Abstract
Programmed cell death 4 (Pdcd4) suppresses neoplastic transformation by inhibiting the activation of c-Jun and consequently AP-1-dependent transcription. We report that Pdcd4 blocks c-Jun activation by inhibiting the expression of mitogen-activated protein kinase kinase kinase kinase 1 (MAP4K1)/hematopoietic progenitor kinase 1, a kinase upstream of Jun N-terminal kinase (JNK). cDNA microarray analysis of Pdcd4-overexpressing RKO human colon carcinoma cells revealed MAP4K1 as the sole target of Pdcd4 on the JNK activation pathway. Cotransfection of a MAP4K1 promoter-reporter with Pdcd4 demonstrated inhibition of transcription from the MAP4K1 promoter. Ectopic expression of Pdcd4 in metastatic RKO cells suppressed invasion. MAP4K1 activity is functionally significant in invasion, as overexpression of a dominant negative MAP4K1 (dnMAP4K1) mutant in RKO cells inhibited not only c-Jun activation but also invasion. Overexpression of a MAP4K1 cDNA in Pdcd4-transfected cells rescued the kinase activity of JNK. Thus, Pdcd4 suppresses tumor progression in human colon carcinoma cells by the novel mechanism of down-regulating MAP4K1 transcription, with consequent inhibition of c-Jun activation and AP-1-dependent transcription.
Programmed cell death 4 (Pdcd4) is a novel tumor suppressor. The Pdcd4 gene was first identified as a differentially expressed mRNA when cells were treated with apoptosis inducers (40). Pdcd4 gene expression is up- or down-regulated in response to apoptosis inducers (3, 9, 40, 51). Despite its name, the role of Pdcd4 in programmed cell death remains unclear.
A differential display of mRNA analysis of mouse epidermal JB6 variants demonstrated that Pdcd4 is highly expressed in JB6 P− (transformation-resistant) but not P+ (transformation-susceptible) cells (11). Reduction of Pdcd4 protein in P− cells by antisense Pdcd4 is accompanied by acquisition of a P+ phenotype (11). Overexpression of sense Pdcd4 in stably transfected P+ cells renders them resistant to tetradecanoyl phorbol acetate (TPA)-induced transformation and inhibits the expression of tumor phenotype in JB6 transformed cells. This indicates that elevated Pdcd4 protein is sufficient to inhibit tumor promoter-induced transformation and to suppress tumor phenotype (47, 48). Moreover, mice transgenic for Pdcd4 are resistant to skin tumorigenesis and tumor progression (22). Pdcd4 suppresses TPA-induced transformation and tumor phenotype as well as tumorigenesis by specifically inhibiting AP-1 but not NF-κB or serum response element (SRE)-dependent transcription (47, 48). Pdcd4 inhibits AP-1-dependent transcription by inhibiting the transactivation but not the synthesis of c-Jun and c-Fos (48). These findings suggest that Pdcd4 might suppress tumor progression by regulating the signal transduction pathways leading to AP-1 transactivation. Although Bitomsky et al. (5) recently showed in an in vitro kinase assay that Pdcd4 inhibits Jun N-terminal kinase (JNK) activity, the mechanism by which Pdcd4 inhibits JNK kinase activity remains unclear.
Yeast two-hybrid, immunoprecipitation, glutathione S-transferase (GST) pull-down, and immunolocalization analyses have established that Pdcd4 binds to eukaryotic translation initiation factor 4A (eIF4A) and eIF4G (18, 24, 46). Pdcd4 inhibits the helicase activity of eIF4A and inhibits translation in both the rabbit reticulocyte lysate system and transfected cells (45, 46, 50). Binding to eIF4A is required for Pdcd4 to inhibit translation. Pdcd4 mutants inactivated for eIF4A binding fail to inhibit translation in transfected cells (45). In addition, binding to eIF4A and consequent inhibition of translation initiation appear to be required for Pdcd4 to inhibit AP-1-dependent transcription, because Pdcd4D418A, a mutant inactivated for binding to eIF4A, fails to inhibit either translation or AP-1-dependent transcription (46). These findings suggest that inhibition of AP-1-dependent transcription by Pdcd4 occurs through inhibiting translation of regulatory factors that control the activation of AP-1-dependent transcription. The mRNAs targeted by Pdcd4 are unknown. AP-1 regulates several events required for cell invasion, including expression of matrix metalloproteinases (MMPs) (4, 13, 14) and cell motility (35, 41). Increased expression or activation of AP-1 component proteins enhances invasion (20, 32). In addition, inhibition of AP-1 activity by dominant negative c-Jun, TAM67, suppresses invasion and other events in tumor progression (17, 26, 49).
Pdcd4 expression shows a progressive decrease with human cancer progression in some disease sites. Analysis of Pdcd4 expression in human lung tumor cell lines and primary lung carcinomas revealed that loss of Pdcd4 expression correlates with tumor progression (10). In addition, protein expression patterns in NCI 60 human cancer cell lines and a set of human breast cancer cell lines showed reduced expression of Pdcd4 protein in the more progressed tumor cell lines (1, 23). These findings suggest that Pdcd4 may act to suppress tumor progression.
In order to elucidate the mechanism by which Pdcd4 inhibits the c-Jun activation important for invasion, we stably overexpressed Pdcd4 in human colon carcinoma RKO cells. The results showed that Pdcd4 inhibits JNK kinase activity, JNK phosphorylation, c-Jun phosphorylation, AP-1-dependent transcription, and RKO cell invasion. Pdcd4 targets the expression of MAP kinase kinase kinase kinase 1 (MAP4K1, also known as hematopoietic progenitor kinase 1 [HPK1]), a kinase upstream of JNK (MAP1K). By targeting MAP4K1, Pdcd4 inhibits events important in driving invasion, namely, JNK activation and consequent AP-1-dependent transcription. Pdcd4 does not target JNK directly but targets upstream activities.
MATERIALS AND METHODS
Antibodies.
The MKK4, phospho-MKK4, JNK, phospho-JNK, c-Jun, phospho-c-Jun (Ser63 or Ser73), and phosph-ERK1/2 antibodies were purchased from Cell Signaling. The MAP4K1 (or HPK1) (N-19) antibody was purchased from Santa Cruz Biotechnology. The Pdcd4 antibody was generated as previously described (47).
Construction of plasmids.
The Pdcd4 lentivirus expression plasmid used to generate lentivirus containing recombinant Pdcd4 was created by using the ViraPower Lentiviral Gateway expression kit (Invitrogen). Briefly, the Pdcd4 cDNA was cloned into pDONR201 vector by using BP Clonase and named pDONR-Pdcd4. The Pdcd4 cDNA was then excised from the pDONR-Pdcd4 plasmid and inserted into the pLenti6/V5-DEST vector by using LR Clonase and termed pLenti-Pdcd4. The dominant negative MAP4K1 cDNA was excised from pCI-HPK1-M46 (29), ligated into NheI and NotI sites of the pcDNA3.1+/Zeo vector (Invitrogen), and named pMAP4K1(m46). The pMAP4K1 (wild-type MAP4K1 expression plasmid) was mutated Met to Lys at position 46 of pMAP4K1(m46). All constructs were sequenced to confirm the DNA sequence.
Virus production and transduction.
Virus was produced using the ViraPower lentiviral expression system (Invitrogen) according to the manufacturer's protocol. Briefly, pLenti-Pdcd4 and pLenti-control (pLenti6/V5-DEST with the ccdB gene deleted) were cotransfected with the mixture of the packaging plasmids (Invitrogen) into 293 FT cells. After 72 h, the viral supernatant was collected by centrifugation at 3,000 rpm for 15 min. The viral titer was determined by transduction of RKO cells with serial dilutions of the viral supernatant and colony counting after blasticidin selection (8 μg/ml).
The viral supernatant was added to RKO cells (1 × 105 cells/well in a six-well plate) at a multiplicity of infection of 5 with 6 μg/ml Polybrene. After 24 h, the medium was replaced with fresh McCoy's medium with 10% fetal bovine serum (FBS). After an additional 24 h, the cells were collected for functional assays or selected by blasticidin.
Cloning the MAP4K1 promoter.
