Abstract
African swine fever virus (ASFV), a complex enveloped DNA virus, expresses two polyprotein precursors, pp220 and pp62, which after proteolytic processing give rise to several major components of the virus particle. We have analyzed the structural role of polyprotein pp62, the precursor form of mature products p35 and p15, in virus morphogenesis. Densitometric analysis of one- and two-dimensional gels of purified virions showed that proteins p35 and p15, as well as the pp220-derived products, are present in equimolecular amounts in the virus particle. Immunoelectron microscopy revealed that the pp62-derived products localize at the core shell, a matrix-like domain placed between the DNA-containing nucleoid and the inner envelope, where the pp220-derived products are also localized. Pulse-chase experiments indicated that the processing of both polyprotein precursors is concomitant with virus assembly. Furthermore, using inducible ASFV recombinants, we show that pp62 processing requires the expression of the pp220 core precursor, whereas the processing of both precursors pp220 and pp62 is dependent on expression of the major capsid protein p72. Interestingly, when p72 expression is blocked, unprocessed pp220 and pp62 polyproteins assemble into aberrant zipper-like elements consisting of an elongated membrane-bound protein structure reminiscent of the core shell. Moreover, the two polyproteins, when coexpressed in COS cells, interact with each other to form zipper-like structures. Together, these findings indicate that the mature products derived from both polyproteins, which collectively account for about 30% of the virion protein mass, are the basic components of the core shell and that polyprotein processing represents a maturational process related to ASFV morphogenesis.
African swine fever virus (ASFV) is a complex enveloped deoxyvirus of the Asfarviridae family that infects domestic and wild pigs as well as soft-bodied ticks of the Ornithodoros genus (16, 17, 29, 37). The viral genome, a double-stranded DNA molecule of 170 to 190 kbp, encodes more than 150 polypeptides, 30 to 50 of which are components of the virus particle (11, 20, 39). Besides the structural proteins, the ASFV genome encodes a variety of enzymes involved in DNA replication and repair, gene transcription, and protein modification and a number of proteins potentially involved in the modulation of the virus-host interaction (29, 36, 39).
Extracellular mature particles are composed of several concentric domains: a viral core consisting of a central DNA-containing nucleoid and a surrounding thick protein coat referred to as core shell, an inner lipid envelope derived from the endoplasmic reticulum, an icosahedral protein capsid, and an additional envelope taken from the plasma membrane (4, 5, 12, 28). ASFV particles assemble within cytoplasmic viral factories from endoplasmic reticulum-derived viral membranes (4, 5, 26, 28), which become icosahedral particles by the gradual formation of the outer capsid layer (4, 22). Concomitantly, the core shell is formed underneath the viral envelope, and subsequently the viral DNA and nucleoproteins are packaged and condensed to form the electron-dense nucleoid (4, 8, 10). Intracellular mature virions made at the assembly sites are infectious (7). A fraction of them, however, are transported by microtubule-mediated transport (1, 2) to the plasma membrane, where they are released by budding (9) to give rise to the infectious extracellular enveloped virions.
ASFV, like other complex DNA viruses, regulates its gene expression by temporal control of transcription. Thus, ASFV genes are classified into different classes: early and late genes are transcribed before and after the onset of DNA replication, respectively, whereas transcription of intermediate genes occurs during the onset of genome replication and is then rapidly halted (27). A striking feature of ASFV gene expression is the synthesis of two polyprotein precursors, named pp220 and pp62, to produce several major components of the virus particle. Polyprotein pp220, encoded by the CP2475L gene, is an N-myristoylated polypeptide that, after proteolytic processing, gives rise to proteins p150, p37, p34, and p14 (31). These proteins are present in equimolecular amounts within the mature virion and account for about one-fourth of its total protein mass (4). On the other hand, the major proteins p35 and p15 are processing products of the precursor pp62, encoded by gene CP530R (32). Both polyproteins are expressed late in infection and are posttranslationally processed by the viral cysteine proteinase pS273R (6). Proteolytic processing takes place through a temporally ordered cascade of cleavage events occurring after the second glycine of the consensus motif Gly-Gly-X (25, 31, 32). We have previously shown that the mature products derived from pp220 precursor, as well as the ASFV proteinase, reside at the core shell (4, 6). Consistent with the matrix-like role of this core domain, we also recently showed that repression of pp220 synthesis leads to the assembly of core-deficient icosahedral particles (8).
Here we analyze the structural role of polyprotein pp62 in virus morphogenesis by using biochemical, genetic, and immunocytochemical approaches. The data indicate that proteins p35 and p15 reside at the core shell, as do the polyprotein pp220 products. Moreover, a core shell-like domain can be assembled by coexpressing the two polyprotein-encoding genes. Our data also indicate that polyprotein processing is a maturational process concomitant with virus assembly.
MATERIALS AND METHODS
Cells and viruses.
Vero and COS-7 cells were obtained from the American Type Culture Collection and grown in Dulbecco's modified Eagle's medium containing 5% fetal calf serum. The serum concentration was lowered to 2% during infections. The ASFV strain BA71V, adapted to grow in Vero cells, and the inducible BA71V-derived recombinants vA72 and v220i have already been described (8, 18, 22). The recombinant vaccinia virus vTF7-3 expressing bacteriophage T7 RNA polymerase (21) was kindly provided by Bernard Moss.
Antibodies.
The monospecific rabbit polyclonal sera against the polyprotein pp62-derived structural proteins p35 and p15, the polyprotein pp220-derived product p150, and the major capsid protein p72 have been described previously (6, 7, 31, 32).
1-D and 2-D gel electrophoresis of purified ASFV particles.
Extracellular virus particles metabolically labeled with [35S]methionine or 14C-amino acids were purified by Percoll equilibrium sedimentation as previously described (11). High-resolution two-dimensional (2-D) gel electrophoresis was performed essentially as described previously (3). Sodium dodecyl sulfate (SDS)-polyacrylamide gradient slab gels (7 to 20%) were used for both second-dimension gels and one-dimensional (1-D) SDS-polyacrylamide gel electrophoresis. Isoelectric points (pI) were estimated with a set of pI markers (Bio-Rad).
Quantification of radiolabeled and Coomassie blue-stained proteins was performed with a Bio-Rad GS710 densitometer and Quantity One software (Bio-Rad). Measurements were normalized by taking into account the number of methionine residues of the corresponding [35S]methionine-labeled proteins or with respect to the theoretical molecular masses in the case of 14C-amino acid-labeled and Coomassie blue-stained polypeptides, as follows: p150, 43 methionines or 181.1 kDa; p37, 10 methionines or 41.5 kDa; p34, 3 methionines or 36.3 kDa; p14, 3 methionines or 17.8 kDa; p35, 6 methionines or 35.1 kDa; p15, 4 methionines or 17.7 kDa; and p72, 8 methionines or 73.1 kDa.
