Abstract
The mechanisms that control cell-to-cell spread of human adenoviruses (Ad) are not well understood. Two early viral proteins, E1B-19K and E3-ADP, appear to have opposing effects since viral mutants that are individually deficient in E1B-19K produce large plaques (G. Chinnadurai, Cell 33:759-766, 1983), while mutants deficient in E3-ADP produce small plaques (A. E. Tollefson et al., J. Virol. 70:2296-2306, 1996) on infected cell monolayers. We have used a genetic strategy to identify different viral genes that influence adenovirus type 5 (Ad5) spread in an epithelial cancer cell line. An Ad5 mutant (dl327; lacking most of the E3 region) with the restricted-spread (small-plaque) phenotype was randomly mutagenized with UV, and 27 large-plaque (lp) mutants were isolated. A combination of analyses of viral proteins and genomic DNA sequences have indicated that 23 mutants contained lesions in the E1B region affecting either 19K or both 19K and 55K proteins. Four other lp mutants contained lesions in early regions E1A and E4, in the early L1 region that codes for the i-leader protein, and in late regions that code for the viral structural proteins, penton base, and fiber. Our results suggest that the requirement of E3-ADP for Ad spread could be readily compensated for by abrogation of the functions of E1B-19K and provide genetic evidence that these two viral proteins influence viral spread in opposing manners. In addition to E1B and E3 proteins, other early and late proteins that regulate viral replication and infectivity also influence lateral viral spread. Our studies have identified novel mutations that could be exploited in designing efficient oncolytic Ad vectors.
The efficiency with which a virus spreads from an infected cell to the neighboring uninfected cells is an important determinant of viral pathogenesis. The mechanisms by which nonenveloped viruses, particularly the adenoviruses, spread laterally are not well understood. It is generally believed that subversion of the cellular macromolecular synthesis machineries and the increased intracellular viral load result in cellular destruction and release of virus particles. The human adenoviruses (Ads) replicate in differentiated epithelial cells. The group C Ads infect these cells in a two-step process. The Ads enter the cells by receptor-mediated endocytosis (12, 70). The viral particles first bind to the primary receptor (CAR [coxsackie-adenovirus receptor]) (7, 67) through interaction with the knob structure of the fiber (73, 80). Subsequently, the viral capsid protein penton base interacts with the V type integrins (αVβ3 and αVβ5) via the RGD motif (78). The interaction of the penton base with the integrins also contributes the characteristic Ad cytopathic effect (CPE) (5). After adsorption and internalization into the endosomes, acidification of the endosomes results in cytosolic penetration of the virus particles which is also aided by the penton base (50, 57). Virus particles are then dismantled in a stepwise manner in the cytosol (24). It is possible that the efficiency of virus adsorption with primary and secondary receptors as well as factors that control cytosolic penetration could influence the extent of viral spread at the primary and reinfection levels. The viral structural proteins (e.g., fiber and penton base) that are critical for efficient infection and Ad CPE might be important in regulating viral spread. Two of the viral mutants with the enhanced-spread phenotype described in the present report have mutations in the genes coding for the penton base or the fiber protein.
After the delivery of the viral DNA core into the nucleus, the viral early genes are expressed, starting with the expression of the immediate-early viral gene E1A. The E1A proteins activate the expression of other viral early (E1B, E2, E3, and E4) transcription units (reviewed in reference 23). The primary task of the various early gene products is to prepare the cells to support efficient viral replication. This is accomplished by deregulation of the cell cycle regulatory mechanisms by removing restrictions imposed by cellular cell cycle regulatory proteins such as pRb and p53 (reviewed in reference 38). This feat is achieved by the proteins encoded by the E1A, E1B, and E4 regions. The proteins encoded by the E2 region are intimately involved in viral DNA replication (reviewed in reference 32). The E4 proteins also play an auxiliary role in viral DNA replication, in addition to other regulatory activities (reviewed in reference 60). The E4 proteins also activate signaling pathways to provide a favorable cellular environment for efficient viral replication (40). Although some functions of the various early proteins are known, others remain to be elucidated. Thus, the potential roles of various early gene products in viral spread and pathogenesis remain unexplored. The results presented here suggest that some early viral proteins may influence viral spread.
The L1 transcription unit is also expressed during the early phase (2) and codes for the 52/55K protein (30, 35, 37). The L1 52/55K protein, in association with a late protein, IVa2, plays a role in encapsidation of the viral DNA (42, 83, 84). Some of the mRNAs initiated from the major late promoter, particularly the L1 mRNAs, contain an additional leader (i-leader) spliced together between the second and third segments of the tripartite leader (17). The i-leader codes for a 16-kDa protein (71) which is expressed at early and late phases of viral replication (58). Adenovirus type 5 (Ad5) mutants deficient in the i-leader protein exhibit modest reductions in virus yield and elevated levels of L1 mRNA accumulation in some cell lines (51). However, mutants selected based on the ability to replicate in a human colon cancer cell line (HT29) contained a C-terminal-truncation mutation in the i-leader protein, in addition to other mutations (81). One of the mutants that we have isolated and described in the present paper also has a C-terminal truncation in the i-leader protein, suggesting a role for the i-leader protein in regulation of viral spread.
The E1B region codes for two major proteins, 19K and 55K. The most-well-known activity of the 55K protein is that it binds with the tumor suppressor protein p53 (31) and, in association with E4-Orf6, targets p53 for degradation during late stages of viral infection (20, 26, 45). The 55K-Orf6 complex also targets components of MRN complex (involved in cellular nonhomologous DNA end joining) for degradation (10, 52). Additionally, 55K modulates viral mRNA export (4, 19, 29, 44). Mutations in 55K produce host range and temperature-sensitive phenotypes in certain cell lines (4, 6, 27, 44). One of the 55K mutants, dl1520 (6) (renamed ONYX-015), was first explored as an oncolytic agent with the rationale that it would not be able to replicate efficiently in normal cells with wild-type (wt) p53 and would replicate in tumor cells that contain p53 mutations (8). This rationale has been intensely debated and challenged. More recently, it has been reported that a restriction in export of viral late mRNA in the absence of 55K may determine the tumor selective replication of 55K mutants (41). Therefore, it is possible that the 55K protein may contribute to viral spread in a cell-type-dependent manner. A number of mutants isolated during the present study are defective in the 55K protein in addition to the E1B-19K protein. The replication and spread phenotypes of such mutants are similar to those of mutants with lesions only in E1B-19K. The E1B-19K protein is a homolog of the cellular antiapoptosis protein BCL-2 (reviewed in references 14 and 75). Ad mutants defective in E1B-19K produce large clear plaques (lp) (13), indicative of accelerated viral spread. These mutants are generally characterized by enhanced cell death activity (43, 55, 59, 77). However, some E1B-19K mutants without enhanced death activity also produce large plaques (13, 55). Thus, E1B-19K may have an inhibitory effect on viral spread.
The E3 region codes for seven different polypeptides and is considered nonessential for productive viral replication in established human cancer cell lines (reviewed in references 36 and 79). Some of the E3 proteins inhibit apoptosis induced by FasL and tumor necrosis factor (36, 62, 66) and appear to play a role in viral persistence in immunocompetent mouse models (72). The E3 protein designated ADP (adenovirus death protein) (64) is required for efficient lysis of infected cells and is therefore important for lateral viral spread. This property has been exploited in designing oncolytic Ad vectors that spread at enhanced rates as a result of ADP overexpression (68). Here, we have used an Ad5 mutant (dl327) (61) that contains a deletion encompassing most of the E3 region and consequently spreads at a restricted rate (64) to isolate compensatory mutants. Most of the compensatory mutants contained lesions in the E1B-19K coding region, suggesting that ADP and E1B-19K may be functional antagonists in group C Ad spread. Additionally, we have identified mutations in other early and late genes that also enhance viral spread.
MATERIALS AND METHODS
Cells and viruses.
Human A549, HeLa, and 293 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum. The secondary cultures of human neonatal kidney (HNK) cells were obtained from Diagnostic Hybrids, Inc., and were grown in DMEM supplemented with 10% fetal bovine serum. The present study employed three different parental viruses, wt Ad5 and Ad5 mutants dl327 (61) and pm383 (51). The stocks of different viruses were prepared in A549 or 293 cells and banded in CsCl gradients. The virus titers were determined by plaque assay with A549 cells.
Mutagenesis and construction of recombinants.
