Abstract
Specific targeting of the protein complexes formed by the Polycomb group of proteins is critically required to maintain the inactive state of a group of developmentally regulated genes. Although the role of DNA binding proteins in this process has been well established, it is still not understood how these proteins target the Polycomb complexes specifically to their response elements. Here we show that the grainyhead gene, which encodes a DNA binding protein, interacts with one such Polycomb response element of the bithorax complex. Grainyhead binds to this element in vitro. Moreover, grainyhead interacts genetically with pleiohomeotic in a transgene-based, pairing-dependent silencing assay. Grainyhead also interacts with Pleiohomeotic in vitro, which facilitates the binding of both proteins to their respective target DNAs. Such interactions between two DNA binding proteins could provide the basis for the cooperative assembly of a nucleoprotein complex formed in vitro. Based on these results and the available data, we propose that the role of DNA binding proteins in Polycomb group-dependent silencing could be described by a model very similar to that of an enhanceosome, wherein the unique arrangement of protein-protein interaction modules exposed by the cooperatively interacting DNA binding proteins provides targeting specificity.
In Drosophila, the bithorax complex (BX-C) is responsible for providing the identity of segments posterior to the mesothorax. This homeotic gene complex contains only three coding genes, while the major part of BX-C is arranged into a complex array of functionally autonomous, segment-specific cis-regulatory elements. During early development, the transiently expressed segmentation gene products establish the active or inactive states of the cis regulators, which is then maintained by the trithorax group (trx-G) and the Polycomb group (Pc-G) of genes, respectively, during the rest of development (31). The protein products of the trx-G and Pc-G genes are recruited as distinct protein complexes to the regulatory elements by specific target sites, called trithorax and Polycomb response elements (TRE and PRE, respectively) (12, 20, 51, 56).
Among the known PREs, the iab-7 PRE is particularly well characterized. In transgenic constructs, the iab-7 PRE induces silencing of reporter genes (23). Silencing is stronger when the transgene is homozygous, a phenomenon observed with most PREs and commonly referred to as pairing-sensitive silencing (PSS) (30). PSS by the iab-7 PRE depends on the products of Pc-G, as mutations in several members of this group, including Scm, Pc, Ph, Sce, Psc, z, etc., relieve PSS (10, 23). Five out of the six above-mentioned genes encode proteins that together form the functional core of the PRC1 protein complex, whereas SCM only substoichiometrically associates with it (18, 49). Transgenic constructs carrying the iab-7 PRE create a new binding site for PC, PSC, and PH, and PC was also shown to bind to the iab-7 PRE in vivo and in situ (10, 45). These data indicate that a PC-G complex, similar or identical in composition to PRC1, is targeted to the iab-7 PRE.
PC-G targeting to PREs involves the function of sequence-specific DNA binding proteins, such as Dorsal Switch Protein 1 (DSP1), C-terminal binding protein (CtBP), Pleiohomeotic (PHO), GAGA factor (GAF), and Pipsqueak (PSQ), which interact with GAF and recognize the same sequence motifs (33, 50). Mutations of these DNA binding proteins interact genetically with PREs, and most of them can also interact directly with PC-G proteins (15, 25, 27, 28, 38, 39, 48, 54). Nevertheless, their multimerized binding sites, or even synthetic combinations of them, failed to confer repression on marker genes in transgenic constructs, indicating that association of these DNA binding proteins with Pc-G members might not be sufficiently stable for generation of efficient silencing (1).
In agreement with this conclusion, DSP1, PHO, GAF, and PSQ are not subunits of PRC1 (49). The only sequence-specific DNA binding protein that associates stably with PRC1 is ZESTE, but unlike PRC1, ZESTE has no mammalian homologue, and even in Drosophila this gene is nonessential (34). These observations make ZESTE unlikely to be a sole targeting factor. Indeed, recent experiments indicated that the function of ZESTE in PRC1 is rather structural and, at least in part, independent of its DNA binding ability (42).
ZESTE can also interact with the SWI/SNF chromatin remodeling complex (29). Similarly to ZESTE, none of the above-mentioned DNA binding proteins are dedicated exclusively to Pc-G silencing. For example, PHO, which was characterized genetically as a bona fide member of the Pc-G, can interact not only with PC and PH but also with BRM, a subunit of the dSWI/SNF nucleosome remodeling complex (7, 40). Recruitment of the PC-PH or BRM interaction domain to the cat reporter gene promoter was shown to repress or activate transcription, respectively. However, since in vitro PRC1 inhibits chromatin remodeling by SWI/SNF, simultaneous recruitment of PRC1 and SWI/SNF by either PHO or ZESTE is unlikely to occur in vivo (51). Supporting this conclusion, it was shown that binding of PC and that of SWI/SNF complexes to chromatin are mutually exclusive events not only on the iab-7 PRE but throughout the genome (2, 14, 41).
How these known DNA binding proteins can efficiently and specifically target PC-G complexes to PREs remains an open question. Here we show that the DNA binding protein Grainyhead (GRH) is involved in this process. Similarly to ZESTE, GAF, PSQ, and PHO, Grainyhead alone is insufficient to efficiently target PC-G complexes, but its specific interaction with the PHO protein appears to contribute to the specificity of PC-G targeting.
MATERIALS AND METHODS
Fly work.
X-radiation was used to mutagenize isogenized rosy or Ore-R males that were crossed to Fab-72 females, and the F1 generation was screened for enhancers of the A6-to-A7 transformation. For descriptions of the mutants mentioned in this paper, see http://flybase.bio.indiana.edu or the appropriate publications cited later in the text. Preparation of abdominal cuticles was done according to reference 37.
DNA work.
The various DNA fragments used in this study were amplified from a genomic subclone of the whole Fab-7 region by PCR with the following primers: P1 (2419), TACCGACTAAGTCCGAGCAG; P2 (2515), CAACTTCCTTCGTCCGTCGG; P3 (2761), CGCACTGTCGTAGGCACG; P4 (2923), GTCGGCAATTCGGATTCCCG; P5 (2696), GTTGTGAAGTTCTTGCGACG; P6 (2677), CGTCGCAAGAACTTCACAAC; P7 (2563), GTCTGGGCGACGACGCAGTC; P8 (2582), GACTGCGTCGTCGCCCAGAC; P9 (2588), GTCCCTCGAAATTCCTCCGC; P10 (2611), GGGAGCGGAGGAATTTCGAG; P11 (2867), GCCGAGCTGAAAATGAAGAAC. Molecular coordinates are given in parentheses according to EMBL X78983.
Transgenic construct I, II, and III were made by insertion of P2-P4, P3-P4, and P2-P5 fragments into pCasPeR4, respectively (see Fig. 4E). Cloning was verified by sequencing the relevant portion of the constructs. Constructs were injected into w1118 embryos.
FIG. 4.
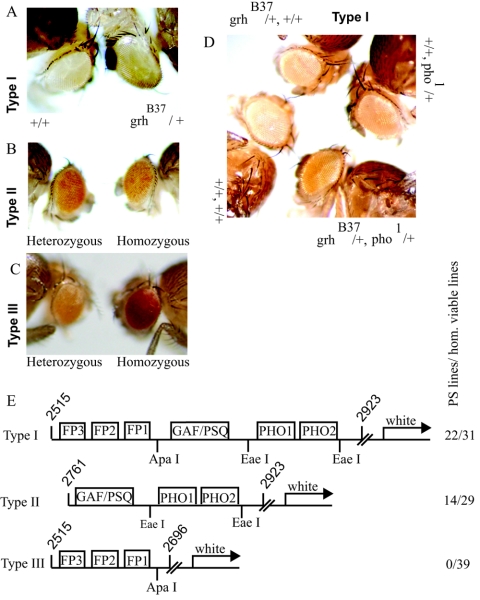
Transgenic constructs carrying iab-7 PRE fragments interact genetically with grh. (A) A typical homozygous type I transformant line that responds to grhB37. Note that in a wild-type background the reporter gene is completely repressed. Such strong silencing was observed in 7 out of the 22 established PS lines. (B) In the case of PS lines carrying a type II construct, the reporter gene is never completely silenced. (C) Type III transformant lines show no signs of pairing sensitivity. (D) In this type I transformant line, neither grhB37 nor pho1 caused a dramatic change in the eye color of the homozygous transgene. However, in the double-mutant background significant derepression of the reporter gene is evident, indicating synergism between grh and pho. (E) Schematic representation of the transgenic constructs. The boxes represent binding sites for DNA binding proteins. Molecular coordinates are given according to EMBL X78983. The number of PS versus homozygous (hom.) viable lines obtained for each construct is indicated on the right.