For immunoblotting with anti-p35 serum, ASFV proteins were separated in 2-D gels and transferred to nitrocellulose membranes (0.2-μm pore size; Renner). Protein detection was carried out with peroxidase-conjugated antibodies and the ECL system (Amersham Pharmacia Biotech) according to the manufacturer's instructions. For immunoprecipitation with anti-p15 serum, [35S]methionine-labeled virions were dissociated in immunoprecipitation buffer (0.01 M Tris-HCl [pH 7.5], 0.15 M NaCl, 1% sodium deoxycholate, 1% IGEPAL CA-630, 0.1% SDS, and protease inhibitors [complete EDTA-free cocktail from Roche]) and incubated with the serum for 2 h at 4°C followed by protein A-Sepharose for 30 min. After extensive washing, immune complexes were solubilized at 37°C in lysis buffer (9.8 M urea, 2% Nonidet P-40, 2% ampholytes [pH 7 to 9], 0.1 M dithiothreitol) for 2-D gel electrophoresis.
Pulse-chase labeling and subcellular fractionation.
Preconfluent monolayers of Vero cells were infected with 10 PFU of BA71V per cell. At 12 h postinfection (hpi), the cells were pulse-labeled for 15 min with 0.5 mCi of [35S]methionine-[35S]cysteine (Promix in vitro cell labeling mix; Amersham Pharmacia Biotech) per ml of methionine- and cysteine-free Dulbecco's modified Eagle's medium. At the end of the labeling period, the medium was removed and the cells were subjected to subcellular fractionation or were incubated with fresh medium for different chase periods. The soluble and the membrane-particulate postnuclear fractions were prepared as described previously (7). The extracellular virus particles were collected from the medium by centrifugation at 20,000 × g for 15 min and mixed with the membrane-particulate fraction. Samples were dissociated at 4°C in immunoprecipitation buffer and incubated with the indicated antibodies immobilized on protein A-Sepharose. Immunoprecipitated proteins were resolved by SDS-12% polyacrylamide gel electrophoresis and detected by autoradiography.
Transient expression in COS cells.
Transfection assays were carried out with plasmids pGEM-CP2475L and KS-CP530R, which contain the genes encoding polyproteins pp220 and pp62, respectively, under the control of the T7 polymerase promoter (6, 32). COS-7 cells were transfected for 1 h at 37°C with 250 ng of DNA per 105 cells by using Lipofectamine Plus reagent (Life Technologies, Inc.) according to the manufacturer's indications. For cotransfection experiments, different ratios of the polyprotein gene-containing plasmids, ranging from 1:3 to 3:1, were tested. The transfected cells were then infected for 1 h at 37°C with recombinant vaccinia virus vTF7-3 expressing T7 RNA polymerase (21) at a multiplicity of infection of 5 PFU per cell. The cells were incubated with 40 μg of cytosine arabinoside (Ara C), an inhibitor of vaccinia virus DNA replication and late protein synthesis, per ml. At 15 hpi, the cells were either subjected to cell fractionation or processed for electron microscopy.
Electron microscopy.
For conventional Epon section analysis, virus-infected Vero cells and transfected COS-7 cells were fixed at the indicated times with 2% glutaraldehyde in 200 mM HEPES (pH 7.4) for 1 h at room temperature. Postfixation, dehydration, and Epon embedding were performed as described previously (8).
For immunoelectron microscopy, the cells were fixed for 1 h with 4% formaldehyde and 0.1% glutaraldehyde in 200 mM HEPES (pH 7.4) on ice. Freeze-substitution, cryosectioning, and immunogold labeling were performed as previously described (5).
RESULTS
Relative abundances and stoichiometries of the polyprotein pp62-derived products p35 and p15 within the virus particle.
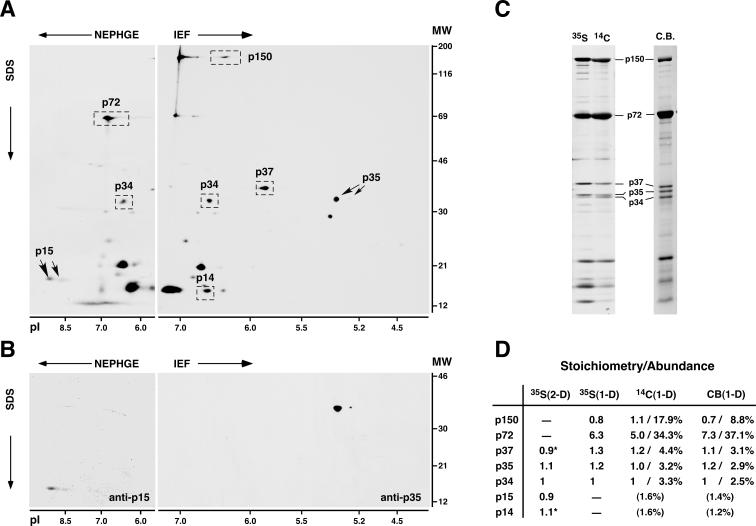
Our first goal was to analyze the contribution of the polyprotein pp62-derived mature products to the protein content of the ASFV particles. Proteins p35 and p15 were first quantified by densitometric analysis of 2-D gels of [35S]methionine-labeled virus particles purified by Percoll gradient sedimentation (11). Figure 1A shows a representative 2-D profile of acidic and basic ASFV structural polypeptides. Proteins p35 and p15 were mapped in 2-D gels by Western immunoblotting or immunoprecipitation, respectively, using monospecific antisera (32). As shown in Fig. 1B, both polyprotein products were resolved as double spots, with the more acidic one being present in a very minor proportion. Protein p35 focused in acidic gels (isoelectric focusing [IEF]) with an estimated pI of 5.4 to 5.3, whereas protein p15 was resolved in basic gels (nonequilibrium pH gradient electrophoresis [NEPHGE]) with an estimated pI of>8.5). These values are in good accordance with the theoretical pIs of proteins p35 (pI 5.4) and p15 (pI 8.8). Since proteins p35 and p15 do not migrate at the same pH range, they were quantified in relation to the polyprotein pp220-derived product p34, which can be detected in the overlapping range of pH 6 to 7 of both IEF and NEPHGE gels (Fig. 1A). The densitometric data were normalized by the number of methionine residues of each protein. As summarized in Fig. 1D [35S(2-D) column], the pp62-derived products p35 and p15, as well as the internal reference p34, were found to be present in the virus particle in similar molar proportions. Since the same stoichiometry has been reported for protein p34 and the other pp220-derived products p150, p37, and p14 (4), these results indicate that equimolecular amounts of all of the polyprotein products compose the ASFV particle.
FIG. 1.