The Ad5 mutant dl327 was subjected to random mutagenesis by exposure to UV. The virus stock was diluted in phosphate-buffered saline and exposed to a germicidal lamp on a shaking platform for 5 min, resulting in a 5-log reduction in virus titer. The mutagenized virus was subjected to a plaque assay with A549 cells (10 to 20 plaques/60-mm dish). The large plaques (compared to those produced by the parental dl327 virus) that arose 8 to 14 days after infection were isolated and purified by further rounds of plaque purification. Recombinants between two different mutants were generated by cotransfection of viral DNA digested with different restriction endonucleases that generated overlapping DNA fragments (15). Two micrograms of each mutant DNA (total digested DNA) was transfected along with 6 μg of the salmon sperm carrier DNA into 293 cells (in 60-mm dishes). After development of detectable CPE (4 to 7 days after transfection), the recombinant viruses were released by freezing and thawing and purified by plaque isolation on A549 cells.
DNA sequence analysis.
The DNA sequences of various mutants were determined by automated sequence analysis of short regions of the viral genomes encompassing the mutations or by sequencing the entire viral genome. DNA sequences of several mutations in the E1B region and in recombinants carrying known mutations were determined by analysis of DNA fragments amplified by PCR using the high-fidelity DNA polymerase Vent (New England Biolabs). The PCR amplifications were carried out using the Hirt supernatant DNA prepared from infected cells. The complete sequences of the viral DNA of some mutants were determined by using primers distributed throughout the genome (∼500 to 900 bp apart).
Antibodies and immunoblot analysis.
The rabbit polyclonal antibodies specific to E1B-19K (25), i-leader (35), L1-52/55K (58), and E3-ADP (64) have been described previously. The rabbit polyclonal antibodies specific to Ad5 virion proteins (ab6982; Abcam) or actin (SC1615; Santa Cruz Biotechnology) were purchased from the indicated commercial sources. The mouse monoclonal antibodies specific to the E1A and E1B-55K proteins were gifts from Peter Yaciuk (Saint Louis University). The proteins were fractionated by 10 to 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The separated proteins were electrotransferred onto a nitrocellulose membrane and probed with a primary antibody followed by a horseradish peroxidase-conjugated secondary antibody (goat anti-rabbit, goat anti-mouse, and donkey anti-goat antibodies were purchased from Santa Cruz Biotechnology) and analyzed with a chemiluminescence detection system (Roche Applied Science) according to manufacturer specifications.
Analysis of intracellular DNA.
For analysis of intracellular viral DNA and fragmented (cellular and viral) DNA, cells were lysed at indicated times after infection and the small-molecular-weight (MW) DNA was prepared by Hirt extraction as described earlier (54) and analyzed by electrophoresis on agarose gels. The gels were stained with either ethidium bromide (EtBr) or Vistra Green. The gels stained with EtBr were analyzed using a gel documentation system (Chem Doc XRS; Bio-Rad) and quantified with Quantity One software. The gels stained with Vistra Green were analyzed with a phosphorimager and quantified using ImageQuant software.
Cell viability assays.
Cell viability was determined by a colorimetric MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] assay (Promega). For this purpose, 8 × 103 cells/well were plated in 96-well plates with DMEM and incubated for 24 h. The cells in triplicate wells were infected with various Ad mutants at 20 PFU/cell in 100 μl of media containing 2% serum. Six days after infection, the media were changed and 20 μl of reaction mixture containing MTS (final concentration, 333 μg/ml) and the electron coupling reagent PES (phenazine ethosulfate; final concentration, 25 μM) was added. The absorbance at 490 nm was measured after a 2-h incubation at 37°C in a 96-well plate reader.
RESULTS
Isolation of compensatory mutants of dl327 and screening.
Although it is generally believed that apoptosis-like cell death programs initiated by viral infection are cellular defense mechanisms to restrict viral replication, we hypothesized that such programs might also play a role in viral release and consequently contribute to viral spread. This hypothesis would predict that a restricted-spread phenotype imposed by the lack of E3 proteins such as ADP could be relieved by mutations that result in enhanced cell death (e.g., in E1B-19K). We took a random-mutagenesis approach to test this hypothesis, since such an approach would identify a spectrum of viral genes that might play a role in viral spread. The Ad5 mutant dl327 contains a deletion of most of the E3 region and produces small plaques on established human cell lines (64). This phenotype of dl327 appears to be predominantly dictated by the absence of ADP. We mutagenized dl327 by exposure to UV and selected 27 mutants that produced lp on A549 cells.
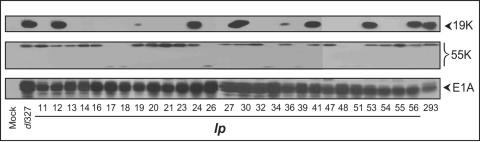
Since mutations in E1B-19K result in the lp phenotype of Ad with the E3 region (13), we first examined if any of the mutants of dl327 were defective in E1B-19K expression. A549 cells were infected with various lp mutants, and the cell extracts were analyzed for expression of E1A and E1B proteins (Fig. 1). Western blot analysis revealed that all mutants expressed comparable levels of E1A. Six mutants (lp12, lp24, lp30, lp41, lp53, and lp56) expressed the E1B-19K protein at levels comparable to that of dl327, while two other mutants (lp19 and lp36) produced much lower levels of E1B-19K. All eight mutants that were positive for E1B-19K expression also expressed E1B-55K. Among the other mutants, 12 were defective in expression of E1B-19K only, while 7 other mutants were defective in expression of both E1B-19K and -55K proteins.
FIG. 1.
Expression of E1A and E1B proteins by lp mutants. Whole-cell lysates of A549 cells were prepared at 24 h postinfection with various lp mutants. The Western blots were probed with monoclonal or polyclonal antibodies specific to E1A or E1B (19K and 55K) proteins.
Since mutations that severely affect E1B-19K functions produce the cyt (cytocidal) phenotype (56, 59), we examined the CPE of all 27 mutants on A549 and secondary HNK cells (not shown). Twenty-one mutants (which included all mutants defective in E1B-19K expression) produced cyt CPE (floating individual and fragmented cells), suggesting that the activity of E1B-19K might be impaired in these mutants. Two other mutants (lp19 and lp36) which produced lower levels of E1B-19K caused less-severe cyt CPE. Four other mutants (lp24, lp30, lp41, and lp53) produced wt Ad CPE. From these studies, several mutants that were either positive or negative for expression of E1B-19K were chosen for direct DNA sequence analysis of the E1B region or the entire viral genome.
DNA sequence analysis of selected lp mutants.
Based on the patterns of expression of E1 proteins (Fig. 1), four lp mutants that were positive for E1B-19K and produced wt CPE (lp24, lp30, lp41, and lp53) were chosen for DNA sequence analysis of the entire viral genome. Additionally, the complete DNA sequences of one of the E1B-19K-negative cyt mutants (lp11) and one mutant that produced low levels of E1B-19K (lp19) were determined. The DNA sequences of the E1A and E1B regions of several mutants that were either positive (lp12 and lp56) or negative (lp20, lp34, and lp48) for E1B-19K were determined. These results are summarized in Table 1. Among the E1B mutants, two (lp12 and lp56) contained mutations that cause one or more amino acid substitutions in E1B-19K. The phenotypes (CPE and plaque morphology) of these two mutants were similar to that of the 19K null mutant (lp11). Both mutants contained a lesion that resulted in a substitution for a Gly(87) residue that is essential for the function of E1B-19K (16). In lp26, lp34, and lp48, insertion of a single G residue at position 2044 results in fusion of the E1B-19K and -55K reading frames. The E1B proteins expressed by these mutants (Fig. 1) contained several lower-MW polypeptides (probed with the E1B-55K antibody), suggesting that the fusion proteins produced by these mutants were labile. Seven mutants induced a similar pattern of E1B protein expression (i.e., lack of both E1B-19K and -55K proteins), but the DNA sequences of only three (E1A and E1B regions) were determined. The mutant lp19 (that produced low levels of E1B-19K) contained two mutations, one within the E1B-19K coding region and the other in the coding region for pV (core protein). Although the mutation within the E1B-19K coding region would result in a frameshift and premature termination, low levels of the E1B-19K protein (full-length) were expressed in cells infected with this mutant (Fig. 1). It is possible that the insertion of an extra T within the T5 repeat of the E1B-19K-coding sequences might not be recognized efficiently by the translational machinery. Mutant lp36, which also produced reduced levels of E1B-19K, contained two silent mutations and two amino acid substitution mutations within the E1B-19K-coding region. These two mutants (lp19 and lp36) were not pursued further. Based on the information gleaned from the DNA sequence analysis and the expression patterns of E1B proteins, two cyt mutants, lp11 (defective only in E1B-19K) and lp26 (defective in both E1B-19K and E1B-55K), and four non-cyt mutants (E1B-19K proficient) that contained mutations elsewhere (lp24, lp30, lp41, and lp53) were chosen for further characterization.
