Abstract
Cytotoxic T-lymphocyte (CTL) responses are crucial for the control of immunodeficiency virus replication. Possible involvement of a dominant single epitope-specific CTL in control of viral replication has recently been indicated in preclinical AIDS vaccine trials, but it has remained unclear if multiple epitope-specific CTLs can be involved in the vaccine-based control. Here, by following up five rhesus macaques that showed vaccine-based control of primary replication of a simian immunodeficiency virus, SIVmac239, we present evidence indicating involvement of multiple epitope-specific CTL responses in this control. Three macaques maintained control for more than 2 years without additional mutations in the provirus. However, in the other two that shared a major histocompatibility complex haplotype, viral mutations were accumulated in a similar order, leading to viral evasion from three epitope-specific CTL responses with viral fitness costs. Accumulation of these multiple escape mutations resulted in the reappearance of plasma viremia around week 60 after challenge. Our results implicate multiple epitope-specific CTL responses in control of immunodeficiency virus replication and furthermore suggest that sequential accumulation of multiple CTL escape mutations, if allowed, can result in viral evasion from this control.
Virus-specific cytotoxic T-lymphocyte (CTL) responses are crucial for the control of immunodeficiency virus infections. The importance of CTLs for control has been indicated by temporal association of CTL appearance with the resolution of primary viremia in human immunodeficiency virus type 1 (HIV-1)-infected humans (9, 24, 33) and by monoclonal anti-CD8 antibody-mediated CD8-depletion experiments in macaque AIDS models (18, 29, 38). Therefore, AIDS vaccine researchers have been making efforts to develop methods efficiently eliciting CTL responses (15, 30), and most of them have used multiple antigens for CTL induction (3, 8). However, it has remained unclear if multiple epitope-specific CTLs can really take part in vaccine-based control of viral replication.
Several preclinical trials of CTL-based AIDS vaccines in macaques have succeeded in the control of replication of a simian-human immunodeficiency virus, SHIV89.6P, that induces acute CD4+ T-cell depletion (3, 8, 27, 37, 40). Unfortunately, most of these vaccine regimens have failed to contain the more realistic challenge of pathogenic simian immunodeficiency viruses (SIVs) that induce chronic disease progression (12, 17). Recently, however, CTL-based control of replication of a pathogenic SIV clone, SIVmac239, has been shown in a preclinical vaccine trial using Burmese rhesus macaques (28). In that study, macaques immunized with a DNA prime/Gag-expressing Sendai virus (SeV-Gag) vector-boost vaccine were challenged intravenously with SIVmac239. Five of eight vaccinees controlled viral replication and had undetectable levels of plasma viremia after 5 weeks of infection. All of the five macaques showed rapid selection of CTL escape mutations in gag, indicating that vaccine-induced CTLs were crucial for the containment of the wild-type, challenge virus. Of the five, three vaccinees that share a major histocompatibility complex class I (MHC-I) haplotype, 90-120-Ia, showed high levels of Gag206-216 (IINEEAADWDL) epitope-specific CTL and rapid selection of a mutant escaping from this CTL. The virus with the CTL escape mutation, GagL216S, leading to an alteration from leucine (L) to serine (S) at the 216th amino acid (aa) in Gag showed diminished replicative ability compared to the wild type. Inoculation of naive macaques with this mutant resulted in persistent viral replication and reversion in the absence of the Gag206-216-specific CTL responses (23). These results have suggested that additional adaptive immune responses as well as Gag206-216-specific CTLs are important for containment of this CTL escape mutant virus with lower viral fitness.
Viral escape from CTL recognition has been frequently observed in HIV-1 and SIV infections, and it may be critical for viral evasion from immune control (5, 6, 10, 15, 16, 32, 35, 36). Indeed, viral evasion from immune control with a single escape mutation from a dominant CTL has been reported in preclinical AIDS vaccine trials, indicating involvement of the single epitope-specific CTL in this control (5, 6). However, these reports have not made it clear whether multiple epitope-specific CTLs can be involved in the vaccine-based control of immunodeficiency virus replication.
In the present study, we have followed, for more than 2 years, the five macaques that showed vaccine-based control of SIVmac239 replication. We have found that three of them maintained control of viral replication for more than 2 years while the other two lost control at approximately week 60 after challenge. Analysis of the latter two has revealed viral evasion from the vaccine-based control by accumulation of multiple CTL escape mutations, indicating involvement of multiple epitope-specific CTLs in this control.
MATERIALS AND METHODS
Animal experiments.
Twelve male Burmese rhesus macaques (Macaca mulatta) used in our previous SIVmac239 challenge experiment (28) were followed up in the present study. These macaques were maintained in accordance with the guidelines for laboratory animals of the National Institute of Infectious Diseases. Blood collection, vaccination, and virus challenge were performed under ketamine anesthesia. Four of the macaques were naive whereas the other eight macaques received a DNA vaccine followed by a single boost with SeV-Gag before an intravenous SIVmac239 challenge. The DNA, CMV-SHIVdEN, used for the vaccination was constructed from an env- and nef-deleted SHIVMD14YE molecular clone DNA (39) and has the genes encoding SIVmac239 Gag, Pol, Vif, and Vpx; SIVmac239-HIV-1DH12 chimeric Vpr; and HIV-1DH12 Tat and Rev as described previously (28). At the DNA vaccination, animals received 5 mg of CMV-SHIVdEN DNA intramuscularly. Six weeks after the DNA prime, animals intranasally received a single boost with 1 × 108 cell infectious units of replication-competent SeV-Gag (V1, V2, V3, and V4) or 6 × 109 cell infectious units of F-deleted replication-defective F(−)SeV-Gag (19, 20, 26, 41). Thirteen weeks after the boost, animals were challenged intravenously with 1,000 50% tissue culture infective doses of SIVmac239 (22).
Quantitation of plasma viral loads.
Plasma RNA was extracted using the High Pure viral RNA kit (Roche Diagnostics, Tokyo, Japan). Serial fivefold dilutions of RNA samples were amplified in quadruplicate by reverse transcription (RT) and nested PCR using SIV gag-specific primers (AGAAACTCCGTCTTGTCAGG and TGATAATCTGCATAGCCGC for the first RT-PCR and GATTAGCAGAAAGCCTGTTGG and TGCAACCTTCTGACAGTGC for the second DNA PCR) to determine the endpoint. Plasma SIV RNA levels were calculated according to the Reed-Muench method as described previously (28, 39). The lower limit of detection in this standard assay is about 4 × 102 copies/ml. For fivefold concentration of plasma, after centrifugation of 1 ml of plasma at 25,000 × g for 2 h, 0.8 ml of its supernatant was discarded and the remaining 0.2 ml was subjected to RNA extraction.
Sequencing.
Fragments corresponding to nucleotides (nt) 1231 to 2958 (containing the entire gag region), nt 2827 to 3960, nt 3811 to 4970, nt 4829 to 5986, nt 5852 to 7000, nt 6843 to 7901, nt 7684 to 8831, nt 8677 to 9723, and nt 9499 to 10196 in the SIVmac239 genome (GenBank accession number M33262) were amplified by nested RT-PCR. Alternatively, genomic DNA was extracted from peripheral blood mononuclear cells (PBMCs) by using the DNeasy kit (QIAGEN K.K., Tokyo, Japan), and the gag fragment was amplified by nested PCR. The PCR products were sequenced using dye terminator chemistry and an automated DNA sequencer (Applied Biosystems, Tokyo, Japan). Alternatively, the PCR products were subcloned into plasmids by using the TOPO cloning system (Invitrogen, Tokyo, Japan) and sequenced.