Gel mapping, stoichiometry, and relative abundances of ASFV polyprotein products in the virus particle. (A) 2-D gel electrophoresis of ASFV labeled with [35S]methionine. Basic proteins were resolved by NEPHGE, while acidic proteins were separated by IEF. The positions of the major capsid protein p72; the polyprotein pp220-derived products p150, p37, p34, and p14; and the polyprotein pp62-derived products p35 and p15 are indicated. The migrations of molecular weight (MW) markers (in thousands) and of pI markers are indicated at the right and at the bottom of the 2-D gels, respectively. (B) 2-D mapping of proteins p35 and p15. Protein p15 was mapped in NEPHGE gels by immunoprecipitation, whereas p35 was identified in IEF gels by immunoblotting. (C) 1-D gel electrophoresis of ASFV particles labeled with [35S]methionine or 14C-amino acids or stained with Coomassie blue (C.B.). The positions of several structural proteins are indicated. (D) Quantitative analysis of the polyprotein products. Polyprotein products and, as a reference, capsid protein p72 were quantified by densitometry analysis of 2-D gels of [35S]methionine-labeled proteins (two experiments) or of 1-D gels of ASFV proteins labeled with [35S]methionine (three experiments) or 14C-amino acids (two experiments) or stained with Coomassie blue (three experiments). Stoichiometry data were normalized by the number of methionine residues of each protein (for 35S-labeled proteins) or by its molecular weight (for 14C-labeled or Coomassie blue-stained proteins) and referred to protein p34. Relative abundances (percentages) were estimated from 1-D gels of 14C-labeled or Coomassie blue-stained proteins. Standard deviations of the means were less than 25% of the means. Proteins p150 and p72 were not quantified in 2-D gels due to deficient focusing, while proteins p15 and p14 were not estimated in 1-D gels because of heterogeneity of the bands. The relative abundances of proteins p15 and p14 are values extrapolated by assuming an equimolar stoichiometry. Stoichiometry data for p37 and p14 [marked with asterisks in the 35S(2-D) column] are from reference 4.
To obtain further evidence for this particular stoichiometry and to determine the contribution of the polyprotein products to the virion mass, we took advantage of the fact that most of them (p150, p37, p35, and p34) can be unambiguously quantified in conventional 1-D gels of ASFV structural proteins. Figure 1C shows typical 1-D profiles of virus polypeptides labeled with [35S]methionine or 14C-amino acids or stained with Coomassie blue. Densitometric data for the polyprotein products were normalized by the number of methionine residues [Fig. 1D, 35S(1-D) column] or by the theoretical molecular mass of each protein [14C(1-D) and CB(1-D) columns]. Quantification of proteins p15 and p14 was not possible due to the band complexity observed in the low range of molecular masses (Fig. 1C). The three measurement methods confirmed the equimolecular stoichiometry between the polyprotein products. The only exception was protein p150, which is underestimated in Coomassie blue-stained gels (4). Also, densitometric analysis of 1-D protein patterns indicated that about 32% of the total protein content of the ASFV particle [data from the 14C(1-D) column] corresponds to the mature products derived from pp220 (about 27%) and pp62 (about 5%). As a reference, the major capsid protein p72 accounts for about 34% of the viral content, and its molar proportion is fivefold higher than that of each polyprotein product.
Proteins p35 and p15 reside at the core shell.
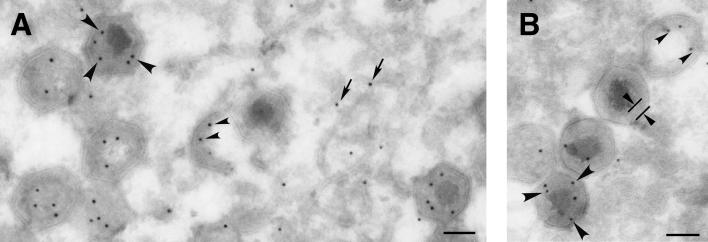
Next we analyzed the localization of polyprotein pp62 and its derived mature proteins in ASFV-infected cells. Previous results based on immunofluorescence microscopy showed that both anti-p35 and anti-p15 antibodies detect the viral factories as well as single virus particles spreading throughout the cytoplasm (32). To study in more detail the distribution of proteins p35 and p15 at the assembly sites, we performed immunogold labeling on thawed cryosections of ASFV-infected cells processed at 20 h postinfection. Figure 2 shows details of virus factories labeled with anti-p35 (Fig. 2A) and anti-p15 (Fig. 2B) antibodies followed by protein A-gold (10-nm diameter). No qualitative difference was found between the labeling patterns with the two antibodies. Significant labeling was detected in close proximity to membranous viral structures, the precursor elements of ASFV particles. However, most of the labeling was associated with both immature and mature virions. Within the assembling particles (without nucleoid), gold particles were detected at the core structure emerging beneath the inner envelope. Within the mature icosahedral virions, gold particles were found predominantly at the core shell surrounding the DNA-containing nucleoid. The same localization has been reported for the polyprotein pp220-derived mature products (4) as well as for ASFV proteinase pS273R (6). Therefore, these results indicate that both ASFV polyproteins are involved in the assembly of the matrix virus domain.
FIG. 2.
Proteins p35 and p15 reside at the core shell. Thawed cryosections of BA71V-infected cells fixed at 20 hpi were incubated with anti-p35 (A) or anti-p15 (B) antibodies followed by protein A-gold (10-nm diameter). Within the virus factory, the pp62 labeling can be observed close to precursor viral membranes (arrows), at the immature core region of assembling virions (small arrowheads), and at the core shell (large arrowheads) surrounding the electron-dense nucleoid of mature particles. The core shell appears delimited in a virus particle in panel B. Bars, 100 nm.
Polyprotein processing is concomitant with virus assembly.
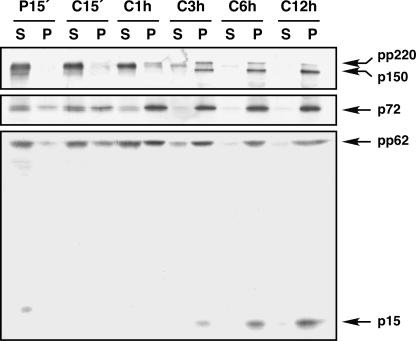
The colocalization of the polyprotein products and the proteinase at the same core domain strongly suggests that proteolytic processing of the ASFV precursors is concomitant with core assembly. To test this hypothesis, we first analyzed the subcellular distribution of the polyproteins in pulse-chase experiments. ASFV-infected cells were pulse-labeled at 12 h postinfection with [35S]methionine-[35S]cysteine for 15 min and then chased for different periods in the presence of cold methionine-cysteine. After each pulse and chase period, the cells were homogenized and the postnuclear supernatants were fractionated into a cytosolic fraction and a membrane-particulate fraction. The fractions were finally immunoprecipitated with anti-pp220/p150 and anti-pp62/p15 antibodies, which detect the first mature products arisen from each polyprotein, and analyzed by SDS-PAGE. As a reference, the major capsid protein p72, a peripheral membrane protein (14), was also monitored. As shown in Fig. 3, protein p72 was initially expressed at the cytosol and rapidly associated (after 1-h chase) with the membrane-particulate fraction, which can be interpreted as being incorporated into assembling virions. Similarly, polyproteins pp220 and pp62 were detected mostly in the soluble fraction after the pulse-labeling and were detected increasingly in the cytoplasmic sediments after the different chase periods. Interestingly, while significant amounts of unprocessed precursors pp220 and pp62 were detected in the membrane-particulate fraction after the 1-h chase, no proteolytic processing was evident until the 3-h chase. Moreover, the polyprotein products p150 and p15 were associated exclusively with the membrane-particulate fraction. These results suggest that polyprotein processing occurs after the incorporation of the core precursors into the assembling virions.
