Abstract
Phosphorylation on Ser/Thr-Pro motifs is a major mechanism regulating many events involved in cell proliferation and transformation, including centrosome duplication, whose defects have been implicated in oncogenesis. Certain phosphorylated Ser/Thr-Pro motifs can exist in two distinct conformations whose conversion in certain proteins is catalyzed specifically by the prolyl isomerase Pin1. Pin1 is prevalently overexpressed in human cancers and is important for the activation of multiple oncogenic pathways, and its deletion suppresses the ability of certain oncogenes to induce cancer in mice. However, little is known about the role of Pin1 in centrosome duplication and the significance of Pin1 overexpression in cancer development in vivo. Here we show that Pin1 overexpression correlates with centrosome amplification in human breast cancer tissues. Furthermore, Pin1 localizes to and copurifies with centrosomes in interphase but not mitotic cells. Moreover, Pin1 ablation in mouse embryonic fibroblasts drastically delays centrosome duplication without affecting DNA synthesis and Pin1 inhibition also suppresses centrosome amplification in S-arrested CHO cells. In contrast, overexpression of Pin1 drives centrosome duplication and accumulation, resulting in chromosome missegregation, aneuploidy, and transformation in nontransformed NIH 3T3 cells. More importantly, transgenic overexpression of Pin1 in mouse mammary glands also potently induces centrosome amplification, eventually leading to mammary hyperplasia and malignant mammary tumors with overamplified centrosomes. These results demonstrate for the first time that the phosphorylation-specific isomerase Pin1 regulates centrosome duplication and its deregulation can induce centrosome amplification, chromosome instability, and oncogenesis.
Centrosomes are major microtubule-organizing structures in animal cells that determine the organization of the mitotic spindle poles that segregate duplicated chromosomes between dividing cells (7, 18, 33, 56, 70). Consequently, defects in either the number or the function of centrosomes can adversely affect mitotic spindle formation, cytokinesis, and genomic stability (19, 56, 70). For example, an increase in the number of centrosomes can result in the organization of multipolar spindles and the eventual missegregation of chromosomes, which contributes to the genetic instability that is often observed during oncogenesis. In fact, centrosome abnormalities and amplifications have been well documented in many human cancers and these changes have been observed at early stages of human cancer development and also correlate with poor clinical outcome in some cancers (12, 17, 26, 38-40, 56, 59-62, 70, 71). In addition, several oncogenes and tumor suppressors have been shown to affect centrosome duplication and/or induce centrosome amplification (6, 14, 25, 34, 38, 51, 52, 56, 58, 69, 76, 83, 89). Therefore, the elucidation of the regulatory mechanisms of centrosome duplication and its abnormal amplification is important for understanding cancer development and may lead to more effective anticancer therapies.
Accurate chromosome segregation to each daughter cell during mitosis requires the duplication of centrosomes once and only once during each cell cycle (7, 18, 33, 56, 70). Centrosome duplication initiates at the G1/S transition and is completed during S phase in mammalian somatic cells. Centrosome duplication must be coupled to the events of the nuclear cell cycle, and their decoupling can result in abnormal centrosome numbers and aberrant mitosis, leading to chromosome instability. This strict coordination has been shown to be regulated by multiple pathways. One major pathway is the activation of Cdk2/cyclin E or A during the G1/S transition (32, 36, 49, 52). Furthermore, E2F activation and Rb phosphorylation by Cdk2 are also required for centrosome duplication (52). Moreover, Cdk2 might be subjected to the regulation of p53-mediated cell cycle checkpoints (13, 22, 28). Finally, several centrosome Cdk substrates have been identified, including BRCA1, nucleophosmin/B23, mMPS1/ESK, and CP110, that play an important role in centrosome duplication (10, 23, 57, 83). These results indicate that Cdk2-mediated protein phosphorylation plays a key role in regulating centrosome duplication during the S phase. However, little is known about whether the coordination between DNA synthesis and centrosome duplication is further regulated after phosphorylation.
Cyclin-dependent protein kinases are Pro-directed kinases that regulate cell cycle progression by phosphorylating exclusively on serine or threonine residues preceding a proline (Ser/Thr-Pro). Although these phosphorylation events have been proposed to function via inducing conformational changes, little was known about the nature and regulation of the conformational changes until recently (45). Recent studies indicate that certain phosphorylated Ser/Thr-Pro (pSer/Thr-Pro) motifs in proteins can exist in the two completely distinct cis and trans conformations; their conversion is normally inhibited by phosphorylation but is specifically catalyzed by the prolyl isomerase Pin1 (44, 45, 63, 84). Pin1 contains an N-terminal WW domain and a C-terminal prolyl isomerase domain. The WW domain binds to specific pSer/Thr-Pro motifs and targets Pin1 to a subset of phosphoproteins, while the isomerase domain induces conformational changes by catalyzing the isomerization of specific pSer/Thr-Pro bonds (47, 84, 90). Such conformational changes have been shown to have profound effects on the function of Pin1 substrates by modulating their catalytic activity, phosphorylation status, protein-protein interaction, subcellular localization, and stability (41, 42, 67, 68, 74, 79, 81, 82, 84, 87, 88, 90). Consequently, Pin1 has been shown to be involved in the regulation of many cellular processes, such as cell proliferation and differentiation (2, 16, 41, 42, 44, 45, 48, 78, 79, 85).
An increasing body of evidence suggests that Pin1 may play an important role in oncogenesis and may be a potential new anticancer target. Pin1 is overexpressed in a large number of human cancers and is also an excellent prognostic marker of poor outcome in some cancers (5, 43, 65, 67, 82). Furthermore, Pin1 can function as a critical catalyst that amplifies multiple oncogenic signaling pathways such as the Neu/Ras/c-Jun, Wnt/β-catenin, and cytokine/NF-κB pathways (67, 68, 82) and its overexpression can transform immortal breast epithelial cells in vitro (66). In contrast, the inhibition of Pin1 in cancer cells via multiple approaches either triggers apoptosis or suppresses transformed phenotypes (44, 46, 64) and Pin1 knockout suppresses the ability of certain oncogenes to induce breast cancer (80). These findings indicate that Pin1 is prevalently overexpressed in human cancers and is important for the activation of multiple oncogenic pathways. However, little is known about the significance of Pin1 overexpression in cancer development in vivo. Furthermore, although cyclin D1, a key molecule in oncogenesis (15, 37, 72, 77, 86) and a major Pin1 downstream target in oncogenesis (41, 67, 68, 80, 82), has been implicated in centrosome amplification in vitro (55), nothing is known about the role of Pin1 in centrosome duplication and chromosome stability.
In this study, we have shown that Pin1 localizes to and copurifies with centrosomes and regulates centrosome duplication. Furthermore, the ablation of Pin1 in mouse embryonic fibroblasts specifically delays centrosome duplication during S phase, whereas overexpression of Pin1 in nontransformed cells induces centrosome amplification, abnormal mitotic spindle formation, chromosome missegregation eventually leading to aneuploidy, and oncogenic transformation. Moreover, the overexpression of Pin1 in transgenic mouse mammary glands also potently causes centrosome amplification, eventually leading to mammary hyperplasia and malignant mammary tumors with overamplified centrosomes. Together with the findings that Pin1 levels strongly correlate with centrosome amplification in human breast cancer tissues, these results indicate that Pin1 is an important regulator of centrosome duplication and that its overexpression can contribute to centrosome abnormality, chromosome instability, and oncogenesis.
MATERIALS AND METHODS
Immunohistochemistry, immunostaining, and immunoblotting.