Peptide-specific CTL responses.
We measured virus-specific T-cell levels by flow cytometric analysis of gamma interferon (IFN-γ) induction after specific stimulation as described previously (28). In brief, PBMCs were cocultured with autologous herpesvirus papio-immortalized B-lymphoblastoid cell lines (B-LCL) (42) pulsed with 1 μM or indicated concentrations of peptides (Sigma Genosys, Ishikari, Japan) for peptide-specific stimulation or unpulsed B-LCL for nonspecific stimulation. Intracellular IFN-γ staining was performed by using the Cytofix-Cytoperm kit (Becton Dickinson, San Jose, California). Peridinin chlorophyll protein-conjugated anti-human CD8, allophycocyanin-conjugated anti-human CD3, and phycoerythrin-conjugated anti-human IFN-γ antibodies (Becton Dickinson) were used. Specific T-cell levels were calculated by subtracting the IFN-γ+ T-cell frequencies after nonspecific stimulation from those after peptide-specific stimulation. Specific T-cell levels less than 100 cells per million PBMCs were considered negative.
Generation of CTL clones and CTL assay.
Gag206-216-specific and Gag241-249-specific CTL clones were obtained from macaque V5 PBMCs cocultured with irradiated, V5-derived B-LCL pulsed with the corresponding peptides. Cytotoxicity was measured in a standard 51Cr release assay. In brief, target cells (5 × 105) were incubated with 150 μCi Na251CrO4 for 1 h, pulsed with the corresponding peptides for 1 h, and cocultured with effector cells for 4 h. The culture supernatants were analyzed with a gamma counter. The spontaneous 51Cr release (cpm spn) was determined by measuring the 51Cr release from the culture containing only target cells. The maximum release (cpm max) was determined by measuring the 51Cr release from target cells in the presence of 2.5% Triton X-100. Percent specific lysis was calculated as follows: percent specific lysis = 100 × (cpm exp − cpm spn)/(cpm max − cpm spn), where cpm exp is the 51Cr release from the culture containing both target and effector cells.
Viral competition assay.
SIV molecular clone DNAs with mutations in gag were constructed by site-directed mutagenesis from the wild-type SIV molecular clone DNA pBRmac239, provided by T. Kodama and R. C. Desrosiers. COS1 cells were transfected with mutant SIV molecular DNAs to obtain mutant SIV stocks. Two million cells of a herpesvirus saimiri-immortalized macaque T-cell (MTC) line (1) were infected with one of the mutant SIVs at the dose of 2 ng of SIV CA (p27), and 1 day later, half of them were cocultured with those infected with another mutant SIV. Two million MTCs were added into the culture on days 8, 12, 16, and 20 after infection. RNA was extracted from the culture supernatant on day 24. The fragment (nt 1231 to nt 3016 in SIVmac239) containing the entire gag region was amplified from the RNA by RT-PCR and was subcloned into plasmids for sequencing to determine dominant sequences.
RESULTS
Reappearance of viremia after 1 year of control in two of the five controllers.
Twelve Burmese rhesus macaques used in our previous SIVmac239 challenge experiment (28) were followed up in the present study (Table 1). Of the 12, eight macaques descended from a male breeder, R-90-120, and four of them shared an MHC-I haplotype, 90-120-Ia. Four macaques were naive whereas eight macaques received a DNA vaccine followed by a single boost with SeV-Gag before an intravenous SIVmac239 challenge. All four naive animals and three of the vaccinees failed to control SIV replication, but five of eight vaccinees controlled SIV replication with undetectable levels of plasma viremia (less than 400 RNA copies/ml) after 5 weeks of infection. We have termed the former seven animals noncontrollers and the latter five controllers in the present study.
TABLE 1.
SIVmac239 challenge experiments
| Macaque | MHC-I haplotypea | Naive or vaccineeb | Set point VLc around wk 12 | CTL escaped at wk 5 | VL around wk 60 |
|---|---|---|---|---|---|
| R-90-120 descendants | |||||
| N2 | 90-120-Ia | Naive | 104-106 | 104-106 | |
| V5 | 90-120-Ia | Vaccinee | <400 | GagL216S | >103 |
| V3 | 90-120-Ia | Vaccinee | <400 | GagL216S | >103 |
| V4 | 90-120-Ia | Vaccinee | <400 | GagL216S | <400 |
| V2 | 90-120-Ib | Vaccinee | 104-106 | Deade | |
| N3 | 90-122-Ie | Naive | 104-106 | 104-106 | |
| V7 | 90-122-Ie | Vaccinee | 104-106 | 104-106 | |
| V6 | 90-122-Ie | Vaccinee | <400 | GagI377T | <400 |
| R-90-088 descendants | |||||
| N1 | 90-088-Ij | Naive | 104-106 | 104-106 | |
| V1 | 90-088-Ij | Vaccinee | 104-106 | 104-106 | |
| R-90-010 descendants | |||||
| N4 | 90-010-Id | Naive | 104-106 | 104-106 | |
| V8 | 90-010-Id | Vaccinee | <400 | GagQ58K | <400 |
MHC-I haplotype was determined by reference strand-mediated conformation analysis (4) as described previously (28). Macaques N2, V3, and V2 are sons of male breeder R-90-120; V5, V4, N3, V7, and V6 are sons of R-94-027; N1 and V1 are sons of R-90-088; N4 and V8 are sons of R-90-010. Breeder R-94-027 is the son of male R-90-120 and female R-90-122 and possesses 90-120-Ia and 90-122-Ie haplotypes. MHC-I haplotypes 90-120-Ia and 90-120-Ib are derived from breeder R-90-120, 90-122-Ie is from R-90-122, 90-088-Ij is from R-90-088, and 90-010-Id is from R-90-010.
All the animals were challenged intravenously with SIVmac239. Vaccinees received a prophylactic DNA prime/SeV-Gag boost vaccine before challenge.
Plasma viral load (RNA copies/ml plasma). VL, viral load.
Rapidly selected CTL escape mutations in Gag as described previously (28).
Macaques N3, V1, V2, and V7 developed AIDS and were euthanized at weeks 104, 105, 42, and 77, respectively.
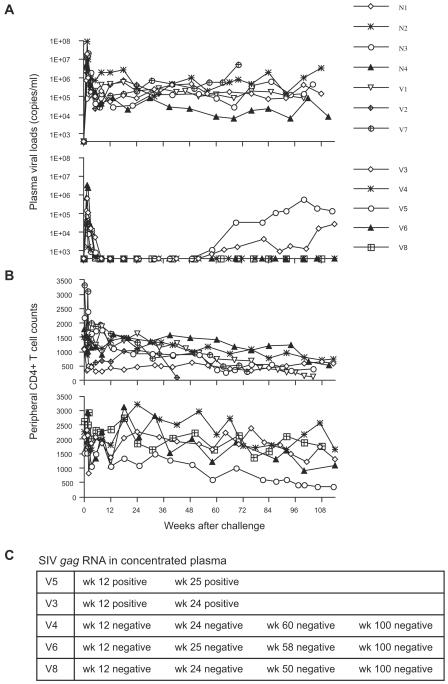
During 2 years of follow-up, all the seven noncontrollers maintained high levels of plasma viremia (Fig. 1A). Four of them developed AIDS and had to be euthanized. By contrast, plasma viremia was undetectable and peripheral CD4+ T-cell counts were maintained even after 2 years of infection in three (V4, V6, and V8) of five controllers (Fig. 1A and B). In the other two controllers (V5 and V3), however, plasma viremia reappeared and was detectable (more than 400 RNA copies/ml) at week 58 after challenge (Fig. 1A). Thus, three of five controllers maintained control of SIV replication for more than 2 years, whereas the other two controllers lost control after 1 year of infection. We have termed the former three animals sustained controllers and the latter two transient controllers in the present study.