FIG. 3.
Processing of ASFV polyproteins occurs after their incorporation into the membrane-particulate fraction. Infected Vero cells were pulse-labeled with [35S]methionine-[35S]cysteine for 15 min (P15′ lanes) at 12 hpi and either immediately processed or subsequently chased for 15 min, 1 h, 3 h, 6 h, and 12 h (C15′, C1 h, C3 h, C6 h, and C12 h lanes, respectively). The postnuclear supernatants were fractionated into membrane-particulate (lanes P) and soluble (lanes S) fractions and immunoprecipitated with anti-pp220/p150, anti-pp62/p15, and anti-p72 antibodies. The proteins detected in the pulse and chases after SDS-polyacrylamide gel electrophoresis are indicated.
Proteolytic processing of precursor pp62 depends on polyprotein pp220 expression.
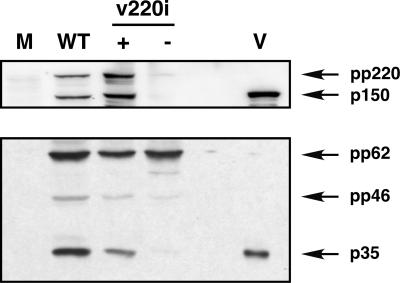
To further explore the relationship between virus assembly and proteolytic processing, an ASFV recombinant, v220i, that inducibly expresses polyprotein pp220 (8) was analyzed. Under nonpermissive conditions, icosahedral particles virtually devoid of the core structure are assembled in the virus factories. These defective particles lack, besides the pp220 products, the p35 and p15 proteins as well as nucleoid components, including the viral genome (8). To investigate whether precursor pp62 is properly processed when pp220 expression is blocked, extracts of v220i-infected cells maintained under permissive or restrictive conditions were analyzed by Western immunoblotting with anti-pp220/p150 and anti-pp62/p35 antibodies. As a control, uninfected cells or cells infected with parental BA71V virus were also analyzed. As shown in Fig. 4, under permissive conditions the proteolytic processing of pp62 precursor occurred to a similar extent as in control infections. In contrast, under restrictive conditions the processing of pp62 was strongly inhibited. This finding indicates that the expression of polyprotein pp220 is required for the processing of pp62 precursor into the mature products p35 and p15, probably because pp220 is also necessary for the incorporation of pp62 precursor into assembling virions (8).
FIG. 4.
Polyprotein pp62 processing depends on polyprotein pp220 expression. Vero cells were mock infected (lane M) or infected with parental BA71V (lane WT) or recombinant v220i virus in the presence (lane +) or absence (lane −) of IPTG (isopropyl-β-d-thiogalactopyranoside). At 20 hpi, the cells were lysed and analyzed, together with purified ASFV particles (lane V), by Western immunoblotting with anti-pp220/p150 and anti-pp62/p35 antibodies. The positions of polyprotein pp220 and its derived protein p150 (upper panel) and of polyprotein pp62, the intermediate processing precursor pp46, and the mature product p35 (lower panel), are indicated.
Expression of the capsid protein p72 is required for processing of polyproteins pp220 and pp62.
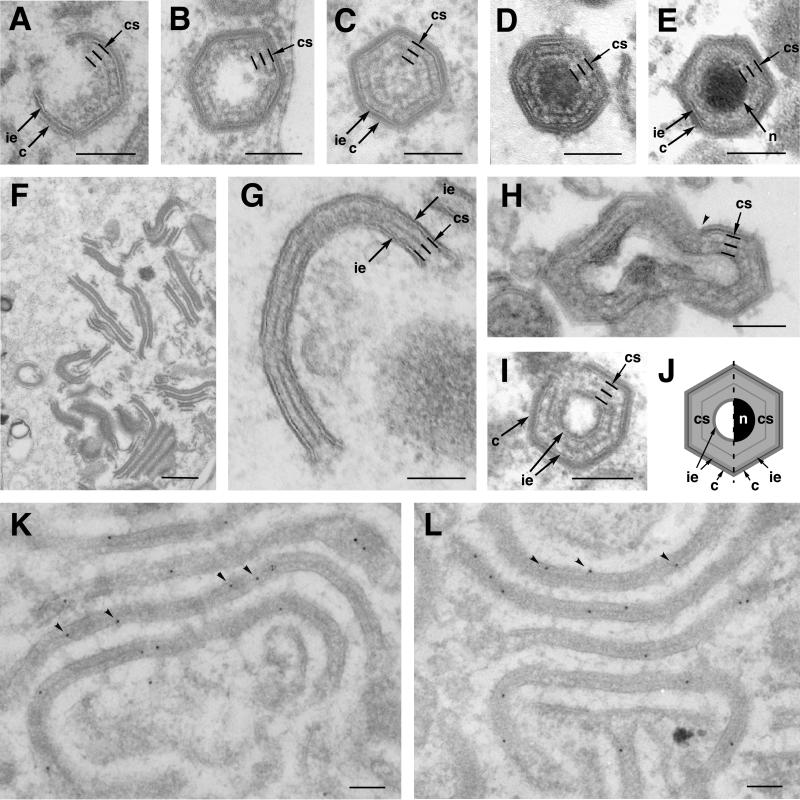
The assembly of the core shell underneath the inner lipid envelope seems to be concomitant with the formation of the outer capsid and appears to occur independently of nucleoid maturation (Fig. 5A to E) (4, 8). As illustrated in Fig. 5A, the emerging core shell is evident when the developing capsid is still open. Before and during the formation of the internal DNA-containing nucleoid (Fig. 5B to D), the surrounding core shell appears like a thick domain (33 ± 3 nm) that is subdivided by a thin electron-dense layer of hexagonal outline. This thin layer, however, is less evident in intracellular (Fig. 5E) and extracellular (not shown) mature virions, which suggests a late structural maturation of this core domain.
FIG. 5.