Formalin-fixed and paraffin-embedded tissue microarrays of human breast cancer tissue were purchased from Imgenex (IMH-364; San Diego, CA). Immunohistochemical staining for Pin1 was performed as described previously (3, 67, 82). For immunofluorescence analysis of centrosome staining, slides were dewaxed in xylene and rehydrated through a series of ethanol to water gradients and 0.3% H2O2 in water was then used to block endogenous peroxidase. The sections were further blocked with 5% normal goat sera (Vector Laboratories, Inc., Burlingame, CA) for 1 h at room temperature, followed by incubation with antipericentrin polyclonal antibodies (BabCo, Denver, CO) at a 1:800 dilution overnight at 4°C. After an extensive wash with phosphate-buffered saline, slides were incubated with biotinylated goat anti-rabbit immunoglobulin G (Vector Laboratories, Inc.) at a 1:200 dilution for 30 min at room temperature, followed by Alexa488-labeled streptavidin (Molecular Probes, Eugene, OR) diluted 1:500 for 30 min at room temperature. After washing with phosphate-buffered saline, the nuclei were stained with propidium iodide (10 μg/ml) for 10 min at room temperature. The ratio of centrosomes to nuclei was scored for each sample of >200 nuclei by using a fluorescence microscope. To detect centrosomes in cultured cells, cells were fixed with 3.7% buffered formaldehyde for 5 min and stained with anti-γ-tubulin (Sigma) or antipericentrin as described previously (67) and immunoblotting was also performed as described previously (67).
Isolation of centrosome fraction.
Isolation of centrosomes was carried out according to a published procedure (8), with the following minor modifications. Cultured cells were incubated with 10 μg/ml nocodazole and 5 μg/ml cytochalasin B for 2 h, rinsed with an isolation buffer (1 mM Tris, pH 8, 0.5 mM EGTA, 0.1% β-mercaptoethanol), and then lysed by swaying the dishes in the isolation buffer containing 0.5% NP-40 at 4°C for 10 min. Next, a 1/50 vol of buffer {0.5 M PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 7.2, 0.1 M EGTA} was added to the extracts, followed by loading onto discontinuous sucrose density gradient set with 3.5 ml of 60% sucrose and 3.5 ml of 40% sucrose prepared in 20 mM PIPES, pH 6.8, 0.5 mM MgCl2, 1 mM EGTA, and 0.1% β-mercaptoethanol and centrifugation for 1 h at 14,500 rpm. Fractions were collected from the bottom and subjected to immunoblotting as described previously (67).
Analysis of centrosome duplication during S phase.
Centrosome duplication assays in Chinese hamster ovary (CHO) cells were performed as described previously (4, 52). Briefly, cells were transfected with the indicated DNA constructs by electroporation (Bio-Rad) according to the manufacturer's instructions. Twelve hours after transfection, hydroxyurea was added at a final concentration of 4 mM for 40 h. Cells were then fixed with formaldehyde for 15 min at room temperature and then with cold methanol for 10 min. Cells were then stained for centrosomes with anti-γ-tubulin or antipericentrin antibodies and analyzed by fluorescent microscopy as described previously (67). DNA synthesis was monitored using the BrdU labeling kit (Roche) according to the manufacturer's instructions with Alexa594-conjugated goat anti-rabbit immunoglobulin G secondary antibodies (Molecular Probes).
To reexpress Pin1 in Pin1−/− mouse embryonic fibroblasts (MEFs), MEFs derived from Pin1−/− mouse embryos and immortalized using the 3T3 protocol as used previously (41, 68) were infected with the pBabe retroviruses expressing Pin1 or control retroviruses; this was followed by selection with puromycin to generate stable rescued Pin1−/− and control Pin1−/− MEFs. To analyze centrosome duplication during the cell cycle, Pin1−/− and rescued Pin1−/− MEFs were incubated with 70 ng/ml nocodazole for 16 h to synchronize cells at late G2 and M phases and then released from the arrest, followed by collecting samples for every 2 h for flow cytometry to analyze cell cycle progression and by immunostaining with anti-γ-tubulin to determine centrosome duplication.
Electron microscopy.
For electron microscopy, cells were fixed for 30 min at room temperature in a buffer containing 0.15 M sodium cacodylate (pH 7.0) and 0.2% glutaraldehyde and processed for electron microscopy as described previously (23, 50)
Analysis of ploidy and cell transformation assays.
Asynchronously growing cells were treated with Colcemid (10 mg/ml; Roche) for 1 h and harvested by trypsinization, followed by resuspension in 10 ml of hypotonic buffer (0.2% KCl/0.2% trisodium citrate) for 12 min at 37°C. After adding 1 ml of an ice-cold fixative (3:1, methanol:acetic acid) for 5 min on ice, cells were harvested and resuspended twice more in 10 ml of the fixative. Finally, cells were dropped onto glass coverslips and at least 100 chromosome spreads per group were analyzed. Cell transformation assays were performed as described previously (66).
Generation of Pin1 transgenic mice.
The human Pin1 cDNA with an NH2-terminal hemagglutinin (HA) epitope tag was subcloned under the control of the mouse mammary tumor virus (MMTV) promoter (provided by P. Leder), followed by microinjection into FVB fertilized eggs to generate transgenic mice as described previously (73). Multiple independent founder mouse lines were obtained and confirmed by Southern and Western blotting to express Pin1 and displayed the similar phenotypes observed.
Primary cultures of mammary epithelial cells and mammary tumor cells and mammary gland whole mounts.
Primary cultures of mammary epithelial cells (MECs) and mammary tumor cells derived from mice were performed as described previously (80), as were mammary gland whole mounts (41, 80). Briefly, mammary glands or tumors were mechanically disaggregated and then subjected to sequential digestions with collagenase and trypsin. Isolated cells were resuspended in mammary epithelial growth medium and plated on collagen-coated culture dishes; this was followed by analyzing centrosome number within a few days to 1 week. To examine the development of the mammary epithelium, the no. 4 mammary glands of nonpregnant MMTV-Pin1 or control mice were dissected, spread onto a glass slide, and fixed, followed by fat removal with acetone. The glands were stained overnight in 0.2% carmine red (Sigma) and 0.5% AlK(SO4)2, dehydrated in graded ethanol solutions, followed by clearing in toluene and methyl salicylate. Photos were taken using a dissecting microscope.
Statistical analysis.
MMTV-Pin1 transgenic and control cohorts were considered for the analysis of the end point, breast cancer-free survival using the Kaplan-Meier method; this was followed by determining the significance of the differences in disease-free survival among the cohorts using a log-rank (Mantel-Cox) test as described previously (80).
RESULTS
Pin1 levels correlate with centrosome amplification in human breast cancer tissues.
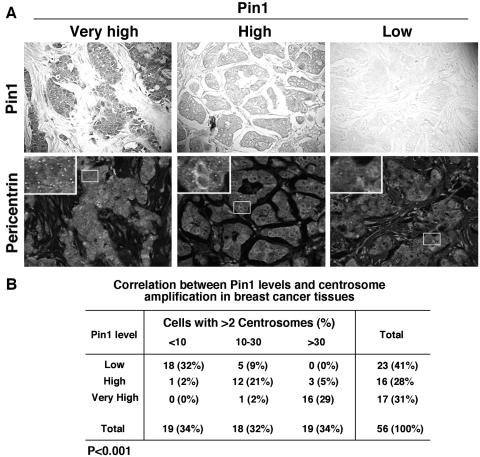
It has been previously shown that Pin1 overexpression and centrosome amplification are often observed in human malignancies, including breast cancer (3, 5, 12, 26, 38-40, 59, 60, 62, 67, 71, 82). Given the critical role of Pro-directed phosphorylation in centrosome duplication, we hypothesized that Pin1 might also play a role in regulating this important cellular process. To test this hypothesis, we first used immunohistochemistry to examine the correlation between Pin1 levels and centrosome amplification in breast cancers. As shown previously (67, 82), Pin1 was overexpressed in various degrees in breast cancer tissues (Fig. 1A). Interestingly, almost all tumors containing very high levels of Pin1 (16 out of 17) had marked centrosome amplification, with more than 30% of cells containing more than two centrosomes per cell (Fig. 1B). In contrast, most tumors containing low Pin1 (18 out of 23) had less than 10% cells containing more than two centrosomes per cell (Fig. 1B). There was a significant correlation between Pin1 expression and centrosome amplification, as determined by the Spearman rank correlation test (P < 0.001). These results suggest that Pin1 levels correlate with centrosome amplification in human breast cancer tissues.
FIG. 1.