The MAP4K1 promoter (1.2 kb) was cloned by PCR. In the first PCR, a fragment was amplified by Pfu polymerase-dependent PCR using forward primer 1, 5′-GGCACGGCGTCCTCAAGGACGGGAACAGG-3′, reverse primer 1, 5′-CCGCTGTAGCAGGTCATAGTGGTCC-3′, and human genomic DNA isolated from RKO cells as template. In the nested PCR, the MAP4K1 promoter (1.2-kb) fragment was amplified by Pfu polymerase-dependent PCR using nested forward primer 2, 5′-GGCTGCCACGTGACTTCTTCCTCGCAGG-3′, reverse primer 2, 5′-GGGTCCACGACGTCCATCCCTGG-3′, and the first PCR product as template. This MAP4K1 promoter (1.2 kb) was then cloned into a pGL3-Basic vector (Promega) and named pMAP4K1-LUC.
Cell culture, transfection, and luciferase activity assay.
The colon carcinoma RKO cells were kindly provided by Douglas Boyd (MD Anderson Cancer Center). Cells were grown in McCoy's medium containing 10% FBS, 2 mM l-glutamine, and 500 U/ml penicillin-streptomycin at 37°C in a humidified atmosphere of 5% CO2 in air. RKO cells (3 × 104 cells/well in a 24-well plate) were transfected with a total of 1.2 μg of DNA (as described in the figure legends) using 2 μl of Fugene 6 (Roche Applied Science). After 4 to 6 h, 1 ml fresh McCoy's medium with 10% FBS was then added into each well. After 48 h, the cells were lysed by 1× passive lysis buffer (Promega) and assayed for luciferase activity as previously described (47).
In vitro migration assay.
Migration assays were performed using 48-well Boyden chambers with 10 μg/ml collagen IV-coated polycarbonate membranes (8-μm pore; Neuro Probe). One hundred thousand cells were resuspended in McCoy-bovine serum albumin (BSA; 0.1%) medium and placed on the upper well. Epidermal growth factor (EGF) was diluted into McCoy-BSA medium at 20 ng/ml and added to the lower well. Chambers were incubated at 37°C for 22 h, after which filters were removed, fixed, and stained with 0.1% (wt/vol) crystal violet and then mounted on glass slides. After removal of nonmigrated cells, cells that had migrated were quantitated by densitometric scanning and image analysis of the fixed and stained cells (Personal Densitometer SI and ImageQuant software; Molecular Dynamics).
Matrigel invasion assay.
The modified Boyden chamber cell invasion assay was carried out according to the manufacturer's instructions (BD Biosciences). Pdcd4-overexpressing RKO (RKO-Pdcd4) cells or control RKO (RKO-vector) cells (1 × 105 cells/ml) were suspended in McCoy-BSA (0.1%) medium and seeded onto Matrigel-coated Transwell filters (8-μm pore size) in Biocoat Matrigel invasion chambers (BD Biosciences). EGF was diluted into McCoy-BSA medium at 20 ng/ml and added to the lower well. Chambers were incubated at 37°C for 22 h, after which filters were removed, fixed, and stained with 0.1% (vol/wt) crystal violet. The number of cells that had invaded through the filter into the lower compartment was determined by counting five random areas.
Gelatin zymography.
RKO cells (5 × 105 cells per well) were plated onto Matrigel-coated (30 μg/ml) six-well plates. After 72 h, the conditioned medium was collected, clarified by centrifugation, concentrated by Centricon YM-30 (10:1 concentration; Millipore), and separated in nonreducing polyacrylamide gels containing 0.1% (wt/vol) gelatin. The gel was incubated with 1× Zymogram renaturing buffer (Invitrogen) for 30 min at room temperature, 1× Zymogram developing buffer (Invitrogen) for 30 min at room temperature, and 1× Zymogram developing buffer at 37°C for overnight. The gel was then stained for protein with See Blue and photographed. Proteolysis was detected as a white zone in a dark field.
JNK kinase assay.
The JNK kinase activity was assayed by using a SAPK/JNK assay kit (Cell Signaling). Briefly, 2 × 106 cells were lysed in 1× cell lysis buffer (Cell Signaling) plus protease inhibitor and incubated on ice for 1 h. After centrifugation, an aliquot (250 μl) of supernatant was added to 2 μg (20 μl) of c-Jun fusion protein beads and incubated with gentle rocking overnight at 4°C. The beads were then washed with 500 μl of 1× cell lysis buffer twice and 1× kinase buffer (Cell Signaling) once. The beads were suspended in 50 μl of kinase buffer and incubated for 30 min at 30°C. The reaction was terminated by adding 25 μl of 3× sodium dodecyl sulfate (SDS) sample buffer (Cell Signaling). The proteins were separated on 10% NuPage bis-Tris polyacrylamide gel (Invitrogen) and Western blotted using phospho-c-Jun (Ser63) antibody.
Preparation of cell lysates and Western blot analysis.
RKO cells were washed with phosphate-buffered saline twice and lysed in lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 1× protease inhibitor mixture [Roche Applied Sciences]). After incubation on ice for 1 h, the supernatant was collected by centrifugation at 14,000 rpm for 10 min. Protein concentration was determined by using the BCA protein assay kit (Pierce). Aliquots containing 15 μg protein were separated on a 10% NuPage bis-Tris polyacrylamide gel and transferred to a nitrocellulose membrane. The protein-bound membrane was incubated with antibody (as indicated in the figure legends), followed by horseradish peroxidase-linked immunoglobulin G, and visualized by chemiluminescence (ECL; Amersham).
Northern blot analysis.
The total RNA was isolated from Pdcd4-overexpressing RKO cells (or control RKO cells) using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. Ten micrograms of RNA was separated on a 1% formaldehyde gel and transferred to a nylon membrane (Roche Applied Sciences) as previously described (47). The membrane was prehybridized with NorthernMax prehybridization/hybridization buffer (Ambion) for 30 min, hybridized by incubation with 32P-labeled MAP4K1 cDNA in NorthernMax prehybridization/hybridization buffer, and then washed according to the manufacturer's instructions.
Microarray analysis.
Fluorescence-labeled cDNA was generated by reverse transcription of 20 μg of total RNA from RKO-Pdcd4 or RKO-vector cells using the Superscript III indirect cDNA labeling system (Invitrogen) and Cy3/Cy5-mono reactive dyes (Amersham Pharmacia Biotech). Cy3-labeled test samples and Cy5-labeled universal control (Stratagene) cDNA samples were combined and concentrated using Microcon YM-30 filter units (Millipore). Samples were diluted in F-hybridization buffer (25% formamide, 5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% SDS containing 10 μg human Cot-1 DNA, 10 μg poly(dA), and 4 μg tRNA) and hybridized to 22K oligonucleotide array slides overnight at 42°C. The slides were then washed with 2× SSC and 0.1% SDS, 1× SSC and 0.1% SDS for 2 min, and 0.2× SSC for 2 min.
Microarrays were scanned and processed using a GenePix 4000A microarray scanner in combination with GenePix Pro 4.0 software (Axon Instruments Inc.). Data analysis was performed using the MADb program suite (http://nciarray.nci.nih.gov) and Microsoft Excel. Data sets representing differentially expressed genes were selected according to the following criteria: (i) mean intensities beyond the threshold set at 500 units and at least two standard deviations above background, (ii) mean signal differences of ≥2.0-fold for duplicate comparisons between cDNAs from RKO-Pdcd4, RKO-Pdcd4 mutant 418, and RKO-vector cells.
RESULTS
Pdcd4 inhibits c-Jun phosphorylation, JNK activity, and AP-1-dependent transcription.
To circumvent the limited efficiency and sensitivity associated with transient transfections, we generated a recombinant lentivirus containing Pdcd4 (lentivirus-Pdcd4). RKO cells were transduced with recombinant lentivirus-Pdcd4 at a multiplicity of infection of 5, selected by blasticidin for 10 to 12 days, and designated as RKO-Pdcd4. As shown in Fig. 1A, the RKO-Pdcd4 cells produced an approximately four- to fivefold-higher Pdcd4 protein level than the cells transduced with lentivirus-empty vector (designated RKO-vector cells). This result indicates that the recombinant lentivirus-Pdcd4 is able to transduce RKO cells and give rise to stable Pdcd4 expression. It is noteworthy that overexpression of Pdcd4 in RKO cells does not induce apoptosis or alter the cell cycle (Fig. 1B). This contrasts with the reported observations of Pdcd4 inhibiting proliferation in human carcinoid cells (19) and inducing apoptosis in human breast cancer cells (1).