Full-length PHO and GRH (NTF-1) cDNAs were gifts from M. Vidal and R. Tjian, respectively (4, 21). These plasmids were used as templates to produce radioactively labeled proteins in a T7/T3 coupled reticulocyte lysate system (Promega). Luciferase cDNA was supplied with the kit.
GRH B/E is a part of the GRH protein which contains the full DNA binding domain and the C-terminal dimerization domain. This protein was used in in vitro DNA binding studies and as bait in a glutathione S-transferase (GST) pulldown assay and is referred to as bacterially expressed GRH in this report. The pGEX-GRH B/E expression plasmid was a gift from S. Bray (59).
DNA-protein and protein-protein interaction studies.
Nuclear extract preparation and gel mobility shift assays were performed according to reference 39. With the same amount of nuclear extract (4 to 5 μg in a 10-μl binding reaction mixture), both qualitative and quantitative features of our experiments are highly reproducible. Specific competitors were used at a 100-fold molar excess before the addition of nuclear extract as the last component of the binding reaction mixture. For the supershifts, 0.5 to 1 μl of the given antibody was added to a binding reaction mixture after a 0.5-h incubation of probe with nuclear extract and incubation was continued for another 30 min. Monoclonal GRH and polyclonal PHO antibodies were provided by S. Bray and P. Verrijzer, respectively (5, 40). For the experiments presented in Fig. 6, GRH B/E and PHO were expressed as GST fusion proteins and liberated from the affinity matrix by thrombin cleavage. Binding reaction mixtures were incubated on ice for 30 min, and electrophoresis was performed in a cold room.
FIG. 6.
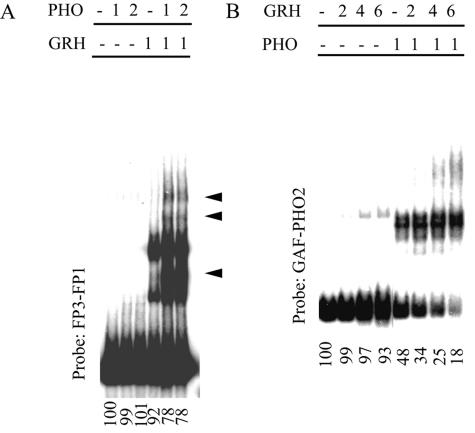
Mutual enhancement of DNA binding between PHO and GRH. (A) A constant amount of GRH is titrated in the presence of increasing amounts of PHO. The probe fragments used here cover the FP3 and FP1 sites but lack the PHO sites (see also Fig. 4E). GRH-PHO complexes are marked by arrowheads. (B) Binding of a constant amount of PHO in the presence of increasing amounts of GRH to a probe which contains only the PHO sites without the FP3 and FP1 sites (see Fig. 4E). A discrete PHO-GRH complex was not detected. Nevertheless, cooperativity can be deduced from the decreasing amount of free DNA, which faithfully reflects the original equilibrium situation even if the specific complexes dissociate during electrophoresis. The amount of free DNA quantified by a PhosphorImager is shown below each lane as a percentage of the input DNA. The amounts of GRH and PHO are marked as relative units above the lanes.
Footprinting reactions were performed in gel shift binding buffer, but less or no nonspecific competitor was used and the buffer was supplemented with polyvinyl alcohol to 2%. It was necessary to use about 30- to 40-fold more DNase I in binding reaction mixtures where nuclear extract was used to achieve comparable digestion of DNA samples.
For the GST pulldown assay, 1.5 to 2 μg of purified GST or GST-GRH B/E was bound to 25 μl of glutathione-Sepharose beads and after washing in gel shift binding buffer radioactively labeled proteins and ethidium bromide to 100 μg/ml were added. After a few hours of incubation at 4°C, beads were washed thoroughly in binding buffer and bound material was eluted in sodium dodecyl sulfate sample buffer.
RESULTS
grainyhead interacts with the iab-7 PRE in vivo.
The iab-7 PRE lies next to the Fab-7 boundary, a chromatin domain insulator element between the neighboring iab-6 and iab-7 cis-regulatory domains of BX-C. Fab-7 ensures the functional autonomy of these cis-regulatory domains, as iab-7 is inactive in the sixth abdominal segment (A6), where iab-6 is active, while iab-7 is activated in segment A7. A large set of internal BX-C deficiencies is available, making this region ideal for genetic studies (37).
Class II deletions, which remove only the boundary region, fuse the otherwise intact cis-regulatory elements iab-6 and iab-7 (Fig. 1F). The consequence of this fusion is that in some A6 cells iab-6 is inactivated by iab-7, while in some other A6 cells iab-6 ectopically activates iab-7. As a result, A6 will become a mixture of cell clones with either A5 or A7 identity. Due to the fact that the Abd-B gene, the expression of which is controlled by these cis regulators, is haploinsufficient, such transformations are evident even under heterozygous conditions (Fig. 1A, left side). Class I deletions, which remove both the Fab-7 boundary and the adjacent iab-7 PRE, transform A6 into a perfect copy of A7 (Fig. 1B, left side), suggesting that in the case of class II deletions it is the iab-7 PRE that mediates the inactivation of iab-6 in A6; thus, the inactivation may depend on Pc-G-mediated silencing. Indeed, if a class II deletion is combined with some, but not all, Pc-G mutations, the resulting phenotype is indistinguishable from that of class I deletions (37). Based on this result, it should be possible to identify mutations in factors that specifically interact with the iab-7 PRE as enhancers of the phenotype of class II deletions.
FIG. 1.
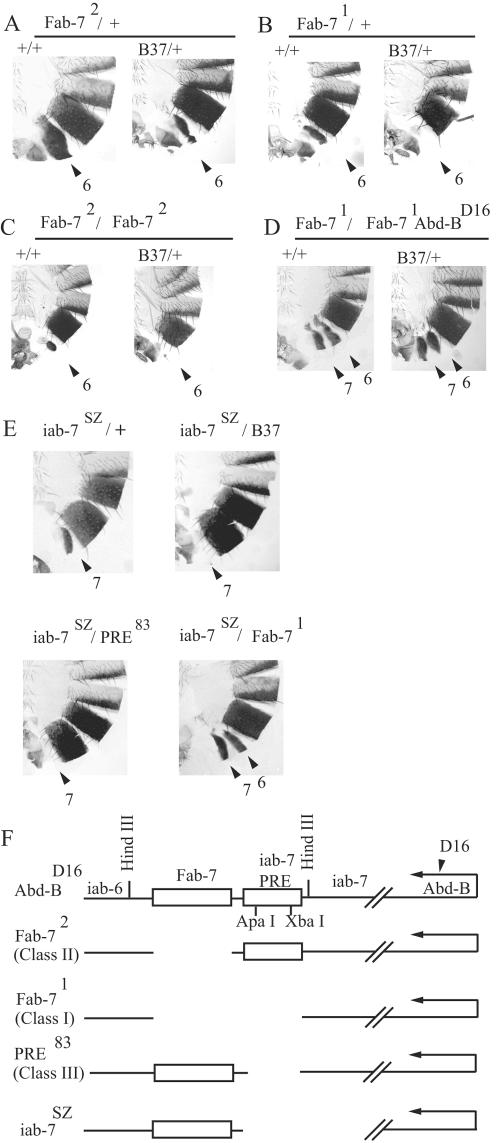
grainyhead interacts with the iab-7 PRE. (A to D) Abdominal cuticle preparations of adult male flies carrying internal BX-C deletions in the wild-type (left side of each panel) or grhB37 mutant (right side of each panel) background. The BX-C deletions are Fab-72/+ (A), Fab-71/+ (B), Fab-72/Fab-72 (C), and Fab-71/Fab-71 Abd-BD16 (D). The numbered arrows show the relevant abdominal segments. A7 normally does not have a visible tergite, and the size reduction of the A6 tergite indicates a partial transformation toward an A7 identity of this segment. Increasing reduction of the tergite size indicates increasingly stronger transformation. (E) grainyhead interaction with the iab-7SZ deletion can be also detected even though the boundary function remains unaffected by this deletion. Note that the phenotype of grhB37/iab-7SZ transheterozygous flies is very similar to that of class III/iab-7SZ flies, while it is markedly different from that of the Fab-71/iab-7SZ combination. (F) Schematic representation of BX-C mutants. For exact molecular coordinates, see reference 37. The Fab-7 boundary and the iab-7 PRE are represented by boxes.