Pin1 levels correlate with centrosome amplification in human breast cancer tissues. (A) Breast cancer tissue sections were immunostained using anti-Pin1 antibodies and visualized by DAB staining (upper panels), and different sections of the same tissues were immunostained with antipericentrin antibodies and visualized by Alexa488-conjugated secondary antibody along with propidium iodide to stain nuclei (lower panels). The ratio of centrosomes to nuclei was scored in each sample of >200 nuclei under a fluorescence microscope. The far left panels show a representative very high Pin1 staining, with >30% of cancer cells containing more than two centrosomes, the middle panels show a representative high Pin1 staining with 10 to 30% of cancer cells containing more than two centrosomes, and the right panels show a representative low Pin1 staining with <10% of cancer cells containing more than two centrosomes. (B) The levels of Pin1 expression and the percentage of cancer cells containing more than two centrosomes were determined in 56 surgical specimens of breast cancer as shown in panel A, and their correlation was analyzed by a Spearman rank correlation test (P < 0.001).
Pin1 localizes to centrosomes in interphase but not mitotic cells.
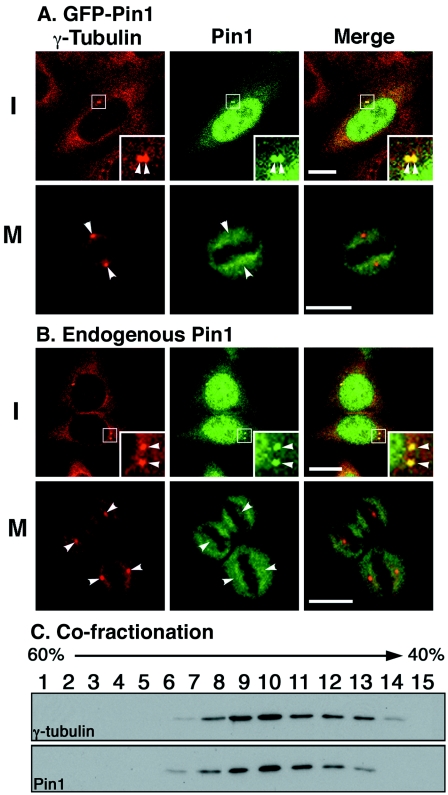
The tight correlation between Pin1 expression and centrosome amplification in human breast cancer suggests a role for Pin1 in regulating centrosome duplication. To explore this possibility, we examined whether Pin1 localizes to centrosomes in a cell cycle-specific manner since centrosomes are duplicated during interphase and separated during mitosis. Exponentially growing NIH 3T3 cells were transfected with either green fluorescent protein (GFP) or GFP-Pin1 followed by immunostaining with γ-tubulin, a centrosome marker. Consistent with previous studies (44, 46), GFP-Pin1 was found to localize primarily in the nucleus of cultured cells but, interestingly, the colocalization of Pin1 and γ-tubulin was also observed as dots adjacent to the nucleus, a characteristic of centrosome staining (Fig. 2A). Importantly, the colocalization of Pin1 and γ-tubulin at the centrosome was not detected in cells in mitotic phase (Fig. 2A), indicating a cell cycle-specific colocalization of exogenously expressed Pin1 with centrosomes.
FIG. 2.
Pin1 localizes to centrosomes in interphase but not mitotic cells. (A) Localization of exogenously expressed Pin1 to centrosomes. NIH 3T3 cells transiently transfected with GFP-Pin1 were immunostained with anti-γ-tubulin antibody, followed by the confocal microscopy. Colocalization of GFP-Pin1 and γ-tubulin was analyzed. Representative transfected cells in interphase and mitotic phase are shown. Bar, 10 μm. Arrowheads, centrosomes. (B) Localization of endogenous Pin1 to centrosomes. Nontransfected NIH 3T3 cells were stained with anti-Pin1 and anti-γ-tubulin antibodies, followed by the confocal microscopy. Bar, 10 μm. Arrowheads, centrosomes. (C) Cofractionation of endogenous Pin1 with centrosomes. Centrosomes were purified NIH 3T3 cells using discontinuous sucrose gradient centrifugation, and different fractions were subjected to immunoblot analysis using anti-γ-tubulin and anti-Pin1 antibodies.
To rule out the possibility that this colocalization is due to overexpression or GFP tag, we examined the localization of endogenous Pin1 using anti-Pin1 antibodies that have extensively been used for staining Pin1 in cultured cells and human tissues (5, 43, 65, 67, 82). Following fixation with paraformaldehyde and methanol, endogenous Pin1 was clearly detected in centrosomes again in interphase cells but not in mitotic cells (Fig. 2B). To independently confirm the association of endogenous Pin1 with centrosomes, centrosomes were purified from NIH 3T3 cells via discontinuous sucrose gradient centrifugation as described previously (8), followed by Western blotting on gradient fractions using anti-Pin1 and anti-γ-tubulin antibodies. Endogenous Pin1 was specifically detected in fractions that contained centrosomes, as determined by cosedimentation with γ-tubulin at the expected sucrose densities (Fig. 2C). Similar colocalization and copurification between Pin1 and centrosomes were also observed in HeLa and CHO cells (data not shown). Taken together, these data indicate that both endogenous and exogenously expressed Pin1 proteins localize to centrosomes during only interphase, not mitotic phase, even although proteins at centrosomes are well known to be phosphorylated during mitosis (18, 53, 56, 70).
Inhibition of Pin1 suppresses centrosome amplification in S-arrested CHO cells.
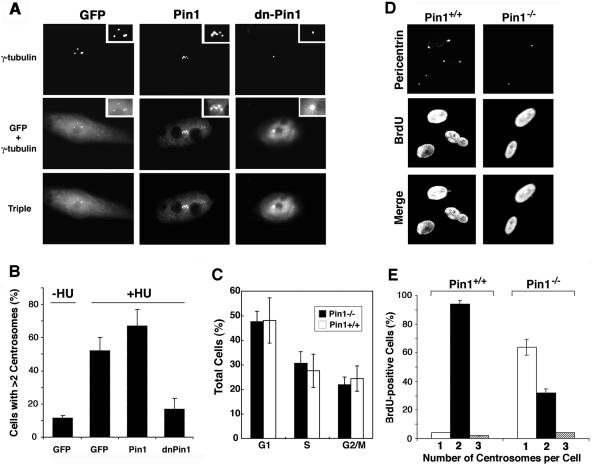
Given that Pin1 localizes to centrosomes in interphase when centrosomes are duplicated, we next examined whether the inhibition of Pin1 function might affect centrosome duplication. For this purpose, we first used CHO cells, a cell model system widely used to study centrosome duplication because their centrosomes undergo multiple rounds of duplication when cells are arrested in S phase with hydroxyurea (4, 35, 49, 52). To inhibit cellular Pin1 function in hamster cells, we used a GFP-Pin1 WW domain mutant which contains an Ala substitution at Ser16, as this mutant has been previously shown to constitutively bind to endogenous Pin1 substrates but fails to catalyze the isomerization, thereby inhibiting endogenous Pin1 function in a dominant-negative manner (dn-Pin1) (46, 66). CHO cells were transfected with plasmid encoding either GFP control vector, GFP-Pin1, or GFP-dn-Pin1 and then treated with hydroxyurea for 40 h; this was followed by evaluating the number of centrosomes after staining with anti-γ-tubulin. As expected, GFP-Pin1 and GFP-dn-Pin1, but not control GFP, localized to the centrosomes (Fig. 3A). Interestingly, relative to nontransfected cells or cells transfected with GFP alone, the number of cells containing abnormal numbers of centrosomes (more than two per cell) was increased in cells overexpressing Pin1 (Fig. 3B). More impressively, such cells containing an abnormal centrosome number were markedly reduced by the expression of dn-Pin1 (Fig. 3B). These results indicate that inhibition of Pin1 suppresses the centrosome amplification that usually occurs in S-arrested CHO cells.
FIG. 3.