The core shell and the aberrant zipper-like structures are structurally and biochemically related. Cells infected with parental BA71V (A, B, C, D, and E) and recombinant vA72 (F, G, H, and I) were processed by conventional Epon embedding at 16 to 24 hpi. (A to E) Different stages of the assembly of an intracellular mature particle. Note that the assembly of the core shell (indicated by three parallel lines) beneath the inner envelope is concomitant with the formation of the outer capsid (A) and precedes the maturation of the DNA-containing nucleoid (B to D). Note also that the fine electron-dense layer that subdivides the core shell is less evident in intracellular mature virions (E). (F and G) Aberrant zipper-like structures assembled at vA72 virus factories at 16 hpi under restrictive conditions. Zipper-like elements consist of a core shell-like structure limited by two inner envelopes (G). (H and I) Icosahedral aberrant viruses formed in vA72-infected cells maintained for 16 h under restrictive conditions and then treated with inducer for an additional 8-h period. Note that the zipper elements become icosahedral, double-enveloped particles after capsid assembly (arrowhead in panel H) on one of the two flanking envelopes. Note also that the core shell of these particles (I) is nearly identical to that of normal assembling virions (B to D). (J) A diagram illustrates the similarities and differences between a normal (right half) and an aberrant (left half) particle. Note that the aberrant particle contains an additional innermost lipid envelope but lacks the nucleoid. (K and L) Immunogold labeling of vA72-infected cells maintained under restrictive conditions for 24 h. Lowicryl sections were labeled with anti-pp62 (K) or anti-pp220 (L) antibodies followed by protein A-gold (10-nm diameter). c, capsid; ie, inner envelope; cs, core shell; n, nucleoid. Bars, 100 nm.
When capsid formation is inhibited by means of an inducible recombinant for protein p72, the core assembly does not take place (22). Instead, zipper-like viral structures accumulate at the assembly sites (Fig. 5F). These aberrant elements consist of an elongated thick (32- ± 2-nm) protein layer resembling the core shell, which is limited by adjacent lipid envelopes (Fig. 5G). The analogy between the core shell and the protein intermediate domain of the zipper-like structures becomes even more evident when p72 expression is induced after the previous accumulation of the aberrant zippers under restrictive conditions (22). As illustrated in Fig. 5H and I, p72 expression leads to the assembly of the capsid layer on one face of the zipper-like structures, which become polyhedral particles. These aberrant virions usually appear empty, as they lack an electron-dense nucleoid; however, they contain an additional innermost envelope (Fig. 5J). The structure of the core shell in these double-enveloped particles is very similar to that found in developing normal virions (compare Fig. 5B and I). In order to explore the composition of the zipper-like viral structures, we performed immunoelectron microscopy with anti-pp220 and anti-pp62 antibodies on sections of vA72-infected cells maintained under restrictive conditions for 24 h. As illustrated in Fig. 5K and L, the labeling of pp220 and pp62 was essentially associated with the zipper-like structures at the virus factories.
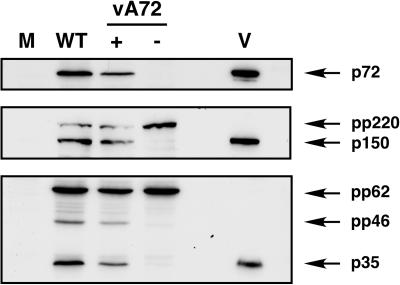
Next we analyzed whether the core precursors are properly cleaved when p72 expression is repressed. Cells infected with recombinant vA72 under permissive and restrictive conditions, as well as control mock- and BA71V-infected cells, were processed at 24 hpi. Cell extracts and purified virions were then analyzed by Western immunoblotting with antibodies against the core precursors pp220 and pp62 and against capsid protein p72 as a reference. As shown in Fig. 6, under permissive conditions the proteolytic processing of both precursors occurred to a similar extent as in control cells infected with parental virus. In contrast, under restrictive conditions the proteolytic processing of pp220 and pp62 precursors into their respective mature products p150 and p35 was drastically inhibited. These results indicate that the expression of capsid protein p72 is needed for the proper assembly of the core and for polyprotein processing.
FIG. 6.
Proteolytic processing of core polyproteins pp220 and pp62 requires the expression of the major capsid protein p72. Vero cells were mock infected (lane M) or infected for 20 h with parental BA71V (lane WT) or recombinant vA72 virus in the presence (lane +) or absence (lane −) of IPTG. Cell extracts were analyzed, together with purified ASFV particles (lane V), by immunoblotting with antibodies against capsid protein p72, polyprotein pp220 and mature product p150, or polyprotein pp62 and mature product p35. The positions of the detected proteins are indicated.
Polyproteins pp220 and pp62 coassemble into a core shell-like structure.
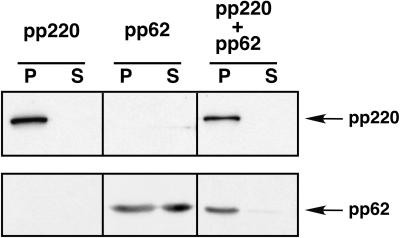
The colocalization of unprocessed polyproteins pp220 and pp62 in the zipper-like structures led us to investigate whether the two precursors were able to interact with each other. To this purpose, we took advantage of a previous result showing that polyprotein pp220, when expressed alone in COS-7 cells, assembles into dense, thick protein coats bound to cell membranes through its myristic moiety (8). As a first approach, the polyprotein pp220- and pp62-encoding genes, under the control of the T7 promoter, were independently transfected or cotransfected into COS-7 cells. After transfection, the cells were infected with recombinant vaccinia virus vTF7-3, expressing the T7 RNA polymerase (21), in the presence of Ara C, an inhibitor of vaccinia virus DNA replication and late gene expression. At 7 hpi the cells were fractionated into a low-speed (500 × g) sediment and a postnuclear supernatant. Finally, the fractions were assayed for polyprotein expression by Western immunoblotting. As shown in Fig. 7, pp220, when expressed alone, was found exclusively in the low-speed sediment, as a consequence of its assembly in the dense membrane-bound coats (8). On the other hand, when precursor pp62 was expressed alone, most of the protein was detected in the postnuclear supernatant, although a significant fraction was also found in the low-speed pellet. Interestingly, when the polyproteins were coexpressed, both of them appeared almost exclusively in the sediment, thus indicating that the pp220 precursor influences the subcellular distribution of pp62 precursor.
FIG. 7.
Coexpression of polyproteins pp220 and pp62 in COS-7 cells. COS-7 cells were transfected with plasmids bearing the pp220- or pp62-encoding genes or cotransfected with both plasmids and infected with vaccinia virus vTF7-3 in the presence of Ara C. At 7 hpi, the cells were fractionated into a low-speed sediment (lanes P) and a postnuclear supernatant (lanes S) and the fractions were analyzed by Western immunoblotting with anti-pp220 or anti-pp62 serum.
