP-binding cassette, subfamily B (Tap1) | U60020 | 3.2, 0.003 | 2.4, 0.008 | 1.3, 0.250 |
| Viral hemorrhagic septicemia virus (VHSV)-induced gene 1 (Vig1) | AA204579 | 2.8, 0.013 | 2.7, 0.007 | 1, 0.972 |
| Metabolism | ||||
| Histidine decarboxylase (Hdc) | X57437 | 5.3, 0.000 | 3.4, 0.001 | 1.6, 0.009 |
| Purine-nucleoside phosphorylase (Pnp) | U35374 | 2.6, 0.001 | 2.5, 0.000 | 1, 0.746 |
| Signaling/communication—apoptosis | ||||
| Fas death domain-associated protein (Daxx) | AF110520 | 2.9, 0.008 | 2.6, 0.002 | 1.1, 0.700 |
| Signaling/communication—ligands | ||||
| Chemokine (C-C motif) ligand 4 (Ccl4) | X62502 | 18, 0.000 | 4.3, 0.000 | 4.2, 0.000 |
| Chemokine (C-C motif) ligand 5 (Ccl5) | AF065947 | 12.5, 0.000 | 6.7, 0.001 | 1.9, 0.021 |
| Lectin, galactose binding, soluble 9 (Lgals9) | U55060 | 2.8, 0.007 | 2.3, 0.001 | 1.2, 0.453 |
| Signaling/communication—receptors | ||||
| Component of Sp100-rs (Csprs) | M55219 | 9.5, 0.001 | 10.3, 0.001 | 0.9, 0.452 |
| M55219 | 3.8, 0.020 | 4.1, 0.013 | 0.9, 0.616 | |
| Membrane-spanning four-domains, subfamily A, member 4D (Ms4a4d) | AI509811 | 9.4, 0.011 | 13.7, 0.005 | 0.7, 0.144 |
| Signaling/communication—transducers | ||||
| Guanylate nucleotide binding protein 3 (GBP3) | AW047476 | 3.3, 0.005 | 4.3, 0.000 | 0.8, 0.218 |
| N-Myc (and STAT) interactor (Nmi) | AF019249 | 3.2, 0.009 | 3.5, 0.001 | 0.9, 0.647 |
| T-cell-specific GTPase (Tgtp) | L38444 | 18.4, 0.000 | 25, 0.000 | 0.7, 0.346 |
| Unknown | ||||
| FLN29 gene product | AW049897 | 2.5, 0.005 | 2, 0.013 | 1.3, 0.167 |
| Macrophage-expressed gene 1 (Mpg-1) | L20315 | 2.3, 0.046 | 3, 0.025 | 0.8, 0.476 |
| Mus musc, 2 d pregnant adult female ovary cDNA | AA822898 | 3.1, 0.007 | 3.1, 0.002 | 1, 0.998 |
| RIKEN cDNA 1300004C08 gene | AW046479 | 5.7, 0.001 | 2.4, 0.007 | 2.3, 0.001 |
Fold change (FC) ≥ 2 and P ≤ 0.05 for c87/mock (m), D/m and c87/c8, D/c8.
Grouped according to functions of encoded proteins. Even though several gene products serve more than one function, each gene is represented in only one functional classification based on the focus of this paper.
GenBank accession number refers to the sequence used to design a gene's probe for the Affymetrix U74Av2 chips.
TABLE 2.
Cellular genes that had significantly higher expression following infection with the host-shutoff-inducing strains of reovirus, c8 and c87, than after infection with strain Dearinga
| Function and gene nameb | Accession no.c | FC, P value for c87/D | FC, P value for c8/D | FC, P value for c87/c8 |
|---|---|---|---|---|
| Cell cycle | ||||
| Growth arrest and DNA-damage-inducible 45 alpha (GADD45α) | U00937 | 2, 0.013 | 2, 0.025 | 1, 0.976fs |
| Gene/protein expression—DNA binding | ||||
| Tumor necrosis factor, alpha-induced protein 3 (Tnfip3) | U19463 | 8.6, 0.007 | 7, 0.041 | 1.2, 0.590 |
| Gene/protein expression—transcription | ||||
| DNA-damage-inducible transcript 3 (GADD153/CHOP) | X67083 | 3.2, 0.041 | 3, 0.028 | 1.1, 0.906 |
| Jun oncogene (Jun) | X12761 | 2.3, 0.018 | 2.3, 0.025 | 1, 0.953 |
| Nuclear receptor subfamily 1, group D, member 1 (Nr1d1) | AI834950 | 2.9, 0.003 | 2.2, 0.014 | 1.3, 0.195 |
| IFN response and other defenses | ||||
| Heat shock 70-kDa protein 1-like (Hsc70t) | L27086 | 3.7, 0.004 | 3.7, 0.012 | 1, 0.927 |
| Heat shock protein, 105 kDa (HSP105) | L40406 | 3.3, 0.028 | 2.2, 0.004 | 1.5, 0.393 |
| Signaling/communication—ligand | ||||
| Growth differentiation factor 15 (Gdf15) | AJ011967 | 4.8, 0.001 | 3.1, 0.039 | 1.5, 0.255 |
| Signaling/communication—phosphatase | ||||
| Dual-specificity phosphatase 1 (Dusp1) | X61940 | 2.6, 0.013 | 2.6, 0.000 | 1, 0.875 |
| Signaling/communication—receptor | ||||
| Inositol 1,4,5-triphosphate receptor 5 (Itpr5) | AF031127 | 6.7, 0.025 | 4.3, 0.012 | 1.5, 0.591 |
| Signaling/communication—transducers | ||||
| GTP binding protein 2 (Gtpbp2) | AW124369 | 2.2, 0.002 | 2, 0.050 | 1.1, 0.640 |
| Myeloid differentiation primary response gene 116 (MyD116/GADD34) | X51829 | 5.2, 0.003 | 3.2, 0.140d | 1.6, 0.286 |
| Unknown | ||||
| cDNA sequence BC037006 | AI850846 | 3.4, 0.043 | 2.1, 0.000 | 1.6, 0.414 |
| Expressed sequence AA407930 | AI841579 | 3.4, 0.000 | 2.2, 0.011 | 1.5, 0.062 |
| RIKEN cDNA 0610012A05 gene | AA815845 | 2.7, 0.000 | 2, 0.028 | 1.4, 0.161 |
FC ≥ 2 and P ≤ 0.05 for c87/m, c8/m and c87/D, c8/D.
Grouped according to functions of encoded proteins. Even though several gene products serve more than one function, each gene is represented in only one functional classification based on the focus of this paper.
GenBank accession number refers to the sequence used to design a gene's probe for the Affymetrix U74Av2 chips.
As a consequence of the stringency of our analysis and multiple P values being taken into account, the P value for c8/D is >0.005.
When the three reovirus strains were compared regarding the ability to alter cellular gene expression (Fig. 2B and C), we saw that c87 had the most dramatic effects. One hundred sixty-nine genes had a ≥3-fold increase in expression after c87 infection relative to mock-infected cells, compared to 94 and 79 genes with increased expression following Dearing and c8 infections, respectively. Only 41 genes were consistently increased in expression ≥3-fold following infection with all three reovirus isolates. In contrast, there was a total of 96 genes that had an increase in expression of at least threefold after infection with only one reovirus isolate (67 genes in c87-infected cells, 14 in Dearing-infected cells, and 15 in c8-infected cells). As might be expected, subsets of cellular genes appeared to be coordinately regulated following infection with reovirus strains that share certain phenotypic consequences of infection. For example, there were 23 genes whose expression increased ≥3-fold only during infection with the host shutoff-inducing reovirus strains c8 and c87. Similarly, the expression of 44 cellular genes was increased only after infection with the strong IFN-β mRNA- and apoptosis-inducing strains, Dearing and c87. Dearing and c8 infections are phenotypically distinct in terms of host shutoff, IFN-β mRNA expression, and apoptosis induction (Fig. 1). Interestingly, there were no cellular genes that had increased expression following only Dearing and c8 infections at a threshold of a threefold change. Even when the threshold was dropped to 2.5- or 2-fold, there were only five and eight genes, respectively, that were induced to that extent by c8 and Dearing infections but not by c87 infection (data not shown).