Accordingly, we performed several X-ray mutagenesis screenings with the class II allele Fab-72. Among the enhancer mutants, one complementation group, represented by five alleles in our collection, is described here. Two alleles are associated with a cytologically visible breakpoint in 54F, and deficiency mapping placed the locus between the proximal breakpoints of the Pcl11b and Pcl7b deletions. Previously, four complementation groups were isolated within this interval (6). Noncomplementation with alleles of one of the four complementation groups showed that we isolated new mutant alleles of the previously described gene grainyhead (grh) (6). The previously isolated grh alleles, including the molecularly characterized amorphic allele B37, are also strong Fab-72 enhancers, indicating that loss-of-function grh mutations affect the function of the iab-7 PRE (Fig. 1A).
Surprisingly, subsequent analysis revealed that the phenotypes of not only class II deficiencies, but those of class I deficiencies, such as Fab-71, are also enhanced by grh alleles (Fig. 1B). Nevertheless, the last result does not conflict with a possible role for grh in the iab-7 PRE. Earlier studies showed that class III deficiencies (which remove the iab-7 PRE only) also enhance the phenotype of both class I and class II deficiencies, suggesting, in accordance with the PSS phenomenon, that in situ the two iab-7 PREs interact with each other. This hypothesis is further supported by the observation that the class III deficiencies show a gain-of-function phenotype under hemizygous or homozygous, but not under heterozygous, conditions (37).
In order to definitely prove that grh acts on the iab-7 PRE and not on the Fab-7 boundary, we tested grhB37 in a homozygous Fab-72 background, where the boundary is deleted in both chromosomes. Tergite clones in A6 of Fab-72 homozygous flies are tiny but still discernible. In a grhB37 background, this phenotype was further enhanced, resulting in complete loss of A6 tergites (Fig. 1C). This shows that the sequence required for GRH to play a role in the ectopic inactivation of iab-6 is not deleted in Fab-72. To test the effect of grh mutations on a homozygous class I allele, we used the Fab-71/Fab-71 Abd-BD16 background. The reason for using this mutant combination is that homozygous class I alleles transform A6 into a perfect copy of A7, resulting in complete absence of the A6 tergite in males. However, in this mutant combination, due to the presence of a point mutation in Abd-B, the regulator capacity of the cis fusion element is lost on the Abd-BD16 chromosome, resulting in visible, similar-size tergites in the A6 and A7 segments (52). This phenotype was not changed in a grhB37 background, showing that all of the sequence that requires GRH is removed by this class I deletion (Fig. 1D). The fact that grh is epistatic over a class I, but not over a class II, deficiency clearly indicates that GRH acts on the iab-7 PRE.
This interpretation also predicts that grh should interact genetically with other Bx-C deficiencies deleting the iab-7 PRE, even if the boundary remains intact. The iab-7SZ deletion removes only the iab-7 cis-regulatory region, while it leaves the Fab-7 boundary unaffected. As anticipated, the loss-of-function phenotype of heterozygous iab-7SZ flies is suppressed in the grhB37 background (Fig. 1E). A genetic combination of a PRE deletion with iab-7SZ gave essentially the same result, while the combination of iab-7SZ with the class I allele Fab-71 resulted in a strikingly different phenotype (Fig. 1E) (52). These results also support the view that the genetic interaction of grh with Fab-7 mutations is fully attributable to the requirement of GRH for proper functioning of the iab-7 PRE.
We do not detect any homeotic transformation in flies heterozygous for grh mutations. This is in contrast to many (but not all) Pc-G gene mutations, which often have weak, dominant homeotic phenotypes such as extra sex combs on the second and third legs. Moreover, mutant grh alleles do not show strong genetic interactions with other Pc-G mutations tested, including alleles of most members of the PRC1 complex. In this respect, the Sce gene, which encodes the PRC1 subunit RING, is a notable exception (19, 22). While only a few Sce1/+ males have sex combs on their posterior legs, we observed extra sex combs on the second and third legs of nearly all male flies with the genotype Sce1/+ grhB37/+ (Fig. 2).
FIG. 2.
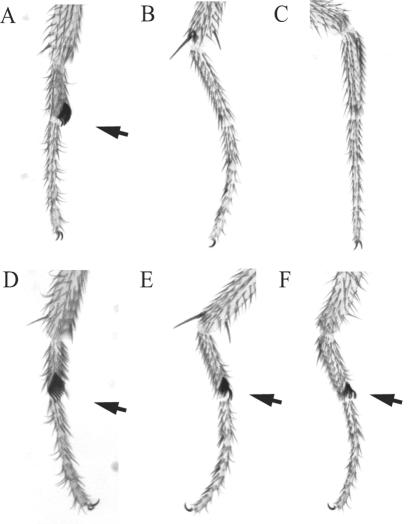
grainyhead interacts genetically with Sex combs extra (Sce). The first, second, and third legs of wild-type (A to C) and grhB37/+ Sce1/+ (D to F) male flies are shown. Arrows point to sex combs, whose presence on the second and third legs indicate partial transformation of the respective thoracic segments (T2 and T3) into a T1 identity, which is a characteristic phenotype of certain Pc-G alleles and allele combinations (8). The average number of legs having at least one sex comb tooth is 2.2 in Sce1 heterozygous male flies (n = 177) and 5.4 in grhB37/+ Sce1/+ transheterozygous males (n = 220). We have never observed ectopic sex combs on grh heterozygous flies.
Lack of a dominant homeotic phenotype and interaction with Sce1 also characterize the mutations of the Pc-G gene pleiohomeotic (pho) (8). The similarity between grh and pho is further strengthened by the finding that pho alleles also enhance the phenotype of both Fab-72/+ and Fab-71/+ (data not shown).
GRH interacts with the iab-7 PRE in vitro.
GRH, also known as NTF-1 and Elf-1, was originally characterized as a transcription factor involved in transcriptional activation by TFIID through direct interaction with the subunits TAF60 and TAF150 (11). In addition, GRH was also shown to physically interact with RING, a subunit of PRC1 in both Drosophila and humans (57). Drosophila and mammalian RING is encoded by a Pc-G gene, linking GRH to Pc-G silencing (16, 19, 22). It is therefore possible that GRH plays a role in the silencing function of the iab-7 PRE by directly binding and recruiting the silencing complex to it. However, another trivial possibility is that grh act as a positive regulator of Pc-G genes and therefore contributes to iab-7 PRE-mediated silencing only indirectly.
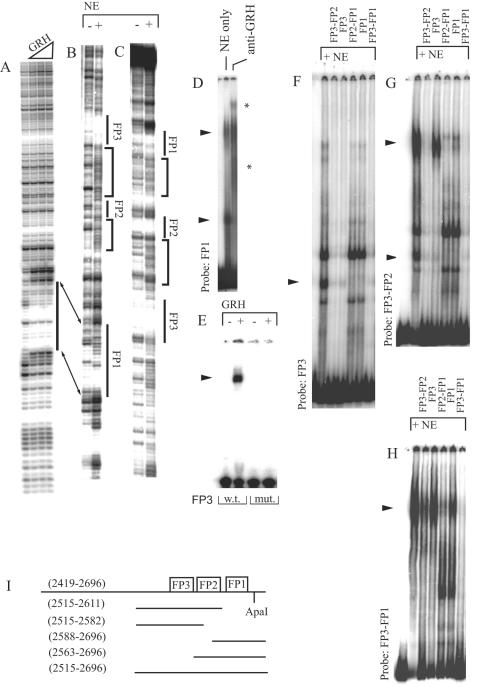
To distinguish between these two possibilities, we used DNase I footprinting in order to test if the bacterially expressed GRH protein can bind to the iab-7 PRE in vitro. These experiments led to the identification of one binding site, FP1, which was also protected when nuclear extract was used as a protein source (Fig. 3A to C). In gel shift experiments with nuclear extract, the formation of two complexes was detected on this binding site and both were recognized by anti-GRH antibody, supporting the idea that the FP1 binding activity corresponds to GRH (Fig. 3D).
FIG. 3.