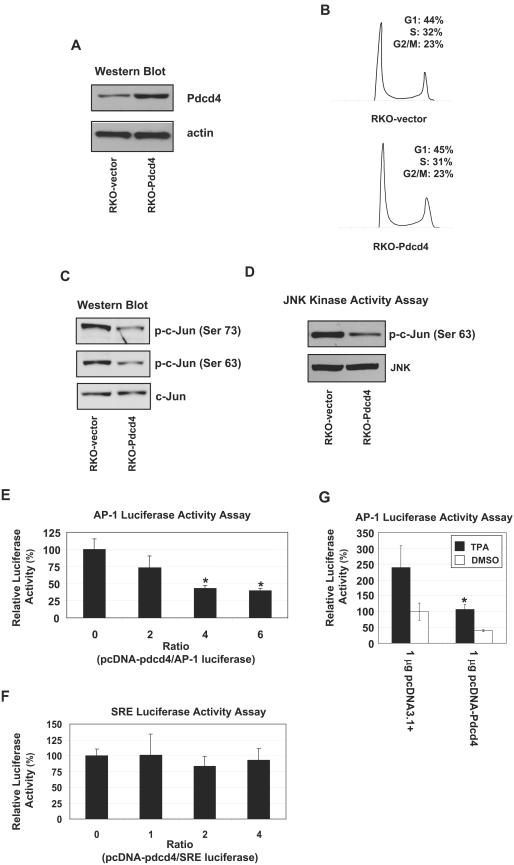
FIG.1.
Stable Pdcd4 expression in RKO cells inhibits JNK and c-Jun activation. (A) Stable overexpression of Pdcd4 in RKO cells by lentivirus transduction. RKO cells were transduced by recombinant lentivirus-Pdcd4 (RKO-Pdcd4) or lentivirus-empty (RKO-vector) at a multiplicity of infection of 5. (B) Pdcd4 does not alter the cell cycle in RKO cells. Cell cycle was assayed by fluorescence-activated cell sorting analysis after propidium iodide staining. (C) Pdcd4 inhibits phosphorylation of c-Jun. Western blotting was performed on cell lysates from RKO-Pdcd4 or RKO-vector cells with phospho-c-Jun (Ser 63) or phospho-c-Jun (Ser 73) antibodies. The corresponding blot was stripped and reprobed with Jun antibody. (D) Pdcd4 inhibits JNK kinase activity. The total JNK was pulled down from the indicated cell lysates by GST-c-Jun and was analyzed for JNK kinase activity. Phospho-c-Jun was visualized by phospho-c-Jun (Ser 63) antibody, and the bead-bound JNK was visualized by JNK antibody. (E and F) Pdcd4 inhibits AP-1-dependent transcription but not SRE-dependent transcription. RKO cells were cotransfected with increasing amounts (0 to 1.2 μg) of pcDNA-Pdcd4 and 0.2 μg of 4× AP-1-LUC (E) or 0.2 μg of SRE-LUC (F). The total DNA was maintained at 1.4 μg by adding pcDNA3.1+. The luciferase activity with 0 μg of pcDNA-Pdcd4 is designated as 100%. These experiments were repeated three times following each of five independent transfections, and representative data are shown. Results are expressed as means ± standard deviations. *, significant difference compared with 0 μg of pcDNA-Pdcd4 as determined by Student's t test (P < 0.01). (G) Pdcd4 inhibits TPA-induced AP-1-dependent transcription. RKO cells were transfected with 1.0 μg of pcDNA-Pdcd4 (pcDNA3.1+) and 0.2 μg of 4× AP-1-LUC. The luciferase activity with 1.0 μg of pcDNA3.1+ is designated as 100%. These experiments were repeated two times following each of five independent transfections, and representative data are shown. Results are expressed as means ± standard deviations. *, significant difference compared with 0 μg of pcDNA-Pdcd4 as determined by Student's t test (P < 0.05).
Previously we demonstrated that Pdcd4 suppresses AP-1-dependent transcription in mouse epidermal JB6 cells (47, 48). The mechanism of inhibition involves suppression of c-Jun transactivation (48). To investigate whether Pdcd4 inhibits the phosphorylation of c-Jun in RKO cells, we performed a Western blot assay using antibodies against phospho-c-Jun (Ser-63) or phospho-c-Jun (Ser-73). The levels of phospho-c-Jun were two- to threefold lower in the RKO-Pdcd4 cells than in the RKO-vector cells (Fig. 1C). The reduction of c-Jun phosphorylation found in the RKO-Pdcd4 cells occurs posttranslationally, because the total protein levels of c-Jun are similar in the RKO-Pdcd4 cells and the RKO-vector cells (Fig. 1C). Although others (5) have found that the amount of ectopically expressed c-Jun is decreased in the presence of Pdcd4, the results shown here demonstrate that Pdcd4 does not inhibit endogenous c-Jun expression, in agreement with our previous report (48). Since phosphorylation of c-Jun is primarily attributable to JNK (44), we asked whether Pdcd4 inhibits JNK kinase activity. As shown in Fig. 1D, the JNK kinase activity in RKO-Pdcd4 cells was approximately 20% to 30% of that seen in RKO-vector cells, whereas the levels of bound JNK protein are similar. This finding suggests that Pdcd4 inhibits JNK kinase activity, resulting in inhibition of c-Jun phosphorylation at serines 63 and 73.
To test whether Pdcd4-inhibited c-Jun phosphorylation produces inhibition of AP-1-dependent transcription in human colon carcinoma RKO cells, we cotransfected a Pdcd4 expression plasmid (pcDNA-Pdcd4) with a 4× AP-1 luciferase reporter gene (4× AP-1-LUC). Pdcd4 inhibited basal AP-1-dependent transcription in a concentration-dependent manner (Fig. 1E). AP-1-dependent transcription was approximately 60% inhibited at a 4:1 ratio of pcDNA-Pdcd4 to 4× AP-1-LUC. Concentrations as high as a 6:1 ratio of pcDNA-Pdcd4 to reporter also showed approximately 60% inhibition. In contrast, Pdcd4 did not inhibit transcription expected to be JNK independent, namely, that regulated through the SRE (Fig. 1F). In addition, the TPA-induced AP-1-dependent transcription was approximately 50% inhibited at a 5:1 ratio of pcDNA-Pdcd4 to 4× AP-1-LUC (Fig. 1G). These findings indicate that Pdcd4 inhibits both basal and TPA-induced AP-1-dependent transcription and that inhibition of AP-1-dependent transcription by Pdcd4 in RKO cells occurs, at least in part, through inhibition of c-Jun phosphorylation.
Pdcd4 specifically regulates the JNK signal transduction pathway.
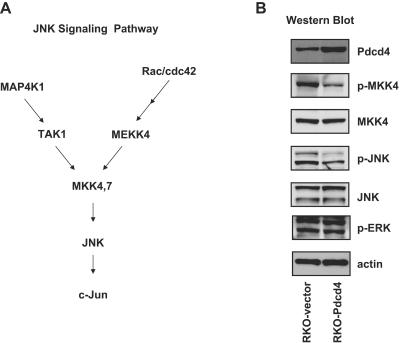
To investigate whether Pdcd4 regulates the JNK signal transduction pathway, cell lysates from RKO-Pdcd4 and RKO-vector cells were separated on SDS-polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, and incubated with various antibodies (Fig. 2B). The levels of phospho-MKK4 and phospho-JNK were two- to threefold lower in the RKO-Pdcd4 cells than in the RKO-vector cells, whereas the levels of total MKK4 and JNK proteins were similar with and without ectopic Pdcd4 expression. This indicates that Pdcd4 regulates MKK4 and JNK activities posttranslationally (Fig. 2B). These results also indicate that Pdcd4 regulates a kinase that is upstream of MKK4 and JNK. In contrast, the levels of phospho-ERK were similar in RKO-Pdcd4 and RKO-vector cells, indicating that Pdcd4 specifically regulates the JNK but not the ERK signal transduction pathway. Phospho-p38 in both RKO-Pdcd4 and RKO-vector cells was undetectable (data not shown).
FIG. 2.