Reovirus infection led to fewer decreases in cellular gene expression than increases (see Supplementary Table 2). Of the 150 genes whose expression decreased at least threefold (P ≤ 0.05), 138 had decreased expression following c87 infection (Fig. 2C). Significantly fewer genes had decreased expression during Dearing and c8 infections, with 17 and 18 genes affected, respectively. Only 3 of the 150 cellular genes had decreased expression, by at least threefold, following infection with each of the three virus strains. Consistent with our analysis of increases in gene expression, we found small subsets of genes whose expression similarly decreased following infection with reovirus strains that result in similar infection phenotypes.
Using a cutoff of a 3-fold change, we identified a gene as differentially regulated if it increased 3- or 3.1-fold in two of the three infections, even if its expression increased 2.9-fold after infection with the third strain. Since our goal was to identify changes in cellular gene expression that might advance our understanding of translational control in reovirus-infected cells, we applied another type of analysis to our data set. First, we identified cellular genes that had at least a twofold change in expression, with a P value of ≤0.05, compared to mock-infected L929 cells. This selected genes whose expression was significantly altered by reovirus infection. Then, to identify sets of genes whose expression changes might be related to particular biological phenotypes, we narrowed that subset to those genes whose expression increased or decreased ≥2-fold (P ≤ 0.05) after infection with two of the reovirus strains relative to the level of expression after infection with the third strain. In this manner, we were able to identify genes whose expression was significantly altered as a consequence of infection with strains that induce strong host shutoff (c8 and c87) or high levels of IFN-β and apoptosis (Dearing and c87).
The extent of IFN-β mRNA induced as a consequence of reovirus infection correlates with ISG expression and is associated with apoptosis induction.
As predicted, this comparative analysis revealed that a number of ISGs, including IFN-β itself and IFN regulatory factor 7 (IRF7), were expressed at higher levels following infection with Dearing and c87, the strains of reovirus that induced more IFN-β mRNA (Table 1; Fig. 1B). Of the 43 genes that had significant increases in expression specific to Dearing and c87, 22 are components of the IFN response or other innate defense systems. Because c8 also induces IFN-β mRNA, albeit to a significantly lower level than strains Dearing and c87, these data argue that the level of type I IFN induced as a consequence of reovirus infection impacts the number of ISGs induced and the extent of their induction. We also identified a subset of genes with increased expression whose protein products have been linked to apoptotic signaling. These include nuclear antigen Sp100, myeloid differentiation response gene 88 (MyD88), and the Fas death domain-associated protein Daxx. The increased expression of IRF7 and Sp100 following infection with c87 has been previously reported (21). Given that both NF-κB activation (18) and JNK activation (14) have been implicated in reovirus-induced apoptosis and that strains Dearing and c87 are associated with high levels of apoptosis, the increased expression of MyD88 and Daxx is not surprising. MyD88 is a general adaptor molecule for the Toll family of receptors that induces NF-κB activation (1), while Daxx activates apoptosis through the JNK signal transduction cascade (11, 78).
Infection with c8 and c87, the host shutoff-inducing strains of reovirus, induces components of the unfolded protein response.
We were particularly interested in those expression changes that were unique to cells infected with the strong host shutoff-inducing strains c8 and c87. Fifteen genes had significant increases in expression following c8 and c87 infections compared to Dearing-infected L929 cells (Table 2). Many of these are stress-responsive genes, including those for heat shock proteins Hsc70t and HSP105, growth arrest and DNA-damage-inducible 45α (GADD45α), GADD34, and the transcription factors GADD153/CHOP and Jun. Increased expression of CHOP is a classic marker of ER stress and the UPR (36). The UPR couples protein synthesis and protein folding within the ER and leads to translational shutoff when the level of total proteins or misfolded proteins within the lumen of the ER exceeds its processing capacity (reviewed in reference 68). Thus, it was surprising that CHOP transcript levels increased significantly following reovirus infection, given that mammalian orthoreoviruses do not encode glycoproteins or proteins with signal sequences (54) that would be expected to increase the ER luminal content during viral infection. Our microarray analysis did not reveal significant increases in the expression of other genes commonly associated with UPR activation, including BiP/GRP78, Herp, and GRP94 (36, 66).
Reovirus infection alters expression of genes whose protein products regulate eIF2α phosphorylation.
The UPR is just one of a number of cellular stresses that leads to eIF2α phosphorylation (30). Others include amino acid deprivation (40), heme deficiency (13), and viral infection/dsRNA (16). eIF2α phosphorylation and subsequent translational inhibition are reversible, enabling the regulated expression of proteins that facilitate recovery from stress. Two genes known to be induced as a consequence of ER stress, P58IPK and GADD34, function to prevent and reverse eIF2α phosphorylation, respectively. P58IPK binds to PKR and the PKR-like ER stress sensor kinase PERK and interferes with their ability to phosphorylate eIF2α (71, 77). In contrast, GADD34 recruits the catalytic subunit of protein phosphatase 1 to eIF2α, leading to its dephosphorylation (9). Our microarray analysis revealed that the expression of both GADD34 and P58IPK was altered after infection with the host shutoff-inducing strains of reovirus. GADD34 expression increased 6- and 9.7-fold (P ≤ 0.05) after infection with reovirus strains c8 and c87, respectively (Table 2; see Supplementary Table 1). This is consistent with the finding that CHOP levels also increased significantly in these infected cells, since CHOP is reported to directly activate GADD34 expression (48). In contrast, P58IPK expression decreased as a consequence of infection with these host shutoff-inducing strains. P58IPK expression was 4.4-fold and 3.5-fold higher in uninfected L929 cells than in c87- and c8-infected cells, respectively (Table 3; see Supplementary Table 2).
TABLE 3.
Cellular genes whose expression decreased as a consequence of c8 and c87 infectionsa
| Function and gene nameb | Accession no.c | FC, P value for D/c87 | FC, P value for D/c8 | FC, P value for c87/c8 |
|---|---|---|---|---|
| Cell cycle | ||||
| Cell division cycle 6 homolog (Cdc6) | AJ223087 | 2.5, 0.002 | 3.1, 0.001 | 1.2, 0.042 |
| Gene/protein expression—helicase | ||||
| DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 (Ddx5) | X65627 | 3.2, 0.045 | 2.4, 0.039 | 0.7, 0.309 |
| Gene/protein expression—transcription | ||||
| Distal-less homeobox 1 (Dlx1) | U51000 | 6.3, 0.001 | 2.5, 0.021 | 2.5, 0.028 |
| Splicing factor, arginine/serine-rich 5 (Sfrs5) | AF020308 | 2.4, 0.028 | 2, 0.006 | 0.8, 0.427 |
| Signaling/communication—transducer | ||||
| DnaJ (Hsp40) homolog, subfamily C, member 3 [P58(IPK)] | AI604013 | 3.9, 0.015 | 3.1, 0.004 | 0.8, 0.407 |
| Unknown | ||||
| Expressed sequence AI326115 | AI049391 | 2.5, 0.004 | 2.4, 0.040 | 1, 0.933 |
FC ≥ 2 and P ≤ 0.05 for m/c87, m/c8 and D/c87, D/c8.
Grouped according to functions of encoded proteins. Even though several gene products serve more than one function, each gene is represented in only one functional classification based on the focus of this paper.
GenBank accession number refers to the sequence used to design a gene's probe for the Affymetrix U74Av2 chips.