TABLE 1.
Mutations in selected lp mutants
| Mutant(s) | nt sequenced | Site | Mutation | Genea | Change (aa)b |
|---|---|---|---|---|---|
| lp11 | 1-35934 | 1797 | G→A | E1B-19K | Trp(28)→Ter |
| lp12 | 333-3614 | 1973 | G→A | E1B-19K | Gly(87)→Glu |
| lp19 | 1-35934 | 1740 | +T | E1B-19K | (E1B-19K-Frameshift-Ter)* |
| 17186 | G→C | L2 pV | Gln(214)→His | ||
| lp20 | 333-3614 | 1861 | G→T | E1B-19K | Glu(50)→Ter |
| lp26 | 159-2578 | 2044 | +G | E1B-19K-55K | E1B-19K(112)-55K(11-496) fusion |
| lp34, lp48 | 333-3614 | 2044 | |||
| lp36 | 333-3614 | 1923 | G→A | E1B-19K | Ala(71)→Thr |
| 1924 | G→A | Silent | |||
| 2001 | G→A | Silent | |||
| 2003 | G→A | Ser(97)→Asn | |||
| lp56 | 333-3614 | 1972-1973 | GG→AA | E1B-19K | Gly(87)→Lys |
| 2059 | G→A | E1B-19K | Asp(116)→Asn | ||
| E1B-55K | Gly(14)→Glu | ||||
| lp24 | 1-35934 | 1317 | G→A | E1A | Silent |
| 1319 | G→A | E1A | Arg(215)→Lys | ||
| 17971 | C→A | (L2-L3) | Silent | ||
| 28517 | A→T | E3 (12.5K-7.1K) | Silent | ||
| 34659 | T→C | E4-Orf3 | Glu(14)→Gly | ||
| 34661 | C→T | E4-Orf3 | Silent | ||
| lp30 | 1-35934 | 14650 | G→T | Penton base | Trp(165)→Leu |
| 23994 | G→T | (L3-E2A) | Silent | ||
| lp41 | 1-35934 | 32777 | G→A | Fiber | Glu(581)→Lys |
| lp53 | 1-35934 | 8299 | C→T | i-Leader | Gln(108)→Ter |
| E2B (Ad Pol) | Silent |
The mutations that are located between two protein coding regions or transcription units are indicated within parentheses. Pol, DNA polymerase.
The amino acid (aa) changes caused by the mutations are indicated within parentheses. *, see text.
CPE and plaque morphology of lp mutants.
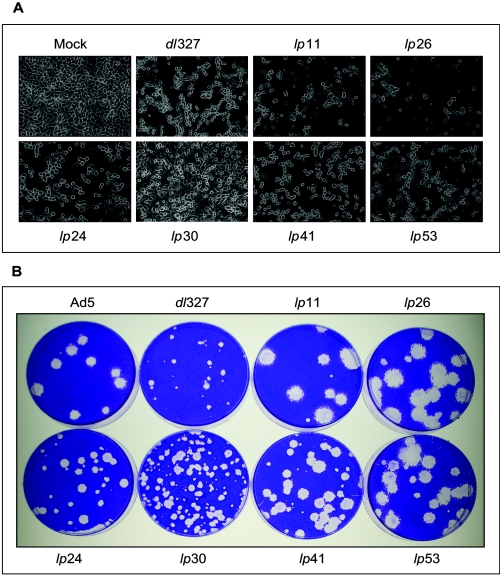
During the initial characterization of the lp mutants, we classified them on the basis of CPE in A549 and HNK cells. Such grouping correlated very well with the functional status of E1B-19K of the mutants, which was learned by subsequent studies of expression of E1B proteins and DNA sequence analysis (Fig. 1 and Table 1). The CPEs of selected lp mutants on A549 cells are shown in Fig. 2A. wt Ad5 and dl327 induced the characteristic Ad CPE (adherent clumped cells), while mutants lp11 and lp26 produced cyt CPE. Among the non-cyt mutants, lp24, lp41, and lp53 induced wt-like CPE while lp30 consistently produced restricted CPE. The plaque morphologies of these mutants are shown in Fig. 2B. As expected, dl327 formed small plaques (compared to the wt) on A549 cells. All lp mutants formed large plaques compared to the parental virus dl327. The cyt mutants (lp11 and lp26) formed much larger plaques than the non-cyt mutants (lp24, lp30, lp41, and lp53). Among the non-cyt mutants, lp41 and lp53 formed larger plaques than lp24 and lp30. Within this group of mutants, lp30 produced more heterogeneous-sized plaques. Thus, mutants such as lp41 and lp53 produce large plaques without causing severe CPE (like lp11 and lp26). It should be noted that the plaque assays for various lp mutants contained a few small plaques. Such variations in plaque size of a genetically homogeneous virus population might be attributed to stochastic fluctuations in virus infection and in viral gene expression. Such a phenomenon has been well documented with bacterial and animal viruses (3, 74).
FIG. 2.
CPE and plaque morphology of lp mutants. (A) CPE. A549 cells were infected with various lp mutants at 20 PFU/cell and photographed under phase contrast at 36 h postinfection. (B) Plaque morphology. Monolayers of A549 cells in 60-mm dishes were infected with various mutants, overlaid with 0.5% agarose in growth medium containing 2% fetal bovine serum, and stained with crystal violet at 10 days postinfection.
Lateral spread of lp mutants.
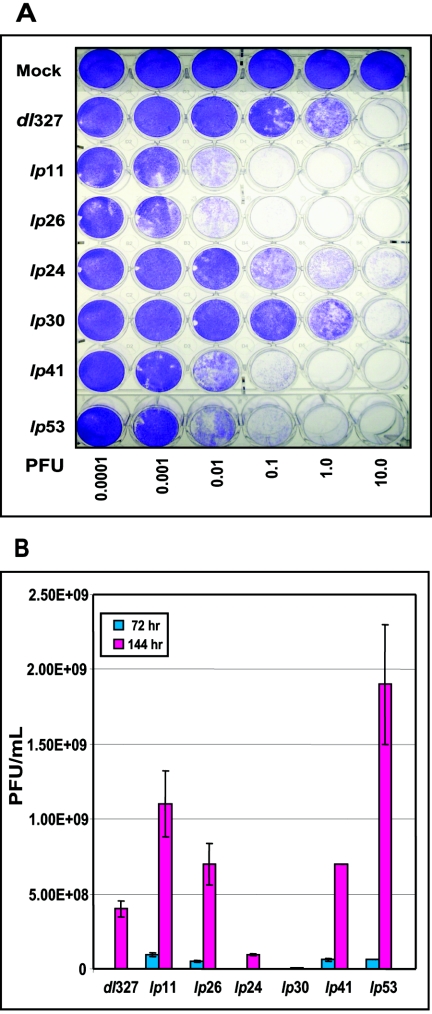
The lateral spread of the selected lp mutants on A549 cell monolayers was determined by infecting cells in 24-well plates with different multiplicities of infection (MOI). The viable cells were stained with crystal violet 6 days after infection (Fig. 3A) and quantified from the dye extracted from the plates (data not shown). This assay revealed that the spread of lp11 and lp26 was at least 100-fold greater than that of dl327, while mutants lp41 and lp53 spread at 10- and 20-fold-greater levels, respectively, than dl327. Mutant lp24 also spread at an enhanced level but was slower than lp41 and lp53. The spread phenotype of lp30 was more or less comparable to that of dl327, despite a significant fraction of large plaques formed by lp30. These results clearly indicate that mutations which obliterate the function of E1B-19K can readily overcome the spread restriction imposed by the lack of E3 proteins such as ADP. Interestingly, mutations in other early and late genes also enhanced the spread of the E3-deleted virus. A 24-well-format spread assay carried out with the HeLa cell line also produced results comparable to that obtained with A549 cells, except the effects of the E1B mutants (lp11 and lp26) and the penton base mutant (lp30) were more pronounced (not shown). A multistep viral-growth assay was also carried out to determine the spread potentials of the various mutants. For this purpose, A549 cells were infected with various mutants at an MOI of 0.1 PFU/cell and the yield of progeny virus at 72 h and 144 h after infection was determined by plaque assay (Fig. 3B). These results were in general agreement with the 24-well-format spread assay (Fig. 3A). However, yields of two mutants, lp24 and lp30, were lower than that of dl327. The reason for the lower virus yield of these mutants after multiple rounds of replication is unclear. However, cellular response such as premature cell death by apoptosis or necrosis may contribute to such a reduction in virus yield. The data for lp11 and lp26 support this hypothesis. Mutants lp11 and lp26 yielded less progeny virus than lp53, despite sharing with lp53 the most pronounced spread and lp phenotype.