Pdcd4 inhibits the JNK pathway but not the ERK activation pathway. (A) JNK signaling pathway. JNK can be activated through a signaling pathway from Rac/cdc42 or, alternatively, through a signaling pathway from MAP4K1. (B) Pdcd4 inhibits phosphorylation of JNK and MKK-4 but not ERK. Western blotting was performed on cell lysates from the indicated cells with phospho-JNK, phospho-MKK-4, and phospho-ERK antibodies. The corresponding blots were stripped and reprobed with Pdcd4, JNK, MKK-4, and actin antibodies.
Pdcd4 inhibits the transcription of MAP4K1.
In order to identify a Pdcd4 target gene that regulates the JNK signal transduction pathway, we performed cDNA microarray assays. The ultimate objective of the microarray analysis is to identify one or more mRNAs whose expression is regulated transcriptionally by proteins whose synthesis is sensitive to Pdcd4 inhibition. The total RNA from RKO-Pdcd4 cells and RKO-Pdcd4 mutant 418 cells was isolated and labeled with Cy3 and cohybridized with Cy5-labeled universal human control on 20K human oligonucleotide arrays. A total of 412 genes were down-regulated more than 2.0-fold in RKO-Pdcd4 cells relative to RKO-Pdcd4 mutant 418 cells. Known AP-1 targets SERPINA1, STAT6, MMP7, HMGA1, and FGFR1 were among the Pdcd4 down-regulated genes. MMP-7 (7) and HMGA1 (16, 37) are known to be involved in tumor progression or metastasis. MAP4K1 was the only one of the 412 down-regulated genes on the JNK signaling pathway (2). MAP4K1 mRNA expression was three- to eightfold suppressed by Pdcd4. MAP4K1, also designated hematopoietic progenitor kinase 1, is a kinase upstream of JNK that regulates the activity of JNK kinase in hematopoietic and other cells (21). Ectopic expression of MAP4K1 activates JNK activity and strongly elevates AP-1-dependent transcription (21). In contrast, expression of dnMAP4K1 mutant inhibits JNK kinase activity (52).
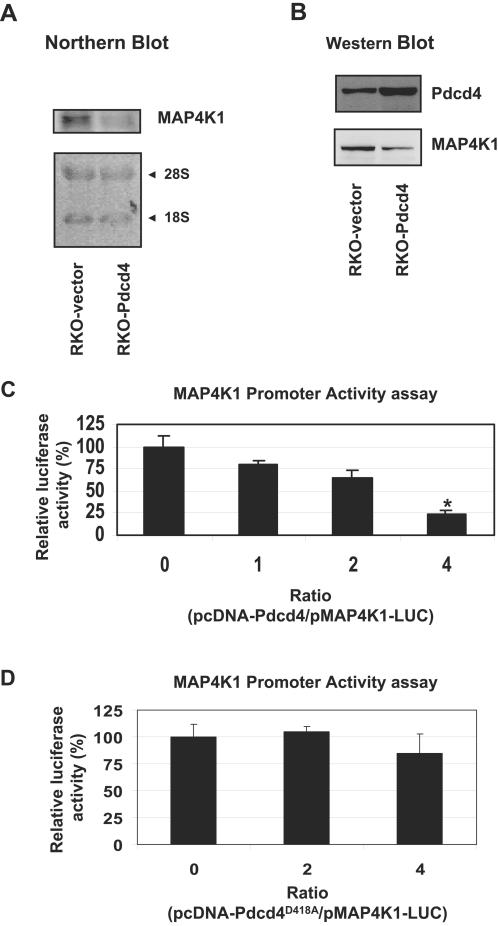
To confirm the results of the microarrays, Northern and Western blot analyses were performed. As shown in Fig. 3A, the MAP4K1 mRNA level in RKO-vector cells was approximately fivefold higher than that in RKO-Pdcd4 cells. The level of MAP4K1 protein in RKO-vector cells was approximately twofold higher than that in the RKO-Pdcd4 cells (Fig. 3B). These findings suggest that Pdcd4 acts pretranslationally to inhibit MAP4K1 mRNA synthesis or stability.
FIG. 3.
Pdcd4 inhibits expression of MAP4K1, a kinase upstream of JNK. (A) Pdcd4 inhibits MAP4K1 mRNA expression. Northern blotting was performed on the total RNA from the indicated cells and visualized by hybridization with 32P-labeled MAP4K1 cDNA. The experiments were repeated twice. (B) Pdcd4 inhibits MAP4K1 protein expression. Western blotting was performed on cell lysates (50 μg) from the indicated cells with MAP4K1 antibody. The corresponding blot was stripped and reprobed with Pdcd4 antibody. The experiments were repeated four times, and representative data are shown. (C) Pdcd4 inhibits transcription from the MAP4K1 promoter. RKO cells were cotransfected with increasing amounts (0 to 0.8 μg) of pcDNA-Pdcd4 and 0.2 μg of MAP4K1-LUC. The assay measures transcription rather than translation of luciferase, as the MAP4K1-Luc construct contains an unstructured 5′UTR in the luciferase gene. The total DNA was maintained at 1.0 μg by adding pcDNA3.1+. The luciferase activity with 0 μg of pcDNA-Pdcd4 is designated as 100%. These experiments were repeated three times following each of five independent transfections, and representative data are shown. Results are expressed as means ± standard deviations. *, significant difference compared with 0 μg of pcDNA-Pdcd4 as determined by Student's t test (P < 0.005).
To test whether Pdcd4 inhibits transcription from the MAP4K1 promoter, we cloned the human MAP4K1 promoter (1,201 bp) and fused it to a luciferase reporter gene (pMAP4K1-LUC). As shown in Fig. 3C, cotransfected Pdcd4 inhibits MAP4K1 promoter activity in a concentration-dependent manner. MAP4K1 promoter activity was approximately 75% inhibited at a 4:1 ratio of pcDNA-Pdcd4 to pMAP4K1-LUC. This transcriptional inhibition is relatively specific for MAP4K1, as luciferase expression driven by other transcriptional promoters, including SRE (Fig. 1F) or NF-κB (47), is not inhibited by Pdcd4. The lack of inhibition of SRE-LUC or NF-κB-LUC expression also eliminates the possibility that Pdcd4 could be inhibiting pMAP4K1-LUC expression at the level of luciferase translation (the luciferase construct lacks a structured 5′ untranslated region [UTR]). Others have reported that Pdcd4 can interfere with transcriptional enhancement by the c-Jun-p300 interaction (5). To ascertain whether c-Jun-p300 enhancement might apply to MAP4K1 transcription, we cotransfected a c-Jun expression plasmid with MAP4K1-LUC; this produced no effect on MAP4K1 promoter activity. Expression of P300 enhanced the luciferase activity driven by the MAP4K1 promoter, but this enhanced luciferase activity was not inhibited when the Pdcd4 expression plasmid was cotransfected (data not shown). Therefore, although other transcriptional promoters may be so regulated, inhibition of transcription from the MAP4K1 promoter by Pdcd4 appears not to occur through attenuation of c-Jun and/or P300 enhancement. Thus, transcription from the MAP4K1 promoter is inhibited by Pdcd4. Because Pdcd4 is a translation inhibitor that preferentially inhibits translation of 5′UTR structured mRNAs such as those encoding oncogenes or transcription factors (45, 46), Pdcd4 may translationally regulate transcription factors or transcription factor activators required for transcription of MAP4K1. To test the hypothesis that Pdcd4-inhibited MAP4K1 mRNA expression occurs as a consequence of Pdcd4-inhibited translation, a Pdcd4 mutant expression plasmid, pcDNA-Pdcd4D418A, which is defective for binding to eIF4A and for inhibition of translation (41, 42), was transfected with MAP4K1-LUC. As shown in Fig. 3D, Pdcd4D418A did not inhibit luciferase activity driven from the MAP4K1 promoter at a 4:1 ratio of pcDNA-Pdcd4D418A to MAP4K1-LUC. This result suggests that Pdcd4 translationally controls MAP4K1 expression.
Inhibition of invasiveness, cell migration, and ECM protease activity by Pdcd4.