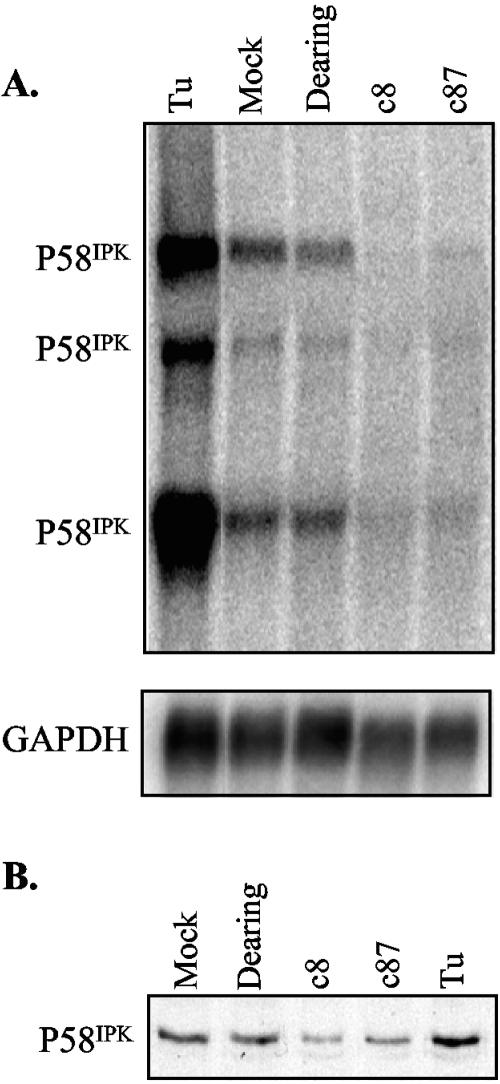
P58IPK is considered important for the ability of influenza virus and some plant viruses to resist the antiviral functions of PKR (7, 71, 72). Given that infection with reovirus strains c8 and c87 increased the expression of CHOP and GADD34, it was somewhat surprising to find that, at the same time, these strains led to a decrease in P58IPK mRNA expression. Like CHOP and GADD34, P58IPK is transcriptionally induced in response to ER stress (75). Since the decrease in P58IPK mRNA expression was unexpected, we confirmed this result by Northern blot analysis. L929 cells were mock infected or infected with the three strains of reovirus. As a positive control, cells were treated with tunicamycin to inhibit protein glycosylation and induce ER stress (77). At 19.5 h p.i., total RNA was extracted and separated by electrophoresis, and P58IPK mRNA was detected using a P58IPK-specific 32P-labeled DNA probe. Consistent with published reports (42), we observed three P58IPK mRNA species (Fig. 3A). As expected, tunicamycin treatment increased P58IPK transcript levels in L929 cells. While the transcript levels were similar in mock- and Dearing-infected L929 cells, P58IPK mRNA decreased in c8- and c87-infected cells to 38% and 44%, respectively, of the level in mock-infected cells. Consistent with the results from our Northern blot analysis, when cell extracts from mock-infected, reovirus-infected, and tunicamycin-treated L929 cells were probed using a P58IPK-specific monoclonal antibody, we found that the level of P58IPK protein decreased in c8- and c87-infected cells to 51% and 68%, respectively, of the level in mock-infected cells (Fig. 3B).
FIG. 3.
P58IPK expression changes in reovirus-infected cells. L929 cells were treated with tunicamycin (Tu), mock infected, or infected with the indicated strains of reovirus at an MOI of 80 PFU/cell. (A) At 19.5 h p.i., total RNAs were isolated, and the amounts of P58IPK mRNA were analyzed by Northern blotting (top panel). The blot was then stripped and reprobed with a GAPDH mRNA-specific probe (bottom panel). (B) At 19.5 h p.i., cell extracts were harvested, and P58IPK protein in 25 μg of cell extract was detected by immunoblot analysis.
Does the ER stress kinase PERK contribute to reovirus-induced host shutoff?
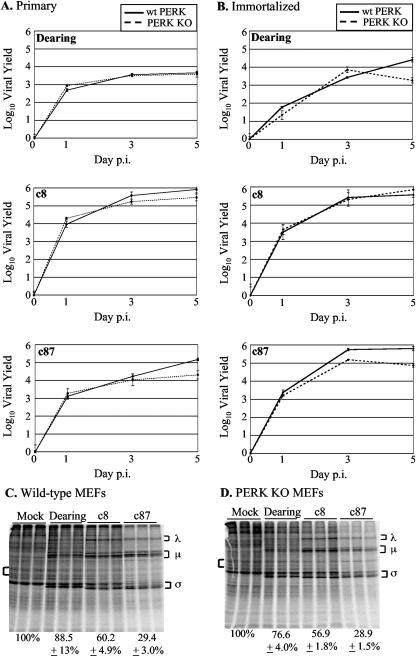
Decreases in the level of P58IPK in c8- and c87-infected cells would be predicted to increase the activity of PERK and/or PKR. Because infection with these strains increased the expression of the UPR marker CHOP, and since c8 and c87 induce host shutoff in the absence of PKR (70), we considered the possibility that PERK might influence reovirus replication and play a role in reovirus-induced host shutoff. Single-cycle growth analyses in MEFs from wt or PERK KO mice (29) revealed that the final yields were as much as 0.5 to 1 log higher in wt MEFs than in PERK KO MEFs (Fig. 4A and B). These data indicate that, like PKR (70), PERK is not detrimental to reovirus replication.
FIG. 4.
Effect of PERK on reovirus replication and role of PERK in reovirus-induced host shutoff. Primary (A) and immortalized (B) wt and PERK KO MEFs were infected with the indicated strains of reovirus at a multiplicity of 2 PFU/cell. Virus yields at 0, 1, 3, and 5 days p.i. were measured by a plaque assay. Each time point represents the mean (± SD) derived from three samples. Primary wt (C) and PERK KO (D) MEFs were mock infected or infected in triplicate with the indicated reovirus strains at a multiplicity of 80 PFU/cell. Cells were metabolically labeled at 19.5 h p.i. with [35S]methionine-cysteine for 2 h, and proteins were separated by SDS-polyacrylamide gel electrophoresis. Reovirus proteins are labeled with brackets to the right of the gel image. Percentages underneath the gel images represent the percentage of cellular translation relative to the level in uninfected cells quantified from the area indicated by the left bracket ± SD.
To determine if PERK contributes to host shutoff in reovirus-infected cells, we analyzed protein synthesis in infected and mock-infected primary MEFs from wt and PERK KO mice. At 19 h p.i., cells were metabolically labeled with [35S]methionine-cysteine, and the level of cellular translation was determined (Fig. 4C and D). Infection with strain Dearing had a minimal effect on cellular translation in either cell type, consistent with previous results (65, 67, 70). Infection of wt MEFs with c8 or c87 resulted in host shutoff (Fig. 4C), although not to the extent typically observed in MEFs (70). The amounts of cellular translation were similar in the presence or absence of PERK. These results suggest that, although the presence of PERK modestly increases reovirus yields, this ER-localized eIF2α kinase does not play a major role in reovirus-induced host shutoff.
Reovirus infection does not lead to IRE1α/β activation.
PERK is one of three identified UPR transducers associated with the ER membrane. Two others, IRE1α/β and activating transcription factor 6 (ATF6), activate transcriptional pathways that regulate the cell's response to stress. An increase in unfolded client proteins within the ER results in the dissociation of the BiP chaperone from the luminal domain of IRE1α/β, enabling it to dimerize. This activates the cytoplasmic endoribonuclease activity of IRE1α/β to initiate an unconventional splicing reaction that converts the X-box protein 1 (XBP-1) transcript to an mRNA encoding a functional transcription factor (10). XBP-1 target genes include those involved in UPR-mediated protein degradation (66). To understand if the induction of GADD34 and CHOP gene expression in reovirus-infected cells reflects an activation of the UPR's transcriptional pathways, we used real-time PCR to quantify the amounts of spliced XBP-1 mRNA (4) in uninfected and reovirus-infected L929 cells. As a positive control, cells were treated with tunicamycin for 6 h (32). As expected, we found that the level of spliced XBP-1 mRNA was increased 10.2-fold in tunicamycin-treated cells compared to that in untreated control cells. At 19.5 h p.i., the time at which we detected increases in CHOP and GADD34 expression by microarray analysis, we did not detect any increase in spliced XBP-1 mRNA in reovirus-infected samples; strains Dearing, c8, and c87 resulted in 0.81-, 0.18-, and 0.45-fold the amount of spliced XBP-1 mRNA in mock-infected cells, respectively. Thus, reovirus infection clearly does not activate the ER-resident endonuclease IRE1α/β to induce the XBP-1 transcriptional branch of the UPR.
Wild-type phosphorylatable eIF2α is associated with more efficient reovirus replication.