DNase I footprinting demonstrates GRH binding to the iab-7 PRE. The probe fragment used here was amplified by the P1 and P5 primers (see Materials and Methods) and labeled at the proximal end (A and B) or at the distal end (C). Bacterially expressed GRH was used as the protein source for the experiment shown in panel A, and nuclear extract (NE) was used for the experiments shown in panels B and C, where the resulting footprints (FP) are numbered. In panel B, instead of clear protection of the sequence, protein binding to FP1 is indicated only by the increased DNase I sensitivity around the FP1 site. Note that the marked changes in the DNase I digestion pattern between FP1-FP2 and FP2-FP3, indicated by brackets on both strands, are highly compatible with the short-range looping model described in the text. FP2 corresponds to the sequence TCGCAGAAAG. (D) Anti-GRH antibody recognizes both complexes formed on a single FP1 site in a gel shift assay. GRH-specific shifts are marked by arrowheads throughout. Supershifts are indicated by asterisks. (E) In a gel shift experiment, bacterially expressed GRH binds to a wild-type (w.t.) FP3 oligonucleotide (CTGCATTTTTTTTGTTTTTGTCT) but not to the mutated (mut.) sequence (CTGCATTTTTTTTCACTTTGTCT[the mutation is underlined]). Gel shifts demonstrate cooperative binding of GRH. (F) GRH binding to the FP3 motif in a gel shift assay can be deduced from the competition of the marked shifts by nonoverlapping GRH binding site FP1 (the applied competitor fragments are referred to by their proximal and distal binding sites and are indicated above the lanes). (G) The upper GRH-specific shift detected with the FP3-FP2 probe is essentially absent from the FP3 probe, which indicates context-dependent, cooperative interaction between FP3 and FP2. (H) Another level of cooperativity is suggested by the experiment with the FP3-FP1 probe, since the formation of the single complex on this probe cannot be interpreted as the sum of the pattern of binding to its subfragments (compare with panels D, F, and G). Moreover, subfragments of the probe could not compete as efficiently for binding as the large fragment itself. (I) Schematic representation of the subfragments used as probes and as competitors in the gel shift experiments. Binding sites are represented by boxes. Molecular coordinates are shown in parentheses according to EMBL X78983.
We found a second DNA sequence within the iab-7 PRE with some degree of similarity to FP1 (Table 1). Interestingly, this sequence, named FP3, was not protected against DNase I cleavage by the bacterially expressed GRH protein but was clearly protected when nuclear extract was used (FP3 in Fig. 3B and C). Nevertheless, an oligonucleotide corresponding to the FP3 sequence was faintly shifted by the bacterially expressed GRH protein in a gel shift assay, indicating that this sequence can serve as a weak binding site for GRH (Fig. 3E).
TABLE 1.
| Name | Sequence |
|---|---|
| FP1 | CACAATCCTGTTTTTT |
| FP3 | GCATTTTTTTTGTTTTTG |
| Ftz I | TGATTCGGTATCGAAA |
| Ftz II | GGGAGCTGTTTTTTCC |
| Ftz IIIa | TCGGATGGTTTTATAT |
| Ftz IIIb | AATAAGGGTGTAGAGT |
| Ftz IV | CTTTCTGGTTTTGCAG |
| Ubx | CAATCTGGTTTTGAGC |
| Ddc be-1 | GAAACCGGTTAT |
| Ddc be-2 | TGAACCGGTCCTGCGG |
We surmised that cooperative interaction with an another binding activity may facilitate the binding of GRH to its otherwise weak FP3 site when nuclear extract is used. In the case of such a short-range interaction, an altered DNase I cleavage pattern is expected between the binding sites of the cooperative partners in footprinting experiments as a result of looping of the intervening DNA (24). Such an effect was indeed observed between FP3 and another binding site, FP2, and also between FP1 and FP2 (marked by brackets in Fig. 3B and C).
We tested the contribution of the FP2 site to FP3 binding in gel shift assays by using different subfragments of the DNA used for the footprinting experiments (these subfragments are referred to by the binding sites they carry; a more detailed description is given in Fig. 3I and Materials and Methods). GRH binding to FP3 and FP3-FP2 was clearly detected with nuclear extract (Fig. 3F and G). The shifts observed with these FP3-containing probes could be competed by the nonoverlapping GRH binding site FP1, indicating that these protein-DNA complexes are indeed the consequences of GRH binding. Importantly, the upper GRH-specific shift in Fig. 3G was detected only with probes that also contain the FP2 site, indicating a context-dependent interaction between this binding activity and FP3 (compare Fig. 3F and G). As could be anticipated from the presumed cooperativity, FP3 alone was a much weaker competitor of this shift than FP3-FP2 (Fig. 3G). Cooperativity between FP2 and FP1 was also observed, but a contribution of FP2 to the strength of GRH binding was less evident (data not shown). These results support the notion that FP2 has a role in GRH binding.
With the FP3-FP2-FP1 subfragment as a probe, only one shift was observed. This shift could also be competed by the FP1 and FP3 subfragments, indicating that this single shift is still the consequence of GRH binding (Fig. 3H). It is also clear that FP3 is a weaker competitor than FP1, in agreement with the finding that FP3 is a lower-affinity binding site for GRH. Weak, if any, competition was observed with an oligonucleotide overlapping only the FP2 site (data not shown). Nevertheless, in concert with the hypothesis that FP2 cooperates with GRH, in the presence of the FP2 site, both FP3 and FP1 compete better for this single shift. This effect was more prominent on the competing ability of the FP3 site (Fig. 3H). Significantly, none of the subfragments of the FP3-FP2-FP1 fragment competed as well as FP3-FP2-FP1 itself, indicating that GRH binding to this fragment is stronger than that to any of its subfragments. This suggests another level of cooperation. GRH can form homodimers, which was shown to increase the affinity of GRH for its binding sites, and we also suspect that dimerization is required for further stabilization of the complex formed on the FP3-FP2-FP1 fragment (4, 59). However, the stretch of DNA between the FP3 and FP1 GRH binding sites is short, and cooperative interactions would require looping of the intervening sequences, which has energy constraints. Looping of a short stretch of DNA would need the aid of DNA binding proteins to bend it (62). As the FP2 site is about halfway between the GRH binding sites, FP1 and FP3, the function of this unidentified protein may not only cooperate with single GRH sites but also promote their interaction by DNA bending. Addressing these possibilities directly would require identification of the FP2 binding factor.
Eukaryotic DNA binding proteins usually recognize short, often degenerated sequence motifs. Thus, for a protein like GRH, tens of thousands of nonfunctional binding sites, which may be called pseudosites, are expected to occur in the Drosophila genome. However, the experiments presented above indicate that several levels of cooperativity are required for efficient in vitro binding of GRH to the iab-7 PRE. This result would not be expected in the case of a pseudosite and supports the hypothesis that GRH plays a direct role, unrelated to its function as a transcription factor, in the functioning of the iab-7 PRE.
grh interacts genetically with pho in transgenic constructs.
In order to further substantiate that GRH binds to the iab-7 PRE in vivo, we have generated transgenic constructs carrying various iab-7 PRE fragments upstream of the mini-white reporter gene and examined the influence of the grhB37 mutant background on pairing-dependent silencing (Fig. 4). We used three constructs for this purpose, a type I construct containing the in vitro GRH binding sites along with the GAF/PSQ, DSP1, and PHO consensus sites; a type II construct containing the GAF/PSQ, DSP1, and PHO consensus sites but lacking the GRH binding sites; and a type III construct containing only the GRH binding sites (Fig. 4E).
The type I transgenic construct responds to the dosage of grh, as the eye color of four out of the eight examined homozygous pairing-sensitive (PS) lines became darker in the grhB37 background (Fig. 4A). Transformant lines containing the type II construct, although frequently PS, do not respond to the presence of grhB37, as revealed bycomparing the eye colors of grhB37/+ type II/type II and +/+ type II/type II flies (seven PS lines were tested). Since reduction of the GRH dosage could influence reporter gene expression only when the GRH binding sites were present in the construct, we concluded that these sites also bind GRH in vivo. We established 39 homozygous viable transformant lines of type III construct, but none of them were PS (Fig. 4C). This led us to conclude that, like other binding sites occurring in PREs, GRH sites on their own are insufficient to generate PSS (1).
An inherent limitation of the transgenic approach is that both the silencing and the expression of the reporter gene could be subject to genomic position effects. Nevertheless, it is noteworthy that the overall degree of silencing observed in type II lines was considerably lower compared to that of type I lines (compare Fig. 4A, B, and C). This may indicate that the contribution of GRH to silencing in type I lines is to promote the stability of the binding of another silencing factor. This is supported by the effect of the double-mutant combination pho1/+ grhB37/+ on the eye color of some of the type I transformant lines. As shown in Fig. 4D, grhB37 alone has no effect on this particular line while pho1 derepresses the transgene only weakly. By contrast, the eye color is much darker in the double-mutant combination, suggesting that GRH and PHO are not only involved in the silencing process but, at least in the case of this particular transgene, interact cooperatively.
GRH interacts with PHO in vitro.