FIG. 3.
Spread and multistep growth of lp mutants. (A) Spread. A549 cells in 24-well plates (5 × 104 cells/well) were infected with various mutants at indicated PFU, maintained in growth media containing 2% fetal bovine serum, and stained with crystal violet at 6 days postinfection. (B) Multistep growth. A549 cells were infected with various mutants at an MOI of 0.1 PFU/cell, and the yield of progeny virus was determined at 72 and 144 h after infection by plaque assay with A549 cells.
DNA synthesis and expression of late proteins.
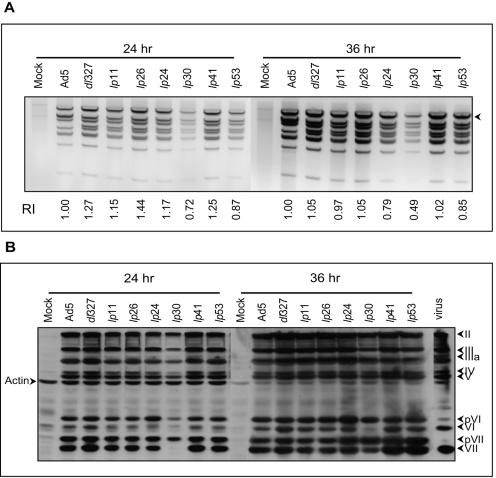
Next, we analyzed the levels of viral DNA replication and expression of late viral proteins to determine if they correlated with the mutant phenotypes. For analysis of viral DNA replication, the low-MW intracellular DNA was extracted from infected cells (at 24 h and 36 h postinfection), digested with the restriction endonuclease HindIII, and analyzed by agarose gel electrophoresis (Fig. 4A). These results suggest that the cells infected with various mutants (except lp30) contained comparable amounts of viral DNA. The analysis of late viral proteins also did not reveal major differences in the levels of expression of these proteins (Fig. 4B). However, lp30, which carries a mutation in the penton base, exhibited a significant lag in processing of the precursors for the core proteins pVI and pVII. It is possible that the reduced levels of viral DNA and late proteins observed with cells infected with lp30 may reflect asynchronous infection by this mutant virus, which might be caused by the mutation in the penton base.
FIG. 4.
Viral DNA replication and late protein expression by lp mutants. (A) DNA replication. A549 cells (1 × 106) were infected with various mutants at 20 PFU/cell, and the Hirt supernatants were prepared, digested with HindIII, and analyzed (equal amounts of extracts) by electrophoresis in 1% agarose gels stained with EtBr. The intensity of fragment A (indicated by the arrowhead) in each lane was quantified with Quantity One (Bio-Rad) software. RI, relative intensities based on wt-infected cells. (B) Expression of late proteins. Whole-cell extracts of infected cells were separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the Western blots were probed first with anti-Ad5 antibody, followed by antiactin antibody. The proteins of wt Ad5 virions purified by CsCl banding are shown as markers at the right.
Viral replication.
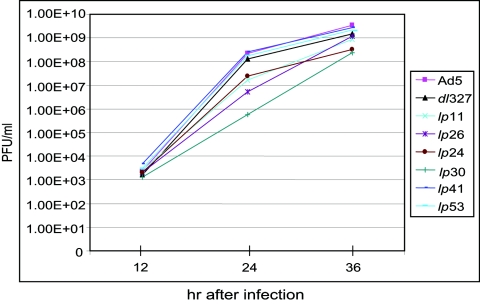
To determine if the spread phenotypes of various mutants might be due to differences in the overall viral replication potentials, we carried out a one-step growth curve analysis. A549 cells were infected with Ad5 mutants at an MOI of 5, and the virus yields at different times after infection were determined (Fig. 5). Mutants dl327, lp53, and lp41 replicated with kinetics comparable to that of wt Ad5. Although the cyt mutants (lp11 and lp26) and the non-cyt mutant lp24 replicated at slower rates, there were only modest reductions in the final virus yield (at 36 h after infection). Mutant lp30 replicated with a significant lag period, and the virus yield at 36 h postinfection was about 10-fold lower than that of the parental virus (dl327). However, the spread phenotype of this mutant in multiple rounds of replication was comparable to that of dl327 (Fig. 3A). Thus, the spread phenotypes of different mutants do not appear to correlate with virus replication. These results further suggest that the modulation of cellular processes by the viral gene products is more critical for efficient viral spread.
FIG. 5.
Replication of lp mutants. The single-step growth curves of various mutants were determined with A549 cells infected at 5 PFU/cell. The average virus titers from three independent experiments are shown. hr, hours.
Cell death activity.
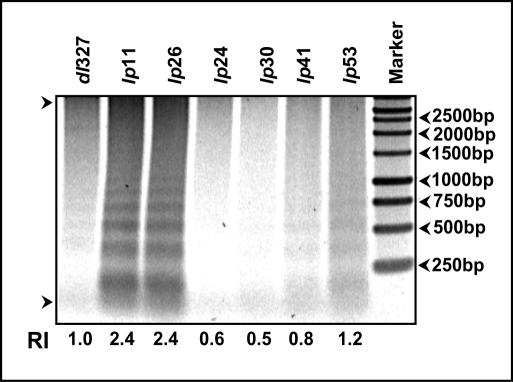
In order to determine if the spread phenotypes of different mutants might be related to cell death activity, we determined the fragmentation pattern of intracellular DNA as a measure of cell death. A549 cells were infected with different mutants, and the low-MW intracellular DNA was extracted at 36 h after infection and analyzed by electrophoresis in agarose gels (Fig. 6). As expected, the intracellular DNA was extensively degraded in cells infected with the cyt mutants (lp11 and lp26). Cells infected with lp53 contained slightly elevated levels of fragmented DNA compared to cells infected with dl327, while cells infected with other lp mutants (lp24, lp30, and lp41) contained lower levels of fragmented DNA. These results suggest that the enhanced-spread phenotype of lp11 and lp26 might be related to the enhanced cell death activity of these mutants. However, the prominent non-cyt mutants, such as lp41 and lp53, might spread at enhanced levels, independently of such cell-killing activity.
FIG. 6.
Cell death activity. The intracellular low-MW DNA was extracted at 36 h postinfection and analyzed by agarose gel (1.5%) electrophoresis as a measure of cell death activity. The intensity of the DNA fragments in each lane was analyzed with a phosphorimager. RI, relative intensities of the DNA fragments (based on dl327) in the areas indicated between the two arrows.
Effect of i-leader C-terminal-truncation and null mutations.