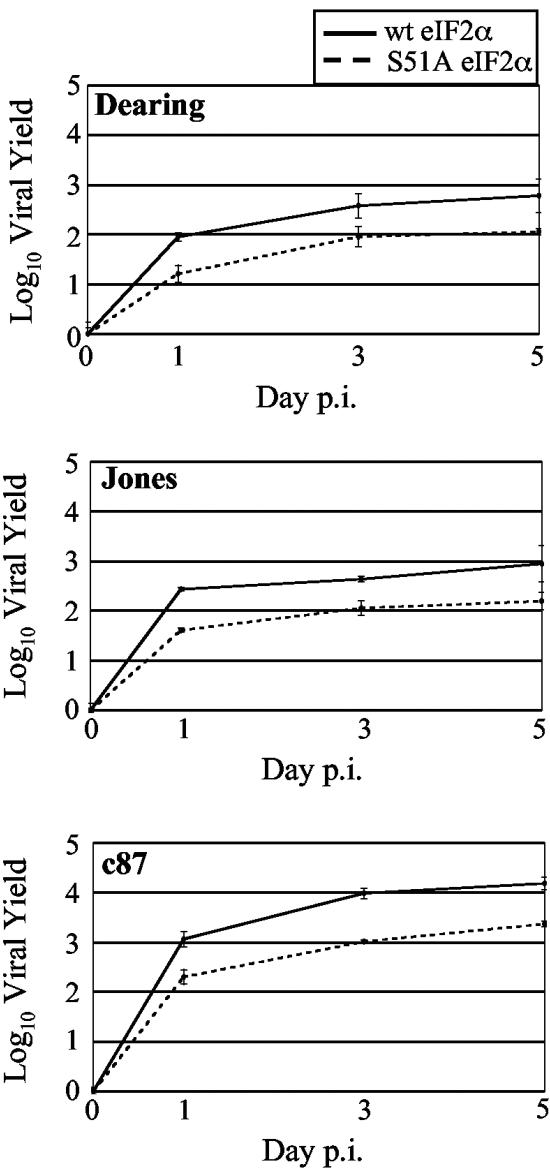
As mentioned above, the capacity of c8 and c87 to decrease P58IPK expression would be predicted to increase eIF2α phosphorylation by enabling PKR and/or PERK dimerization and activation (71, 77). When eIF2α is phosphorylated on serine 51, eIF2’s affinity for the guanine nucleotide exchange factor eIF2B increases. This results in a general inhibition of translation initiation by preventing the recycling of eIF2-GDP to the GTP-bound form that recruits the initiator methionine tRNA to the 40S ribosomal subunit. eIF2α phosphorylation is detrimental to the replication of most viruses (5, 79), consistent with its induction by IFN-induced pathways and viral dependence on the host translational machinery. In contrast, our results for wt and PKR or PERK KO cells indicate that reovirus yields were not improved in the KO cells; rather, the presence of these eIF2α kinases modestly increased viral yields (70) (Fig. 4A and B). To directly analyze the effect of eIF2α phosphorylation on reovirus replication, we measured viral yields in knock-in MEFs in which serine 51 of eIF2α was mutated to a nonphosphorylatable alanine (S51A) (64). We found that all strains of reovirus replicated more slowly and reached lower yields in MEFs expressing the constitutively active S51A mutant eIF2α than in MEFs expressing wt eIF2α (Fig. 5). Together, our data support a model in which eIF2α phosphorylation promotes more efficient reovirus replication.
FIG. 5.
Reovirus replication in MEFs that have constitutively active eIF2α. wt MEFs and MEFs expressing S51A mutant eIF2α were infected with the indicated strains of reovirus at a multiplicity of 2 PFU/cell. Amounts of infectious virus present at 0, 1, 3, and 5 days p.i. were determined as described in the legend to Fig. 4.
The phospho-eIF2α-dependent transcription factor ATF4 increases the efficiency of reovirus replication.
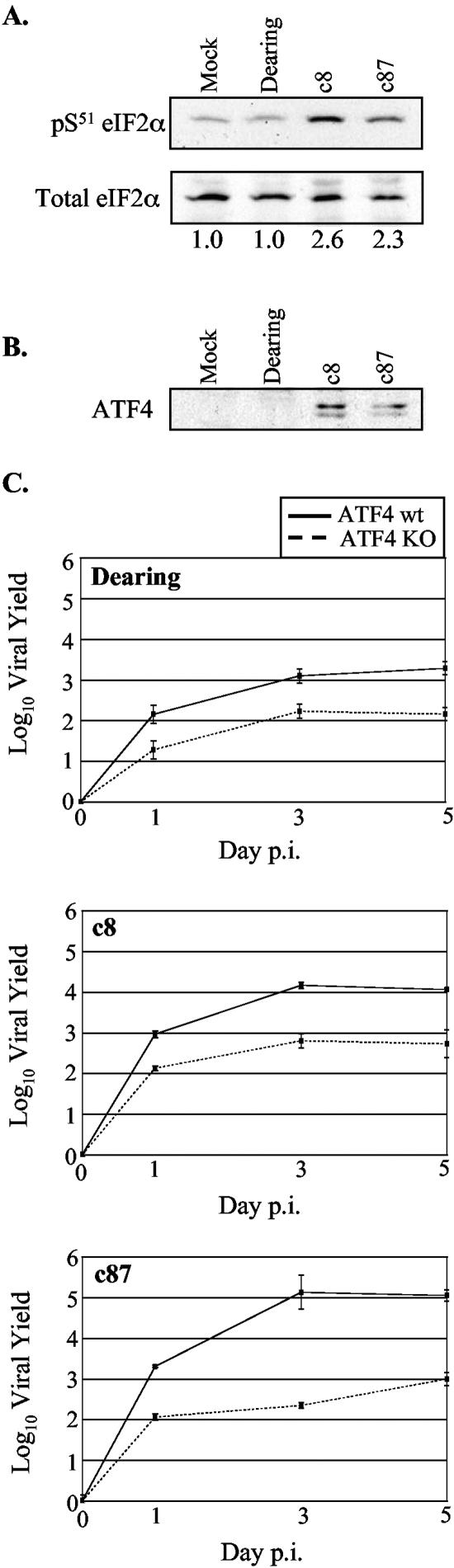
Our growth data clearly demonstrate that eIF2α phosphorylation is not required for reovirus replication but that its capacity to be phosphorylated on S51 provides a replicative advantage. Interestingly, it has previously been reported that the extent of eIF2α phosphorylation varies in reovirus-infected cells, with a higher degree of phosphorylation following infection with the host shutoff-inducing strain Jones than either strain Dearing or strain Lang (45). Given that reovirus yields were higher in cells expressing wt, phosphorylatable eIF2α and that strain Dearing consistently reached lower final yields than either c8 or c87, we compared the extents of eIF2α phosphorylation in L929 cells infected with these three strains. Cell extracts were prepared at 20 h p.i., and proteins were subjected to immunoblotting using total and phospho-eIF2α (S51)-specific rabbit polyclonal antibodies (Fig. 6A). Quantitative analysis revealed that in c8- and c87-infected cells, levels of phospho-eIF2α were 2.6- and 2.3-fold higher, respectively, than those in mock-infected samples. In contrast, eIF2α phosphorylation was not increased after infection with strain Dearing.
FIG. 6.
ATF4 expression in reovirus-infected cells and its impact on reovirus replication. (A and B) L929 cells were mock infected or infected with the indicated strains of reovirus at an MOI of 80 PFU/cell, and cell extracts were harvested at 19.5 h p.i. Samples were adjusted to contain 20 μg of protein for eIF2α analysis and 25 μg of protein for ATF4 analysis. The levels of phosphorylated eIF2α (A, top), total eIF2α (A, bottom), and ATF4 (B) were examined by immunoblot analysis. The band intensities for phosphorylated and total eIF2α were determined using NIH Image. The ratio of phosphorylated eIF2α to total eIF2α was set at 1 for mock-infected cells, and relative ratios for the infected samples are displayed beneath panel A. (C) Primary wt and ATF4 KO MEFs were infected with the indicated strains of reovirus at an MOI of 2 PFU/cell. The amounts of infectious virus present at 0, 1, 3, and 5 days p.i. were determined as described in the legend to Fig. 4.
How might phosphorylation of eIF2α on S51 promote more efficient viral replication? Although it is associated with general translation inhibition, eIF2α phosphorylation leads to the preferential translation of certain mRNAs. One such transcript encodes ATF4. When eIF2α is phosphorylated, rather than initiating at an upstream open reading frame, the ribosome scans the ATF4 transcript until it reaches the authentic open reading frame for ATF4 initiation (28, 46, 64). ATF4 then induces the transcription of several genes, many of which are involved in amino acid metabolism and redox regulation; this gene expression program has been referred to as the integrated stress response (ISR) (31). Both CHOP and GADD34, stress response genes that are induced following c8 and c87 infections, are transcriptionally activated by ATF4 (28, 56). Our finding that infection with the host shutoff-inducing strains c8 and c87 led to increased eIF2α phosphorylation predicted that ATF4 protein levels would also be higher in c8- and c87-infected cells. This was confirmed when ATF4 levels were analyzed in cell extracts from uninfected and reovirus-infected L929 cells (Fig. 6B). While we did not detect ATF4 in Dearing-infected L929 cells, given the fact that strain Dearing has been documented to induce a low level of eIF2α phosphorylation (45), we cannot rule out the possibility that a small quantity of ATF4 was synthesized but was below the level of detection in our experiments.