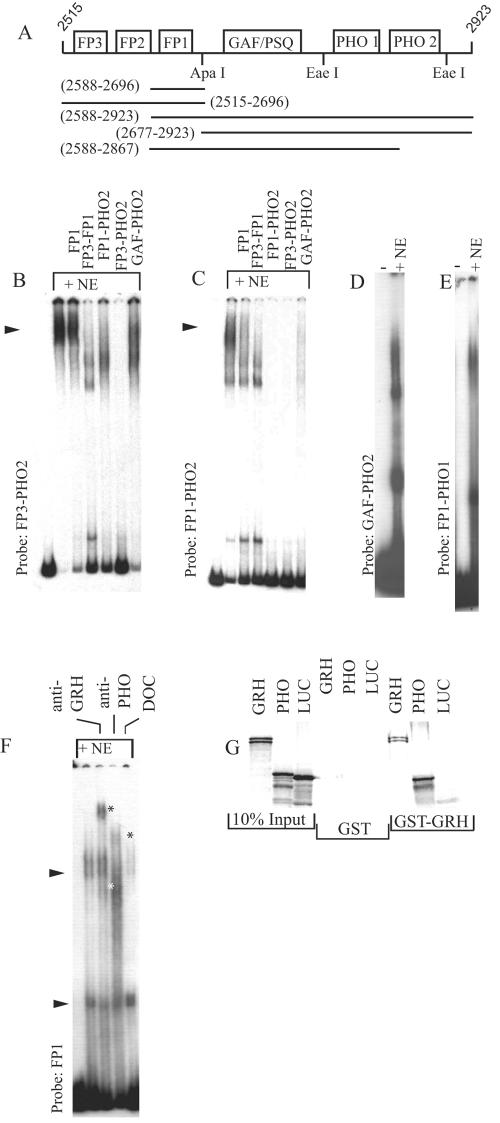
In order to establish that GRH and PHO cooperate in vitro, we employed the gel shift assay with a fragment in which the FP3-FP2-FP1 sites were present along with the GAF/PSQ, DSP1, and PHO consensus sites (Fig. 5A). Two signs of cooperativity are evident: (i) in the presence of the same amount of nuclear extract used in the gel shift experiments described above, almost all of this probe DNA is complexed, leaving only a small amount unbound, and (ii) the resulting DNA-protein complex is refractory to competition by subfragments of the probe DNA (Fig. 5B; compare the competition ability, for example, of a single FP1 site in the gel shift experiments presented in Fig. 3F and G with that shown in Fig. 5B). With a shorter probe that lacks the FP3 and FP2 sites, relatively high-affinity binding was still observed, although cooperativity is less spectacular in this case (Fig. 5C). Deleting either the FP1 or the distal PHO site from the probe fragment resulted in an apparent loss of cooperative binding (Fig. 5D and E). These experiments indicate that the presence of various binding sites on the longer DNA probes facilitates and reinforces interactions in such a way that a more stable complex is formed.
FIG. 5.
A highly cooperative nucleoprotein complex is formed on the iab-7 PRE in vitro. (A) Schematic representation of probes and subfragments. Binding sites for DNA binding proteins are represented by boxes. Molecular coordinates are given in parentheses according to EMBL X78983. These subfragments are referred to by their proximal and distal binding sites throughout. (B) When the FP3-PHO2 template is used in gel shift experiments, almost all of the probe is bound and the resulting complex is highly resistant to competition either by the single binding site FP1 or even by the GAF/PSQ-PHO2 subfragment, indicating cooperative assembly of a protein complex. (C) Signs of cooperativity are still evident, although to a lesser extent, when the FP1-PHO2 template is used. Further truncation of this probe to remove the FP1 or the PHO2 site results in loss of cooperative binding (panels D and E, respectively). (F) The protein complex formed on the FP1 GRH site is supershifted by both anti-GRH and anti PHO, indicating that GRH and PHO are members of the same protein complex. Supershifts are indicated by asterisks. The effect of deoxycholate (DOC) is also shown; this detergent eliminated the upper, but not the lower, shift at a concentration of 0.1%, indicating that the upper shift is the result of a detergent-sensitive protein-protein interaction. (G) In a GST pulldown experiment, PHO interacts with GRH. GRH homodimerization (4, 59) was used as a positive and luciferase (LUC) as a negative control for binding. PHO binds to the GST-GRH matrix but not to GST alone.
In another set of gel shift experiments, we used the FP1 site as a probe and detected the formation of two GRH-specific shifts with nuclear extract (Fig. 3D). Of these two shifts, the upper one was recognized by both anti-GRH and anti-PHO antibodies (Fig. 5F). This result clearly indicates that GRH and PHO are components of the same complex. We tested if the two proteins can interact with each other directly in a pulldown experiment. PHO binding to the GST-GRH, but not to the GST, matrix was evident, demonstrating that these proteins can interact directly (Fig. 5G). Since this interaction is conserved from Drosophila to humans, it is very likely that their interaction domains, not yet identified, are also conserved (47).
Interaction of GRH and PHO increases affinity for their binding sites.
To see the functional consequence of the GRH-PHO interaction, we tested the binding of bacterially expressed GRH to its binding site in the presence of increasing amounts of bacterially expressed PHO. A GRH-PHO complex was clearly observed, along with a concomitant decrease in the unbound probe DNA, indicating that binding of PHO to GRH increases its affinity for DNA (Fig. 6A). In a reciprocal experiment, we tested the effect of GRH on PHO binding to its DNA target. Once again, the sharp decrease in the amount of free DNA indicated that GRH increases the affinity of PHO for its binding sites (Fig. 6B). Interestingly, a concomitant increase in a discrete PHO-GRH complex was not observed in this experiment. Instead, we observed a smear above the discrete PHO-specific shifts in the simultaneous presence of GRH, indicating that the PHO-GRH-specific complex is unstable and gradually dissociates during electrophoresis.
If GRH and PHO mutually increase each other's ability to bind DNA, their effect would be expected to be more prominent when a probe containing their binding sites in cis is used (62). Contrary to this expectation, we were unable to convincingly reproduce cooperative binding in vitro with such DNA probes and bacterially expressed GRH and PHO (data not shown). Conceivably, the DNA between the GRH and PHO binding sites prevents the expected interaction. Thus, GRH-PHO interaction is very likely to be necessary but insufficient to recapitulate the cooperative assembly observed when nuclear extract is used, and other, as yet unidentified, proteins may be involved in this process.
DISCUSSION
Genome-wide prediction indicated that the occurrence of the same limited set of consensus motifs can fairly accurately predict the PRE function of a DNA sequence (46). This observation suggests that many, if not all, PREs use the same set of DNA binding proteins. One of the frequently occurring consensus sequences within PREs is a poly-T motif. Many, although not all, GRH binding sites are T rich, and our studies indicate that at least in some cases the poly-T consensus sequence may be a binding site for this protein. However, like other DNA binding proteins involved in PRE function, GRH alone cannot explain the specificity of targeting, since its function is not limited to PREs. In other contexts, GRH acts as a transcriptional activator (11). The fact that an array of distinct sequence motifs is required to accurately predict PREs probably means that there is no single major targeting activity. Indeed, in the case of the engrailed PRE it was demonstrated that all binding sites of DNA binding proteins are equally important for silencing activity (1). Identification of GRH as a PRE-related DNA binding protein and, in particular, its cooperative interaction with another member of this group both in vivo and in vitro may help in understanding the targeting of PC-G to PREs during development.
Recently, a cooperative interaction between GAF and PHO has been demonstrated (35). In contrast to the case of GRH and PHO, cooperation between GAF and PHO is independent of the physical interaction between the two proteins and requires a nucleosomal context. Although the physical basis of this cooperative interaction is not understood, it also suggests that cooperativity may be an important principle in the organization of nucleoprotein assembly at PREs.
Plurality and diversity of components but unity of purpose.