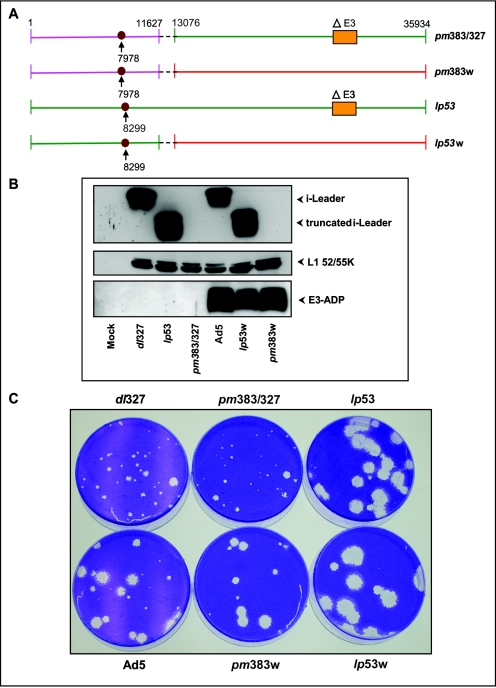
Among the various non-cyt mutants, the phenotype of lp53 is interesting considering that an i-leader null mutant (pm383) was reported to exhibit only modest phenotypes (51). Therefore, it was of interest to determine if the phenotype of lp53 is due to a dominant function conferred by the truncated i-leader polypeptide or due to a general functional defect of the protein. For this purpose, we generated a recombinant virus (pm383/327) by transferring the i-leader null mutation of pm383 (51) to dl327 by overlap recombination. The recombinant was obtained by cotransfection of the DNA of pm383 (mutation at nucleotide [nt] 7978; ATG→GTG) digested with AvrII and the DNA of dl327 digested with NsiI in 293 cells. This restriction digestion strategy was designed to generate recombinants via the overlapping sequences located between nucleotide positions 11627 and 13076 (Fig. 7A) of the largest left-end fragment of pm383 generated by AvrII (nt 1 to 13076) and the largest right-end fragment of dl327 generated by NsiI (nt 11627 to 35934). A similar strategy was also used to generate recombinants of pm383 (pm383w) or lp53 (lp53w) with the wt E3 region (Fig. 7A). The recombinant viruses were isolated, and the mutations in the recombinant genome were ascertained by DNA sequence analysis of the i-leader and the E3 regions. The mutational effect on i-leader protein expression was determined by Western blot analysis (Fig. 7B). Mutants lp53 and lp53w produced copious amounts of the C-terminally truncated i-leader protein, while recombinants pm383/327 and pm383w did not produce detectable levels of the i-leader protein. The Western blot analysis did not show major differences in expression of L1 52/55K proteins. The analysis of E3-ADP also revealed the expected patterns. The recombinants in the dl327 background did not express ADP, while the recombinants under a wt E3 background expressed it (Fig. 7B). The effect of the i-leader null mutation was then compared with that of the C-terminal-truncation mutant under ΔE3 (dl327) and wt E3 backgrounds by a plaque assay with A549 cells (Fig. 7C). While the lp53 mutation caused large plaques under both backgrounds, the null mutation (pm383) did not significantly change the plaque morphology under both backgrounds. These results suggest that the C-terminally truncated version of the i-leader may contribute to the phenotype of lp53.
FIG. 7.
Effect of i-leader null and C-terminal-truncation mutants. (A) Genome structures of recombinants containing the i-leader mutations (pm383 or lp53). The i-leader mutations (at nt 7978 and 8299) are indicated. The regions of overlap recombination in recombinant viruses are indicated by dashed lines. (B) Expression of i-leader, L1 52/55K, and E3-ADP proteins. Western blot analysis was carried out with whole-cell lysates prepared at 24 h postinfection. (C). Plaque morphologies of recombinant and parent viruses. Cells were stained 10 days postinfection and photographed.
Effect of E3 region on E1B-19K mutation.
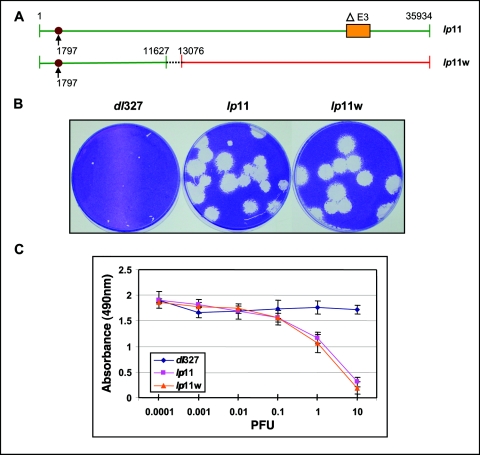
Most of the compensatory mutants that we have isolated affected the function of E1B-19K. These results suggested an agonist/antagonist relationship between E3 and E1B-19K. We reasoned that if E3 contributes to viral spread independently of E1B-19K, the effect of the E3 region would be additive with that of the E1B-19K mutation. Also, if E3 proteins function primarily to neutralize the activity of E1B-19K, the effect may not be additive. To test this hypothesis, we constructed a recombinant (lp11w) by introducing the lp11 mutation into a wt Ad5 background (Fig. 8A). The recombinant virus was generated by the overlap recombination strategy described above (using viral DNA fragments generated by AvrII and NsiI), and the expected genomic structure (E1B and E3 regions) was confirmed by DNA sequence analysis. The plaque sizes of lp11 and lp11w were more or less similar, and no additive increase in plaque size was discernible (Fig. 8B). To compare the cytolytic potentials of lp11 and lp11w, we carried out an MTS-based cytolysis assay. A549 cells in 96-well plates were infected with dl327, lp11, or lp11w at different MOI, and cell viability was determined at 144 h after infection (Fig. 8C). These results revealed no significant differences in the cytolytic potentials of lp11 and lp11w. The cytolytic activities of lp11 and lp11w were also similar in HeLa cells (data not shown). Thus, our results suggest that the presence of the E3 region and the E1B-19K mutation (lp11) may not have a significant additive effect on cytolysis of Ad-infected cells. Although our results suggest that E3 might primarily antagonize the effect of E1B-19K, they cannot fully rule out that inclusion of the E3 region may have a small enhancing effect on the cytolytic potential of E1B-19K mutants.
FIG. 8.
Effect of E3 region on E1B-19K mutation. (A) Genome structures of lp11 and lp11w. Recombinant lp11w was constructed by overlap recombination, and the region of recombination is indicated by dashed lines. (B) Plaque morphologies of the recombinant and parents. (C) Cytolysis by Ad mutants. A549 cells in 96-well plates were infected with indicated viruses at various MOI, and cell viability was determined by the MTS assay at 144 h after infection.
DISCUSSION
We have used a genetic strategy to identify Ad5 genes that play a role in lateral viral spread. We have exploited the finding by Tollefson et al. that E3 mutants lacking ADP are deficient in lateral spread of group C Ads (64). Many of the compensatory lp mutants (23 out of 27) of the ADP null mutant (dl327) that we isolated by random mutagenesis contain mutations that affect the function of the E1B-19K protein. These results provide strong genetic evidence that ADP and E1B-19K play opposing roles in Ad spread. While more is known about the function of E1B-19K, little is known about ADP. Even though ADP is an early gene, it is expressed at high levels during the late phase of the viral cycle (64). Interestingly, E1B-19K is also expressed at high levels during late phases of the viral life cycle (25, 69), suggesting both proteins may function in stoichiometric manners. Our present results have revealed that E1B-19K mutants form large plaques in the absence of ADP. The presence of the E3 region in an E1B-19K mutant (lp11w) (Fig. 8) does not significantly enhance plaque size or cytolysis. These results suggest that ADP may antagonize the function of E1B-19K.
ADP is an integral membrane glycoprotein that is predominantly localized in the nuclear envelope/Golgi regions (65). It appears that ADP may cause virus release by disrupting nuclear membrane integrity, since cells infected with ADP-negative mutants appear to contain an intact nuclear membrane with accumulated virus load inside the nucleus (63). The anchoring of ADP in the inner nuclear membrane appears to be critically important for the activity of ADP (65). ADP has not been implicated in any proapoptotic response during normal viral infection. However, overexpression of ADP has been reported to induce a unique pattern of cell death that resembles neither apoptosis nor necrosis. Overexpression of ADP under the control of the cytomegalovirus or Ad major late promoter has been reported to cause enhanced cell death that exhibited some characteristics of apoptosis (82, 85). However, comparison of the cell death patterns of two conditionally replicative vectors (containing a deletion in the CR2 region of E1A) that differed in the presence or absence of E3 has implicated ADP in induction of necrosis-like cell death (1). Although the mechanism by which ADP causes nuclear membrane destabilization and viral egress remains to be clarified, the lack of functional E1B-19K can overcome the restriction in virus release imposed by the lack of ADP to a large extent. It is possible that ADP and E1B-19K may function as an agonist and an antagonist, respectively, in the same pathway or that they may function through distinct mechanisms to achieve a balanced viral replication and spread.
How does the lack of functional E1B-19K enhance viral spread? The E1B-19K protein appears to be a viral homolog of Bcl-2 and functions as a potent inhibitor of apoptosis (14, 75). It is known that E1B-19K suppresses an Ad-induced cell death program (which resembles apoptosis) during viral infection (43, 55, 77). A fraction of E1B-19K has been reported to be localized in the nuclear envelope region in physical association with nuclear lamins (46, 76). The E1B-19K protein appears to protect against proteolysis of the lamins during apoptosis (47). A simple explanation for the enhanced viral spread in the absence of E1B-19K would be that disruption of the nuclear envelope mediated by proteolysis of the components of the nuclear lamina may release the virus load. However, whether E1B-19K has a viral-spread inhibitory function independent of its antiapoptosis function also merits further investigation. This is relevant since some E1B-19K mutants (e.g., lp3) that do not exhibit the deg (fragmentation of chromosomal DNA) phenotype (associated with apoptosis) also produce large plaques (13).