Pin1 inhibition or ablation suppresses centrosome duplication in CHO cells and mouse embryonic fibroblasts. (A) and (B) Inhibition of Pin1 suppresses centrosome amplification in S-arrested CHO cells. CHO cells were transfected with plasmids encoding GFP, GFP-Pin1, or GFP-dnPin1 and then treated with hydroxyurea (HU) for 40 h to trigger abnormal centrosome amplification. (A) Centrosome numbers were analyzed quantitatively by immunofluorescence analysis with anti-γ-tubulin antibody and 4′,6′-diamidino-2-phenylindole (DAPI). (B) The percentage of cells containing >2 centrosomes was scored in 300 transfected cells. Error bars, standard deviations from three independent experiments. (C) Pin1+/+ and Pin1−/− MEFs have similar cell cycle profiles at early passages. Exponentially growing primary MEFs derived from Pin1+/+ and Pin1−/− mouse embryos at early passages (below four) were subjected to flow cytometry analysis. Error bars, standard deviations from three independent experiments (D) and (E) Pin1 deletion delays centrosome duplication during S phase in primary fibroblasts. (D) Pin1+/+ and Pin1−/− MEFs were labeled with BrdU for 3 h and coimmunostained with anti-BrdU monoclonal and antipericentrin polyclonal antibodies to analyze centrosome numbers in S-phase cells. (E) For each sample, the number of centrosomes was counted in 200 BrdU-positive cells. Error bars, standard deviations from three independent experiments.
Pin1 ablation in MEFs drastically delays centrosome duplication without affecting DNA synthesis.
The above results suggest that endogenous Pin1 may be important for centrosome duplication. To examine this possibility, we examined the effects of the loss of Pin1 function on centrosome duplication by using MEFs derived from Pin1 knockout (Pin1−/−) and wild-type (Pin1+/+) mice (24, 41). At early passages, both Pin1−/− and Pin1+/+ MEFs grew well and did not have any obvious differences. However, unlike Pin1+/+ MEFs, which continued to grow well for at least a dozen passages, Pin1−/− MEFs usually began to exhibit reduced cell growth after 6 to 8 passages and eventually entered senescence by ∼9 to 10 passages. Therefore, we used Pin1+/+ and Pin1−/− MEFs at passage numbers lower than four, which displayed similar cell cycle profiles based on flow cytometry analysis (Fig. 3C). In order to monitor centrosome duplication in S-phase cells, MEFs were pulse-chased with BrdU for 3 h, followed by coimmunostaining with antipericentrin and anti-BrdU antibodies to determine the coordination between DNA synthesis and centrosome duplication. Approximately 30% of Pin1+/+ and Pin1−/− cells were BrdU-positive (Fig. 3D and E). Importantly, almost all BrdU-positive Pin1+/+ cells contained two centrosomes per cell (Fig. 3D and E), indicating that centrosomes are already duplicated as expected. Under the same conditions, only ∼30% of BrdU-positive Pin1−/− cells, however, contained duplicated centrosomes, with ∼65% of the cells containing only a single centrosome (Fig. 3D and E). These results indicate that centrosome duplication is defective during S phase in most Pin1−/− cells.
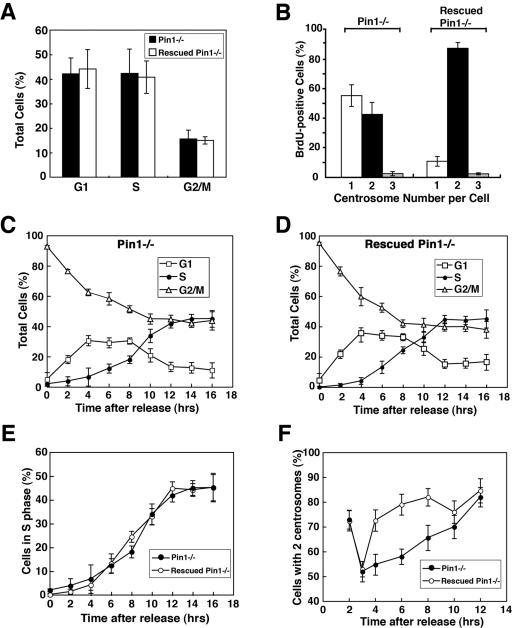
To ensure that this defective centrosome duplication in Pin1−/− MEFs is due to loss of Pin1 function, we needed to stably reexpress Pin1 in Pin1−/− MEFs. For this purpose, Pin1−/− MEFs immortalized using the 3T3 protocol as described previously (41, 68) were infected with the pBabe retroviruses expressing Pin1 or control retroviruses, followed by selection with puromycin to generate stable rescued Pin1−/− and control Pin1−/− MEFs. Expression of Pin1 in rescued Pin1−/− MEFs was comparable to Pin1+/+ MEFs, as confirmed by immunoblotting and immunostaining (data not shown). Furthermore, cell cycle profiles of the immortalized control Pin1−/− and rescued Pin1−/− MEFs were similar (Fig. 4A), although they were slightly different from those of primary MEFs at early passages (Fig. 3C). Importantly, only ∼40% of vector control Pin1−/− MEFs that were BrdU-positive had duplicated their centrosomes (Fig. 4B), resembling early passages of primary Pin1−/− MEFs (Fig. 3E). However, ∼90% rescued Pin1−/− MEFs that were BrdU-positive duplicated their centrosomes (Fig. 4B), which was indistinguishable from those in Pin1+/+ MEFs at early passages (Fig. 3E). These results indicate that the defect in centrosome duplication in Pin1−/− MEFs is specifically due to loss of Pin1 function.
FIG. 4.
Pin1 ablation inhibits centrosome duplication during the S phase in mouse embryonic fibroblasts. (A and B) Rescuing the centrosome duplication defeat in Pin1−/− MEFs by reexpression of Pin1. Pin1−/− MEFs immortalized using the 3T3 protocol and infected with the pBabe control retroviruses or retroviruses expressing Pin1, followed by selection with puromycin, resulting in stable control Pin1−/− and rescued Pin1−/− MEFs. Error bars, standard deviations. (A) Cell cycle profiles of these stable Pin1−/− and rescued Pin1−/− MEFs were determined by flow cytometry analysis. (B) These Pin1−/− and rescued Pin1−/− MEFs were labeled with BrdU for 3 h and coimmunostained with anti-BrdU monoclonal and antipericentrin polyclonal antibodies; this was followed by counting the number of centrosomes in 200 BrdU-positive cells. (C to F) Inhibition of centrosome duplication during the S phase by Pin1 ablation. Pin1−/− and rescued Pin1−/− MEFs were incubated with 70 ng/ml nocodazole for 16 h to synchronize cells at late G2 and M phases and then released from the arrest; this was followed by collecting samples for every 2 h for flow cytometry to analyze cell cycle progression (C to E) and by immunostaining with anti-γ-tubulin to determine centrosome duplication (F).
The fact that Pin1−/− MEFs are able to undergo cell division suggests that centrosome duplication is delayed but not completely blocked. To directly examine this possibility, control Pin1−/− and rescued Pin1−/− MEFs were incubated with nocodazole for 16 h to synchronize cells at late G2 and M phases and then released from the arrest; this was followed by collecting samples for every 2 h for flow cytometry to analyze cell cycle progression and immunostaining with anti-γ-tubulin to determine centrosome duplication. After the release, both control Pin1−/− and rescued Pin1−/− MEFs entered the cell cycle (Fig. 4C and D) and S phase with similar kinetics (Fig. 4E), indicating that these cells have similar cell cycle profiles. However, relative to rescued Pin1−/− cells, centrosome duplication was significantly delayed for many hours in Pin1−/− MEFs, although they were able to eventually duplicate their centrosomes when reaching G2 phase (Fig. 4F). Interestingly, the inhibition of CP110 has also been shown to delay but not completely block centrosome duplication in U2OS cells (10). These results indicate that loss of Pin1 function drastically delays centrosome duplication without affecting DNA synthesis in mouse embryonic fibroblasts.
Overexpression of Pin1 drives centrosome duplication.