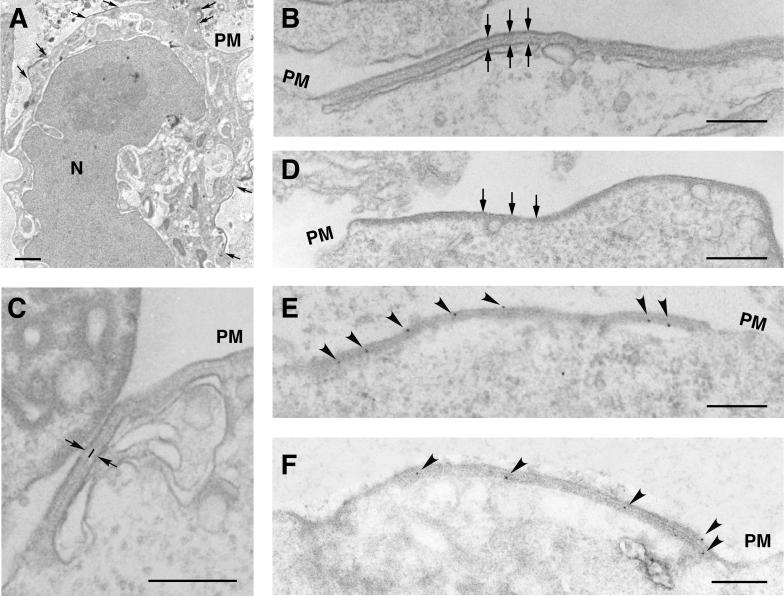
To analyze in more detail a possible interaction between the two polyproteins, we performed electron microscopy on the cotransfected COS-7 cells (Fig. 8). As previously described (8), dense, membrane-bound, thick coats were observed when polyprotein pp220 was expressed alone (see below). In contrast, no identifiable pp62-induced structure could be detected after expression of the pp62 precursor (not shown) despite the finding that a significant amount of the pp62 pool sedimented at low speed in the fractionation experiments. However, when both genes were cotransfected, we observed, besides the characteristic pp220-induced coats (Fig. 8D), a novel type of membrane-bound structure (Fig. 8A). As shown in detail in Fig. 8B and C, these pp220- and pp62-induced structures consist of an elongated trilaminar domain of symmetrical appearance bound to two adjacent lipid membranes. The central domain of these structures is very similar to that of the aberrant zipper-like structures assembled in ASFV infections (5, 22) (Fig. 5G and H) and, therefore, to the core shell of ASFV particles (4) (Fig. 5A to E). In fact, it is composed of two similar layers separated by a thinner electron-dense layer. With regard to the limiting membranes, the pp220- and pp62-induced structures were mainly associated with the plasma membrane and intracellular endosomal-lysosomal vesicles, as in the case of the pp220-induced coats (8). A comparison between the pp220- and pp62-induced zippers (Fig. 8B) and the pp220-induced coats (Fig. 8D) revealed clear differences: the pp220-containing coats were significantly thinner than the zippers (24 ± 4 nm versus 32 ± 3 nm), lacked the symmetrical trilaminar organization of the zipper-like elements, and were bound to only one membrane. The abundance of pp220 coats was always higher than that of pp220- and pp62-induced zippers, in a broad spectrum of tested pp220-to-pp62 DNA ratios (from 1:3 to 3:1). This might be due to an inefficient expression of complete pp220 copies, which are 2,475 amino acids long, in the COS cell expression system, which would affect the interaction with pp62. Alternatively, the formation of pp220 coats might be kinetically favored with respect to the assembly of pp220- and pp62-induced zipper elements. To confirm that the zipper-like structures induced in COS-7 cells were made of both polyproteins, Lowicryl sections of freeze-substituted cotransfected cells were incubated with anti-pp220 and anti-pp62 antibodies followed by protein A-gold. As shown in Fig. 8E and F, both pp220 and pp62 labeling was associated with the zipper-like structures. Together, these results show that the two polyprotein precursors interact with each other to form a core shell-like viral domain.
FIG. 8.
Polyproteins pp220 and pp62 assemble into zipper-like structures. COS-7 cells were cotransfected with plasmids bearing the pp220- and pp62-encoding genes and infected with vaccinia virus vTF7-3 in the presence of Ara C. At 12 hpi, the cells were processed for conventional Epon embedding (B, C, and D) or freeze-substitution and Lowicryl embedding (A, E, and F). (A) A cotransfected cell containing pp220- and pp62-induced zipper-like structures in different areas of the cell surface (arrows). (B and C) At a higher magnification, the zipper-like structures reveal a symmetrical organization consisting of a trilaminar cytosolic domain of about 32 nm limited by lipid membranes (arrows). Two thick layers separated by a thin electron-dense layer (line in panel C) compose the protein domain. (D) For comparison, a pp220-induced coat, which is bound to only one membrane and is approximately 24 nm thick, is shown. (E and F) The Lowicryl sections of cotransfected cells were incubated with anti-pp220 (E) or anti-pp62 (F) serum followed by protein A-gold (10-nm diameter). Note that both antibodies label zipper-like structures associated with the plasma membrane (PM) and intracellular cisternae. Bars, 100 nm (A) and 200 nm (B to F).
DISCUSSION
A striking feature of ASFV is that it expresses several major structural proteins as polyprotein precursors (31, 32). Previous work showed that the polyprotein pp220 products p150, p37, p34, and p14 are major components of the viral matrix-like domain referred to as the core shell, which is placed between the inner envelope and the DNA-containing nucleoid (4, 8). In the present study, we have examined the structural role of the other polyprotein, pp62, and its mature products p35 and p15 and have further characterized the significance of polyprotein processing in ASFV assembly.
We have provided evidence that polyprotein pp62 is also involved in the construction of the core shell. Thus, the data indicate that the mature proteins p35 and p15 reside at the core shell and that the two unprocessed polyproteins pp220 and pp62 are able to interact with each other to form a core shell-like structure. The products of the two polyproteins account for nearly one-third of the total virus mass (another one-third corresponds to the capsid protein p72) and are present in similar molar proportions. This equimolecular stoichiometry does not appear to be an obvious consequence of their initial expression as polyprotein precursors. Thus, the temporal regulation of polyprotein processing, the cleavage efficiencies of the different Gly-Gly-X signals, or the relative stabilities of the resulting products could cause certain products to accumulate faster and to a greater extent than others. Indeed, the appearance of the latest pp220 products, p37 and p14, is delayed for several hours with respect to that of the earliest protein, p150, as judged by pulse-chase experiments (31). Rather, our data indicate that the equimolecular stoichiometry is the consequence of a certain ordered arrangement of the polyproteins in the assembling virions, as occurs, for example, with the processing products of picornavirus structural polyproteins (24).