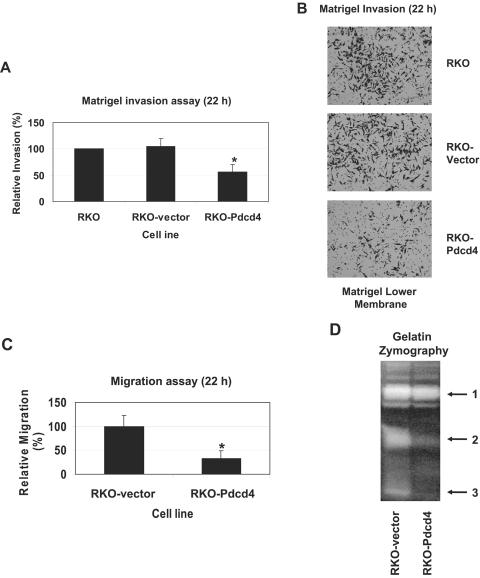
Down-regulation of JNK activity and AP-1-dependent transcription inhibits tumor cell invasion (17, 34, 42). Because Pdcd4 inhibits activation of JNK and AP-1-dependent transcription in metastatic RKO cells (Fig. 1), elevating Pdcd4 would be expected to suppress RKO cells' invasive ability. To examine whether Pdcd4 inhibits cell invasion, we assayed the capacity to pass through a Matrigel barrier in a modified Boyden chamber assay. RKO-Pdcd4 cells (or RKO-vector cells) were placed in the upper well of the invasion chamber, and the attractant (EGF, 20 ng/ml) was placed in the lower well. After 22 h, the unmigrated cells were removed and the cells that had migrated to the lower face of the membrane were stained with 0.1% crystal violet. As shown in Fig. 4A and B, the ability of RKO-Pdcd4 cells to invade through Matrigel was approximately 50 to 60% reduced compared to RKO-vector cells, while RKO-vector and wild-type cells had a similar invasive ability. This change in invasion led us to examine the effects of Pdcd4 on cell migration and extracellular matrix (ECM) protease activity, two components of the invasion process. The cell migration assays were performed as the Matrigel invasion assays were, except in the absence of Matrigel. The ability of RKO-Pdcd4 cells to migrate through the membrane was reduced approximately twofold relative to the RKO-vector cells (Fig. 4C). To investigate whether Pdcd4 suppresses ECM protease activity in Pdcd4-overexpressing RKO cells, we performed Zymogram assays. The conditioned medium from RKO-Pdcd4 cells (or RKO-vector cells) was separated on a polyacrylamide gel containing 0.1% gelatin and stained with 0.2% Coomassie blue. As shown in Fig. 4D, the conditioned medium from the RKO-Pdcd4 cells had less protease activity than that from control cells (Fig. 4D), indicating that Pdcd4 inhibits the expression or activity of ECM protease. The first band was identified as MMP2 (data not shown), whereas the second and third bands were unidentified MMPs. These findings indicate that Pdcd4 inhibits RKO cell invasion at least in part through inhibition of cell migration and ECM protease activity.
FIG. 4.
Pdcd4 inhibits RKO cell invasion. (A) Pdcd4 inhibits Matrigel invasion. Matrigel invasion assays were performed with the indicated cells for 22 h using EGF (20 ng/ml) as stimulus. Each value is the mean ± standard deviation of four independent experiments. *, significant difference compared with wild-type RKO cells as determined by Student's t test (P < 0.01). (B) Representative images of the lower membrane surface from Matrigel invasion assays. The stained cells can be distinguished from the 8-μm membrane pores. (C) Pdcd4 inhibits RKO cell migration. Boyden chamber cell migration assays were performed with the indicated cells for 22 h using EGF (20 ng/ml) as stimulus. Each value is the mean ± standard deviation of four independent experiments. *, significant difference compared with wild-type RKO cells as determined by Student's t test (P < 0.01). (D) Pdcd4 inhibits ECM protease activity. ECM protease activity was determined by using gelatin zymography. Conditioned media from indicated cells were separated on a 0.1% gelatin acrylamide gel. After activation of ECM protease, the gel was then stained for protein with See Blue and photographed. Arrows indicate the different protease activities between RKO-Pdcd4 and RKO-vector cells. Arrow 1 indicates MMP-2, while the enzymes designated by arrows 2 and 3 are unknown.
Inactivation of MAP4K1 activity contributes to inhibited invasiveness.
MAP4K1 is a kinase upstream of JNK that regulates the activation of JNK (52). Ectopic expression of a dnMAP4K1 in which methionine is substituted in place of lysine 46 inhibits MAP4K1 kinase and JNK activities (21, 52). In order to determine whether inhibition of MAP4K1 activity by Pdcd4 contributes to suppression of the invasive ability in RKO cells, we stably transfected a dnMAP4K1 expression plasmid. The pooled cells were selected for zeocin resistance for 14 days. Figure 5A shows that dnMAP4K1 [pMAP4K1(m46)] was expressed in stably transfected cells designated m46-1 and m46-2. The level of phospho-c-Jun in m46-1 and m46-2 cells was three- to fourfold lower than that in vector control cells. In agreement with previous findings (52), this result indicates that overexpression of dnMAP4K1 inhibits transactivation of c-Jun. It is noteworthy that inhibiting c-Jun activation by dnMAP4K1 is specific, because MAP4K1 has been demonstrated to inhibit transforming growth factor β-induced but not UVC-induced JNK activation (48). In addition, dominant negative MAP4K5 but not dnMAP4K1 blocks the UVC-induced JNK activation.
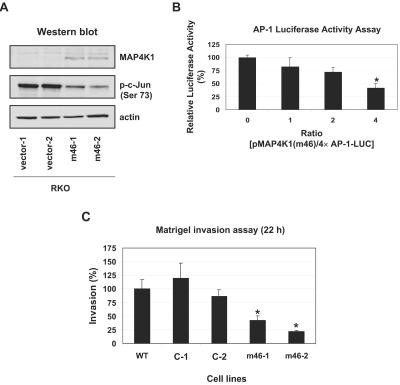
FIG. 5.
Dominant negative MAP4K1 inhibits RKO cell invasion. (A) Western blotting was performed on cell lysates (10 μg) from stably expressed pMAP4K1(m46) (or empty vector) with phospho-c-Jun (Ser 73), c-Jun, MAP4K1, and actin antibodies. (B) dnMAP4K1 inhibits AP-1-dependent transcription in RKO cells. RKO cells were cotransfected with increasing amounts (0 to 0.8 μg) of pMAP4K1(m46) and 0.2 μg of 4× AP-1-LUC. The total DNA was maintained at 1.0 μg by adding pcDNA3.1+. The luciferase activity with 0 μg of pMAP4K1(m46) is designated as 100%. These experiments were repeated three times following each of five independent transfections, and representative data are shown. Results are expressed as means ± standard deviations. *, significant difference compared with 0 μg of pMAP4K1 as determined by Student's t test (P < 0.01). (C) dnMAP4K1 inhibits RKO cell Matrigel invasion. Matrigel invasion assays were performed with the indicated cells for 22 h using EGF (20 ng/ml) as stimulus. Each value is the mean ± standard deviation of three independent experiments. *, significant difference compared with wild-type RKO cells as determined by Student's t test (P < 0.01).
To further test whether overexpression of dnMAP4K1 inhibits AP-1-dependent transcription, the dnMAP4K1 expression plasmid [pMAP4K1(m46)] was cotransfected with the 4× AP-1 luciferase reporter gene (4× AP-1-LUC) into RKO cells. Pdcd4 inhibited AP-1-dependent transcription in a concentration-dependent manner (Fig. 5B). AP-1-dependent transcription was approximately 60% inhibited at a 4:1 ratio of pMAP4K1(m46) to 4× AP-1-LUC. These dnMAP4K1-expressing cells (m46-1 and m46-2 cells) were further analyzed for Matrigel invasion. The ability of m46-1 and m46-2 cells to invade through Matrigel was approximately 60 to 80% reduced relative to wild-type RKO cells (Fig. 5C), whereas the vector control cells (C-1 and C-2 cells) showed invasive abilities similar to wild-type RKO cells. The finding that inactivation of MAP4K1 suppresses RKO cell invasion demonstrates that MAP4K1 activity is required for invasion.