In addition to GADD34 and CHOP, reovirus infection increased the expression of other ATF4 target genes (31), including 4EBP-1, tRNA synthetases, amino acid transporters, and CCAAT/enhancer binding proteins. These findings led us to hypothesize that the expression of ATF4 target genes might mitigate some of the stresses of viral infection and that the presence of ATF4 would increase viral yields. To test this, we analyzed the replication of reovirus strains Dearing, c8, and c87 in primary wt and ATF4 KO MEFs (33). Our analysis revealed that c87 replication was profoundly decreased in MEFs that lacked an intact ATF4 gene; final yields were >2 logs lower in the KO cells than in wt cells (Fig. 6C). Dearing and c8 replication was also decreased in ATF4 KO MEFs, with final yields approximately 1 log lower than those in wt cells. These results argue that efficient reovirus replication depends on one or more cellular genes whose transcription is induced by ATF4 and reveal that the relative impact of ATF4 depends on the infecting viral strain.
Reovirus infection induces formation of stress granules.
Phosphorylation of eIF2α leads not only to the synthesis of ATF4 but also to the formation of “stalled” incomplete 48S preinitiation complexes that are incapable of translation initiation (reviewed in references 2 and 3). Although these complexes contain mRNAs, they lack the 60S ribosomal subunit and may also lack essential constituents of the 48S preinitiation complex, including eIF2 and eIF5 (37, 41). Two cellular RNA-binding proteins, TIA-1 and TIAR, bind to mRNAs associated with the stalled complexes, self-oligomerize, and aggregate into cytoplasmic SGs (38). Because reovirus infection induces eIF2α phosphorylation (45) (Fig. 6A), we hypothesized that SGs would form as a consequence of reovirus infection.
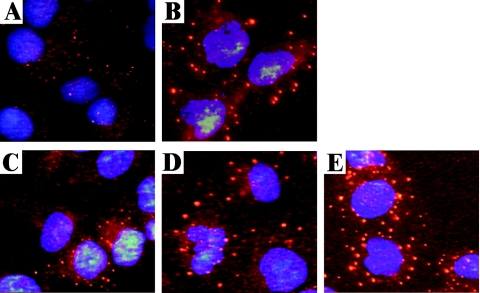
To test this hypothesis, we mock infected or infected DU145 cells with Dearing, c8, or c87. We chose DU145 cells for this analysis because they have been used in other analyses of SG formation (37, 38) and because c8- and c87-induced host shutoff occurs in this cell line (data not shown). As a positive control, cells were treated with sodium arsenite (49). At 19.5 h p.i., cells were fixed and stained for TIAR by indirect immunofluorescence. Consistent with reports in the literature (37, 38), some TIAR was localized within punctate cytoplasmic structures, characteristic of SGs, in sodium arsenite-treated DU145 cells (Fig. 7B). This contrasts with its more diffuse cytoplasmic localization in untreated, mock-infected cells (Fig. 7A). In cells infected with reovirus strains c8 and c87, we detected TIAR in large SGs that were similar in size to those observed in sodium arsenite-treated cells (Fig. 7D and E). Dearing-infected cells displayed an intermediate phenotype, with less abundant and smaller SGs than those observed in sodium arsenite-treated cells or in those infected with the host shutoff-inducing strains c8 and c87 (Fig. 7C). Thus, reovirus-induced SG formation varies in a strain-specific manner that correlates with the extent of host translational shutoff and eIF2α phosphorylation.
FIG. 7.
Stress granule formation in reovirus-infected cells. DU145 cells were mock infected (A), treated with sodium arsenite (B), or infected with Dearing (C), c8 (D), or c87 (E) at a multiplicity of 80 PFU/cell. At 19.5 h p.i., cells were fixed, and the intracellular localization of TIAR and nuclei was determined by indirect immunofluorescence.
DISCUSSION
Reovirus induces dramatic strain-dependent alterations in cellular gene expression.
Only a few studies have provided links between alterations in cellular gene expression and the phenotypic changes that are a consequence of mammalian reovirus infection (21, 57) and, to date, no expression analysis has focused on virus-induced host shutoff. We utilized oligonucleotide microarray technology to analyze cellular transcript levels 19.5 h after infection with reovirus strains Dearing, c8, and c87. These isolates differ in their capacities to induce type I IFN, apoptosis, and host translational shutoff, with strains Dearing and c87 inducing high levels of IFN-β mRNA and apoptosis and strains c8 and c87 sharing the property of host shutoff. Our analysis revealed that although the strains have similar replication kinetics in L929 cells, cellular gene expression is profoundly impacted by the infecting reovirus strain. Although some genes (62 of 251 whose expression was altered) were similarly impacted by infection with all three strains (see Supplementary Tables 3 and 4), expression of the majority of the genes (189 of 251) was only significantly altered after infection with one or two reovirus strains. As expected, virus pairs that shared phenotypes altered the expression of similar subsets of cellular genes. Remarkably, we found very few shared effects of Dearing and c8 infections on cellular gene expression. While these reovirus isolates are distinct in terms of the three phenotypes we measured, one might have imagined that they would induce other shared biological consequences in L929 cells that would similarly alter cellular gene expression. It was also notable that c87, which strongly induces IFN-β mRNA, apoptosis, and host shutoff, had the largest effect on cellular gene expression. We are currently working to determine if these changes in c87-infected cells are due to transcriptional or posttranscriptional mechanisms.
Genes whose expression increased twofold or more after infection with all strains included some encoding proteins involved in cell cycle control (GADD45β/Myd118 and Cyr61), protein degradation (autophagy 12-like), gene expression (C/EBPγ, Ets2, Taf1a, and Trim 25), and IFN regulatory pathways (PKR, ISG15, Usp18, and Trif). Only two of these transcripts, those for Cyr61 and C/EBPγ, were also found to be increased in human embryonic kidney 293 cells infected with reovirus strains c87 (T3 Abney) and Lang (21), further emphasizing the distinct responses mammalian cells have to infections with different reovirus isolates. The finding that all three strains in our analysis increased expression (more than threefold) of the autophagy 12-like gene (52) is intriguing in light of emerging evidence that some RNA viruses subvert components of cellular autophagic pathways for replication (34, 58).
Although reovirus strain Dearing is unique in its capacity to spare the cell from host translational shutoff, we failed to identify alterations in gene expression unique to Dearing-infected cells that helped to explain why infection with this reovirus isolate does not lead to host shutoff. Dearing infection only uniquely increased the expression of paired related homeobox 1, IFN regulatory factor 1, and myxomavirus resistance 1 and decreased the expression of 3-hydroxy-3-methylglutaryl-coenzyme A lyase.
Some ER stress response genes are induced in reovirus-infected cells, but infection does not induce a typical UPR.
We found that infection with the host shutoff-inducing reovirus strains c8 and c87 altered the expression of genes whose protein products impact translation during cellular stress responses. DeBiasi and colleagues also noted that cellular genes involved in the ER stress response (GADD34, GADD45, and HSP70t genes) were induced as a consequence of reovirus infection (21) and concluded that such changes were part of a generalized stress response to infection with all strains of reovirus. However, both of the isolates used in their experiments, c87 and Lang, induce host shutoff (65). In light of our analysis, we believe this stress response is enhanced when the infecting strain induces host translational shutoff.
The significant increases in expression of CHOP, GADD34, and GADD45α as well as the heat shock proteins Hsc70t and HSP105 suggested that infection with host-shutoff-inducing strains of reovirus might cause ER stress and induce an UPR. Upon ER stress, the chaperone BiP dissociates from the luminal domains of PERK, ATF6, and IRE1α/β in order to bind to misfolded proteins and prevent their aggregation (47). This release enables each of the three UPR sensors to become activated, either through dimerization (PERK and IRE1α/β) or via trafficking to the Golgi apparatus (ATF6). Emerging evidence indicates that PERK, ATF6, and IRE1α/β can be activated independent of one another (8, 62, 80). Since we failed to detect any increase in XBP-1 splicing following reovirus infection and did not observe an increased expression of genes downstream of IRE1α/β or ATF6, we do not believe the transcriptional arms of the UPR are activated in reovirus-infected L929 cells. The fact that P58IPK levels decrease upon infection with c8 and c87 supports this conclusion, since P58IPK is a target of XBP-1 (43). The lack of a transcriptional UPR in reovirus-infected cells makes teleological sense, as typical targets of ATF6 and genes downstream of IRE1α/β include ER chaperones and proteins involved in ER-associated degradation, and reovirus infection would not be expected to increase the load of either folded or misfolded proteins within the ER.