FIG. 1.
Follow-up of 12 macaques after SIVmac239 challenge. (A) Plasma viral loads. Top, noncontrollers; bottom, controllers. (B) Peripheral CD4+ T-cell counts (per μl). Top, noncontrollers; bottom, controllers. (C) Detection of viral genomes in concentrated plasma obtained from the controllers. Positive, detected (>8 × 101 copies/ml); negative, undetectable.
Of four macaques possessing the MHC-I haplotype 90-120-Ia, all of the three vaccinees, V5, V3, and V4, successfully controlled SIV replication, although one naive macaque, N2, failed. Remarkably, two of the three controllers possessing 90-120-Ia lost control around week 60.
We examined viral loads in the controllers by detection of viral genomes in concentrated plasma (Fig. 1C). The cutoff line of this assay is about 80 RNA copies/ml whereas that of our standard assay for quantitation of plasma viral RNA is approximately 400 RNA copies/ml. In both of the transient controllers, viral RNA was detected in the concentrated plasma during the period of control although it was undetectable by our standard assay. In contrast, viral RNA was undetectable even in the concentrated plasma in all of the sustained controllers. These results indicate that SIV replication was contained to much lower levels in the sustained controllers compared to the rather high levels in the transient controllers.
Viral mutations in the transient controllers.
The previous study (28) showed rapid selection of CTL escape mutations in gag in all of the controllers (Table 1), indicating the importance of the CTL responses in the control of SIV replication. We then examined gag sequences to see if additional viral mutations were involved in the loss of control in the transient-controllers (Table 2). In a sustained controller (V4) possessing the MHC-I haplotype 90-120-Ia, we observed rapid selection of the GagL216S mutation leading to escape from Gag206-216-specific CTL responses (referred to as Gag206-216-CTL-escape mutation) both in plasma viral RNA and in proviral DNA of PBMCs. This mutation was maintained, but no other mutation became dominant even at week 85. In the other two sustained controllers (V6 and V8), the rapidly selected CTL escape mutations were observed in viral RNA but not in proviral DNA. This may reflect the possibility that accumulated mutant copies were too small for their detection in provirus compared to the wild type in these two macaques.
TABLE 2.
Dominant sequences in Gag in the five controllers
| Macaque | Wk | Sample | Amino acid change(s) in Gaga |
|---|---|---|---|
| V5 | 5 | Plasma viral RNA | L216S |
| 58 | Plasma viral RNA | L216S, D244E, I247L, A312V, A373T | |
| V3 | 5 | Plasma viral RNA | L216S |
| 64 | Plasma viral RNA | (V145A), (P172S), L216S, D244E, (V375A), (P376S) | |
| V4 | 5 | Plasma viral RNA | L216S |
| 12 | PBMC proviral DNA | L216S | |
| 85 | PBMC proviral DNA | L216S | |
| V6 | 5 | Plasma viral RNA | I377T |
| 12 | PBMC proviral DNA | No mutation | |
| 100 | PBMC proviral DNA | No mutation | |
| V8 | 5 | Plasma viral RNA | Q58K |
| 12 | PBMC proviral DNA | No mutation | |
| 100 | PBMC proviral DNA | No mutation |
Fragments containing the SIV gag region were amplified by nested RT-PCR and subjected to sequencing. Dominant mutations leading to amino acid changes are shown. The parentheses indicate that both the wild-type and the mutant sequences were detected clearly at the position.
In both of the transient controllers (V5 and V3) possessing the MHC-I haplotype 90-120-Ia, the Gag206-216-CTL-escape mutation was rapidly selected and still maintained at approximately week 60. In contrast to the sustained controllers, we found multiple additional mutations in the reemerged viruses in both of these macaques. In macaque V5, viral genomes with GagL216S, GagD244E (aspartic acid [D]-to-glutamic acid [E] alteration at the 244th aa in Gag), GagI247L (isoleucine [I] to L at the 247th aa), GagA312V (alanine [A] to valine [V] at the 312th aa), and GagA373T (A to threonine [T] at the 373rd aa) mutations were dominant at week 58. In macaque V3, viral genomes with GagV145A (V to A at the 145th aa), GagL216S, GagD244E, and GagP376S (proline [P] to serine[S] at the 376th aa) mutations were dominant, but those with GagP172S (P to S at the 172nd aa), GagL216S, GagD244E, and GagV375A (V to A at the 375th aa) mutations were also detected at week 64.
We then examined gag sequences during control in both of the transient controllers (Tables 3 and 4). This analysis showed that, in addition to the GagL216S mutation, the GagD244E mutation was initially selected, followed by selection of the mutations leading to alterations around the 375th aa in Gag in both of these macaques. In this regard, the two transient controllers showed similar patterns of sequential accumulation of mutations.
TABLE 3.
Accumulation of mutations in macaque V5
| Wk | Sample | Frequencya | Amino acid change(s) in Gagb |
|---|---|---|---|
| 5 | Plasma | 10/10 | L216S |
| Viral RNA | |||
| 18 | PBMC | 7/10 | L216S, D244E |
| Proviral DNA | 3/10 | L216S, D244E, A373T | |
| 32 | PBMC | 6/11 | L216S, D244E, A373T |
| Proviral DNA | 5/11 | L216S | |
| 58 | Plasma | 8/10 | L216S, D244E, I247L, A312V, A373T |
| Viral RNA | 2/10 | V145A, L216S, D244E, I247L, A312V, A373T |
Number of clones with change(s)/total number of clones.
Amplified gag fragments were subcloned into plasmids for sequencing. In general, mutations detected more than once are shown.
TABLE 4.
Accumulation of mutations in macaque V3
| Wk | Sample | Frequencya | Amino acid change(s) in Gagb |
|---|---|---|---|
| 5 | Plasma | 10/10 | L216S |
| Viral RNA | |||
| 24 | Concentrated plasma | 2/9 | L216S |
| Viral RNAc | 1/9 | L216S, D244E | |
| 3/9 | L216S, D244E, V375A | ||
| 2/9 | L216S, D244E, V375M | ||
| 1/9 | L216S, D244E, V375I | ||
| 64 | Plasma | 8/10 | V145A, L216S, D244E, P376S |
| Viral RNA | 2/10 | P172S, L216S, D244E, V375A |
Number of clones with change(s)/total number of clones.
Amplified gag fragments were subcloned into plasmids for sequencing. In general, mutations detected more than once are shown.
We successfully obtained the gag fragments for sequencing from concentrated plasma in macaque V3 although we failed to amplify them in macaque V5 during the period of viral control.
Accumulation of CTL escape mutations in the transient controllers.