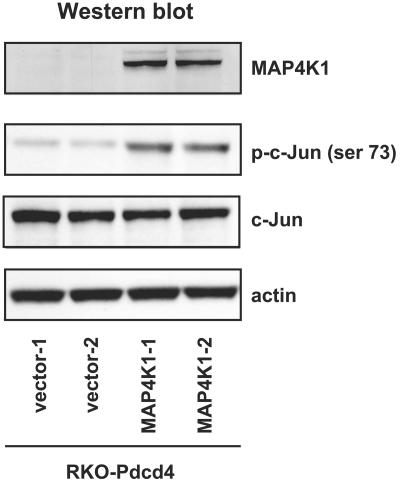
Expression of MAP4K1 protein rescues c-Jun phosphorylation from Pdcd4 inhibition.
To determine whether MAP4K1 is a functionally significant target of Pdcd4 when it inhibits JNK signaling, we performed a stable transfection of pMAP4K1 (or empty vector) into RKO-Pdcd4 cells. After selection for zeocin resistance for 14 days, the pooled cells were assayed for expression of MAP4K1 and phosphorylated c-Jun. As shown in Fig. 6, MAP4K1 introduced into RKO-Pdcd4 cells (MAP4K1-1 and MAP4K1-2) was successfully overexpressed at the protein level. The levels of phosphorylated c-Jun in MAP4K1-overexpressing RKO-Pdcd4 (MAP4K1-1 and MAP4K1-2) cells were approximately four- to sixfold higher than those in control Pdcd4-only cells (vector 1 and vector 2), whereas the levels of total c-Jun in MAP4K1-overexpressing and control cells were similar. This finding indicates that ectopic expression of MAP4K1 protein in RKO-Pdcd4 cells relieves the inhibition of c-Jun activation by Pdcd4. Thus, MAP4K1 expression is a critical target of Pdcd4 when it inhibits JNK signaling.
FIG. 6.
MAP4K1 reverses Pdcd4-inhibited c-Jun phosphorylation. Cell lysates (10 μg) from wild-type RKO, stably transfected pMAP4K1 RKO, and stably transfected vector RKO cells were prepared as described in Materials and Methods. Western blotting was performed on cell lysates with phospho-c-Jun (ser 73), c-Jun, MAP4K1, and actin antibodies.
DISCUSSION
Pdcd4 suppresses colon carcinoma cell invasion by a novel mechanism. The suppression occurs at least in part through inhibition of MAP4K1 expression. Pdcd4 suppresses transformation and tumor phenotype by inhibiting c-Jun and c-Fos transactivation and, consequently, attenuating AP-1-dependent transcription (48). The present study extends this mechanistic understanding to now implicate the JNK kinase activation pathway as a Pdcd4 target (Fig. 1). Inhibition of JNK kinase activity by Pdcd4 is attributable to attenuated expression of MAP4K1, a kinase three steps upstream of JNK (Fig. 2 and 3). Because JNK is the major kinase responsible for activating phosphorylation of c-Jun, these findings provide a mechanistic explanation for the inhibition by Pdcd4 of c-Jun transactivation and consequently of the AP-1 transactivation important to tumor progression.
Pdcd4 decreases the level of endogenous phospho-c-Jun but not of c-Jun protein in colon carcinoma RKO cells (Fig. 1). This contrasts with the observations of others (5) who reported attenuated levels of ectopically introduced c-Jun protein in quail fibroblast QT6 cells expressing Pdcd4. It is possible that QT6 cells may have higher protease activity than RKO cells or that ectopic and endogenous c-Jun are expressed or activated differently.
Elevation of the Pdcd4 protein level in RKO cells is not sufficient to induce apoptosis or to alter the cell cycle (Fig. 1B). Moreover, transgenic expression of Pdcd4 in mouse epidermis does not induce apoptosis in vivo (22). Although Pdcd4 was initially identified as being up-regulated in response to apoptosis inducers (40), cytotoxic chemotherapeutics have been found to inhibit Pdcd4 expression (3, 33). Recently, Afonja et al. (1) showed that transient transfection of Pdcd4 cDNA induces apoptosis in breast cancer T-47D and MCF-7 cells (1). Our results dissociating Pdcd4 expression from apoptosis are in agreement with previous findings that overexpression of Pdcd4 cDNA does not induce apoptosis in NIH 3T3 cells (40). A possible explanation for the discrepancies is that an additional factor present in breast cancer cell lines may be required for inducing apoptosis by Pdcd4, while this factor is deficient in RKO and NIH 3T3 cells as well as mouse epidermis.
Elevation of Pdcd4 protein in RKO cells inhibits Matrigel invasion, cell motility, and ECM protease activity (Fig. 3). The suppression of RKO cell invasion by Pdcd4 appears to be attributable at least in part to inhibition of AP-1-dependent transcription (Fig. 1). AP-1 is a transcription factor complex comprised of Jun-Jun homodimers or Jun-Fos heterodimers. The Jun protein family includes c-Jun, JunB, and JunD. The Fos protein family contains c-Fos, Fra-1, Fra-2, and FosB. AP-1 has been shown to regulate several events required for cell invasion, including expression of MMPs (4, 13, 14) and cell motility (35, 41). Increased expression or activation of AP-1 component proteins enhances invasion (20, 32). Cells transformed by oncogenic forms of Fos or Jun proteins are invasive (20, 26). In addition, inhibition of AP-1 activity by dominant negative c-Jun, TAM67, suppresses invasive ability in keratinocytes (17), fibroblasts (26), and squamous carcinomas (49) and blocks papilloma-to-carcinoma conversion (12). The inhibition of AP-1-dependent transcription by Pdcd4 (Fig. 1) is likely to contribute to suppression of RKO cell invasion (Fig. 3). Transactivation of the AP-1 protein complex occurs principally through the MAPK cascade (38). The MAPK signaling cascade includes three signal transduction pathways, i.e., JNK, ERK, and p38 signaling. JNK regulates the activation of c-Jun by phosphorylating it at Ser-63 and Ser-73. Suppression of JNK activity by blockade of the receptor for advanced glycation end products decreases tumor metastasis (43). Moreover, the JNK inhibitor SP600125 represses c-Jun activation and type IV collagenase expression, required events for invasion. These findings indicate that suppression of JNK activity resulting in inhibition of AP-1 transactivation is an important mechanism for suppression of tumor cell invasion (42).
The finding that Pdcd4 inhibits the expression of MAP4K1 mRNA and protein (Fig. 3) indicates that MAP4K1 is a molecular target of Pdcd4. MAP4K1 is a mammalian STE20-like protein serine/threonine kinase which activates the JNK signaling pathway (31). MAP4K1 activates JNK through the signaling pathway MAP4K1 → TAK1 → MKK4 → JNK (52) and does not affect other MAPK signaling pathways, including ERK and p38 signaling (25). Therefore, MAP4K1 specifically enhances AP-1-dependent transcription through activation of c-Jun (29, 31). Some reports suggest that MAP4K1 may inhibit AP-1 activity (30, 39). It is possible that, by binding to different associated proteins, MAP4K1 may become a positive or a negative regulator of AP-1-dependent transcription (6). MAP4K1 has been shown to play a critical role in T-cell activation (28, 39). Although MAP4K1 is constitutively expressed in several tissues (21), the function of MAP4K1 is still unclear. The observation that down-regulation of MAP4K1 suppresses invasive ability in RKO cells may suggest a motility-related function of MAP4K1 in normal tissues.
Previously, we have demonstrated that Pdcd4 is a binding partner of eIF4A that inhibits eIF4A's helicase activity (46). Pdcd4 preferentially inhibits translation of mRNAs having secondary structure in the 5′UTR (45). So-called “translationally repressed” mRNAs have been described which contain G- or C-rich regions in the 5′UTR with potential to form a stable secondary structure(s) in the 5′UTR (8, 36). Typically, the products of such “translationally repressed” mRNAs are transcription factors, growth factors, or proto-oncogenes (15). The mechanism by which Pdcd4 inhibits transcription from the MAP4K1 promoter (Fig. 4) may involve inhibiting translation of a transcription factor(s) or of a transcription factor activator(s) that is required for MAP4K1 transcription. Lankat-Buttgereit et al. (27) have recently reported that overexpression of Pdcd4 cDNA in HEK293 cells decreases the protein but not the mRNA level of carbonic anhydrase, suggesting that carbonic anhydrase may be a candidate translational target of Pdcd4. Direct translational targets of Pdcd4 have not yet been demonstrated.