An analysis of viral growth in wt and PERK KO MEFs revealed that reovirus yields are slightly higher in the presence of PERK. While not as dramatic, these findings are consistent with what we observed with PKR wt and KO MEFs, in which the presence of the eIF2α kinase has the paradoxical effect of increasing viral yields (70). Although we do not have direct evidence for PERK activation, we know that eIF2α is phosphorylated in reovirus-infected PKR KO MEFs, indicating that at least one other eIF2α kinase is activated upon infection.
Reovirus infection decreases expression of P58IPK, a PKR/PERK inhibitor.
Our microarray analysis indicated that infection with host shutoff-inducing strains of reovirus decreased the expression of P58IPK. We confirmed this result by Northern blot analysis and demonstrated that P58IPK protein levels were also decreased in c8- and c87-infected cells. This is the first example, to our knowledge, of a viral infection that specifically decreases P58IPK expression. Our findings are interesting in light of the fact that P58IPK plays a positive role in the pathogenesis of other RNA viruses (influenza, tobacco mosaic, and tobacco etch viruses). In plant virus infections, P58IPK is required for virulence and functions as a susceptibility factor (7). P58IPK activity is increased in influenza virus-infected cells; this is proposed to limit PKR-mediated eIF2α phosphorylation (44, 50). In reovirus-infected cells, we suspect that decreased P58IPK expression is a cellular response that benefits viral replication, given that reovirus yields are increased in the presence of the eIF2α kinases and decreased in cells expressing a constitutively active, nonphosphorylatable eIF2α. Consistent with this, we found that c8 and c87 replicate to higher yields than strain Dearing, which does not induce a decrease in the level of P58IPK.
Reovirus induces, and benefits from, an integrated stress response.
Our data demonstrate that infections with the host-shutoff-inducing strains of reovirus result in increased eIF2α phosphorylation, perhaps as a consequence of decreased P58IPK levels. This enables the synthesis of ATF4, which activates the integrated stress response, a gene expression pathway geared towards promoting survival of stressed cells (28, 56). ISR genes, such as those encoding CHOP and GADD34, that depend on the synthesis of ATF4 (28, 56) were clearly activated in c8- and c87-infected cells. Future experiments with knockout cells will allow us to determine which of the stress kinases activate this gene expression pathway in reovirus-infected cells. There is evidence for the involvement of PKR in reovirus-induced eIF2α phosphorylation (19, 20, 26, 51, 55, 63). However, our observation that eIF2α is phosphorylated in reovirus-infected PKR KO MEFs (data not shown) suggests that PERK and/or GCN2 may also contribute to the activation of an ISR in infected cells.
Our results demonstrate that reovirus replicates to lower yields in the absence of PKR, PERK, or ATF4 and in MEFs with nonphosphorylatable S51A mutant eIF2α. The effect of the ATF4 KO mutation is greater than that of either of the single kinase KO mutations, consistent with the conclusion that more than one stress kinase is activated in reovirus-infected cells. Together, our results strongly argue that eIF2α kinase activation and subsequent induction of the ISR enable more efficient reovirus replication. We can envision a number of different, but not mutually exclusive, mechanisms that could explain this surprising result. First, we have previously shown that the capsid protein σ3 localizes to the perinuclear region of cells infected with host shutoff-inducing strains of reovirus (65). Because σ3 has the capacity to bind dsRNA, we hypothesized that PKR activation is prevented in the vicinity of the perinuclear viral factories (65). Because the majority of viral transcripts are synthesized from secondary transcriptase particles that assemble in this area, such a phenomenon might put viral transcripts at a selective advantage for limited translational machinery. Second, viral translation may be facilitated because the phosphorylation of eIF2α promotes polyribosome disassembly (35), enabling ribosomal subunits to initiate translation on other mRNAs. This would disrupt the continuous translation of some cellular mRNAs and might enable greater translation of abundant reovirus transcripts. Third, we have demonstrated that reovirus infection induces the formation of stress granules. These could increase the competitive advantage of reovirus transcripts by sequestering cellular mRNA, leaving the available translational machinery free to interact with abundant viral mRNAs. Finally, eIF2α phosphorylation induces the expression of cell survival pathways through both ATF4 and NF-κB (23, 31). Our work (Fig. 6) and that of our colleagues (18) suggest that these antiapoptotic gene expression programs may improve the intracellular environment to promote viral replication.
In conclusion, we have demonstrated that reovirus infection induces striking changes in cellular gene expression and that these vary significantly depending on the infecting strain. Infections with the host-shutoff-inducing strains of reovirus lead to the expression of genes involved in cellular stress responses. The trigger(s) for these responses has yet to be defined but may include dsRNA, ER stress, and/or nutrient deprivation. The surprising finding that reovirus replicates most efficiently under stressful conditions that compromise host protein synthesis raises interesting questions about the mechanisms involved in viral translation and the relationship between apoptosis and viral growth.
Acknowledgments
We thank Heather Harding and David Ron for wt and PERK KO MEFs and James McCarthy for DU145 cells. We are indebted to Martin Prlic, who assisted with fluorescence-activated cell sorting analysis, and Kezhong Zhang, who analyzed XBP-1 splicing for us. We are grateful to Alan Goodman, Nancy Kedersha, John Parker, and David Ron for helpful discussions and feedback on various aspects of this study. Finally, we thank Rebecca Duerst, Joseph Golden, and Stephen Rice for critical reviews of the manuscript and Jessica Bahe and Tim Leonard for technical assistance.
This work was supported by NIH grants AI45990 (to L.A.S.), AI49494 (to P.R.B.), AI22646 (to M.G.K.), and DK42394 (to R.J.K.). L.A.S. and P.R.B. were each also supported by seed grants from the University of Minnesota Academic Health Center.