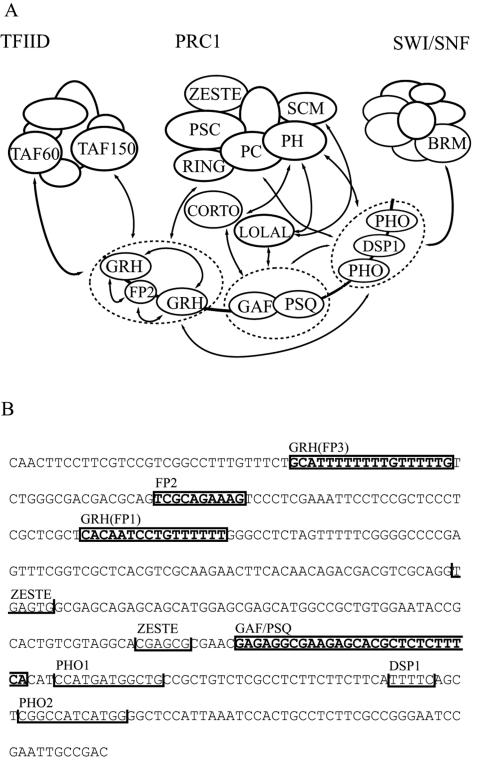
What could be the impact of cooperativity on PC-G targeting? Theoretically, one of the most significant problems encountered by a DNA binding protein is the huge excess of potential binding sites in the genome, including both functional sites and pseudosites. It can be assumed that if any of the DNA binding proteins involved in targeting are present in limited amounts in the nucleus, then their binding occurs only at the highest-affinity sites, where a combination of certain binding sites facilitates their cooperative binding. Several observations contradict this simple model. First, if the amount of these DNA binding proteins were limited, their mutations would be expected to result in strong haploinsufficient phenotypes, which is not the case. Second, studies on the DNA binding proteins EVE, FTZ, and GAF demonstrated that in vivo they also bind to genes that are not controlled by them. These functionally irrelevant sequences may represent pseudosites, and the relatively low level of binding at these sites may indicate a low binding affinity (44, 61). Thus, it appears that restricted binding site occupancy of DNA binding proteins is not necessary for specificity in gene regulation. Likewise, even though the DNA binding proteins present on PREs may bind to nonfunctional sites, it is likely that the functionally relevant high-affinity sites are distinguished from pseudosites in vivo by the unique arrangement of distinct, stably bound cooperative partners. However, although in our model of targeting of PRC1 to the iab-7 PRE, cooperativity at the level of the DNA binding proteins is critically required for binding stability, by itself it is insufficient to provide the required specificity of the targeting process (Fig. 7A).
FIG. 7.
(A) Model of PRC1 targeting to iab-7 PRE. For simplicity, similar DNA binding proteins are circled and treated as single targeting domains. Cooperative interactions between DNA binding proteins and subunits of PRC1 are indicated by arrows. These interactions are described in the text and in references 15, 38, 40, 48, and 57. Based on the available data, the interaction of GAF with PRC1 subunits requires the adaptor proteins LOLA-LIKE and CORTO (38, 48). The possible roles of other proteins, such as RYBP and CtBP, should also be considered (3, 21, 54). The putative GAF-PHO interaction would require a nucleosomal context (35). TFIID can interact with GRH (11), whereas SWI/SNF can interact with PHO (40). Therefore, it would be conceivable that these two complexes can also be recruited to the iab-7 PRE. According to our model, however, stable recruitment of TFIID and SWI/SNF is unlikely to occur because their binding is not supported by other interacting modules in the context of the iab-7 PRE. (B) Locations of binding sites for regulatory proteins within the largest iab-7 PRE fragment used in this study are shown. Closed boxes indicate footprinted sequences. The boundaries of the FP3, FP2, and FP1 footprints are derived from experiments with nuclear extract, and the FP1 site was also verified by using purified protein. Protection of the GAF/PSQ motif was detected with nuclear extract (data not shown; 39). Consensus sites for the PHO, ZESTE, and DSP1 proteins are indicated by open boxes (14, 15, 39).
In contrast to the DNA binding components, other constituents of the silencing complex appear to be limiting factors. This is suggested by the fact that most Pc-G genes were identified either on the basis of their characteristic haploinsufficient phenotypes or on the basis of their dominant genetic interaction with other known Pc-G members (8). The number of potential PRE sequences is also relatively small, as a genome-wide survey estimated it to be not more than a few hundred in Drosophila (46). This brings us to the question of how the abundant DNA binding proteins link the limited amount of PC-G complexes to the low-frequency target sites with high specificity.
The first clue comes from studies showing that all of the PRE DNA binding proteins have the ability to interact with various PC-G proteins that are all subunits of the same preformed protein complex, PRC1 (summarized in Fig. 7A). These interactions appear to be weak by themselves, as illustrated by the fact that although the occurrence of these interactions can be demonstrated by using short protocols like immunoprecipitation, the resulting complexes do not survive nonequilibrium methods used for traditional biochemical purification of protein complexes. The consequence of the cooperativity at the level of DNA binding proteins is that the otherwise weak interaction surfaces are integrated into a stable composite surface that can serve as a high-affinity docking site for the limited amount of PRC1 complex. In our model, this second level of cooperativity would provide targeting specificity.
Notably, the same DNA binding proteins involved in PC-G targeting can separately participate in weak interactions with various other protein complexes involved in processes unrelated to, or the opposite of, Pc-G-dependent silencing, such as TFIID-dependent transcription or chromatin remodeling by SWI/SNF (11, 40). Based on the available data, interaction surfaces of any such complex are not shared by these DNA binding proteins, and according to our model, their concerted recruitment to PREs is unlikely. Also, in agreement with the experimental data, this model predicts that in the absence of DNA none of the DNA binding proteins will be able to interact stably with the complex to be recruited. The integration of several weak protein-protein interaction modules into a single entity is a prerequisite for the complex to dock on chromatin (Fig. 7A).
It has been shown that transcription through the iab-7 PRE displaces PC-G proteins and results in concomitant recruitment of the TRX and BRM proteins (10, 14, 26). Thus, iab-7 PRE appears to be a switchable element and the potential, for example, of PHO to interact with protein partners having a function that is the opposite of PC-G silencing might be realized under certain circumstances. There is insufficient data to explain the mechanism underlying this switch. One possibility is that binding of some DNA binding proteins to DNA or to their interacting partners is modified by posttranslational modifications, as it was shown in the case of the human homologue of Grh (60). According to our model, even the modification of a single actor (e.g., GRH) can radically influence the overall assembly configuration of the targeting complex and might be responsible for the dynamic nature of the iab-7 PRE.
Similarity to the targeting of transcriptional activator complexes.
Our model shows remarkable similarity to the functional and structural organization of enhanceosomes (9). For example, multimerization of the binding sites of any of the DNA binding proteins involved in beta interferon (IFN-β) enhanceosome formation does not reproduce faithfully the virus inducibility of the intact enhancer. Instead, these synthetic enhancers respond promiscuously to inducers that are normally not involved in regulation of the IFN-β gene. The molecular basis of the selective inducer response of the enhanceosome is established by the following cooperative interactions. First, in their original context, the mutually cooperative interactions at the level of DNA binding proteins promote binding stability. Second, on the resulting spatially arranged protein surface, each DNA binding protein contributes to the recruitment of a protein complex through interactions with one of its subunits (36, 55). We conclude that the integration of different, hierarchical levels of cooperativity could be a general principle in the targeting of protein complexes to chromatin.
The validity of the enhanceosome model has already been demonstrated by in vitro reassembly of the IFN-β enhanceosome with well-defined recombinant components (32). In vitro studies with a nucleosomal template have provided valuable insights into the role of PRC1 in regulation of the chromatin structure (18, 51). However, in this experimental system the excess of PRC1 and nonspecific DNA binding of PRC1 complex members overcomes the problem of targeting. Our initial attempt to reconstitute cooperativity at the level of DNA binding proteins failed, possibly because the simultaneous presence of several other DNA binding proteins is required for cooperative assembly. Until these components of PREs are identified, it is likely that PC-G targeting cannot be faithfully reconstituted in vitro. The identification of as-yet-unknown DNA binding protein components of PREs, together with the conceptual framework presented here, will hopefully facilitate these studies.
Direct versus indirect targeting.
Recent results showed that in vivo stable recruitment of PC to the Ubx PRE critically depends on the presence of the E(Z) protein (63). E(Z) is a member of a PC-G complex, which is distinct from PRC1, and possesses histone methyltransferase activity (13, 20, 43). These findings led to a model wherein, upon binding of the EZ complex, its enzymatic activity could provide the mark for the specific targeting of PRC1. Hence, recruiting of PRC1 would only indirectly depend on sequence-specific DNA binding proteins, as they primarily act as recruiters of the E(Z) complex, but not PRC1 (63). Contrary to the predictions of this model, we found that although mutations in PRC1 complex members are similarly strong dominant enhancers of the Fab-72 phenotype as grh and pho, amorphic E(z) alleles in heterozygous condition are not (our unpublished data). Thus, our results indicate a rather intimate link between these DNA binding proteins and PRC1 complex members. However, it is still possible that in a nucleosomal context the histone mark could provide an additional constituent for binding whose presence can be critical in vivo in certain tissues. Certain PC-G group members have a tissue-specific phenotype, and GRH is also not ubiquitously expressed, which supports this notion (5, 6, 53).