On the basis of the transient-expression experiments with pp220 and pp62 in COS cells, it is tempting to speculate that both precursors self-assemble to form the protein scaffold of the core shell. We have previously shown that polyprotein pp220, when expressed alone, binds to a variety of cellular membranes through its N-terminal myristic moiety to form long (tens of micrometers) and thick (24-nm) coats (8). In the present paper we show that both polyproteins pp220 and pp62 form a thicker (about 32-nm) distinct structure that binds to one lipid membrane at each side. The symmetrical appearance of these zipper-like structures suggests that the central domain resembling the core shell is composed of two membrane-bound equivalent sheets separated by a thinner electron-dense layer. Among other possibilities, one simple model to explain this particular architecture would consist of two pp220-containing opposite layers interconnected by precursor pp62. A similar organization might also occur at the developing core shell of normal ASFV particles. However, unlike the zipper-like structure, the bona fide core shell is an asymmetrical domain limited by a lipid envelope and an inner DNA-containing nucleoid. This major difference could be due to other events occurring during the virus assembly where core maturation is concomitant with capsid formation (4). In relation to this, we have shown by using a conditional lethal mutant affected in the major capsid protein p72 that the core assembly depends on p72 expression (22). When p72 expression and capsid formation are eliminated, both polyprotein precursors remain unprocessed and assemble into aberrant zipper-like structures (reference 22 and this report) similar to those observed in the transient-expression experiments with pp220 and pp62 precursors. Under these restrictive conditions, no DNA-containing nucleoids are formed at the virus factories. From these results it seems probable that the construction of the capsid on the inner envelope determines in some way the assembly of an asymmetrical core shell beneath this envelope. For example, the progressive bending of the assembling capsid could prevent interaction of the enclosed membrane-attached core shell with a second adjacent envelope, which would lead to the formation of aberrant double-enveloped particles such as those obtained from zipper-like structures upon p72 expression (Fig. 5H and I). Another possibility is that capsid formation triggers polyprotein processing in such a way that it enables the inner face of the assembling core shell to interact with the nucleoid components. In line with this idea, a maturational role has also been proposed for the proteolytic processing of the core precursor proteins of poxviruses, which use cleavage motifs (Ala-Gly-X) similar to those of ASFV (Gly-Gly-X). In the case of vaccinia virus, conditional lethal mutations of several genes have been shown to affect both the proteolytic cleavage of the precursor proteins and the assembly of the core, including the DNA uptake (13, 19, 23).
The specific role of polyprotein processing in ASFV morphogenesis is not fully understood at present. The available data strongly suggest that polyprotein processing is a maturational process concomitant with the core assembly. First, ASFV proteinase has been detected in the assembling core of immature virions and in the core shell of mature particles (6). On the other hand, proteolytic processing probably takes place upon the incorporation and assembly of the major capsid protein p72 and the core precursors pp220 and pp62 into the developing particles, as deduced from pulse-chase experiments (reference 15 and this report). In support of this view is the finding that proteolytic processing is inhibited when p72 expression is repressed. Furthermore, the analysis of recombinant v220i revealed that when pp220 expression is blocked, the precursor pp62 remains virtually uncleaved. This suggests that only after its interaction with polyprotein pp220 inside the developing particle would the precursor pp62 be properly processed.
ASFV is structurally related to the iridoviruses and to other large icosahedral DNA viruses such as the phycodnaviruses (4, 12, 33, 34). Despite this, no significant sequence similarity has been reported at present for the genes encoding structural proteins, with the sole exception of the major capsid protein p72 (30). Also, there is no evidence for polyprotein expression in these related DNA viruses. Interestingly, however, the occurrence of aberrant virus forms very similar to the ASFV zipper-like structures has been reported for HincV-1, a brown algal DNA virus (38), and FV3, the prototype of iridoviruses (35). Regarding FV3, such aberrant forms arise at the virus factories at very late times of infection (Don B. Stoltz, personal communication) or when the infected cells are treated with metabolic inhibitors such as actinomycin D (35). In the case of ASFV, zipper-like elements are frequently found at very late times in normal infections (our unpublished observations) or when the expression of the capsid proteins p72 and pE120R is eliminated by using conditional lethal mutants (7, 22). In our view, the existence of these aberrant forms reinforces the structural resemblance between ASFV, the iridoviruses and related viruses and suggests that all of these viruses share equivalent structural motifs in their core proteins.
Acknowledgments
We thank Donald B. Stoltz for critical reading of the manuscript and helpful discussions. We thank M. Guerra, F. Ocaña, and M. Rejas for technical assistance.
This study was supported by grants from the Dirección General de Investigación Científica y Técnica (PB96-0902-C02-01), the European Community (FAIR-CT97-3441), and the Ministerio de Educación y Cultura (AGF98-1352-CE) and by an institutional grant from Fundación Ramón Areces.
REFERENCES
- 1.Alonso, C., J. Miskin, B. Hernáez, P. Fernández-Zapatero, L. Soto, C. Cantó, I. Rodríguez-Crespo, L. Dixon, and J. M. Escribano. 2001. African swine fever virus protein p54 interacts with the microtubular motor complex through direct binding to light-chain dynein. J. Virol. 75:9819-9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alves de Matos, A. P., and Z. G. Carvalho. 1993. African swine fever virus interaction with microtubules. Biol. Cell 78:229-234. [DOI] [PubMed] [Google Scholar]
- 3.Andrés, G., C. Simón-Mateo, and E. Viñuela. 1993. Characterization of two African swine fever virus 220-kDa proteins: a precursor of the major structural protein p150 and an oligomer of phosphoprotein p32. Virology 194:284-293. [DOI] [PubMed] [Google Scholar]
- 4.Andrés, G., C. Simón-Mateo, and E. Viñuela. 1997. Assembly of African swine fever virus: role of polyprotein pp220. J. Virol. 71:2331-2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrés, G., R. García-Escudero, C. Simón-Mateo, and E. Viñuela. 1998. African swine fever virus is enveloped by a two-membraned collapsed cisterna derived from the endoplasmic reticulum. J. Virol. 72:8988-9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andrés, G., A. Alejo, C. Simón-Mateo, and M. L. Salas. 2001. African swine fever virus protease: a new viral member of the SUMO-1-specific protease family. J. Biol. Chem. 276:780-787. [DOI] [PubMed] [Google Scholar]
- 7.Andrés, G., R. García-Escudero, E. Viñuela, M. L. Salas, and J. M. Rodríguez. 2001. African swine fever virus structural protein pE120R is essential for virus transport from the assembly sites to the plasma membrane but not for infectivity. J. Virol. 75:6758-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andrés, G., R. García-Escudero, M. L. Salas, and J. M. Rodríguez. 2002. Repression of African swine fever virus polyprotein pp220-encoding gene leads to the assembly of icosahedral core-less particles. J. Virol. 76:2654-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breese, S. S., Jr., and C. J. DeBoer. 1966. Electron microscope observation of African swine fever virus in tissue culture cells. Virology 28:420-428. [DOI] [PubMed] [Google Scholar]
- 10.Brookes, S. M., A. D. Hyatt, T. Wise, and R. M. E. Parkhouse. 1998. Intracellular virus DNA distribution and the acquisition of the nucleoprotein core during African swine fever virus particle assembly: ultrastructural in situ hybridisation and DNase-gold labelling. Virology 249:175-188. [DOI] [PubMed] [Google Scholar]
- 11.Carrascosa, A. L., M. del Val, J. F. Santarén, and E. Viñuela. 1985. Purification and properties of African swine fever virus. J. Virol. 54:337-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carrascosa, J. L., J. M. Carazo, A. L. Carrascosa, N. García, A. Santisteban, and E. Viñuela. 1984. General morphology and capsid fine structure of African swine fever virus particles. Virology 132:160-172. [DOI] [PubMed] [Google Scholar]
- 13.Cassetti, M. C., M. Merchlinsky, E. J. Wolffe, A. S. Weisberg, and B. Moss. 1998. DNA packaging mutant: repression of the vaccinia virus A32 gene results in noninfectious, DNA-deficient, spherical, enveloped particles. J. Virol. 72:5769-5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cobbold, C., J. T. Whittle, and T. Wileman. 1996. Involvement of the endoplasmic reticulum in the assembly and envelopment of African swine fever virus. J. Virol. 70:8382-8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cobbold, C., and T. Wileman. 1998. The major structural protein of African swine fever virus, p73, is packaged into large structures, indicative of viral capsid or matrix precursors, on the endoplasmic reticulum. J. Virol. 72:5215-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Costa, J. V. 1990. African swine fever virus, p. 247-270. In G. Darai (ed.), Molecular biology of iridoviruses. Kluwer Academic Publishers, Boston, Mass.