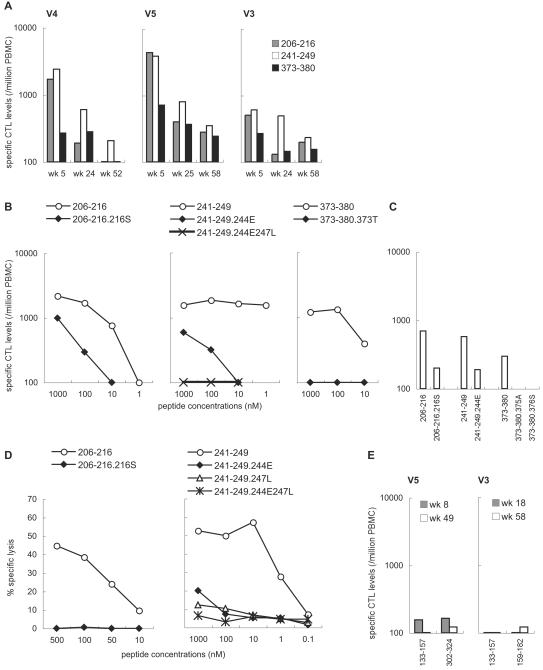
To see if the mutations observed in the transient controllers were CTL escape mutations, we examined IFN-γ induction after stimulation with peptides corresponding to the regions around the mutation sites. In addition to the Gag206-216 epitope, we mapped two CTL epitopes, Gag241-249 (SSVDEQIQW) and Gag373-380 (APVPIPFA). High levels of these three epitope-specific (Gag206-216-specific, Gag241-249-specific, and Gag373-380-specific) CTL responses were observed in all the three controllers possessing MHC-I haplotype 90-120-Ia in the early phase of infection (Fig. 2A). The Gag206-216-specific and Gag241-249-specific CTL responses were especially dominant. These CTL levels were considerably reduced in the chronic phase, probably reflecting diminished SIV replication during the control. Reduction in Gag206-216-specific CTL responses was faster, consistent with the fastest selection of the Gag206-216-CTL-escape mutation.
FIG. 2.
CTL responses in the controllers (V4, V5, and V3) possessing MHC-I haplotype 90-120-Ia. (A) Gag206-216-specific, Gag241-249-specific, and Gag373-380-specific CTL levels in the macaques V4, V5, and V3. (B) IFN-γ induction in macaque V5 after stimulation with the wild-type or the mutant peptides. In the left panel, PBMCs obtained at 2 weeks after SeV-Gag boost were stimulated by coculture with B-LCL pulsed with indicated concentrations of the wild-type Gag206-216-epitope peptide (206-216, IINEEAADWDL) or the mutant peptide with an L216S alteration (206-216.216S, IINEEAADWDS) corresponding to the 206th to 216th aa in Gag. In the middle panel, PBMCs at 2 weeks after SeV-Gag boost were stimulated by coculture with B-LCL pulsed with the wild-type Gag241-249-epitope peptide (241-249, SSVDEQIQW), the mutant peptide with a D244E alteration (241-249.244E, SSVEEQIQW), or the mutant peptide with D244E and I247L alterations (241-249.244E247L, SSVEEQLQW) corresponding to the 241st to 249th aa in Gag. In the right panel, PBMCs at 1 week after SeV-Gag boost were stimulated by coculture with B-LCL pulsed with the wild-type Gag373-380 epitope peptide (373-380, APVPIPFA) or the mutant peptide with an A373T alteration (373-380.373T, TPVPIPFA) corresponding to the 373rd to 380th aa in Gag. (C) IFN-γ induction in macaque V3 after stimulation with the wild-type or the mutant peptides. PBMCs at week 5 (206-216, 206-216.216S, 373-380, 373-380.375A, and 373-380.376S) or week 8 (241-249 and 241-249.244E) after challenge were used. (D) Recognition of wild-type and mutant epitope peptides by Gag206-216-specific and Gag241-249-specific CTL clones. In the left panel, the cytotoxic activities of a Gag206-216-specific CTL clone for target cells pulsed with the wild-type Gag206-216 epitope (206-216) or the L216S mutant epitope (206-216.216S) peptide were measured at an effector-to-target ratio (E:T) of 2:1. In the right panel, the cytotoxic activities of a Gag241-249-specific CTL clone for target cells pulsed with the wild-type Gag241-249 epitope (241-249) or mutant epitope peptides with D244E (241-249.244E), I247L (241-249.247L), or D244E-I247L (241-249.244E247L) alterations were measured at an E:T of 2:1. (E) CTL responses to the peptides corresponding to the region around the sites of GagV145A, GagP172S, and GagA312V mutations. PBMCs were cocultured with B-LCL pulsed with a mixture of peptides corresponding to the 133rd to 147th, 137th to 153rd, and 143rd to 157th aa in Gag; those corresponding to the 159th to 174th, 164th to 178th, and 168th to 182nd aa in Gag; or those corresponding to the 302nd to 316th, 306th to 320th, and 310th to 324th aa in Gag for Gag133-157-specific (133-157), Gag159-182-specific (159-182), or Gag302-324-specific (302-324) stimulation.
Both of the transient controllers (V5 and V3) showed diminished recognition of the peptide with the GagD244E mutation, Gag241-249.244E (SSVEEQIQW), by Gag241-249-specific CTL responses (Fig. 2B and 2C). The peptide with the GagI247L mutation in addition to the GagD244E (SSVEEQLQW) showed further-reduced sensitivity to CTL recognition. This indicates that the GagD244E and GagI247L mutations were selected for by Gag241-249-specific CTLs (referred to as Gag241-249-CTL-escape mutations). Furthermore, the GagA373T, GagV375A, and GagP376S mutations in the Gag373-380 peptide (APVPIPFA) resulted in diminished recognition by Gag373-380-specific CTL responses (Fig. 2B and 2C), indicating that the GagA373T, GagV375A, and GagP376S mutations were selected for by Gag373-380-specific CTLs (referred to as Gag373-380-CTL-escape mutations). Thus, viruses in both of the transient controllers accumulated the Gag241-249-CTL-escape mutation and the Gag373-380-CTL-escape mutation in addition to the Gag206-216-CTL-escape mutation. Additionally, we obtained a Gag206-216-specific CTL clone and a Gag241-249-specific CTL clone and confirmed these escapes (Fig. 2D).
To determine if the remaining mutations, GagV145A, GagP172S, and GagA312V, that were observed in the reemerged viruses were within CTL epitope regions, we further examined IFN-γ induction after stimulation with peptide mixtures corresponding to the 133rd to 157th aa, the 159th to 182nd aa, and the 302nd to 324th aa, respectively. The responses were at marginal levels (Fig. 2E), and we were unable to determine whether these mutations were selected for by CTLs.
Loss of viral fitness by the accumulated mutations.
Next, we examined the effect of the mutations observed in viruses from the transient controllers on viral fitness. We constructed three groups of mutant SIV clones from an SIVmac239 molecular clone by site-directed mutagenesis as shown in Table 5. The group P virus (P1), SIVmac239Gag216S, contains a single CTL escape mutation selected in 5 weeks in both macaques V5 and V3 and has diminished replicative ability compared to the wild-type SIVmac239 as described previously (28). The group Q viruses have the Gag206-216-CTL-escape, Gag241-249-CTL-escape, and Gag373-380-CTL-escape mutations. The group R viruses contain the four or five mutations dominant in the reemerged viruses.
TABLE 5.
List of SIV mutants
| Group and abbreviationa | Name | Amino acid change(s) in Gag | Macaque(s) in which selected |
|---|---|---|---|
| P | |||
| P1 | SIVmac239Gag216S | L216S | V5 and V3 |
| Q | |||
| Q1 | SIVmac239Gag216S244E373T | L216S, D244E, A373T | V5 |
| Q2 | SIVmac239Gag216S244E375A | L216S, D244E, V375A | V3 |
| Q3 | SIVmac239Gag216S244E376S | L216S, D244E, P376S | V3 |
| R | |||
| R1 | SIVmac239Gag216S244E247L312V373T | L216S, D244E, I247L, A312V, A373T | V5 |
| R2 | SIVmac239Gag172S216S244E375A | L216S, D244E, V375A, P172S | V3 |
| R3 | SIVmac239Gag145A216S244E376S | L216S, D244E, P376S, V145A | V3 |
Group P, Gag206-216-CTL-escape mutant rapidly selected in 5 weeks; group Q, Gag206-216-, Gag241-249-, and Gag373-380-CTL escape mutants; group R, mutants selected in the reemerged viruses.