In conclusion, in addition to inhibiting tumor promoter-induced transformation and tumor phenotype (47, 48) as well as tumorigenesis (22), Pdcd4 suppresses human tumor cell invasion. MAP4K1 appears to be a critical target of Pdcd4 when it inhibits invasion. Inhibited MAP4K1 expression leads to inhibition of JNK activation and consequently to inhibition of AP-1 activity and invasion. Elevating or mimicking Pdcd4 expression may be a promising therapeutic strategy.
Acknowledgments
We thank Mike Brattain (Roswell Park Cancer Center) and Douglas Boyd (MD Anderson Cancer Center) for providing RKO cells and the members of the Gene Regulation Section, Laboratory of Cancer Prevention, for helpful discussions.
This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
REFERENCES
- 1.Afonja, O., D. Juste, S. Das, S. Matsuhashi, and H. H. Samuels. 2004. Induction of PDCD4 tumor suppressor gene expression by RAR agonists, antiestrogen and HER-2/neu antagonist in breast cancer cells. Evidence for a role in apoptosis. Oncogene 23:8135-8145. [DOI] [PubMed] [Google Scholar]
- 2.Anonymous. 2005. KEGG: Kyoto encyclopedia of genes and genomes. 36.0 ed. Kanehisa Laboratories. [Online.] http://www.genome.jp/dbget-bin/show_pathway?hsa04010+11184.
- 3.Azzoni, L., O. Zatsepina, B. Abede, I. M. Bennett, P. Kanakaraj, and B. Perussia. 1998. Differential transcriptional regulation of CD161 and a novel gene, 197/15a, by IL-2, IL-15, and IL-12 in NK and T cells. J. Immunol. 161:3493-3500. [PubMed] [Google Scholar]
- 4.Benbow, U., and C. E. Brinckerhoff. 1997. The AP-1 site and MMP gene regulation: what is all the fuss about? Matrix Biol. 15:519-526. [DOI] [PubMed] [Google Scholar]
- 5.Bitomsky, N., M. Bohm, and K. H. Klempnauer. 2004. Transformation suppressor protein Pdcd4 interferes with JNK-mediated phosphorylation of c-Jun and recruitment of the coactivator p300 by c-Jun. Oncogene 23:7484-7493. [DOI] [PubMed] [Google Scholar]
- 6.Boomer, J. S., and T. H. Tan. 2005. Functional interactions of HPK1 with adaptor proteins. J. Cell Biochem. 95:34-44. [DOI] [PubMed] [Google Scholar]
- 7.Bramhall, S. R., J. P. Neoptolemos, G. W. Stamp, and N. R. Lemoine. 1997. Imbalance of expression of matrix metalloproteinases (MMPs) and tissue inhibitors of the matrix metalloproteinases (TIMPs) in human pancreatic carcinoma. J. Pathol. 182:347-355. [DOI] [PubMed] [Google Scholar]
- 8.Brown, E. J., and S. L. Schreiber. 1996. A signaling pathway to translational control. Cell 86:517-520. [DOI] [PubMed] [Google Scholar]
- 9.Chauhan, D., D. Auclair, E. K. Robinson, T. Hideshima, G. Li, K. Podar, D. Gupta, P. Richardson, R. L. Schlossman, N. Krett, L. B. Chen, N. C. Munshi, and K. C. Anderson. 2002. Identification of genes regulated by dexamethasone in multiple myeloma cells using oligonucleotide arrays. Oncogene 21:1346-1358. [DOI] [PubMed] [Google Scholar]
- 10.Chen, Y. H., T. Knosel, G. Kristiansen, A. Pietas, M. E. Garber, S. Matsuhashi, I. Ozaki, and I. Petersen. 2003. Loss of PDCD4 expression in human lung cancer correlates with tumour progression and prognosis. J. Pathol. 200:640-646. [DOI] [PubMed] [Google Scholar]
- 11.Cmarik, J. L., H. Min, G. Hegamyer, S. Zhan, M. Kulesz-Martin, H. Yoshinaga, S. Matsuhashi, and N. H. Colburn. 1999. Differentially expressed protein Pdcd4 inhibits tumor promoter-induced neoplastic transformation. Proc. Natl. Acad. Sci. USA. 96:14037-14042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooper, S. J., J. MacGowan, J. Ranger-Moore, M. R. Young, N. H. Colburn, and G. T. Bowden. 2003. Expression of dominant negative c-jun inhibits ultraviolet B-induced squamous cell carcinoma number and size in an SKH-1 hairless mouse model. Mol. Cancer Res. 1:848-854. [PubMed] [Google Scholar]
- 13.Crawford, H. C., and L. M. Matrisian. 1996. Mechanisms controlling the transcription of matrix metalloproteinase genes in normal and neoplastic cells. Enzyme Protein 49:20-37. [DOI] [PubMed] [Google Scholar]
- 14.Curran, S., and G. I. Murray. 2000. Matrix metalloproteinases: molecular aspects of their roles in tumour invasion and metastasis. Eur. J. Cancer 36:1621-1630. [DOI] [PubMed] [Google Scholar]
- 15.De Benedetti, A., and A. L. Harris. 1999. eIF4E expression in tumors: its possible role in progression of malignancies. Int. J. Biochem. Cell Biol. 31:59-72. [DOI] [PubMed] [Google Scholar]
- 16.Dhar, A., J. Hu, R. Reeves, L. M. Resar, and N. H. Colburn. 2004. Dominant-negative c-Jun (TAM67) target genes: HMGA1 is required for tumor promoter-induced transformation. Oncogene 23:4466-4476. [DOI] [PubMed] [Google Scholar]
- 17.Dong, Z., H. C. Crawford, V. Lavrovsky, D. Taub, R. Watts, L. M. Matrisian, and N. H. Colburn. 1997. A dominant negative mutant of jun blocking 12-O-tetradecanoylphorbol-13-acetate-induced invasion in mouse keratinocytes. Mol. Carcinog. 19:204-212. [DOI] [PubMed] [Google Scholar]
- 18.Goke, A., R. Goke, A. Knolle, H. Trusheim, H. Schmidt, A. Wilmen, R. Carmody, B. Goke, and Y. H. Chen. 2002. DUG is a novel homologue of translation initiation factor 4G that binds eIF4A. Biochem. Biophys. Res. Commun. 297:78-82. [DOI] [PubMed] [Google Scholar]
- 19.Goke, R., C. Gregel, A. Goke, R. Arnold, H. Schmidt, and B. Lankat-Buttgereit. 2004. Programmed cell death protein 4 (PDCD4) acts as a tumor suppressor in neuroendocrine tumor cells. Ann. N. Y. Acad. Sci. 1014:220-221. [DOI] [PubMed] [Google Scholar]
- 20.Hennigan, R. F., K. L. Hawker, and B. W. Ozanne. 1994. Fos-transformation activates genes associated with invasion. Oncogene 9:3591-3600. [PubMed] [Google Scholar]
- 21.Hu, M. C., W. R. Qiu, X. Wang, C. F. Meyer, and T. H. Tan. 1996. Human HPK1, a novel human hematopoietic progenitor kinase that activates the JNK/SAPK kinase cascade. Genes Dev. 10:2251-2264. [DOI] [PubMed] [Google Scholar]
- 22.Jansen, A. P., C. E. Camalier, and H. N. Colburn. 2005. Epidermal expression of the translation inhibitor Pdcd4 suppresses tumorigenesis. Cancer Res. 65:6034-6041. [DOI] [PubMed] [Google Scholar]
- 23.Jansen, A. P., C. E. Camalier, C. Stark, and N. H. Colburn. 2004. Characterization of programmed cell death 4 in multiple human cancers reveals a novel enhancer of drug sensitivity. Mol. Cancer Ther. 3:103-110. [PubMed] [Google Scholar]
- 24.Kang, M. J., H. S. Ahn, J. Y. Lee, S. Matsuhashi, and W. Y. Park. 2002. Up-regulation of PDCD4 in senescent human diploid fibroblasts. Biochem. Biophys. Res. Commun. 293:617-621. [DOI] [PubMed] [Google Scholar]
- 25.Kiefer, F., L. A. Tibbles, M. Anafi, A. Janssen, B. W. Zanke, N. Lassam, T. Pawson, J. R. Woodgett, and N. N. Iscove. 1996. HPK1, a hematopoietic protein kinase activating the SAPK/JNK pathway. EMBO J. 15:7013-7025. [PMC free article] [PubMed] [Google Scholar]