REFERENCES
- 1.Aliprantis, A. O., R. B. Yang, D. S. Weiss, P. Godowski, and A. Zychlinsky. 2000. The apoptotic signaling pathway activated by Toll-like receptor-2. EMBO J. 19:3325-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, P., and N. Kedersha. 2002. Stressful initiations. J. Cell Sci. 115:3227-3234. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, P., and N. Kedersha. 2002. Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones 7:213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Back, S. H., M. Schroder, K. Lee, K. Zhang, and R. J. Kaufman. 2005. ER stress signaling by regulated splicing: IRE1/HAC1/XBP1. Methods 35:395-416. [DOI] [PubMed] [Google Scholar]
- 5.Balachandran, S., P. C. Roberts, L. E. Brown, H. Truong, A. K. Pattnaik, D. R. Archer, and G. N. Barber. 2000. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity 13:129-141. [DOI] [PubMed] [Google Scholar]
- 6.Barber, G. N., S. Thompson, T. G. Lee, T. Strom, R. Jagus, A. Darveau, and M. G. Katze. 1994. The 58-kilodalton inhibitor of the interferon-induced double-stranded RNA-activated protein kinase is a tetratricopeptide repeat protein with oncogenic properties. Proc. Natl. Acad. Sci. USA 91:4278-4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bilgin, D. D., Y. Liu, M. Schiff, and S. P. Dinesh-Kumar. 2003. P58(IPK), a plant ortholog of double-stranded RNA-dependent protein kinase PKR inhibitor, functions in viral pathogenesis. Dev. Cell 4:651-661. [DOI] [PubMed] [Google Scholar]
- 8.Brewer, J. W., and L. M. Hendershot. 2005. Building an antibody factory: a job for the unfolded protein response. Nat. Immunol. 6:23-29. [DOI] [PubMed] [Google Scholar]
- 9.Brush, M. H., D. C. Weiser, and S. Shenolikar. 2003. Growth arrest and DNA damage-inducible protein GADD34 targets protein phosphatase 1 alpha to the endoplasmic reticulum and promotes dephosphorylation of the alpha subunit of eukaryotic translation initiation factor 2. Mol. Cell. Biol. 23:1292-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calfon, M., H. Zeng, F. Urano, J. H. Till, S. R. Hubbard, H. P. Harding, S. G. Clark, and D. Ron. 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415:92-96. [DOI] [PubMed] [Google Scholar]
- 11.Chang, H. Y., H. Nishitoh, X. Yang, H. Ichijo, and D. Baltimore. 1998. Activation of apoptosis signal-regulating kinase 1 (ASK1) by the adapter protein Daxx. Science 281:1860-1863. [DOI] [PubMed] [Google Scholar]
- 12.Chawla-Sarkar, M., D. J. Lindner, Y. F. Liu, B. R. Williams, G. C. Sen, R. H. Silverman, and E. C. Borden. 2003. Apoptosis and interferons: role of interferon-stimulated genes as mediators of apoptosis. Apoptosis 8:237-249. [DOI] [PubMed] [Google Scholar]
- 13.Chen, J. 2000. Heme-regulated eIF2alpha kinase, p. 529-546. In N. Sonenberg, J. W. Hershey, and M. B. Mathews (ed.), Translational control of gene expression, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 14.Clarke, P., S. M. Meintzer, C. Widmann, G. L. Johnson, and K. L. Tyler. 2001. Reovirus infection activates JNK and the JNK-dependent transcription factor c-Jun. J. Virol. 75:11275-11283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clarke, P., and K. L. Tyler. 2003. Reovirus-induced apoptosis: a minireview. Apoptosis 8:141-150. [DOI] [PubMed] [Google Scholar]
- 16.Clemens, M. J. 1996. Protein kinases that phosphorylate eIF2 and eIF2B, and their role in eukaryotic cell translational control, p. 139-172. In J. Hershey, M. Mathews, and N. Sonenberg (ed.), Translational control. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 17.Clemens, M. J., M. Bushell, I. W. Jeffrey, V. M. Pain, and S. J. Morley. 2000. Translation initiation factor modifications and the regulation of protein synthesis in apoptotic cells. Cell Death Differ. 7:603-615. [DOI] [PubMed] [Google Scholar]
- 18.Connolly, J. L., S. E. Rodgers, P. Clarke, D. W. Ballard, L. D. Kerr, K. L. Tyler, and T. S. Dermody. 2000. Reovirus-induced apoptosis requires activation of transcription factor NF-kappaB. J. Virol. 74:2981-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Benedetti, A., G. J. Williams, and C. Baglioni. 1985. Inhibition of binding to initiation complexes of nascent reovirus mRNA by double-stranded RNA-dependent protein kinase. J. Virol. 54:408-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Benedetti, A., G. J. Williams, L. Comeau, and C. Baglioni. 1985. Inhibition of viral mRNA translation in interferon-treated L cells infected with reovirus. J. Virol. 55:588-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeBiasi, R. L., P. Clarke, S. Meintzer, R. Jotte, B. K. Kleinschmidt-Demasters, G. L. Johnson, and K. L. Tyler. 2003. Reovirus-induced alteration in expression of apoptosis and DNA repair genes with potential roles in viral pathogenesis. J. Virol. 77:8934-8947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Debiasi, R. L., M. K. Squier, B. Pike, M. Wynes, T. S. Dermody, J. J. Cohen, and K. L. Tyler. 1999. Reovirus-induced apoptosis is preceded by increased cellular calpain activity and is blocked by calpain inhibitors. J. Virol. 73:695-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deng, J., P. D. Lu, Y. Zhang, D. Scheuner, R. J. Kaufman, N. Sonenberg, H. P. Harding, and D. Ron. 2004. Translational repression mediates activation of nuclear factor kappa B by phosphorylated translation initiation factor 2. Mol. Cell. Biol. 24:10161-10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feduchi, E., M. Esteban, and L. Carrasco. 1988. Reovirus type 3 synthesizes proteins in interferon-treated HeLa cells without reversing the antiviral state. Virology 164:420-426. [DOI] [PubMed] [Google Scholar]
- 25.Furlong, D. B., M. L. Nibert, and B. N. Fields. 1988. Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. J. Virol. 62:246-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.George, C. X., and C. E. Samuel. 1988. Mechanism of interferon action. Expression of reovirus S3 gene in transfected COS cells and subsequent inhibition at the level of protein synthesis by type I but not by type II interferon. Virology 166:573-582. [DOI] [PubMed] [Google Scholar]
- 27.Golden, J. W., J. Linke, S. Schmechel, K. Thoemke, and L. A. Schiff. 2002. Addition of exogenous protease facilitates reovirus infection in many restrictive cells. J. Virol. 76:7430-7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harding, H. P., I. Novoa, Y. Zhang, H. Zeng, R. Wek, M. Schapira, and D. Ron. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6:1099-1108. [DOI] [PubMed] [Google Scholar]
- 29.Harding, H. P., H. Zeng, Y. Zhang, R. Jungries, P. Chung, H. Plesken, D. D. Sabatini, and D. Ron. 2001. Diabetes mellitus and exocrine pancreatic dysfunction in perk−/− mice reveals a role for translational control in secretory cell survival. Mol. Cell 7:1153-1163. [DOI] [PubMed] [Google Scholar]
- 30.Harding, H. P., Y. Zhang, and D. Ron. 1999. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397:271-274. [DOI] [PubMed] [Google Scholar]
- 31.Harding, H. P., Y. Zhang, H. Zeng, I. Novoa, P. D. Lu, M. Calfon, N. Sadri, C. Yun, B. Popko, R. Paules, D. F. Stojdl, J. C. Bell, T. Hettmann, J. M. Leiden, and D. Ron. 2003. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11:619-633. [DOI] [PubMed] [Google Scholar]
- 32.Helenius, A. 1994. How N-linked oligosaccharides affect glycoprotein folding in the endoplasmic reticulum. Mol. Biol. Cell 5:253-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hettmann, T., K. Barton, and J. M. Leiden. 2000. Microphthalmia due to p53-mediated apoptosis of anterior lens epithelial cells in mice lacking the CREB-2 transcription factor. Dev. Biol. 222:110-123. [DOI] [PubMed] [Google Scholar]
- 34.Jackson, W. T., T. H. Giddings, Jr., M. P. Taylor, S. Mulinyawe, M. Rabinovitch, R. R. Kopito, and K. Kirkegaard. 2005. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 3:e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaufman, R. J. 2004. Regulation of mRNA translation by protein folding in the endoplasmic reticulum. Trends Biochem. Sci. 29:152-158. [DOI] [PubMed] [Google Scholar]
- 36.Kaufman, R. J. 1999. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 13:1211-1233. [DOI] [PubMed] [Google Scholar]
- 37.Kedersha, N., S. Chen, N. Gilks, W. Li, I. J. Miller, J. Stahl, and P. Anderson. 2002. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell 13:195-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kedersha, N. L., M. Gupta, W. Li, I. Miller, and P. Anderson. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 147:1431-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kedl, R., S. Schmechel, and L. Schiff. 1995. Comparative sequence analysis of the reovirus S4 genes from 13 serotype 1 and serotype 3 field isolates. J. Virol. 69:552-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kimball, S. R., M. J. Clemens, V. J. Tilleray, R. C. Wek, R. L. Horetsky, and L. S. Jefferson. 2001. The double-stranded RNA-activated protein kinase PKR is dispensable for regulation of translation initiation in response to either calcium mobilization from the endoplasmic reticulum or essential amino acid starvation. Biochem. Biophys. Res. Commun. 280:293-300. [DOI] [PubMed] [Google Scholar]