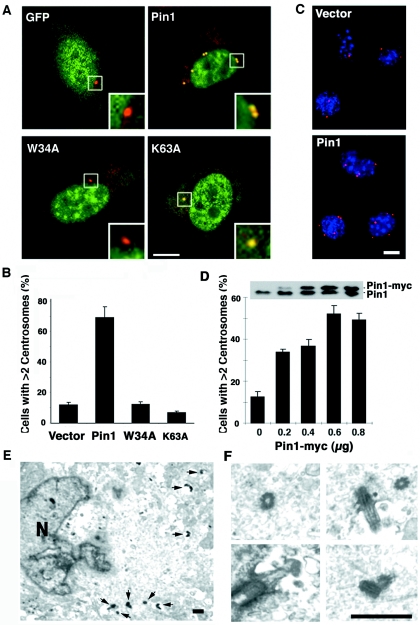
The above results that inhibition or ablation of Pin1 suppresses centrosome duplication and that Pin1 overexpression correlates with centrosome amplification in human breast cancer suggest that Pin1 overexpression might drive centrosome duplication. Centrosome duplication initiates at the G1/S transition in mammalian cells and is completed during S phase. In contrast to CHO cells, where multiple rounds of centrosome duplication are observed under prolonged S-phase arrest (Fig. 3A and B), such extra rounds of centrosome duplication have not been reported in nontransformed NIH 3T3 cells. However, it has been shown that these S-arrested NIH 3T3 cells are permissive for centrosome duplication and have been easy assays for identifying the role of a specific protein in centrosome duplication (23, 33, 75). Therefore, we examined the effects of Pin1 overexpression on centrosome duplication by transfecting NIH 3T3 cells with GFP-Pin1 or control vector and arresting them in G1/S phase by aphidicolin, followed by examining centrosome number as described previously (10, 23, 75). The S arrest was confirmed by flow cytometry (data not shown), indicating that overexpression of Pin1 is not sufficient to overcome the S arrest. Surprisingly, however, more than 60% of GFP-Pin1 transfected cells contained more than two centrosomes (Fig. 5A and B). In contrast, such centrosome amplification was rarely observed in GFP-transfected cells (Fig. 5A and B). These results demonstrate that overexpression of GFP-Pin1 induces centrosome duplication and accumulation in S-arrested NIH 3T3 cells.
FIG. 5.
Overexpression of Pin1 drives centrosome duplication. (A and B) GFP-Pin1, but not its WW domain or PPIase domain mutant, induces centrosome duplication. NIH 3T3 cells were transfected with plasmid encoding GFP, GFP-Pin1, or Pin1 WW domain (W34A) or PPIase domain (K63A) mutant, and then arrested at the G1/S boundary by 10 μg/ml aphidicolin. (A) Cells were stained with anti-γ-tubulin antibody (red) and DAPI (blue). Bar, 10 μm. (B) Cells containing more than two centrosomes were scored in 300 transfected cells. Error bars, standard deviations. (C and D) Pin1-myc induces centrosome duplication. NIH 3T3 cells transfected with different amounts of Pin1-myc or control vector were arrested at the G1/S boundary. (C) Cells were immunostained with anti-myc epitope antibodies, anti-γ-tubulin antibody, and DAPI. Bar, 10 μm. (D) Percentage of cells containing more than two centrosomes was scored in 300 transfected cells. Error bars, standard deviations. (E and F) Confirmation of centrosome amplification by electron microscopy. NIH 3T3 cells expressing Pin1-myc were subjected to electron microscopy. (E) The electron microscopic image shows that one cell contained at least six centrioles in a single section, with high magnification images of four centrioles being shown in panel F. Bar, 400 nm. Arrowheads, centinoles. N, nucleus.
It has been shown that Pin1 binds to and isomerizes specific pSer/Thr-Pro motifs via its WW domain and PPIase domain, respectively, and both of these activities are required for Pin1 to act on its phosphorylated substrates (42, 45, 47, 90). Therefore, we examined whether the effects of Pin1 on centrosome duplication depend on its phosphorylation-specific binding and isomerase activities using a Pin1 point mutant in the WW domain (W34A) or in the PPIase domain (K63A), which cannot bind to or isomerize pSer/Thr-Pro motifs, respectively, as described previously (47, 90). As expected, the K63A mutant localized to the centrosomes but the W34A mutant failed to localize to centrosomes (Fig. 5A), confirming that the centrosome localization of Pin1 is mediated by its WW domain, as shown for other subcellular localization (45, 64). More importantly, neither the WW domain nor the PPIase mutant could induce centrosome duplication in S-arrested NIH 3T3 cells (Fig. 4B). These results indicate that both binding and isomerizing activities of Pin1 are required for its ability to induce centrosome duplication.
To rule out the possibility that the GFP tag affects the function of Pin1 in centrosome regulation, we utilized a plasmid encoding Pin1 fused to a C-terminal myc tag (Pin1-myc) and repeated the experiments. Pin1-myc again potently increased cells containing more than two centrosomes per cell in a dose-dependent fashion (Fig. 5C and D). To confirm that Pin1-myc overexpression causes centrosome amplification, we used electron microscopy to visualize centrioles, which are definitive evidence for centrosome amplification, in Pin1-myc overexpression (56). Indeed, these cells contained more than four centrioles per cells. Figure 5E and F show electron microscopic images from one such cell containing at least six centrioles in a single section. These results together indicate that Pin1 overexpression drives multiple rounds of centrosome duplication in S-arrested NIH 3T3 cells.
Pin1-induced centrosome duplication causes abnormal spindle formation, aneuploidy, and cell transformation in NIH 3T3 cells.
It has been shown that Pin1 is widely overexpressed in most human cancers and correlates with poor clinical outcome (3, 5, 67, 82). Given that Pin1 is an important regulator of centrosome duplication and that its overexpression causes centrosome duplication and accumulation (often referred to as centrosome amplification) in S-arrested NIH 3T3 cells, an important question was whether Pin1-induced centrosome amplification has any pathological consequences. Centrosome abnormalities have been shown to cause aberrant spindle formation and chromosome segregation in mitotic phase (7, 18, 33, 56, 70). We therefore first examined whether Pin1 overexpression affects spindle formation and chromosome segregation during mitosis. To address this question, NIH 3T3 cells were stably transfected with either control vector or Pin1-myc and then arrested in S phase by aphidicolin. Twenty-four hours after release from the arrest (about one cell cycle), both spindle formation and cytokinesis were analyzed during the initial mitotic phase (Fig. 6A). Interestingly, most Pin1-expressing cells, but not vector control cells, contained more than two centrosomes (Fig. 6B). Many of these cells with supernumerary centrosomes underwent bipolar divisions, with multiple numbers of centrosomes coalescing at the two broad spindle poles as reported previously (25). However, ∼20% of these Pin1-expressing cells containing supernumerary centrosomes displayed multipolar spindle formation and nucleation (Fig. 6B and C). These cells subsequently underwent abnormal chromosome segregation and cytokinesis, with one cell being divided into three daughter cells (Fig. 6D). These data indicate that Pin1-induced centrosome amplification leads to chromosome missegregation and abnormal cell division.
FIG. 6.
Pin1-induced centrosome amplification results in aneuploidy and cell transformation. (A) Experimental scheme. NIH 3T3 cells were transfected with Pin1-myc or control vector and selected with G418 to generate pools of stable transfectants. After confirming the expression of Pin1-Myc in cells with anti-Myc antibodies, cells were arrested at the G1/S boundary for 24 h with aphidicolin and released into fresh medium; this was followed by various assays. (B to D) Pin1 overexpression causes the formation of multipolar spindles. At each point following the release from cell cycle arrest, cells were fixed and costained with anti-α-tubulin and anti-γ-tubulin antibodies followed by DAPI staining. (B) Representative mitotic cells with bipolar or multipolar spindle poles were shown. (C) Cells containing more than two mitotic spindle poles were scored in 200 mitotic cells. A representative Pin1-myc transfected cell with three-directional cell division is shown in panel D. Error bars, standard deviations. (E and F) Pin1 overexpression induces aneuploidy. Stable NIH 3T3 cells were released from the aphidicolin block, cultured for the indicated number of passages and then harvested for metaphase spreads. Representative chromosome preparations are shown in panel E, and the chromosome number per cell was calculated from over 100 individual metaphase spreads in each group (F). (G and H) Pin1 overexpression induces cell transformation. Stable NIH 3T3 cells were released from the aphidicolin block and seeded on plastic plates for 3 weeks, followed by crystal violet staining (G), or cultured in soft agar for 3 weeks and the number of colonies formed per 1,000 cells was scored (H). Colony numbers are the means ± standard deviations of three independent experiments (right panel). Error bars, standard deviations.