- 26.Lamb, R. F., R. F. Hennigan, K. Turnbull, K. D. Katsanakis, E. D. MacKenzie, G. D. Birnie, and B. W. Ozanne. 1997. AP-1-mediated invasion requires increased expression of the hyaluronan receptor CD44. Mol. Cell. Biol. 17:963-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lankat-Buttgereit, B., C. Gregel, A. Knolle, A. Hasilik, R. Arnold, and R. Goke. 2004. Pdcd4 inhibits growth of tumor cells by suppression of carbonic anhydrase type II. Mol. Cell. Endocrinol. 214:149-153. [DOI] [PubMed] [Google Scholar]
- 28.Ling, P., C. F. Meyer, L. P. Redmond, J. W. Shui, B. Davis, R. R. Rich, M. C. Hu, R. L. Wange, and T. H. Tan. 2001. Involvement of hematopoietic progenitor kinase 1 in T cell receptor signaling. J. Biol. Chem. 276:18908-18914. [DOI] [PubMed] [Google Scholar]
- 29.Ling, P., Z. Yao, C. F. Meyer, X. S. Wang, W. Oehrl, S. M. Feller, and T. H. Tan. 1999. Interaction of hematopoietic progenitor kinase 1 with adapter proteins Crk and CrkL leads to synergistic activation of c-Jun N-terminal kinase. Mol. Cell. Biol. 19:1359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liou, J., F. Kiefer, A. Dang, A. Hashimoto, M. H. Cobb, T. Kurosaki, and A. Weiss. 2000. HPK1 is activated by lymphocyte antigen receptors and negatively regulates AP-1. Immunity 12:399-408. [DOI] [PubMed] [Google Scholar]
- 31.Ma, W., C. Xia, P. Ling, M. Qiu, Y. Luo, T. H. Tan, and M. Liu. 2001. Leukocyte-specific adaptor protein Grap2 interacts with hematopoietic progenitor kinase 1 (HPK1) to activate JNK signaling pathway in T lymphocytes. Oncogene 20:1703-1714. [DOI] [PubMed] [Google Scholar]
- 32.Marconcini, L., S. Marchio, L. Morbidelli, E. Cartocci, A. Albini, M. Ziche, F. Bussolino, and S. Oliviero. 1999. c-Fos-induced growth factor/vascular endothelial growth factor D induces angiogenesis in vivo and in vitro. Proc. Natl. Acad. Sci. USA. 96:9671-9676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Onishi, Y., and H. Kizaki. 1996. Molecular cloning of the genes suppressed in RVC lymphoma cells by topoisomerase inhibitors. Biochem. Biophys. Res. Commun. 228:7-13. [DOI] [PubMed] [Google Scholar]
- 34.Ozanne, B. W., L. McGarry, H. J. Spence, I. Johnston, J. Winnie, L. Meagher, and G. Stapleton. 2000. Transcriptional regulation of cell invasion: AP-1 regulation of a multigenic invasion programme. Eur. J. Cancer 36:1640-1648. [DOI] [PubMed] [Google Scholar]
- 35.Pilcher, B. K., J. A. Dumin, B. D. Sudbeck, S. M. Krane, H. G. Welgus, and W. C. Parks. 1997. The activity of collagenase-1 is required for keratinocyte migration on a type I collagen matrix. J. Cell Biol. 137:1445-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Proud, C. G. 1994. Translation. Turned on by insulin. Nature 371:747-748. [DOI] [PubMed] [Google Scholar]
- 37.Reeves, R., D. D. Edberg, and Y. Li. 2001. Architectural transcription factor HMGI(Y) promotes tumor progression and mesenchymal transition of human epithelial cells. Mol. Cell. Biol. 21:575-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson, M. J., S. A. Stippec, E. Goldsmith, M. A. White, and M. H. Cobb. 1998. A constitutively active and nuclear form of the MAP kinase ERK2 is sufficient for neurite outgrowth and cell transformation. Curr. Biol. 8:1141-1150. [DOI] [PubMed] [Google Scholar]
- 39.Sauer, K., J. Liou, S. B. Singh, D. Yablonski, A. Weiss, and R. M. Perlmutter. 2001. Hematopoietic progenitor kinase 1 associates physically and functionally with the adaptor proteins B cell linker protein and SLP-76 in lymphocytes. J. Biol. Chem. 276:45207-45216. [DOI] [PubMed] [Google Scholar]
- 40.Shibahara, K., M. Asano, Y. Ishida, T. Aoki, T. Koike, and T. Honjo. 1995. Isolation of a novel mouse gene MA-3 that is induced upon programmed cell death. Gene 166:297-301. [DOI] [PubMed] [Google Scholar]
- 41.Shin, E. Y., S. Y. Kim, and E. G. Kim. 2001. c-Jun N-terminal kinase is involved in motility of endothelial cell. Exp. Mol. Med. 33:276-283. [DOI] [PubMed] [Google Scholar]
- 42.Shin, M., C. Yan, and D. Boyd. 2002. An inhibitor of c-jun aminoterminal kinase (SP600125) represses c-Jun activation, DNA-binding and PMA-inducible 92-kDa type IV collagenase expression. Biochim. Biophys. Acta 1589:311-316. [DOI] [PubMed] [Google Scholar]
- 43.Taguchi, A., D. C. Blood, G. del Toro, A. Canet, D. C. Lee, W. Qu, N. Tanji, Y. Lu, E. Lalla, C. Fu, M. A. Hofmann, T. Kislinger, M. Ingram, A. Lu, H. Tanaka, O. Hori, S. Ogawa, D. M. Stern, and A. M. Schmidt. 2000. Blockade of RAGE-amphoterin signalling suppresses tumour growth and metastases. Nature 405:354-360. [DOI] [PubMed] [Google Scholar]
- 44.Vasilevskaya, I., and P. J. O'Dwyer. 2003. Role of Jun and Jun kinase in resistance of cancer cells to therapy. Drug Resist. Update 6:147-156. [DOI] [PubMed] [Google Scholar]
- 45.Yang, H. S., M. H. Cho, H. Zakowicz, G. Hegamyer, N. Sonenberg, and N. H. Colburn. 2004. A novel function of the MA-3 domains in transformation and translation suppressor Pdcd4 is essential for its binding to eukaryotic translation initiation factor 4A. Mol. Cell. Biol. 24:3894-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang, H. S., A. P. Jansen, A. A. Komar, X. Zheng, W. C. Merrick, S. Costes, S. J. Lockett, N. Sonenberg, and N. H. Colburn. 2003. The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol. Cell. Biol. 23:26-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang, H. S., A. P. Jansen, R. Nair, K. Shibahara, A. K. Verma, J. L. Cmarik, and N. H. Colburn. 2001. A novel transformation suppressor, Pdcd4, inhibits AP-1 transactivation but not NF-κB or ODC transactivation. Oncogene 20:669-676. [DOI] [PubMed] [Google Scholar]
- 48.Yang, H. S., J. L. Knies, C. Stark, and N. H. Colburn. 2003. Pdcd4 suppresses tumor phenotype in JB6 cells by inhibiting AP-1 transactivation. Oncogene 22:3712-3720. [DOI] [PubMed] [Google Scholar]
- 49.Yuspa, S. H. 1998. The pathogenesis of squamous cell cancer: lessons learned from studies of skin carcinogenesis. J. Dermatol. Sci. 17:1-7. [DOI] [PubMed] [Google Scholar]
- 50.Zakowicz, H., H. S. Yang, C. Stark, A. Wlodawer, N. Laronde-Leblanc, and N. H. Colburn. 2005. Mutational analysis of the DEAD-box RNA helicase eIF4AII characterizes its interaction with transformation suppressor Pdcd4 and eIF4GI. RNA 11:261-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang, Z., and R. N. DuBois. 2001. Detection of differentially expressed genes in human colon carcinoma cells treated with a selective COX-2 inhibitor. Oncogene 20:4450-4456. [DOI] [PubMed] [Google Scholar]
- 52.Zhou, G., S. C. Lee, Z. Yao, and T. H. Tan. 1999. Hematopoietic progenitor kinase 1 is a component of transforming growth factor beta-induced c-Jun N-terminal kinase signaling cascade. J. Biol. Chem. 274:13133-13138. [DOI] [PubMed] [Google Scholar]