Among the other compensatory mutants that we have isolated, lp53 (which results in C-terminal truncation of the i-leader protein) exhibits the most pronounced effect on relieving the E3 deficiency in viral spread. Additionally, the lp53 mutation has an additive effect with the E3 region (Fig. 7) and with a deletion within the E1B-19K region (T. Subramanian and G. Chinnadurai, unpublished data), suggesting that C-terminal truncation of the i-leader protein may augment viral spread independently of the E1B-19K/ADP mechanisms. The mutant lp53 contains a single base pair change (C→T) in the Ad5 genome resulting in truncation of the C-terminal 38 amino acids of the i-leader protein. In striking contrast, an i-leader null mutation does not appear to influence viral spread under both E3 (plus or minus) contexts (Fig. 7). A previous genetic screen of Ad5 mutants for enhanced replication in a colon cancer cell line (HT29) identified two mutants (81). Both mutants contained multiple lesions in the Ad5 genome, including a C→T transition that resulted in truncation of the C-terminal 21 amino acids of the i-leader protein. Recombinant viruses that contained only the i-leader mutation also exhibited enhanced cytolysis of HT29 cells. Although Yan et al. (81) ascribed this property to enhanced viral DNA replication, the mutants used for such analysis contained multiple mutations. Since the DNA replication pattern of lp53 does not appear to be significantly different from that of the parental virus dl327, the enhanced-spread phenotype of lp53 may not be ascribed to any effect on DNA replication. The observation that the i-leader null mutation (pm383) does not influence the spread phenotype is intriguing. It is possible that the full-length i-leader protein may be in an inactive conformation and may be functionally activated by proteolysis of the C-terminal region under specific conditions. The mechanism by which the i-leader mutation enhances viral spread would have to await future characterization of full-length and C-terminal-truncation proteins.
The mutations in lp41 (fiber knob) and lp30 (penton base) are located in viral structural proteins that mediate interaction of the virus particles with primary and secondary cellular Ad receptors. These mutations may enhance virus infection during primary and secondary infection cycles. Although it is expected that viral structural proteins may play critical roles in viral spread, the use of the genetic strategy (dl327) has enabled the identification of some of these genes. Our studies have pointed out that some of the early genes, such as E1A and E4, in addition to viral late proteins, may play a role in viral spread. Attempts to resolve the mutations in lp24 (which contains amino acid substitution mutations in the E1A and E4 regions) have suggested that the lp phenotype of this mutant may segregate predominantly with the E4-Orf3 mutation (results not shown). The E4 proteins encoded by the Orf3 and Orf6 regions play overlapping roles in Ad replication (9, 28). The Orf3 protein is highly conserved among various Ad serotypes and plays a role in efficient viral DNA replication by inhibition of concatemerization of the viral DNA by targeting components of the cellular nonhomologous end-joining pathway (52, 53). The Orf3 protein is tightly associated with the nuclear matrix (48) and reorganizes nuclear structures known as promyelocytic leukemia oncogenic domains (11, 22). It is possible that Orf3 may play a role in viral spread through its role in viral DNA replication or through other functions associated with promyelocytic leukemia oncogenic domains. Thus, our genetic screening of random mutants has identified the role of several viral early and late genes that influence the extent of viral spread.
Our present results have important implications in designing efficient oncolytic Ad vectors. The use of these vectors has become a promising platform of cancer therapy (18, 39). The vector designs that include mutations in the E1B-19K gene would offer two advantages, enhanced lateral viral spread and chemosensitivity to anticancer drugs (due to abrogation of the antiapoptosis activity) used in combination therapies. Additionally, such vectors may have a reduction in the overall virus load due to the apoptotic response of infected cells. Although E1B-19K mutant viruses have been used in some studies (33, 34, 49), their utility is underappreciated. A strategy based on overexpression of ADP to achieve enhanced viral spread has been used for oncolytic Ad vector designs (21). Our results presented here suggest that viruses containing the E1B-19K mutation (lp11) in the presence or absence of ADP exhibit similar spread potentials. Thus, the ADP coding region could be advantageously used for expression of heterologous therapeutic genes in large amounts rather than to achieve enhanced viral spread in the presence of an E1B-19K mutation. Some of the other mutants that we have identified could also be incorporated into the E1B-19K mutant background. For example, we have observed that incorporation of the lp53 mutation (i-leader) and an E1B-19K mutation (dl250 [55]) in a single recombinant causes additive enhancement in viral spread (Subramanian and Chinnadurai, unpublished). The possibility that some of the mutants identified here may have enhanced tropism for different tumor cells also remains to be explored.
Acknowledgments
This work was supported by a research grant, CA-33616, from the National Cancer Institute.
We thank Maurice Green, Tom Shenk, Bill Wold, and Ann Tollefson for antibodies and mutant viruses.
REFERENCES
- 1.Abou El Hassan, M. A., I. van der Meulen-Muileman, S. Abbas, and F. A. Kruyt. 2004. Conditionally replicating adenoviruses kill tumor cells via a basic apoptotic machinery-independent mechanism that resembles necrosis-like programmed cell death. J. Virol. 78:12243-12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akusjarvi, G., and H. Persson. 1981. Controls of RNA splicing and termination in the major late adenovirus transcription unit. Nature 292:420-426. [DOI] [PubMed] [Google Scholar]
- 3.Arkin, A., J. Ross, and H. H. McAdams. 1998. Stochastic kinetic analysis of developmental pathway bifurcation in phage lambda-infected Escherichia coli cells. Genetics 149:1633-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babiss, L. E., H. S. Ginsberg, and J. E. Darnell, Jr. 1985. Adenovirus E1B proteins are required for accumulation of late viral mRNA and for effects on cellular mRNA translation and transport. Mol. Cell. Biol. 5:2552-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai, M., B. Harfe, and P. Freimuth. 1993. Mutations that alter an Arg-Gly-Asp (RGD) sequence in the adenovirus type 2 penton base protein abolish its cell-rounding activity and delay virus reproduction in flat cells. J. Virol. 67:5198-5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barker, D. D., and A. J. Berk. 1987. Adenovirus proteins from both E1B reading frames are required for transformation of rodent cells by viral infection and DNA transfection. Virology 156:107-121. [DOI] [PubMed] [Google Scholar]
- 7.Bergelson, J. M., J. A. Cunningham, G. Droguett, E. A. Kurt-Jones, A. Krithivas, J. S. Hong, M. S. Horwitz, R. L. Crowell, and R. W. Finberg. 1997. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320-1323. [DOI] [PubMed] [Google Scholar]
- 8.Bischoff, J. R., D. H. Kirn, A. Williams, C. Heise, S. Horn, M. Muna, L. Ng, J. A. Nye, A. Sampson-Johannes, A. Fattaey, and F. McCormick. 1996. An adenovirus mutant that replicates selectively in p53-deficient human tumor cells. Science 274:373-376. [DOI] [PubMed] [Google Scholar]
- 9.Bridge, E., and G. Ketner. 1989. Redundant control of adenovirus late gene expression by early region 4. J. Virol. 63:631-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carson, C. T., R. A. Schwartz, T. H. Stracker, C. E. Lilley, D. V. Lee, and M. D. Weitzman. 2003. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 22:6610-6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carvalho, T., J. S. Seeler, K. Ohman, P. Jordan, U. Pettersson, G. Akusjarvi, M. Carmo-Fonseca, and A. Dejean. 1995. Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J. Cell Biol. 131:45-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chardonnet, Y., and S. Dales. 1970. Early events in the interaction of adenoviruses with HeLa cells. I. Penetration of type 5 and intracellular release of the DNA genome. Virology 40:462-477. [DOI] [PubMed] [Google Scholar]