We then compared viral fitness of the mutant viruses by determination of dominant viruses in the coculture of mutant virus-infected cells with cells infected by another mutant (Table 6). The competitions between groups P and Q revealed that the group Q viruses with Gag206-216-CTL-escape, Gag241-249-CTL-escape, and Gag373-380-CTL-escape mutations showed lower viral fitness than did group P with a single Gag206-216-CTL-escape mutation, indicating that additions of Gag241-249-CTL-escape and Gag373-380-CTL-escape mutations reduced viral fitness. The competitions between groups Q and R did not show recovery of viral fitness by the GagI247L, GagA312V, GagP172S, or GagV145A mutation. Consistent with these results, the group R viruses showed lower viral fitness than did the group P virus. Thus, CTLs from both of the transient controllers (V5 and V3) selected for Gag241-249-CTL-escape and Gag373-380-CTL-escape mutations in addition to the Gag206-216-CTL-escape mutation with viral fitness costs. Viruses with the Gag mutations observed at viremia reappearance showed lower viral fitness than did the SIVmac239Gag216S selected in 5 weeks of infection.
TABLE 6.
Competition between SIV mutantsa
| Competi- tion no. | SIV mutant used | Amino acid mutation(s) | Frequencyb |
|---|---|---|---|
| 1 | P1 | L216S | 13/17 |
| Q1 | L216S, D244E, A373T | 2/17 | |
| L216S, D244E, | 1/17 | ||
| L216S, A373T | 1/17 | ||
| 2 | P1 | L216S | 15/15 |
| R1 | L216S, D244E, I247L, A312V, A373T | 0/15 | |
| 3 | Q1 | L216S, D244E, A373T | 12/14 |
| R1 | L216S, D244E, I247L, A312V, A373T | 1/14 | |
| L216S, D244E, A312V, A373T | 1/14 | ||
| 4 | P1 | L216S | 11/12 |
| Q2 | L216S, D244E, V375A | 0/12 | |
| L216S, V375A | 1/12 | ||
| 5 | P1 | L216S | 11/15 |
| R2 | P172S, L216S, D244E, V375A | 0/15 | |
| L216S, V375A | 3/15 | ||
| P172S, L216S, V375A | 1/15 | ||
| 6 | Q2 | L216S, D244E, V375A | 8/12 |
| R2 | P172S, L216S, D244E, V375A | 4/12 | |
| 7 | P1 | L216S | 12/12 |
| Q3 | L216S, D244E, P376S | 0/12 | |
| 8 | P1 | L216S | 7/12 |
| R3 | V145A, L216S, D244E, P376S | 0/12 | |
| V145A, L216S | 1/12 | ||
| L216S, D244E | 1/12 | ||
| L216S, P376S | 1/12 | ||
| V145A, L216S, D244E | 1/12 | ||
| L216S, D244E, P376S | 1/12 | ||
| 9 | Q3 | L216S, D244E, P376S | 7/12 |
| R3 | V145A, L216S, D244E, P376S | 5/12 |
MTCs infected with one SIV mutant were cocultured with those infected with another SIV mutant. RNA was extracted from the culture supernatant on day 24 after infection, and the gag fragment amplified from the RNA was subcloned into plasmids for sequencing.
Number of clones with mutation(s)/total number of clones.
DISCUSSION
In the present study, we have followed five rhesus macaques that showed vaccine-based control of SIVmac239 replication in a preclinical trial of a CTL-based AIDS vaccine (28). Two of them showed increases in plasma viral loads after 1 year of control, but the other three maintained the control without detectable plasma viremia for more than 2 years. This result suggests that vaccine induction of CTLs can result in sustained control of immunodeficiency virus replication.
Among the five macaques we followed, three (V5, V3, and V4) shared an MHC-I haplotype, 90-120-Ia, and rapidly selected for a Gag206-216-specific CTL-escape mutant by 5 weeks after challenge. Among these three, one macaque (V4) maintained this control without additional mutations in the provirus, while the other two (V5 and V3) accumulated viral mutations and lost control with reappearance of plasma viremia (more than 400 RNA copies/ml). Because the rapidly selected Gag206-216-CTL-escape mutant virus with the GagL216S mutation showed diminished replicative ability, it was expected that the additional mutations accumulated in macaques V5 and V3 might contribute to recovery of viral fitness. Indeed, some CTL escape mutant viruses with lower viral fitness are known to require additional compensatory mutations to restore their replicative competence (13, 21, 34, 43). However, our results have revealed that mutations accumulated in macaques V5 and V3 did not result in recovery of viral fitness. Viruses accumulated the Gag241-249-CTL-escape mutation (GagD244E) and the Gag373-380-CTL-escape mutation (GagA373T, GagV375A, or GagP376S) with viral fitness costs. Therefore, escape from Gag241-249-specific and Gag373-380-specific CTLs as well as Gag206-216-specific CTLs was essential in the process of viral evasion from the control. This suggests that these three epitope-specific (Gag206-216-specific, Gag241-249-specific, and Gag373-380-specific) CTL responses were crucial for the control in these macaques. This is the first evidence indicating multiple epitope-specific CTL-based control of SIV replication.
It remains unclear what determines the time and the order of appearance of CTL escape mutations. These may be influenced by CTL levels and selective pressure, viral fitness costs by mutations, and mutation rates (T-to-C change in L216S mutation, T-to-G in D244E, G-to-A in A373T, T-to-C in V375A, and C-to-T in P376S). In macaques V5 and V3, Gag206-216-specific and Gag241-249-specific CTL responses were detected dominantly in the early phase of SIV infection, and the Gag206-216-CTL-escape and Gag241-249-CTL-escape mutations were selected for first. These results might suggest that Gag206-216-specific and Gag241-249-specific CTL responses played a central role in the control of SIV replication in both of these macaques. Interestingly, the SIV Gag241-249 epitope (SSVDEQIQW) is homologous to the HLA-B57/5801-restricted CTL epitope, TW10 (TSTLQEQIAW), in HIV-1 Gag (Gag240-249). Like the D244E mutation within the SIV Gag241-249 epitope, an escape mutation within the HIV-1 Gag TW10 epitope has been reported to be selected for with viral fitness costs by this TW10-specific CTL (25). Thus, this region in Gag CA could be a promising epitope candidate for CTL-based AIDS vaccines.
The viruses that reemerged around week 60 in macaques V5 and V3 had other Gag mutations (GagA312V in V5 and GagV145A or GagP172S in V3) in addition to the Gag206-216-CTL-escape, the Gag241-249-CTL-escape, and the Gag373-380-CTL-escape mutations. Our results did not show recovery of viral fitness by these mutations, either, although we failed to determine whether these mutations might result in evasion from another epitope-specific CTL response. Importantly, viruses with the Gag mutations observed at viremia reappearance showed lower replicative ability than did the SIVmac239Gag216S selected around week 5. Therefore, it is inferred that the viruses with lower viral fitness can replicate to detectable levels in plasma because of their evasion from multiple epitope-specific CTL responses essential for this control. Whereas Barouch et al. (5, 6) reported a single CTL escape mutation followed by viral breakthrough (viremia recrudescence) in SHIV89.6P and SIVsmE660 infection, our results indicate that accumulation of multiple CTL escape mutations can result in viral breakthrough from the vaccine-based control of SIVmac239 replication.