Since aneuploidy is a cellular consequence that is often associated with centrosome amplification and mitotic spindle defects (7, 18, 33, 56, 70), we next examined whether the centrosome abnormalities rendered by Pin1 overexpression result in aneuploidy by continuously culturing the above cells and analyzing their chromosome numbers. As expected, the majority of vector-transfected NIH 3T3 cells contained 2N content DNA, with a smaller subset of cells having 4N content DNA, and similar results were obtained at early (P2) or later (P8) passages (Fig. 6E and F). Interestingly, in early passages (P2), cells expressing Pin1-myc had almost a regular number of chromosomes, with only small variations (Fig. 6E and F). However, following eight passages, the distribution of chromosome numbers was dramatically broadened to include many cells containing fewer than 40 chromosomes of less than 2N or more than 4N (Fig. 6E and F). These findings indicate that Pin1 overexpression results in increased chromosome instability and aneuploidy.
Centrosome defects and resulting chromosome instability have been suggested to play an important role in oncogenesis (7, 18, 33, 56, 70). Given that Pin1 overexpression results in increased genetic instability and aneuploidy in nontransformed NIH 3T3 cells, we next explored whether these Pin1-overexpressing cells have any transforming phenotypes by subjecting them to focus formation and soft agar colony formation assays. Cells expressing Pin1-myc exhibited a significantly higher number of foci in plastic plates than vector control cells (Fig. 6G). Furthermore, Pin1-myc-expressing cells, but not vector control cells, were capable of forming colonies in soft agar (Fig. 6H). These results demonstrate that Pin1-overexpressing NIH 3T3 cells display various cell transformation properties. Taken together, these results indicate that Pin1 overexpression induces centrosome amplification, chromosome instability, and cell transformation in NIH 3T3 cells.
Overexpression of Pin1 induces centrosome amplification in mouse mammary glands.
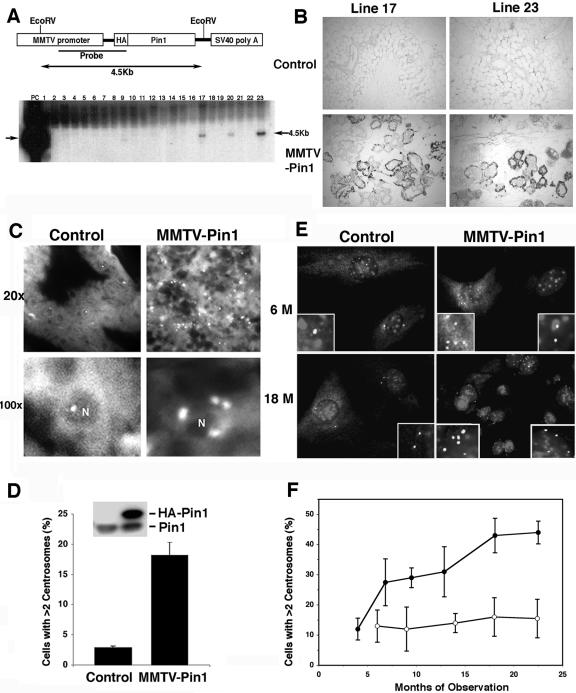
Given that Pin1 overexpression induces centrosome amplification, chromosome instability, and cell transformation in vitro, a key question is whether it has a similar function in vivo. This question is also critical since nothing is known regarding whether Pin1 overexpression is sufficient to induce cancer development in vivo, even though Pin1 has been shown to overexpress in many human cancers and to be important for several oncogenic pathways. To address this question, we generated transgenic mice expressing human Pin1 cDNA under the transcriptional control of the MMTV long terminal repeat promoter/enhancer (Fig. 7A), an approach widely used to access the tumorigenicity of individual oncogenes in breast cancer in vivo (30, 54, 73). To distinguish exogenous Pin1 from endogenous protein, we tagged Pin1 with an HA tag that does not affect Pin1 function in vivo (67, 82). Transgenic mice were first identified by PCR-based genotyping (Fig. 7A) and then confirmed by immunoblotting and immunostaining analyses using anti-HA and anti-Pin1 antibodies (Fig. 7B and D). Pin1 was overexpressed in many, but not all, mammary epithelial cells in both transgenic lines (Fig. 7B and D), as shown for many other MMTV-driven transgenes (9, 11, 31). Two independent transgenic mouse lines, lines 17 and 23, overexpressed Pin1 at similar levels with similar distribution patterns in mammary epithelial cells (Fig. 7B and D). They were further selected to generate homozygous females, which were bred to produce litters two to three times. The phenotypes below were observed in both MMTV-Pin1 transgenic mouse lines at similar frequencies.
FIG. 7.
Pin1 overexpression induces centrosome amplification in mammary epithelial cells in mice. (A) Generation of MMTV-Pin1 transgenic mice. The upper panel is a schematic representation of HA-Pin1 transgene under the control of an MMTV promoter. The lower panel shows the result of Southern blot analysis of the genomic DNA after EcoRI digestion from 23 founder lines obtained from original microinjection. PC, positive control. Note that two independent lines, 17 and 23, expressed HA-Pin1 proteins (inset in panel D) and displayed similar phenotypes. (B) Expression of HA-Pin1 in mammary epithelial cells. Mammary tissue sections from two MMTV-Pin1 transgenic founder mice and control mice were stained with anti-HA 12CA5 monoclonal antibody. (C and D) Centrosome amplification in Pin1-transgenic mouse mammary tissues. Mammary tissue sections from 6-month-old MMTV-Pin1 transgenic or control mice were immunostained with anti-γ-tubulin antibody, followed by immunofluorescence microscopy (C), with the percentage of cells containing more than two centrosomes being scored (D). Error bars, standard deviations. Expression of endogenous Pin1 and exogenous HA-Pin1 in mammary glands was detected by anti-Pin1 antibodies (inset). (E and F) Age-dependent increase in centrosome amplification in primary MEC cultures derived from MMTV-Pin1 transgenic mice. Primary MECs were cultured from different ages of control mice and MMTV-Pin1 transgenic mice without tumors and immunostained with antipericentrin antibody and DAPI staining (E), followed by scoring cells containing more than two centrosomes in 200 cells (F). Results are the means ± standard deviations of three to five mice for each group. Error bars, standard deviations.
To examine whether overexpression of Pin1 in transgenic mice affects centrosome duplication in mice, mammary tissue sections from 6-month-old MMTV-Pin1 transgenic and control mice were fixed and immunostained with antipericentrin antibody. Strikingly, the number of centrosomes in Pin1 transgenic mammary tissues was markedly increased compared to that for the controls (Fig. 7C). About 20% of MECs in MMTV-Pin1 transgenic mice had more than two centrosomes per cell, but only ∼2% of MECs had similar centrosome amplification in control mice (Fig. 7D), indicating that centrosomes are amplified in MMTV-Pin1 transgenic mammary glands. However, no detectable increase in the centrosome number was detected in control mice. These results indicate that overexpression of Pin1 in mammary glands causes centrosome amplification.
To more accurately evaluate the frequency of MECs with centrosome amplification relative to aging in transgenic mice, we isolated primary MECs from mouse mammary glands without tumors in MMTV-Pin1 transgenic and control mice at different ages and then cultured them for a few days to a week as described previously (80); this was followed by immunostaining with antipericentrin antibodies. Under these primary culture conditions, ∼10% of MECs derived from 5-month-old control mice contained more than two centrosomes per cell and this number did not significantly change with aging (Fig. 7E and F). In contrast, the percentage of primary MECs with centrosome amplification significantly increased with aging in MMTV-Pin1 transgenic mice. The frequency of MECs with centrosome amplification in Pin1 transgenic mice that were 4 months old was close to that for controls; however, this frequency was significantly increased to 30% by 6 months of age and then further increased slowly to ∼45% by 18 to 24 months of age (Fig. 7E and F). Taken together, these immunostaining results on tissue sections and primary cell cultures all indicate that overexpression of Pin1 causes centrosome amplification in mouse mammary glands and the effect appears to be more obvious with aging.
Overexpression of Pin1 induces mammary hyperplasia and malignant mammary tumors with overamplified centrosomes in transgenic mice.