- 17.Dixon, L. K., J. V. Costa, J. M. Escribano, D. L. Rock, E. Viñuela, and P. J. Wilkinson. 2000. The Asfarviridae, p. 159-165. In M. H. V. Van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carsten, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Seventh report of the International Committee for the Taxonomy of Viruses. Academic Press, New York, N.Y.
- 18.Enjuanes, L., A. L. Carrascosa, M. A. Moreno, and E. Viñuela. 1976. Titration of African swine fever virus. J. Gen. Virol. 32:471-477. [DOI] [PubMed] [Google Scholar]
- 19.Ericsson, M., S. Cudmore, S. Shuman, R. C. Condit, G. Griffiths, and J. Krijnse Locker. 1995. Characterization of ts16, a temperature-sensitive mutant of vaccinia virus. J. Virol. 69:7072-7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Esteves, A., M. I. Marques, and J. V. Costa. 1986. Two-dimensional analysis of African swine fever virus proteins and proteins induced in infected cells. Virology 152:192-206. [DOI] [PubMed] [Google Scholar]
- 21.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.García-Escudero, R., G. Andrés, F. Almazán, and E. Viñuela. 1998. Inducible gene expression from African swine fever virus recombinants: analysis of the major capsid protein p72. J. Virol. 72:3185-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heljasvaara, R., D. Rodríguez, C. Risco, J. L. Carrascosa, M. Esteban, and J. R. Rodríguez. 2001. The major core protein P4a (A10L gene) of vaccinia virus is essential for correct assembly of viral DNA into the nucleoprotein complex to form immature viral particles. J. Virol. 75:5778-5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hellen, C. U. T., and E. Wimmer. 1992. Maturation of poliovirus capsid proteins. Virology 187:391-397. [DOI] [PubMed] [Google Scholar]
- 25.López-Otín, C., C. Simón-Mateo, L. Martínez, and E. Viñuela. 1989. Gly-Gly-X, a novel consensus sequence for the proteolytic processing of viral and cellular proteins. J. Biol. Chem. 264:9107-9110. [PubMed] [Google Scholar]
- 26.Moura Nunes, J. F., J. D. Vigario, and A. M. Terrinha. 1975. Ultrastructural study of African swine fever virus replication in cultures of swine bone marrow cells. Arch. Virol. 49:59-66. [DOI] [PubMed] [Google Scholar]
- 27.Rodríguez, J. M., M. L. Salas, and E. Viñuela. 1996. Intermediate class of mRNAs in African swine fever virus. J. Virol. 70:8584-8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roullier, I., S. M. Brookes, A. D. Hyatt, M. Windsor, and T. Wileman. 1998. African swine fever virus is wrapped by the endoplasmic reticulum. J. Virol. 72:2373-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salas, J., M. L. Salas, and E. Viñuela. 1999. African swine fever virus: a missing link between poxviruses and iridoviruses?, p. 467-480. In E. Domingo, R. G. Webster, and J. J. Holland (ed.), Origin and evolution of viruses. Academic Press, London, United Kingdom.
- 30.Schnitzler, P., and G. Darai. 1993. Identification of the gene encoding the major capsid protein of fish lymphocystis disease virus. J. Gen. Virol. 74:2143-2150. [DOI] [PubMed] [Google Scholar]
- 31.Simón-Mateo, C., G. Andrés, and E. Viñuela. 1993. Polyprotein processing in African swine fever virus: a novel gene expression strategy for a DNA virus. EMBO J. 12:2977-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simón-Mateo, C., G. Andrés, F. Almazán, and E. Viñuela. 1997. Proteolytic processing in African swine fever virus: evidence for a new structural polyprotein, pp62. J. Virol. 71:5799-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stoltz, D. B. 1971. The structure of icosahedral cytoplasmic deoxyriboviruses. J. Ultrastruct. Res. 37:219-239. [DOI] [PubMed] [Google Scholar]
- 34.Stoltz, D. B. 1973. The structure of icosahedral cytoplasmic deoxyriboviruses. II. An alternative model. J. Ultrastruct. Res. 43:58-74. [DOI] [PubMed] [Google Scholar]
- 35.Tripier, F., J. Braunwald, L. Markovic, and A. Kirn. 1977. Frog virus 3 morphogenesis: effect of temperature and metabolic inhibitors. J. Gen. Virol. 37:39-52. [DOI] [PubMed] [Google Scholar]
- 36.Tulman, E. R., and D. L. Rock. 2001. Novel virulence and host range genes of African swine fever virus. Curr. Opin. Microbiol. 4:456-461. [DOI] [PubMed] [Google Scholar]
- 37.Viñuela, E. 1987. Molecular biology of African swine fever virus, p. 31-49. In Y. Becker (ed.), African swine fever. Nijhoff, Boston, Mass.
- 38.Wolf, S., I. Maier, C. Katsaros, and D. G. Müller. 1998. Virus assembly in Hincksia hincksiae (Ectocarpales, Phaeophyceae), an electron and fluorescent microscopy study. Protoplasma 203:153-167. [Google Scholar]
- 39.Yáñez, R. J., J. M. Rodríguez, M. L. Nogal, L. Yuste, C. Enríquez, J. F. Rodríguez, and E. Viñuela. 1995. Analysis of the complete nucleotide sequence of African swine fever virus. Virology 208:249-278. [DOI] [PubMed] [Google Scholar]