In a sustained controller (V4) sharing the MHC-I haplotype 90-120-Ia with macaques V5 and V3, Gag206-216-specific CTL responses are considered to be involved in the sustained control even at week 85, because the GagL216S mutation was maintained without reversion (7, 11, 14, 23, 25). In addition, Gag241-249-specific and Gag373-380-specific CTLs are expected to play an important role in this control, and failure in accumulating Gag241-249-CTL-escape and Gag373-380-CTL-escape mutations may be associated with the sustained control. In contrast, it is inferred that, in macaques V5 and V3, viruses were allowed to accumulate CTL escape mutations leading to reappearance of plasma viremia. The magnitude of Gag206-216-specific, Gag241-249-specific, Gag373-380-specific, or total Gag-specific CTL responses did not appear to correlate with the level of control (Fig. 2) (25). It may be that, in macaque V4, additional effective CTLs that were not induced in V3 or V5 contributed to sustained control of SIV replication together with Gag206-216-specific, Gag241-249-specific, and Gag373-380-specific CTLs.
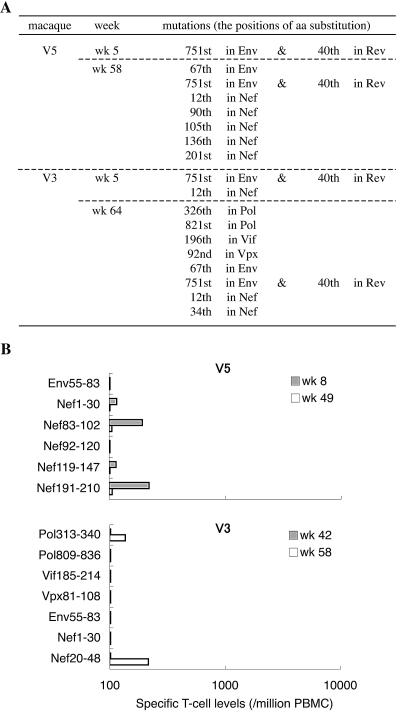
We focused on SIV gag sequences because we used a Gag-expressing vector for the boost in our vaccine system and because vaccine-induced CTL responses were detectable only to Gag (28). In macaques V5 and V3, however, we examined sequences of all of the viral protein coding regions in the SIV genomes at week 5 and around week 60 (Fig. 3). We found that a mutation leading to an arginine (R)-to-glycine (G) alteration at the 751st aa in Env and a lysine (K)-to-R alteration at the 40th aa in Rev was dominant at week 5 in both of them. The wild-type sequence at this position in the SIVmac239 molecular clone is considered to be a suboptimal nucleotide that frequently reverts to an alternative sequence in vivo (2, 31). Indeed, we found this mutation also in the noncontrollers, indicating no association of this mutation with viral control or evasion in the present study. At week 5, no other nonsynonymous mutation became dominant in macaque V5, while one additional mutation in nef was found in macaque V3. Around week 60, several additional mutations were dominant in both macaques. Positions of some of the mutations were within or around epitopes for CTLs, but those CTL responses were only at marginal levels. Even considering the possible contribution of some of these mutations in the viral genome outside gag to the loss of control, it is reasonable to conclude that escape from Gag206-216-specific, Gag241-249-specific, and Gag373-380-specific CTL responses was crucial for the viral evasion in macaques V5 and V3.
FIG. 3.
Mutations in viral genomes encoding SIV proteins other than Gag. (A) Viral mutations in macaques V5 and V3. Dominant mutations leading to amino acid changes are shown. (B) CTL responses to the peptides corresponding to the region around the mutation sites. PBMCs derived from macaque V5 at week 8 or 49 were stimulated by coculture with B-LCL pulsed with a mixture of peptides corresponding to the 55th to 83rd aa in Env (Env55-83), the 1st to 30th aa in Nef (Nef1-30), the 83rd to 102nd aa in Nef (Nef83-102), the 92nd to 120th aa in Nef (Nef92-120), the 119th to 147th aa in Nef (Nef119-147), or the 191st to 210th aa in Nef (Nef191-210). PBMCs from V3 at week 42 or 58 were stimulated by coculture with B-LCL pulsed with a mixture of peptides corresponding to the 313th to 340th aa in Pol (Pol313-340), the 809th to 836th aa in Pol (Pol809-836), the 185th to 214th aa in Vif (Vif185-214), the 81st to 108th aa in Vpx (Vpx81-108), Env55-83, Nef1-30, or the 20th to 48th aa in Nef (Nef20-48).
In summary, our follow-up study of macaques that showed vaccine-based control of primary SIV replication has revealed that sequential accumulation of multiple CTL escape mutations, if allowed, can result in viral evasion from this control. This finding indicates, for the first time, that multiple epitope-specific CTLs can be involved in control of immunodeficiency virus replication. This has an important implication for vaccine design, suggesting the rationale for eliciting multiple epitope-specific CTL responses to contain HIV replication.
Acknowledgments
This work was supported by a grant from the Ministry of Education, Culture, Sports, Science, and Technology; grants from the Japan Health Sciences Foundation; and grants from the Ministry of Health, Labor, and Welfare in Japan.
We thank T. Kodama and R. C. Desrosiers for providing an SIVmac239 molecular clone; Y. Ami, F. Ono, K. Komatsuzaki, A. Hiyaoka, A. Oyama, H. Ogawa, K. Hanari, K. Oto, H. Oto, H. Akari, and K. Terao for assistance in the animal experiments; and DNAVEC Corp., A. Kato, M. Kano, M. Miyazawa, M. Yasunami, A. Kimura, K. Mori, N. Yamamoto, T. Takemori, T. Sata, T. Kurata, A. Nomoto, and Y. Nagai for their help.