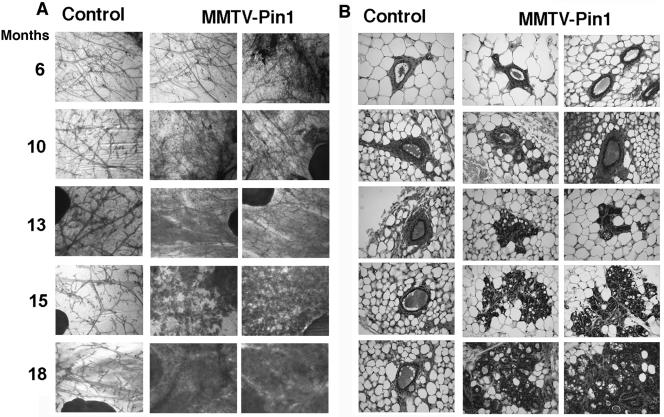
To examine whether Pin1 overexpression results in any pathological changes in mammary glands, we first performed whole mount analysis and histological examination of mammary glands for MMTV-Pin1 and control mice at various ages. MMTV-Pin1 mice showed progressive hyperplastic changes in an age-dependent manner. At an age of 6 months, when centrosome amplification was detected (Fig. 7), there were no detectable histological changes in MMTV-Pin1 mice relative to controls (Fig. 8). Mammary epithelial hyperplasia, detected as early as 10 months of age, was multilayered and often appeared to originate from ducts (Fig. 8). By an age of 18 months, numerous hyperplastic lesions were observed in the majority of mammary glands in most of the transgenic mice that were examined. In contrast, such hyperplastic changes were rarely observed in control mice (Fig. 8). These results indicate that Pin1 overexpression induces mammary hyperplasia.
FIG. 8.
Pin1 overexpression causes mammary epithelial hyperplasia in mice. The whole-mount (A) and histological (B) appearance of mammary glands derived from different ages of nonpregnant MMTV-Pin1 or control mice. The whole mounts of inguinal mammary glands were prepared, and the epithelial component was stained with carmine red. Histological sections were stained with hematoxylin and eosin.
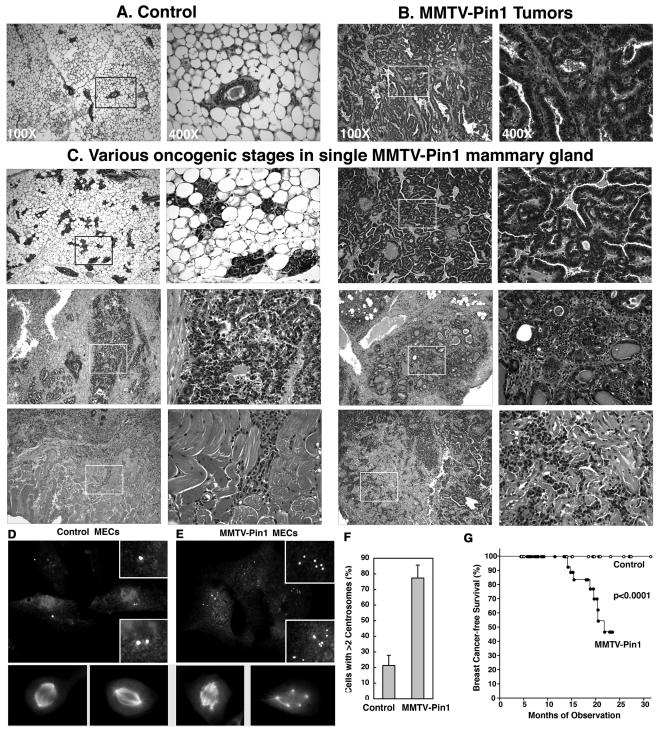
In order to evaluate tumor incidence and age of onset, 54 MMTV-Pin1 and 46 control mice were maintained for aging. Although none of control mice developed mammary tumors, many MMTV-Pin1 mice developed one or several obvious mammary tumor masses, as first detected by palpation and subsequently confirmed by pathological examinations (Fig. 9). These mammary tumors were detected at ages as early as 13 months, and their incidences increased with aging, followed by an apparent plateau at 22 to 24 months. Both the rate and incidence of total mammary gland tumors were highly significant between MMMTV-Pin1 transgenic and control animals (P < 0.0001) (Fig. 9G). These results indicate that MMTV-Pin1 mice develop mammary gland tumors.
FIG. 9.
Pin1 overexpression induces mammary hyperplasia and malignant tumors with overamplified centrosomes in mice. (A to C and G) MMTV-Pin1 mammary glands display various stages of breast cancer development even in a single mammary gland. Histological appearance of 18-month-old nontransgenic control mice (A), MMTV-Pin1 mammary tumors (B), and various oncogenic stages in a single MMTV-Pin1 mammary gland showing hyperplasia (upper left panels), well-differentiated cancer (upper right panels) and a pair of poorly differentiated tumors (middle panels) and invasive tumors (lower panels) (C). (D to F) Centrosome amplification in primary MECs derived from MMTV-Pin1 transgenic mice. Primary MECs were cultured from 18-month-old nontransgenic control mice (D) or MMTV-Pin1 transgenic breast tumors (E); this was followed by immunostaining with anti-γ-tubulin antibody and anti-α-tubulin antibodies and then by DAPI staining. (F) Cells containing more than two centrosomes were scored in 300 mitotic cells. Error bars, standard deviations. (G) Kaplan-Meier analysis of mammary tumors of MMTV-Pin1 and control mice, based on the appearance of obvious tumor masses as detected by palpation and subsequently confirmed by pathological examinations.
Histopathological examinations revealed that Pin1-transgenic mammary glands displayed various stages of breast cancer development, even in a single mammary gland, from hyperplasia, well-differentiated tumors, and poorly differentiated tumors to highly invasive tumors into nearby skeletal muscle and tissues (Fig. 9A to C). These results indicate that tumors were likely developed at multiple foci, which is consistent with widespread centrosome abnormalities in MMTV-Pin1 transgenic mammary glands. To analyze centrosomes in these tumor cells, primary cell cultures were derived from MMTV-Pin1 transgenic mouse tumors and control normal mammary glands, followed by immunostaining with anti-γ-tubulin and anti-α-tubulin antibodies. Centrosome amplification was strikingly obvious in these tumor cells (Fig. 9D and E); close to 80% of tumor cells contained more than two centrosomes per cell, whereas only 20% of MECs from 18-month-old control mice had more than two centrosomes per cell (Fig. 9F). Moreover, as is the case in NIH 3T3 cells overexpressing Pin1 (Fig. 6C to D), although many of these tumor cells with supernumerary centrosomes underwent bipolar divisions, cells displaying multipolar spindle formation and nucleation were readily detected, resulting in chromosome missegregation and aneuploidy (Fig. 9E). Taken together, these results indicate that overexpression of Pin1 leads to centrosome amplification, chromosome instability, and oncogenesis both in cell cultures and in transgenic mice.
DISCUSSION
We here report a novel role for the phosphorylation-specific prolyl isomerase Pin1 in centrosome duplication during the cell cycle and in centrosome amplification and genomic instability in tumorigenesis. First, Pin1 overexpression correlates with centrosome amplification in human breast cancer tissues. Furthermore, Pin1 localizes to and copurifies with centrosomes in interphase cells but not in mitotic cells. Moreover, Pin1 ablation in mouse embryonic fibroblasts drastically delays centrosome duplication without affecting DNA synthesis, and Pin1 inhibition also suppresses centrosome amplification in S-arrested CHO cells. In contrast, overexpression of Pin1 in nontransformed NIH 3T3 cells induces multiple rounds of centrosome duplication under S-arrest conditions, causing the formation of multipolar mitotic spindles and the missegregation of chromosomes, which eventually leads to aneuploidy and cell transformation. The significance of these findings is further substantiated by demonstrations that transgenic overexpression of Pin1 in mouse mammary glands also potently induces centrosome amplification, eventually leading to mammary hyperplasia and malignant mammary tumors with overamplified centrosomes. These results have demonstrated for the first time that Pin1 plays an important role in regulating centrosome duplication and that Pin1 deregulation contributes to centrosome amplification, chromosome instability, and oncogenesis.