- 13.Chinnadurai, G. 1983. Adenovirus 2 Ip+ locus codes for a 19 kd tumor antigen that plays an essential role in cell transformation. Cell 33:759-766. [DOI] [PubMed] [Google Scholar]
- 14.Chinnadurai, G. 1998. Control of apoptosis by human adenovirus genes. Semin. Virol. 8:399-408. [Google Scholar]
- 15.Chinnadurai, G., S. Chinnadurai, and J. Brusca. 1979. Physical mapping of a large-plaque mutation of adenovirus type 2. J. Virol. 32:623-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiou, S. K., C. C. Tseng, L. Rao, and E. White. 1994. Functional complementation of the adenovirus E1B 19-kilodalton protein with Bcl-2 in the inhibition of apoptosis in infected cells. J. Virol. 68:6553-6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chow, L. T., T. R. Broker, and J. B. Lewis. 1979. Complex splicing patterns of RNAs from the early regions of adenovirus-2. J. Mol. Biol. 134:265-303. [DOI] [PubMed] [Google Scholar]
- 18.Dobbelstein, M. 2004. Replicating adenoviruses in cancer therapy. Curr. Top. Microbiol. Immunol. 273:291-334. [DOI] [PubMed] [Google Scholar]
- 19.Dobbelstein, M., J. Roth, W. T. Kimberly, A. J. Levine, and T. Shenk. 1997. Nuclear export of the E1B 55-kDa and E4 34-kDa adenoviral oncoproteins mediated by a rev-like signal sequence. EMBO J. 16:4276-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dobner, T., N. Horikoshi, S. Rubenwolf, and T. Shenk. 1996. Blockage by adenovirus E4orf6 of transcriptional activation by the p53 tumor suppressor. Science 272:1470-1473. [DOI] [PubMed] [Google Scholar]
- 21.Doronin, K., K. Toth, M. Kuppuswamy, P. Krajcsi, A. E. Tollefson, and W. S. Wold. 2003. Overexpression of the ADP (E3-11.6K) protein increases cell lysis and spread of adenovirus. Virology 305:378-387. [DOI] [PubMed] [Google Scholar]
- 22.Doucas, V., A. M. Ishov, A. Romo, H. Juguilon, M. D. Weitzman, R. M. Evans, and G. G. Maul. 1996. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 10:196-207. [DOI] [PubMed] [Google Scholar]
- 23.Flint, J., and T. Shenk. 1997. Viral transactivating proteins. Annu. Rev. Genet. 31:177-212. [DOI] [PubMed] [Google Scholar]
- 24.Greber, U. F., M. Willetts, P. Webster, and A. Helenius. 1993. Stepwise dismantling of adenovirus 2 during entry into cells. Cell 75:477-486. [DOI] [PubMed] [Google Scholar]
- 25.Green, M., K. H. Brackmann, L. A. Lucher, J. S. Symington, and T. A. Kramer. 1983. Human adenovirus 2 E1B-19K and E1B-53K tumor antigens: antipeptide antibodies targeted to the NH2 and COOH termini. J. Virol. 48:604-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harada, J. N., A. Shevchenko, A. Shevchenko, D. C. Pallas, and A. J. Berk. 2002. Analysis of the adenovirus E1B-55K-anchored proteome reveals its link to ubiquitination machinery. J. Virol. 76:9194-9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrison, T., F. Graham, and J. Williams. 1977. Host-range mutants of adenovirus type 5 defective for growth in HeLa cells. Virology 77:319-329. [DOI] [PubMed] [Google Scholar]
- 28.Huang, M. M., and P. Hearing. 1989. Adenovirus early region 4 encodes two gene products with redundant effects in lytic infection. J. Virol. 63:2605-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leppard, K. N., and T. Shenk. 1989. The adenovirus E1B 55 kd protein influences mRNA transport via an intranuclear effect on RNA metabolism. EMBO J. 8:2329-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis, J. B., and M. B. Mathews. 1980. Control of adenovirus early gene expression: a class of immediate early products. Cell 21:303-313. [DOI] [PubMed] [Google Scholar]
- 31.Linzer, D. I., and A. J. Levine. 1979. Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell 17:43-52. [DOI] [PubMed] [Google Scholar]
- 32.Liu, H., J. H. Naismith, and R. T. Hay. 2003. Adenovirus DNA replication. Curr. Top. Microbiol. Immunol. 272:131-164. [DOI] [PubMed] [Google Scholar]
- 33.Liu, T. C., G. Hallden, Y. Wang, G. Brooks, J. Francis, N. Lemoine, and D. Kirn. 2004. An E1B-19 kDa gene deletion mutant adenovirus demonstrates tumor necrosis factor-enhanced cancer selectivity and enhanced oncolytic potency. Mol. Ther. 9:786-803. [DOI] [PubMed] [Google Scholar]
- 34.Liu, T. C., Y. Wang, G. Hallden, G. Brooks, J. Francis, N. R. Lemoine, and D. Kirn. 2005. Functional interactions of antiapoptotic proteins and tumor necrosis factor in the context of a replication-competent adenovirus. Gene Ther. 12:1333-1346. [DOI] [PubMed] [Google Scholar]
- 35.Lucher, L. A., J. S. Symington, and M. Green. 1986. Biosynthesis and properties of the adenovirus 2 L1-encoded 52,000- and 55,000-Mr proteins. J. Virol. 57:839-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McNees, A. L., and L. R. Gooding. 2002. Adenoviral inhibitors of apoptotic cell death. Virus Res. 88:87-101. [DOI] [PubMed] [Google Scholar]
- 37.Miller, J. S., R. P. Ricciardi, B. E. Roberts, B. M. Paterson, and M. B. Mathews. 1980. Arrangement of messenger RNAs and protein coding sequences in the major late transcription unit of adenovirus 2. J. Mol. Biol. 142:455-488. [DOI] [PubMed] [Google Scholar]
- 38.Moran, E. 1993. Interaction of adenoviral proteins with pRB and p53. FASEB J. 7:880-885. [DOI] [PubMed] [Google Scholar]
- 39.Nettelbeck, D. M., and D. T. Curiel. 2003. Tumor-busting viruses. Sci. Am. 289:68-75. [DOI] [PubMed] [Google Scholar]
- 40.O'Shea, C., K. Klupsch, S. Choi, B. Bagus, C. Soria, J. Shen, F. McCormick, and D. Stokoe. 2005. Adenoviral proteins mimic nutrient/growth signals to activate the mTOR pathway for viral replication. EMBO J. 24:1211-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Shea, C. C., L. Johnson, B. Bagus, S. Choi, C. Nicholas, A. Shen, L. Boyle, K. Pandey, C. Soria, J. Kunich, Y. Shen, G. Habets, D. Ginzinger, and F. McCormick. 2004. Late viral RNA export, rather than p53 inactivation, determines ONYX-015 tumor selectivity. Cancer Cell 6:611-623. [DOI] [PubMed] [Google Scholar]
- 42.Ostapchuk, P., J. Yang, E. Auffarth, and P. Hearing. 2005. Functional interaction of the adenovirus IVa2 protein with adenovirus type 5 packaging sequences. J. Virol. 79:2831-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pilder, S., J. Logan, and T. Shenk. 1984. Deletion of the gene encoding the adenovirus 5 early region 1B 21,000-molecular-weight polypeptide leads to degradation of viral and host cell DNA. J. Virol. 52:664-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pilder, S., M. Moore, J. Logan, and T. Shenk. 1986. The adenovirus E1B-55K transforming polypeptide modulates transport or cytoplasmic stabilization of viral and host cell mRNAs. Mol. Cell. Biol. 6:470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Querido, E., P. Blanchette, Q. Yan, T. Kamura, M. Morrison, D. Boivin, W. G. Kaelin, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 15:3104-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rao, L., D. Modha, and E. White. 1997. The E1B 19K protein associates with lamins in vivo and its proper localization is required for inhibition of apoptosis. Oncogene 15:1587-1597. [DOI] [PubMed] [Google Scholar]
- 47.Rao, L., D. Perez, and E. White. 1996. Lamin proteolysis facilitates nuclear events during apoptosis. J. Cell Biol. 135:1441-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sarnow, P., P. Hearing, C. W. Anderson, N. Reich, and A. J. Levine. 1982. Identification and characterization of an immunologically conserved adenovirus early region 11,000 Mr protein and its association with the nuclear matrix. J. Mol. Biol. 162:565-583. [DOI] [PubMed] [Google Scholar]
- 49.Sauthoff, H., S. Heitner, W. N. Rom, and J. G. Hay. 2000. Deletion of the adenoviral E1b-19kD gene enhances tumor cell killing of a replicating adenoviral vector. Hum. Gene Ther. 11:379-388. [DOI] [PubMed] [Google Scholar]