REFERENCES
- 1.Akari, H., K. Mori, K. Terao, I. Otani, M. Fukasawa, R. Mukai, and Y. Yoshikawa. 1996. In vitro immortalization of old world monkey T lymphocytes with herpesvirus saimiri: its susceptibility to infection with simian immunodeficiency viruses. Virology 218:382-388. [DOI] [PubMed] [Google Scholar]
- 2.Alexander, L., L. Denekamp, S. Czajak, and R. C. Desrosiers. 2001. Suboptimal nucleotides in the infectious, pathogenic simian immunodeficiency virus clone SIVmac239. J. Virol. 75:4019-4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amara, R. R., F. Villinger, J. D. Altman, S. L. Lydy, S. P. O'Neil, S. I. Staprans, D. C. Montefiori, Y. Xu, J. G. Herndon, L. S. Wyatt, M. A. Candido, N. L. Kozyr, P. L. Earl, J. M. Smith, H. L. Ma, B. D. Grimm, M. L. Hulsey, J. Miller, H. M. McClure, J. M. McNicholl, B. Moss, and H. L. Robinson. 2001. Control of a mucosal challenge and prevention of AIDS in rhesus macaques by a multiprotein DNA/MVA vaccine. Science 292:69-74. [DOI] [PubMed] [Google Scholar]
- 4.Arguello, J. R., A. M. Little, A. L. Pay, D. Gallardo, I. Rojas, S. G. Marsh, J. M. Goldman, and J. A. Madrigal. 1998. Mutation detection and typing of polymorphic loci through double-strand conformation analysis. Nat. Genet. 18:192-194. [DOI] [PubMed] [Google Scholar]
- 5.Barouch, D. H., J. Kunstman, J. Glowczwskie, K. J. Kunstman, M. A. Egan, F. W. Peyerl, S. Santra, M. J. Kuroda, J. E. Schmitz, K. Beaudry, G. R. Krivulka, M. A. Lifton, D. A. Gorgone, S. M. Wolinsky, and N. L. Letvin. 2003. Viral escape from dominant simian immunodeficiency virus epitope-specific cytotoxic T lymphocytes in DNA-vaccinated rhesus monkeys. J. Virol. 77:7367-7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barouch, D. H., J. Kunstman, M. J. Kuroda, J. E. Schmitz, S. Santra, F. W. Peyerl, G. R. Krivulka, K. Beaudry, M. A. Lifton, D. A. Gorgone, D. C. Montefiori, M. G. Lewis, S. M. Wolinsky, and N. L. Letvin. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature 415:335-339. [DOI] [PubMed] [Google Scholar]
- 7.Barouch, D. H., J. Powers, D. M. Truitt, M. G. Kishko, J. C. Arthur, F. W. Peyerl, M. J. Kuroda, D. A. Gorgone, M. A. Lifton, C. I. Lord, V. M. Hirsch, D. C. Montefiori, A. Carville, K. G. Mansfield, K. J. Kunstman, S. M. Wolinsky, and N. L. Letvin. 2005. Dynamic immune responses maintain cytotoxic T lymphocyte epitope mutations in transmitted simian immunodeficiency virus variants. Nat. Immunol. 6:247-252. [DOI] [PubMed] [Google Scholar]
- 8.Barouch, D. H., S. Santra, J. E. Schmitz, M. J. Kuroda, T. M. Fu, W. Wagner, M. Bilska, A. Craiu, X. X. Zheng, G. R. Krivulka, K. Beaudry, M. A. Lifton, C. E. Nickerson, W. L. Trigona, K. Punt, D. C. Freed, L. Guan, S. Dubey, D. Casimiro, A. Simon, M. E. Davies, M. Chastain, T. B. Strom, R. S. Gelman, D. C. Montefiori, M. G. Lewis, E. A. Emini, J. W. Shiver, and N. L. Letvin. 2000. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science 290:486-492. [DOI] [PubMed] [Google Scholar]
- 9.Borrow, P., H. Lewicki, B. H. Hahn, G. M. Shaw, and M. B. Oldstone. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 68:6103-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borrow, P., H. Lewicki, X. Wei, M. S. Horwitz, N. Peffer, H. Meyers, J. A. Nelson, J. E. Gairin, B. H. Hahn, M. B. Oldstone, and G. M. Shaw. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTL) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3:205-211. [DOI] [PubMed] [Google Scholar]
- 11.Brander, C., and B. D. Walker. 2003. Gradual adaptation of HIV to human host populations: good or bad news? Nat. Med. 9:1359-1362. [DOI] [PubMed] [Google Scholar]
- 12.Feinberg, M. B., and J. P. Moore. 2002. AIDS vaccine models: challenging challenge viruses. Nat. Med. 8:207-210. [DOI] [PubMed] [Google Scholar]
- 13.Friedrich, T. C., C. A. Frye, L. J. Yant, D. H. O'Connor, N. A. Kriewaldt, M. Benson, L. Vojnov, E. J. Dodds, C. Cullen, R. Rudersdorf, A. L. Hughes, N. Wilson, and D. I. Watkins. 2004. Extra-epitopic compensatory substitutions partially restore fitness to simian immunodeficiency virus variants that escape from an immunodominant cytotoxic T-lymphocyte response. J. Virol. 78:2581-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedrich, T. C., E. J. Dodds, L. J. Yant, L. Vojnov, R. Rudersdorf, C. Cullen, D. T. Evans, R. C. Desrosiers, B. R. Mothe, J. Sidney, A. Sette, K. Kunstman, S. Wolinsky, M. Piatak, J. Lifson, A. L. Hughes, N. Wilson, D. H. O'Connor, and D. I. Watkins. 2004. Reversion of CTL escape-variant immunodeficiency viruses in vivo. Nat. Med. 10:275-281. [DOI] [PubMed] [Google Scholar]
- 15.Goulder, P. J., and D. I. Watkins. 2004. HIV and SIV CTL escape: implications for vaccine design. Nat. Rev. Immunol. 4:630-640. [DOI] [PubMed] [Google Scholar]
- 16.Goulder, P. J., R. E. Phillips, R. A. Colbert, S. McAdam, G. Ogg, M. A. Nowak, P. Giangrande, G. Luzzi, B. Morgana, A. Edwards, A. J. McMichael, and S. Rowland-Jones. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212-217. [DOI] [PubMed] [Google Scholar]
- 17.Horton, H., T. U. Vogel, D. K. Carter, K. Vielhuber, D. H. Fuller, T. Shipley, J. T. Fuller, K. J. Kunstman, G. Sutter, D. C. Montefiori, V. Erfle, R. C. Desrosiers, N. Wilson, L. J. Picker, S. M. Wolinsky, C. Wang, D. B. Allison, and D. I. Watkins. 2002. Immunization of rhesus macaques with a DNA prime/modified vaccinia virus Ankara boost regimen induces broad simian immunodeficiency virus (SIV)-specific T-cell responses and reduces initial viral replication but does not prevent disease progression following challenge with pathogenic SIVmac239. J. Virol. 76:7187-7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin, X., D. E. Bauer, S. E. Tuttleton, S. Lewin, A. Gettie, J. Blanchard, C. E. Irwin, J. T. Safrit, J. Mittler, L. Weinberger, L. G. Kostrikis, L. Zhang, A. S. Perelson, and D. D. Ho. 1999. Dramatic rise in plasma viremia after CD8+ T cell depletion in simian immunodeficiency virus-infected macaques. J. Exp. Med. 189:991-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kano, M., T. Matano, A. Kato, H. Nakamura, A. Takeda, Y. Suzaki, Y. Ami, K. Terao, and Y. Nagai. 2002. Primary replication of a recombinant Sendai viral vector in macaques. J. Gen. Virol. 83:1377-1386. [DOI] [PubMed] [Google Scholar]
- 20.Kato, A., Y. Sakai, T. Shioda, T. Kondo, M. Nakanishi, and Y. Nagai. 1996. Initiation of Sendai virus multiplication from transfected cDNA or RNA with negative or positive sense. Genes Cells 1:569-579. [DOI] [PubMed] [Google Scholar]