- 41.Kimball, S. R., R. L. Horetsky, D. Ron, L. S. Jefferson, and H. P. Harding. 2003. Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am. J. Physiol. Cell Physiol. 284:C273-C284. [DOI] [PubMed] [Google Scholar]
- 42.Korth, M. J., C. N. Lyons, M. Wambach, and M. G. Katze. 1996. Cloning, expression, and cellular localization of the oncogenic 58-kDa inhibitor of the RNA-activated human and mouse protein kinase. Gene 170:181-188. [DOI] [PubMed] [Google Scholar]
- 43.Lee, A. H., N. N. Iwakoshi, and L. H. Glimcher. 2003. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 23:7448-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee, T. G., J. Tomita, A. G. Hovanessian, and M. G. Katze. 1992. Characterization and regulation of the 58,000-dalton cellular inhibitor of the interferon-induced, dsRNA-activated protein kinase. J. Biol. Chem. 267:14238-14243. [PubMed] [Google Scholar]
- 45.Lloyd, R. M., and A. J. Shatkin. 1992. Translational stimulation by reovirus polypeptide sigma 3: substitution for VAI RNA and inhibition of phosphorylation of the alpha subunit of eukaryotic initiation factor 2. J. Virol. 66:6878-6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu, P. D., H. P. Harding, and D. Ron. 2004. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J. Cell Biol. 167:27-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma, Y., and L. M. Hendershot. 2002. The mammalian endoplasmic reticulum as a sensor for cellular stress. Cell Stress Chaperones 7:222-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marciniak, S. J., C. Y. Yun, S. Oyadomari, I. Novoa, Y. Zhang, R. Jungreis, K. Nagata, H. P. Harding, and D. Ron. 2004. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 18:3066-3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McEwen, E., N. Kedersha, B. Song, D. Scheuner, N. Gilks, A. Han, J. J. Chen, P. Anderson, and R. J. Kaufman. 2005. Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J. Biol. Chem. 280:16925-16933. [DOI] [PubMed] [Google Scholar]
- 50.Melville, M. W., M. G. Katze, and S. L. Tan. 2000. P58IPK, a novel cochaperone containing tetratricopeptide repeats and a J-domain with oncogenic potential. Cell Mol. Life Sci. 57:311-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miyamoto, N. G., B. L. Jacobs, and C. E. Samuel. 1983. Mechanism of interferon action. Effect of double-stranded RNA and the 5′-O-monophosphate form of 2′,5′-oligoadenylate on the inhibition of reovirus mRNA translation in vitro. J. Biol. Chem. 258:15232-15237. [PubMed] [Google Scholar]
- 52.Mizushima, N., T. Noda, T. Yoshimori, Y. Tanaka, T. Ishii, M. D. George, D. J. Klionsky, M. Ohsumi, and Y. Ohsumi. 1998. A protein conjugation system essential for autophagy. Nature 395:395-398. [DOI] [PubMed] [Google Scholar]
- 53.Nibert, M. L., and B. N. Fields. 1992. A carboxy-terminal fragment of protein mu 1/mu 1C is present in infectious subvirion particles of mammalian reoviruses and is proposed to have a role in penetration. J. Virol. 66:6408-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nibert, M. L., and L. A. Schiff. 2001. Reoviruses and their replication, p. 1679-1728. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 55.Nilsen, T. W., P. A. Maroney, and C. Baglioni. 1982. Inhibition of protein synthesis in reovirus-infected HeLa cells with elevated levels of interferon-induced protein kinase activity. J. Biol. Chem. 257:14593-14596. [PubMed] [Google Scholar]
- 56.Novoa, I., H. Zeng, H. P. Harding, and D. Ron. 2001. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J. Cell Biol. 153:1011-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Poggioli, G. J., R. L. DeBiasi, R. Bickel, R. Jotte, A. Spalding, G. L. Johnson, and K. L. Tyler. 2002. Reovirus-induced alterations in gene expression related to cell cycle regulation. J. Virol. 76:2585-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prentice, E., W. G. Jerome, T. Yoshimori, N. Mizushima, and M. R. Denison. 2004. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 279:10136-10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rice, J. 1995. Mathematical statistics and data analysis. Duxbury Press, Belmont, Calif.
- 60.Rosen, L., J. F. Hovis, F. M. Mastrota, J. A. Bell, and R. J. Huebner. 1960. Observations on a newly recognized virus (Abney) of the reovirus family. Am. J. Hyg. 71:258-265. [DOI] [PubMed] [Google Scholar]
- 61.Rutkowski, D. T., and R. J. Kaufman. 2003. All roads lead to ATF4. Dev. Cell 4:442-444. [DOI] [PubMed] [Google Scholar]
- 62.Rutkowski, D. T., and R. J. Kaufman. 2004. A trip to the ER: coping with stress. Trends Cell Biol. 14:20-28. [DOI] [PubMed] [Google Scholar]
- 63.Samuel, C. E., R. Duncan, G. S. Knutson, and J. W. Hershey. 1984. Mechanism of interferon action. Increased phosphorylation of protein synthesis initiation factor eIF-2 alpha in interferon-treated, reovirus-infected mouse L929 fibroblasts in vitro and in vivo. J. Biol. Chem. 259:13451-13457. [PubMed] [Google Scholar]
- 64.Scheuner, D., B. Song, E. McEwen, C. Liu, R. Laybutt, P. Gillespie, T. Saunders, S. Bonner-Weir, and R. J. Kaufman. 2001. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell 7:1165-1176. [DOI] [PubMed] [Google Scholar]
- 65.Schmechel, S., M. Chute, P. Skinner, R. Anderson, and L. Schiff. 1997. Preferential translation of reovirus mRNA by a sigma3-dependent mechanism. Virology 232:62-73. [DOI] [PubMed] [Google Scholar]
- 66.Schroder, M., and R. J. Kaufman. 2005. The mammalian unfolded protein response. Annu. Rev. Biochem. 74:739-789. [DOI] [PubMed] [Google Scholar]
- 67.Sharpe, A. H., and B. N. Fields. 1982. Reovirus inhibition of cellular RNA and protein synthesis: role of the S4 gene. Virology 122:381-391. [DOI] [PubMed] [Google Scholar]
- 68.Shen, X., K. Zhang, and R. J. Kaufman. 2004. The unfolded protein response—a stress signaling pathway of the endoplasmic reticulum. J. Chem. Neuroanat. 28:79-92. [DOI] [PubMed] [Google Scholar]
- 69.Sherry, B., J. Torres, and M. A. Blum. 1998. Reovirus induction of and sensitivity to beta interferon in cardiac myocyte cultures correlate with induction of myocarditis and are determined by viral core proteins. J. Virol. 72:1314-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smith, J. A., S. C. Schmechel, B. R. Williams, R. H. Silverman, and L. A. Schiff. 2005. Involvement of the interferon-regulated antiviral proteins PKR and RNase L in reovirus-induced shutoff of cellular translation. J. Virol. 79:2240-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tan, S. L., M. J. Gale, Jr., and M. G. Katze. 1998. Double-stranded RNA-independent dimerization of interferon-induced protein kinase PKR and inhibition of dimerization by the cellular P58IPK inhibitor. Mol. Cell. Biol. 18:2431-2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tan, S. L., and M. G. Katze. 1998. Biochemical and genetic evidence for complex formation between the influenza A virus NS1 protein and the interferon-induced PKR protein kinase. J. Interferon Cytokine Res. 18:757-766. [DOI] [PubMed] [Google Scholar]
- 73.Tyler, K. L. 2001. Reoviruses, p. 1729-1746. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 74.Tyler, K. L., M. K. Squier, S. E. Rodgers, B. E. Schneider, S. M. Oberhaus, T. A. Grdina, J. J. Cohen, and T. S. Dermody. 1995. Differences in the capacity of reovirus strains to induce apoptosis are determined by the viral attachment protein sigma 1. J. Virol. 69:6972-6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van Huizen, R., J. L. Martindale, M. Gorospe, and N. J. Holbrook. 2003. P58IPK, a novel endoplasmic reticulum stress-inducible protein and potential negative regulator of eIF2alpha signaling. J. Biol. Chem. 278:15558-15564. [DOI] [PubMed] [Google Scholar]
- 76.Virgin, H. W. T., R. Bassel-Duby, B. N. Fields, and K. L. Tyler. 1988. Antibody protects against lethal infection with the neurally spreading reovirus type 3 (Dearing). J. Virol. 62:4594-4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yan, W., C. L. Frank, M. J. Korth, B. L. Sopher, I. Novoa, D. Ron, and M. G. Katze. 2002. Control of PERK eIF2alpha kinase activity by the endoplasmic reticulum stress-induced molecular chaperone P58IPK. Proc. Natl. Acad. Sci. USA 99:15920-15925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang, X., R. Khosravi-Far, H. Y. Chang, and D. Baltimore. 1997. Daxx, a novel Fas-binding protein that activates JNK and apoptosis. Cell 89:1067-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang, Y. L., L. F. Reis, J. Pavlovic, A. Aguzzi, R. Schafer, A. Kumar, B. R. Williams, M. Aguet, and C. Weissmann. 1995. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J. 14:6095-6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang, K., H. N. Wong, B. Song, C. N. Miller, D. Scheuner, and R. J. Kaufman. 2005. The unfolded protein response sensor IRE1alpha is required at 2 distinct steps in B cell lymphopoiesis. J. Clin. Investig. 115:268-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zweerink, H. J., and W. K. Joklik. 1970. Studies on the intracellular synthesis of reovirus-specified proteins. Virology 41:501-518. [DOI] [PubMed] [Google Scholar]