Alternatively, a histone mark might be critical for maintenance of the repressed state through cell divisions but may not be required for targeting in resting cells. Here it should be noted that the proliferation rate of the imaginal disk and cultured cells used in the study described in reference 63 is different from that of histoblasts, wherefrom abdominal tergites are derived, a difference that may affect the outcome of experiments and conclusions profoundly. Taken together, it is possible that the two models may describe different aspects of Pc-G silencing or different targeting strategies used by different PREs, and they should be treated as complementary, and not concurrent, models.
Acknowledgments
We are indebted to S. Bray, M. Vidal, R. Tjian, and P. Verrijzer for sharing their fly stocks, cDNA clones, and antibodies. We also thank Anita Kiss and Ildikó Krausz for excellent technical assistance and M. Erdélyi, T. Török, and I. Bajusz for critical reading of the manuscript.
This work was supported by grants from the Hungarian National Science Foundation (T038076) and the National Institutes of Health (3 R01GM043432, subcontract 00000870; H.G.), by the Swiss National Foundation, by the State of Geneva (F.K.), and by a young investigator grant from the Human Frontier Science Program (R.K.M.). A.B. was supported by an EMBO short-term fellowship (ASTF 9705).
We have no competing financial interests.
REFERENCES
- 1.Americo, J., M. Whiteley, J. L. Brown, M. Fujioka, J. B. Jaynes, and J. A. Kassis. 2002. A complex array of DNA-binding proteins required for pairing-sensitive silencing by a polycomb group response element from the Drosophila engrailed gene. Genetics 160:1561-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong, J. A., O. Papoulas, G. Daubresse, A. S. Sperling, J. T. Lis, M. P. Scott, and J. W. Tamkun. 2002. The Drosophila BRM complex facilitates global transcription by RNA polymerase II. EMBO J. 21:5245-5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atchison, L., A. Ghias, F. Wilkinson, N. Bonini, and M. L. Atchison. 2003. Transcription factor YY1 functions as a PcG protein in vivo. EMBO J. 22:1347-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Attardi, L. D., and R. Tjian. 1993. Drosophila tissue-specific transcription factor NTF-1 contains a novel isoleucine-rich activation motif. Genes Dev. 7:1341-1353. [DOI] [PubMed] [Google Scholar]
- 5.Bray, S. J., B. Burke, N. H. Brown, and J. Hirsh. 1989. Embryonic expression pattern of a family of Drosophila proteins that interact with a central nervous system regulatory element. Genes Dev. 3:1130-1145. [DOI] [PubMed] [Google Scholar]
- 6.Bray, S. J., and F. C. Kafatos. 1991. Developmental function of Elf-1: an essential transcription factor during embryogenesis in Drosophila. Genes Dev. 5:1672-1683. [DOI] [PubMed] [Google Scholar]
- 7.Brown, J. L., D. Mucci, M. Whiteley, M. L. Dirksen, and J. A. Kassis. 1998. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol. Cell 1:1057-1064. [DOI] [PubMed] [Google Scholar]
- 8.Campbell, R. B., D. A. Sinclair, M. Couling, and H. W. Brock. 1995. Genetic interactions and dosage effects of Polycomb group genes of Drosophila. Mol. Gen. Genet. 246:291-300. [DOI] [PubMed] [Google Scholar]
- 9.Carey, M. 1998. The enhanceosome and transcriptional synergy. Cell 92:5-8. [DOI] [PubMed] [Google Scholar]
- 10.Cavalli, G., and R. Paro. 1999. Epigenetic inheritance of active chromatin after removal of the main transactivator. Science 286:955-958. [DOI] [PubMed] [Google Scholar]
- 11.Chen, J. L., L. D. Attardi, C. P. Verrijzer, K. Yokomori, and R. Tjian. 1994. Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell 79:93-105. [DOI] [PubMed] [Google Scholar]
- 12.Chiang, A., M. B. O'Connor, R. Paro, J. Simon, and W. Bender. 1995. Discrete Polycomb-binding sites in each parasegmental domain of the bithorax complex. Development 121:1681-1689. [DOI] [PubMed] [Google Scholar]
- 13.Czermin, B., R. Melfi, D. McCabe, V. Seitz, A. Imhof, and V. Pirrotta. 2002. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111:185-196. [DOI] [PubMed] [Google Scholar]
- 14.Dejardin, J., and G. Cavalli. 2004. Chromatin inheritance upon Zeste-mediated Brahma recruitment at a minimal cellular memory module. EMBO J. 23:857-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dejardin, J., A. Rappailles, O. Cuvier, C. Grimaud, M. Decoville, D. Locker, and G. Cavalli. 2005. Recruitment of Drosophila Polycomb group proteins to chromatin by DSP1. Nature 434:533-538. [DOI] [PubMed] [Google Scholar]
- 16.del Mar Lorente, M., C. Marcos-Gutierrez, C. Perez, J. Schoorlemmer, A. Ramirez, T. Magin, and M. Vidal. 2000. Loss- and gain-of-function mutations show a polycomb group function for Ring1A in mice. Development 127:5093-5100. [DOI] [PubMed] [Google Scholar]
- 17.Dynlacht, B. D., L. D. Attardi, A. Admon, M. Freeman, and R. Tjian. 1989. Functional analysis of NTF-1, a developmentally regulated Drosophila transcription factor that binds neuronal cis elements. Genes Dev. 3:1677-1688. [DOI] [PubMed] [Google Scholar]
- 18.Francis, N. J., A. J. Saurin, Z. Shao, and R. E. Kingston. 2001. Reconstitution of a functional core polycomb repressive complex. Mol. Cell 8:545-556. [DOI] [PubMed] [Google Scholar]
- 19.Fritsch, C., D. Beuchle, and J. Muller. 2003. Molecular and genetic analysis of the Polycomb group gene Sex combs extra/Ring in Drosophila. Mech. Dev. 120:949-954. [DOI] [PubMed] [Google Scholar]
- 20.Furuyama, T., F. Tie, and P. J. Harte. 2003. Polycomb group proteins ESC and E(Z) are present in multiple distinct complexes that undergo dynamic changes during development. Genesis 35:114-124. [DOI] [PubMed] [Google Scholar]
- 21.Garcia, E., C. Marcos-Gutierrez, M. del Mar Lorente, J. C. Moreno, and M. Vidal. 1999. RYBP, a new repressor protein that interacts with components of the mammalian Polycomb complex, and with the transcription factor YY1. EMBO J. 18:3404-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorfinkiel, N., L. Fanti, T. Melgar, E. Garcia, S. Pimpinelli, I. Guerrero, and M. Vidal. 2004. The Drosophila Polycomb group gene Sex combs extra encodes the ortholog of mammalian Ring1 proteins. Mech. Dev. 121:449-462. [DOI] [PubMed] [Google Scholar]
- 23.Hagstrom, K., M. Muller, and P. Schedl. 1997. A Polycomb and GAGA dependent silencer adjoins the Fab-7 boundary in the Drosophila bithorax complex. Genetics 146:1365-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hochschild, A., and M. Ptashne. 1986. Cooperative binding of lambda repressors to sites separated by integral turns of the DNA helix. Cell 44:681-687. [DOI] [PubMed] [Google Scholar]
- 25.Hodgson, J. W., B. Argiropoulos, and H. W. Brock. 2001. Site-specific recognition of a 70-base-pair element containing d(GA)(n) repeats mediates bithoraxoid polycomb group response element-dependent silencing. Mol. Cell. Biol. 21:4528-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hogga, I., and F. Karch. 2002. Transcription through the iab-7 cis-regulatory domain of the bithorax complex interferes with maintenance of Polycomb-mediated silencing. Development 129:4915-4922. [DOI] [PubMed] [Google Scholar]
- 27.Horard, B., C. Tatout, S. Poux, and V. Pirrotta. 2000. Structure of a polycomb response element and in vitro binding of polycomb group complexes containing GAGA factor. Mol. Cell. Biol. 20:3187-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang, D.-H., Y.-L. Chang, C.-C. Yang, I.-C. Pan, and B. King. 2002. pipsqueak encodes a factor essential for sequence-specific targeting of a Polycomb group protein complex. Mol. Cell. Biol. 22:6261-6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kal, A. J., T. Mahmoudi, N. B. Zak, and C. P. Verrijzer. 2000. The Drosophila brahma complex is an essential coactivator for the trithorax group protein zeste. Genes Dev. 14:1058-1071. [PMC free article] [PubMed] [Google Scholar]