- 21.Kelleher, A. D., C. Long, E. C. Holmes, R. L. Allen, J. Wilson, C. Conlon, C. Workman, S. Shaunak, K. Olson, P. Goulder, C. Brander, G. Ogg, J. S. Sullivan, W. Dyer, I. Jones, A. J. McMichael, and S. Rowland-Jones. 2001. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J. Exp. Med. 193:375-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kestler, H. W., III, D. J. Ringler, K. Mori, D. L. Panicali, P. K. Sehgal, M. D. Daniel, and R. C. Desrosiers. 1991. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell 65:651-662. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi, M., H. Igarashi, A. Takeda, M. Kato, and T. Matano. 2005. Reversion in vivo after inoculation of a molecular proviral DNA clone of simian immunodeficiency virus with a cytotoxic-T-lymphocyte escape mutation. J. Virol. 79:11529-11532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koup, R. A., J. T. Safrit, Y. Cao, C. A. Andrews, G. McLeod, W. Borkowsky, C. Farthing, and D. D. Ho. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68:4650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leslie, A. J., K. J. Pfafferott, P. Chetty, R. Draenert, M. M. Addo, M. Feeney, Y. Tang, E. C. Holmes, T. Allen, J. G. Prado, M. Altfeld, C. Brander, C. Dixon, D. Ramduth, P. Jeena, S. A. Thomas, A. St. John, T. A. Roach, B. Kupfer, G. Luzzi, A. Edwards, G. Taylor, H. Lyall, G. Tudor-Williams, V. Novelli, J. Martinez-Picado, P. Kiepiela, B. D. Walker, and P. J. Goulder. 2004. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 10:282-289. [DOI] [PubMed] [Google Scholar]
- 26.Li, H. O., Y. F. Zhu, M. Asakawa, H. Kuma, T. Hirata, Y. Ueda, Y. S. Lee, M. Fukumura, A. Iida, A. Kato, Y. Nagai, and M. Hasegawa. 2000. A cytoplasmic RNA vector derived from nontransmissible Sendai virus with efficient gene transfer and expression. J. Virol. 74:6564-6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matano, T., M. Kano, H. Nakamura, A. Takeda, and Y. Nagai. 2001. Rapid appearance of secondary immune responses and protection from acute CD4 depletion after a highly pathogenic immunodeficiency virus challenge in macaques vaccinated with a DNA prime/Sendai virus vector boost regimen. J. Virol. 75:11891-11896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matano, T., M. Kobayashi, H. Igarashi, A. Takeda, H. Nakamura, M. Kano, C. Sugimoto, K. Mori, A. Iida, T. Hirata, M. Hasegawa, T. Yuasa, M. Miyazawa, Y. Takahashi, M. Yasunami, A. Kimura, D. H. O'Connor, D. I. Watkins, and Y. Nagai. 2004. Cytotoxic T lymphocyte-based control of simian immunodeficiency virus replication in a preclinical AIDS vaccine trial. J. Exp. Med. 199:1709-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matano, T., R. Shibata, C. Siemon, M. Connors, H. C. Lane, and M. A. Martin. 1998. Administration of an anti-CD8 monoclonal antibody interferes with the clearance of chimeric simian/human immunodeficiency virus during primary infections of rhesus macaques. J. Virol. 72:164-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McMichael, A. J., and T. Hanke. 2003. HIV vaccines 1983-2003. Nat. Med. 9:874-880. [DOI] [PubMed] [Google Scholar]
- 31.O'Connor, D. H., A. B. McDermott, K. C. Krebs, E. J. Dodds, J. E. Miller, E. J. Gonzalez, T. J. Jacoby, L. Yant, H. Piontkivska, R. Pantophlet, D. R. Burton, W. M. Rehrauer, N. Wilson, A. L. Hughes, and D. I. Watkins. 2004. A dominant role for CD8+-T-lymphocyte selection in simian immunodeficiency virus sequence variation. J. Virol. 78:14012-14022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Connor, D. H., T. M. Allen, T. U. Vogel, P. Jing, I. P. DeSouza, E. Dodds, E. J. Dunphy, C. Melsaether, B. Mothe, H. Yamamoto, H. Horton, N. Wilson, A. L. Hughes, and D. I. Watkins. 2002. Acute phase cytotoxic T lymphocyte escape is a hallmark of simian immunodeficiency virus infection. Nat. Med. 8:493-499. [DOI] [PubMed] [Google Scholar]
- 33.Ogg, G. S., X. Jin, S. Bonhoeffer, P. R. Dunbar, M. A. Nowak, S. Monard, J. P. Segal, Y. Cao, S. L. Rowland-Jones, V. Cerundolo, A. Hurley, M. Markowitz, D. D. Ho, D. F. Nixon, and A. J. McMichael. 1998. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science 279:2103-2106. [DOI] [PubMed] [Google Scholar]
- 34.Peyerl, F. W., D. H. Barouch, W. W. Yeh, H. S. Bazick, J. Kunstman, K. J. Kunstman, S. M. Wolinsky, and N. L. Letvin. 2003. Simian-human immunodeficiency virus escape from cytotoxic T-lymphocyte recognition at a structurally constrained epitope. J. Virol. 77:12572-12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phillips, R. E., S. Rowland-Jones, D. F. Nixon, F. M. Gotch, J. P. Edwards, A. O. Ogunlesi, J. G. Elvin, J. A. Rothbard, C. R. Bangham, C. R. Rizza, and A. J. McMichael. 1991. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature 354:453-459. [DOI] [PubMed] [Google Scholar]
- 36.Price, D. A., P. J. Goulder, P. Klenerman, A. K. Sewell, P. J. Easterbrook, M. Troop, C. R. Bangham, and R. E. Phillips. 1997. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc. Natl. Acad. Sci. USA 94:1890-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rose, N. F., P. A. Marx, A. Luckay, D. F. Nixon, W. J. Moretto, S. M. Donahoe, D. Montefiori, A. Roberts, L. Buonocore, and J. K. Rose. 2001. An effective AIDS vaccine based on live attenuated vesicular stomatitis virus recombinants. Cell 106:539-549. [DOI] [PubMed] [Google Scholar]
- 38.Schmitz, J. E., M. J. Kuroda, S. Santra, V. G. Sasseville, M. A. Simon, M. A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B. J. Scallon, J. Ghrayeb, M. A. Forman, D. C. Montefiori, E. P. Rieber, N. L. Letvin, and K. A. Reimann. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283:857-860. [DOI] [PubMed] [Google Scholar]
- 39.Shibata, R., F. Maldarelli, C. Siemon, T. Matano, M. Parta, G. Miller, T. Fredrickson, and M. A. Martin. 1997. Infection and pathogenicity of chimeric simian-human immunodeficiency viruses in macaques: determinants of high virus loads and CD4 cell killing. J. Infect. Dis. 176:362-373. [DOI] [PubMed] [Google Scholar]
- 40.Shiver, J. W., T. M. Fu, L. Chen, D. R. Casimiro, M. E. Davies, R. K. Evans, Z. Q. Zhang, A. J. Simon, W. L. Trigona, S. A. Dubey, L. Huang, V. A. Harris, R. S. Long, X. Liang, L. Handt, W. A. Schleif, L. Zhu, D. C. Freed, N. V. Persaud, L. Guan, K. S. Punt, A. Tang, M. Chen, K. A. Wilson, K. B. Collins, G. J. Heidecker, V. R. Fernandez, H. C. Perry, J. G. Joyce, K. M. Grimm, J. C. Cook, P. M. Keller, D. S. Kresock, H. Mach, R. D. Troutman, L. A. Isopi, D. M. Williams, Z. Xu, K. E. Bohannon, D. B. Volkin, D. C. Montefiori, A. Miura, G. R. Krivulka, M. A. Lifton, M. J. Kuroda, J. E. Schmitz, N. L. Letvin, M. J. Caulfield, A. J. Bett, R. Youil, D. C. Kaslow, and E. A. Emini. 2002. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature 415:331-335. [DOI] [PubMed] [Google Scholar]
- 41.Takeda, A., H. Igarashi, H. Nakamura, M. Kano, A. Iida, T. Hirata, M. Hasegawa, Y. Nagai, and T. Matano. 2003. Protective efficacy of an AIDS vaccine, a single DNA-prime followed by a single booster with a recombinant replication-defective Sendai virus vector, in a macaque AIDS model. J. Virol. 77:9710-9715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Voss, G., S. Nick, C. Stahl-Hennig, K. Ritter, and G. Hunsmann. 1992. Generation of macaque B lymphoblastoid cell lines with simian Epstein-Barr-like viruses: transformation procedure, characterization of the cell lines and occurrence of simian foamy virus. J. Virol. Methods 39:185-195. [DOI] [PubMed] [Google Scholar]
- 43.Yang, O. O., P. T. Sarkis, A. Ali, J. D. Harlow, C. Brander, S. A. Kalams, and B. D. Walker. 2003. Determinant of HIV-1 mutational escape from cytotoxic T lymphocytes. J. Exp. Med. 197:1365-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]