- 50.Seth, P., D. J. P. Fitzgerald, M. C. Willingham, and I. Pastan. 1984. Role of a low-pH environment in adenovirus enhancement of the toxicity of a Pseudomonas exotoxin-epidermal growth factor conjugate. J. Virol. 51:650-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Soloway, P. D., and T. Shenk. 1990. The adenovirus type 5 i-leader open reading frame functions in cis to reduce the half-life of L1 mRNAs. J. Virol. 64:551-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stracker, T. H., C. T. Carson, and M. D. Weitzman. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418:348-352. [DOI] [PubMed] [Google Scholar]
- 53.Stracker, T. H., D. V. Lee, C. T. Carson, F. D. Araujo, D. A. Ornelles, and M. D. Weitzman. 2005. Serotype-specific reorganization of the Mre11 complex by adenoviral E4orf3 proteins. J. Virol. 79:6664-6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Subramanian, T., and G. Chinnadurai. 2003. Pro-apoptotic activity of transiently expressed BCL-2 occurs independent of BAX and BAK. J. Cell. Biochem. 89:1102-1114. [DOI] [PubMed] [Google Scholar]
- 55.Subramanian, T., M. Kuppuswamy, J. Gysbers, S. Mak, and G. Chinnadurai. 1984. 19-kDa tumor antigen coded by early region E1b of adenovirus 2 is required for efficient synthesis and for protection of viral DNA. J. Biol. Chem. 259:11777-11783. [PubMed] [Google Scholar]
- 56.Subramanian, T., M. Kuppuswamy, S. Mak, and G. Chinnadurai. 1984. Adenovirus cyt+ locus, which controls cell transformation and tumorigenicity, is an allele of lp+ locus, which codes for a 19-kilodalton tumor antigen. J. Virol. 52:336-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Svensson, U. 1985. Role of vesicles during adenovirus 2 internalization into HeLa cells. J. Virol. 55:442-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Symington, J. S., L. A. Lucher, K. H. Brackmann, A. Virtanen, U. Pettersson, and M. Green. 1986. Biosynthesis of adenovirus type 2 i-leader protein. J. Virol. 57:848-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takemori, N., C. Cladaras, B. Bhat, A. J. Conley, and W. S. M. Wold. 1984. cyt gene of adenoviruses 2 and 5 is an oncogene for transforming function in early region E1B and encodes the E1B 19,000-molecular-weight polypeptide. J. Virol. 52:793-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tauber, B., and T. Dobner. 2001. Molecular regulation and biological function of adenovirus early genes: the E4 ORFs. Gene 278:1-23. [DOI] [PubMed] [Google Scholar]
- 61.Thimmappaya, B., C. Weinberger, R. J. Schneider, and T. Shenk. 1982. Adenovirus VAI RNA is required for efficient translation of viral mRNAs at late times after infection. Cell 31:543-551. [DOI] [PubMed] [Google Scholar]
- 62.Tollefson, A. E., T. W. Hermiston, D. L. Lichtenstein, C. F. Colle, R. A. Tripp, T. Dimitrov, K. Toth, C. E. Wells, P. C. Doherty, and W. S. Wold. 1998. Forced degradation of Fas inhibits apoptosis in adenovirus-infected cells. Nature 392:726-730. [DOI] [PubMed] [Google Scholar]
- 63.Tollefson, A. E., J. S. Ryerse, A. Scaria, T. W. Hermiston, and W. S. Wold. 1996. The E3-11.6-kDa adenovirus death protein (ADP) is required for efficient cell death: characterization of cells infected with adp mutants. Virology 220:152-162. [DOI] [PubMed] [Google Scholar]
- 64.Tollefson, A. E., A. Scaria, T. W. Hermiston, J. S. Ryerse, L. J. Wold, and W. S. M. Wold. 1996. The adenovirus death protein (E3-11.6K) is required at very late stages of infection for efficient cell lysis and release of adenovirus from infected cells. J. Virol. 70:2296-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tollefson, A. E., A. Scaria, B. Ying, and W. S. M. Wold. 2003. Mutations within the ADP (E3-11.6K) protein alter processing and localization of ADP and the kinetics of cell lysis of adenovirus-infected cells. J. Virol. 77:7764-7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tollefson, A. E., K. Toth, K. Doronin, M. Kuppuswamy, O. A. Doronina, D. L. Lichtenstein, T. W. Hermiston, C. A. Smith, and W. S. M. Wold. 2001. Inhibition of TRAIL-induced apoptosis and forced internalization of TRAIL receptor 1 by adenovirus proteins. J. Virol. 75:8875-8887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tomko, R. P., R. Xu, and L. Philipson. 1997. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc. Natl. Acad. Sci. USA 94:3352-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Toth, K., H. Djeha, B. Ying, A. E. Tollefson, M. Kuppuswamy, K. Doronin, P. Krajcsi, K. Lipinski, C. J. Wrighton, and W. S. Wold. 2004. An oncolytic adenovirus vector combining enhanced cell-to-cell spreading, mediated by the ADP cytolytic protein, with selective replication in cancer cells with deregulated wnt signaling. Cancer Res. 64:3638-3644. [DOI] [PubMed] [Google Scholar]
- 69.Turnell, A. S., R. J. Grand, and P. H. Gallimore. 1999. The replicative capacities of large E1B-null group A and group C adenoviruses are independent of host cell p53 status. J. Virol. 73:2074-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Varga, M. J., C. Weibull, and E. Everitt. 1991. Infectious entry pathway of adenovirus type 2. J. Virol. 65:6061-6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Virtanen, A., P. Alestrom, H. Persson, M. G. Katze, and U. Pettersson. 1982. An adenovirus agnogene. Nucleic Acids Res. 10:2539-2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang, Y., G. Hallden, R. Hill, A. Anand, T. C. Liu, J. Francis, G. Brooks, N. Lemoine, and D. Kirn. 2003. E3 gene manipulations affect oncolytic adenovirus activity in immunocompetent tumor models. Nat. Biotechnol. 21:1328-1335. [DOI] [PubMed] [Google Scholar]
- 73.Watson, G., M. G. Burdon, and W. C. Russell. 1988. An antigenic analysis of the adenovirus type 2 fibre polypeptide. J. Gen. Virol. 69:525-535. [DOI] [PubMed] [Google Scholar]
- 74.Weinberger, L. S., J. C. Burnett, J. E. Toettcher, A. P. Arkin, and D. V. Schaffer. 2005. Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 Tat fluctuations drive phenotypic diversity. Cell 122:169-182. [DOI] [PubMed] [Google Scholar]
- 75.White, E. 2001. Regulation of the cell cycle and apoptosis by the oncogenes of adenovirus. Oncogene 20:7836-7846. [DOI] [PubMed] [Google Scholar]
- 76.White, E., S. H. Blose, and B. W. Stillman. 1984. Nuclear envelope localization of an adenovirus tumor antigen maintains the integrity of cellular DNA. Mol. Cell. Biol. 4:2865-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.White, E., T. Grodzicker, and B. W. Stillman. 1984. Mutations in the gene encoding the adenovirus early region 1B 19,000-molecular-weight tumor antigen cause the degradation of chromosomal DNA. J. Virol. 52:410-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wickham, T. J., P. Mathias, D. A. Cheresh, and G. R. Nemerow. 1993. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell 73:309-319. [DOI] [PubMed] [Google Scholar]
- 79.Wold, W. S., A. E. Tollefson, and T. W. Hermiston. 1995. E3 transcription unit of adenovirus. Curr. Top. Microbiol. Immunol. 199:237-274. [DOI] [PubMed] [Google Scholar]
- 80.Xia, D., L. J. Henry, R. D. Gerard, and J. Deisenhofer. 1994. Crystal structure of the receptor-binding domain of adenovirus type 5 fiber protein at 1.7 A resolution. Structure 2:1259-1270. [DOI] [PubMed] [Google Scholar]
- 81.Yan, W., G. Kitzes, F. Dormishian, L. Hawkins, A. Sampson-Johannes, J. Watanabe, J. Holt, V. Lee, T. Dubensky, A. Fattaey, T. Hermiston, A. Balmain, and Y. Shen. 2003. Developing novel oncolytic adenoviruses through bioselection. J. Virol. 77:2640-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yun, C. O., E. Kim, T. Koo, H. Kim, Y. S. Lee, and J. H. Kim. 2005. ADP-overexpressing adenovirus elicits enhanced cytopathic effect by induction of apoptosis. Cancer Gene Ther. 12:61-71. [DOI] [PubMed] [Google Scholar]
- 83.Zhang, C. T., Z. S. Lin, Z. Zhang, and M. Yan. 1998. Prediction of the helix/strand content of globular proteins based on their primary sequences. Protein Eng. 11:971-979. [DOI] [PubMed] [Google Scholar]
- 84.Zhang, W., and M. J. Imperiale. 2003. Requirement of the adenovirus IVa2 protein for virus assembly. J. Virol. 77:3586-3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zou, A., I. Atencio, W. M. Huang, M. Horn, and M. Ramachandra. 2004. Overexpression of adenovirus E3-11.6K protein induces cell killing by both caspase-dependent and caspase-independent mechanisms. Virology 326:240-249. [DOI] [PubMed] [Google Scholar]