- 30.Kassis, J. A. 2002. Pairing-sensitive silencing, polycomb group response elements, and transposon homing in Drosophila. Adv. Genet. 46:421-438. [DOI] [PubMed] [Google Scholar]
- 31.Kennison, J. A. 1995. The Polycomb and trithorax group proteins of Drosophila: trans-regulators of homeotic gene function. Annu. Rev. Genet. 29:289-303. [DOI] [PubMed] [Google Scholar]
- 32.Kim, T. K., and T. Maniatis. 1997. The mechanism of transcriptional synergy of an in vitro assembled interferon-beta enhanceosome. Mol. Cell 1:119-129. [DOI] [PubMed] [Google Scholar]
- 33.Lehmann, M., T. Siegmund, K. G. Lintermann, and G. Korge. 1998. The pipsqueak protein of Drosophila melanogaster binds to GAGA sequences through a novel DNA-binding domain. J. Biol. Chem. 273:28504-28509. [DOI] [PubMed] [Google Scholar]
- 34.Levine, S. S., A. Weiss, H. Erdjument-Bromage, Z. Shao, P. Tempst, and R. E. Kingston. 2002. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol. Cell. Biol. 22:6070-6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahmoudi, T., L. M. Zuijderduijn, A. Mohd-Sarip, and C. P. Verrijzer. 2003. GAGA facilitates binding of Pleiohomeotic to a chromatinized Polycomb response element. Nucleic Acids Res. 31:4147-4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maniatis, T., J. V. Falvo, T. H. Kim, T. K. Kim, C. H. Lin, B. S. Parekh, and M. G. Wathelet. 1998. Structure and function of the interferon-beta enhanceosome. Cold Spring Harbor Symp. Quant. Biol. 63:609-620. [DOI] [PubMed] [Google Scholar]
- 37.Mihaly, J., I. Hogga, J. Gausz, H. Gyurkovics, and F. Karch. 1997. In situ dissection of the Fab-7 region of the bithorax complex into a chromatin domain boundary and a Polycomb-response element. Development 124:1809-1820. [DOI] [PubMed] [Google Scholar]
- 38.Mishra, K., V. S. Chopra, A. Srinivasan, and R. K. Mishra. 2003. Trl-GAGA directly interacts with lola like and both are part of the repressive complex of Polycomb group of genes. Mech. Dev. 120:681-689. [DOI] [PubMed] [Google Scholar]
- 39.Mishra, R. K., J. Mihaly, S. Barges, A. Spierer, F. Karch, K. Hagstrom, S. E. Schweinsberg, and P. Schedl. 2001. The iab-7 Polycomb response element maps to a nucleosome-free region of chromatin and requires both GAGA and Pleiohomeotic for silencing activity. Mol. Cell. Biol. 21:1311-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mohd-Sarip, A., F. Venturini, G. E. Chalkley, and C. P. Verrijzer. 2002. Pleiohomeotic can link Polycomb to DNA and mediate transcriptional repression. Mol. Cell. Biol. 22:7473-7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mohrmann, L., K. Langenberg, J. Krijgsveld, A. J. Kal, A. J. Heck, and C. P. Verrijzer. 2004. Differential targeting of two distinct SWI/SNF-related Drosophila chromatin-remodeling complexes. Mol. Cell. Biol. 24:3077-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mulholland, N. M., I. F. King, and R. E. Kingston. 2003. Regulation of Polycomb group complexes by the sequence-specific DNA binding proteins Zeste and GAGA. Genes Dev. 17:2741-2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muller, J., C. M. Hart, N. J. Francis, M. L. Vargas, A. Sengupta, B. Wild, E. L. Miller,M. B. O'Connor, R. E. Kingston, and J. A. Simon. 2002. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111:197-208. [DOI] [PubMed] [Google Scholar]
- 44.O'Brien, T., R. C. Wilkins, C. Giardina, and J. T. Lis. 1995. Distribution of GAGA protein on Drosophila genes in vivo. Genes Dev. 9:1098-1110. [DOI] [PubMed] [Google Scholar]
- 45.Orlando, V., E. P. Jane, V. Chinwalla, P. J. Harte, and R. Paro. 1998. Binding of trithorax and Polycomb proteins to the bithorax complex: dynamic changes during early Drosophila embryogenesis. EMBO J. 17:5141-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ringrose, L., M. Rehmsmeier, J. M. Dura, and R. Paro. 2003. Genome-wide prediction of Polycomb/Trithorax response elements in Drosophila melanogaster. Dev. Cell 5:759-771. [DOI] [PubMed] [Google Scholar]
- 47.Romerio, F., M. N. Gabriel, and D. M. Margolis. 1997. Repression of human immunodeficiency virus type 1 through the novel cooperation of human factors YY1 and LSF. J. Virol. 71:9375-9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salvaing, J., A. Lopez, A. Boivin, J. S. Deutsch, and F. Peronnet. 2003. The Drosophila Corto protein interacts with Polycomb-group proteins and the GAGA factor. Nucleic Acids Res. 31:2873-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saurin, A. J., Z. Shao, H. Erdjument-Bromage, P. Tempst, and R. E. Kingston. 2001. A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature 412:655-660. [DOI] [PubMed] [Google Scholar]
- 50.Schwendemann, A., and M. Lehmann. 2002. Pipsqueak and GAGA factor act in concert as partners at homeotic and many other loci. Proc. Natl. Acad. Sci. USA 99:12883-12888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shao, Z., F. Raible, R. Mollaaghababa, J. R. Guyon, C. T. Wu, W. Bender, and R. E. Kingston. 1999. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 98:37-46. [DOI] [PubMed] [Google Scholar]
- 52.Sipos, L., J. Mihaly, F. Karch, P. Schedl, J. Gausz, and H. Gyurkovics. 1998. Transvection in the Drosophila Abd-B domain: extensive upstream sequences are involved in anchoring distant cis-regulatory regions to the promoter. Genetics 149:1031-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Soto, M. C., T. B. Chou, and W. Bender. 1995. Comparison of germline mosaics of genes in the Polycomb group of Drosophila melanogaster. Genetics 140:231-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Srinivasan, L., and M. L. Atchison. 2004. YY1 DNA binding and PcG recruitment requires CtBP. Genes Dev. 18:2596-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thanos, D., and T. Maniatis. 1995. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell 83:1091-1100. [DOI] [PubMed] [Google Scholar]
- 56.Tillib, S., S. Petruk, Y. Sedkov, A. Kuzin, M. Fujioka, T. Goto, and A. Mazo. 1999. Trithorax- and Polycomb-group response elements within an Ultrabithorax transcription maintenance unit consist of closely situated but separable sequences. Mol. Cell. Biol. 19:5189-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tuckfield, A., D. R. Clouston, T. M. Wilanowski, L. L. Zhao, J. M. Cunningham, and S. M. Jane. 2002. Binding of the RING polycomb proteins to specific target genes in complex with the grainyhead-like family of developmental transcription factors. Mol. Cell. Biol. 22:1936-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Uv, A. E., E. J. Harrison, and S. J. Bray. 1997. Tissue-specific splicing and functions of the Drosophila transcription factor Grainyhead. Mol. Cell. Biol. 7:6727-6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Uv, A. E., C. R. Thompson, and S. J. Bray. 1994. The Drosophila tissue-specific factor Grainyhead contains novel DNA-binding and dimerization domains which are conserved in the human protein CP2. Mol. Cell. Biol. 14:4020-4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Volker, J. L., L. E. Rameh, Q. Zhu, J. DeCaprio, and U. Hansen. 1997. Mitogenic stimulation of resting T cells causes rapid phosphorylation of the transcription factor LSF and increased DNA-binding activity. Genes Dev. 11:1435-1446. [DOI] [PubMed] [Google Scholar]
- 61.Walter, J., C. A. Dever, and M. D. Biggin. 1994. Two homeo domain proteins bind with similar specificity to a wide range of DNA sites in Drosophila embryos. Genes Dev. 8:1678-1692. [DOI] [PubMed] [Google Scholar]
- 62.Wang, J. C., and G. N. Giaever. 1988. Action at a distance along a DNA. Science 240:300-304. [DOI] [PubMed] [Google Scholar]
- 63.Wang, L., J. L. Brown, R. Cao, Y. Zhang, J. A. Kassis, and R. S. Jones. 2004. Hierarchical recruitment of polycomb group silencing complexes. Mol. Cell 14:637-646. [DOI] [PubMed] [Google Scholar]