Protein phosphorylation by Pro-directed kinases such as Cdk2 has been shown to play a key role in regulating centrosome duplication in mammalian cells (14, 27, 56). However, little is known about whether centrosome duplication is further regulated after phosphorylation. Recent studies indicate that Pin1-mediated phosphorylation-specific prolyl isomerization regulates the conformation and function of a subset of proteins at the postphosphorylation level (42, 45, 79). Our current study has uncovered a novel role for Pin1 in regulating centrosome duplication and amplification based on the following observations.
Pin1 localizes to and copurifies with centrosomes during interphase when they are duplicated, but not during mitosis when they are separated, even though proteins at centrosomes are heavily phosphorylated during mitosis (18, 56, 70). This dynamic association might provide an explanation for why Pin1 is not identified in the recent proteomic identification of core centrosome proteins (1). Functionally, Pin1 ablation in mouse embryonic fibroblasts drastically delays centrosome duplication without affecting DNA synthesis, indicating a requirement of endogenous Pin1 for the coordination between centrosome duplication and DNA synthesis. Furthermore, the inhibition of Pin1 function in CHO cells is able to suppress the ability of these cells to undergo multiple rounds of centrosome duplication under S-arrest conditions, suggesting a role for Pin1 in centrosome amplification. Indeed, overexpression of Pin1 in nontransformed NIH 3T3 cells potently drives multiple rounds of centrosome duplication under S-arrest conditions. Importantly, the ability of Pin1 to induce centrosome amplification is completely abolished by point mutations that disrupt the ability of Pin1 to bind to or isomerize pSer/Thr-Pro motifs. Moreover, transgenic overexpression of Pin1 in mouse mammary epithelial cells also potently induces centrosome amplification. Together with the previous findings that Pin1 is significantly elevated at the G1/S transition (66) and is also subjected to phosphorylation during the cell cycle (46), these results suggest that Pin1 function is normally needed for the coordination of centrosome duplication and DNA synthesis and its aberration can lead to centrosome amplification.
Our findings also suggest that Pin1-induced centrosome abnormalities may contribute to chromosome instability and oncogenesis and further support the idea of Pin1 as a new anticancer target (43). Centrosome amplifications are often found in many human cancers such as breast, prostate, colon, and lung cancers, where Pin1 is often overexpressed (3, 5, 12, 26, 38-40, 59, 60, 62, 65, 67, 71, 82). Like Pin1 overexpression (3), centrosome defects have been shown to correlate with poor clinical outcome (12, 61, 71), suggesting a possible functional interaction between Pin1 overexpression and centrosome abnormalities. Indeed, we have found that Pin1 levels are significantly correlated with the degree of centrosome amplification in human breast cancer tissues. Furthermore, the overexpression of Pin1 in nontransformed NIH 3T3 cells induces centrosome amplification and chromosome missegregation, eventually leading to aneuploidy and oncogenic transformation. Moreover, transgenic overexpression of Pin1 in mouse mammary glands potently induces centrosome amplification, eventually leading to mammary hyperplasia and malignant mammary tumors with overamplified centrosomes. Given these marked effects of Pin1 overexpression on centrosome amplification in vitro and in vivo and given the well-documented centrosome defects in human cancers, it is likely that Pin1-induced centrosome amplification promotes and/or contributes to oncogenesis. These results also suggest that inhibition of Pin1 might offer an attractive new option for inhibiting centrosome amplification and chromosome instability in cancer cells.
How Pin1 regulates centrosome duplication and how its deregulation leads to centrosome amplification remain to be defined. Pin1 has been shown to increase cyclin D1 protein stability and its transcription in collaboration with Ras/JNK, Wnt/β-catenin and NF-κB pathways (41, 65, 67, 82). Since cyclin D1 plays an essential role in breast cancer development (15, 37, 72, 77, 86) and can increase centrosome duplication in vitro (55), it is possible that Pin1 might regulate centrosome duplication and breast cancer through cyclin D1 and its upstream regulators. In addition, our preliminary results showed that Pin1 binds to several important proteins that are located at centrosomes and phosphorylated Cdk2 substrates and that inhibition of Cdk2 function can suppress the ability of Pin1 to induce centrosome amplification (data not shown), suggesting that Pin1 might also regulate centrosome duplication via Cdk2 substrates on centrosomes. Notably, it has been shown that the Pin1 gene is an Rb/E2F downstream target gene (66) and that G1 Cdks and their downstream Rb/E2F pathway play an important role in centrosome duplication during S phase (20, 21, 32, 36, 49, 52). Therefore, it is possible that during the G1/S transition in normal cells, growth signals activate Cdk kinases which turn on the Rb/E2F pathway, thereby increasing Pin1 expression. This increase in Pin1 levels may in turn promote centrosome duplication by controlling the function of the proteins at centrosomes or their upstream regulators that have been phosphorylated by Cdks in a positive feedback mechanism. However, deregulation of this mechanism due to the constitutively active Rb/E2F pathway and Pin1 overexpression might contribute to centrosome amplification in cancer cells. Therefore, it would be interesting to identify all Pin1-binding proteins at the G1/S transition in normal and cancer cells and then to elucidate their function in centrosome duplication and amplification.
In summary, we have shown that Pin1 plays a critical role in regulating centrosome duplication during the cell cycle and its overexpression causes centrosome amplification and chromosome missegregation, which leads to chromosome instability and oncogenesis in vitro and in vivo. These results provide the first evidence for novel postphosphorylation regulation of centrosome duplication during the cell cycle and its significance in centrosome abnormalities and oncogenesis. These results also suggest that Pin1 may be a new anticancer target for inhibiting centrosome defects and chromosome instability, which are common events in cancer cells.
Acknowledgments
We thank Lew Cantley, Ichiro Aoki, and Hideki Kobayashi for constructive advice; P. Leder for MMTV constructs; Masafumi Nakamura, Yih-Cherng Liou, and Kilian Perrem for constructive discussions and technical assistance; and Kilian Perrem for editing the manuscript.
A.R. and J.L. are a Leukemia and Lymphoma Society Special Fellow and a Human Frontier Science Program Fellow, respectively. This study was supported by the NIH grant GM58556 to K.P.L.
REFERENCES
- 1.Andersen, J. S., C. J. Wilkinson, T. Mayor, P. Mortensen, E. A. Nigg, and M. Mann. 2003. Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426:570-574. [DOI] [PubMed] [Google Scholar]
- 2.Atchison, F. W., B. Capel, and A. R. Means. 2003. Pin1 regulates the timing of mammalian primordial germ cell proliferation. Development 130:3579-3586. [DOI] [PubMed] [Google Scholar]
- 3.Ayala, G., D. Wang, G. Wulf, A. Frolov, R. Le, T. Wheeler, J. M. Sowadski, K. P. Lu, and L. Bao. 2003. Pin1 is a novel prognostic marker in prostate cancer. Cancer Res. 63:6244-6251. [PubMed] [Google Scholar]
- 4.Balczon, R., L. Bao, W. E. Zimmer, K. Brown, R. P. Zinkowski, and B. R. Brinkley. 1995. Dissociation of centrosome replication events from cycles of DNA synthesis and mitotic division in hydroxyurea-arrested Chinese hamster ovary cells. J. Cell Biol. 130:105-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bao, L., G. Sauter, J. Sowadski, K. P. Lu, and D. Wang. 2004. Prevalent overexpression of prolyl isomerase Pin1 in human cancers. Am. J. Pathol. 164:1727-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borel, F., O. D. Lohez, F. B. Lacroix, and R. L. Margolis. 2002. Multiple centrosomes arise from tetraploidy checkpoint failure and mitotic centrosome clusters in p53 and RB pocket protein-compromised cells. Proc. Natl. Acad. Sci. USA 99:9819-9824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bornens, M. 2002. Centrosome composition and microtubule anchoring mechanisms. Curr. Opin. Cell Biol. 14:25-34. [DOI] [PubMed] [Google Scholar]
- 8.Bornens, M., M. Paintrand, J. Berges, M. C. Marty, and E. Karsenti. 1987. Structural and chemical characterization of isolated centrosomes. Cell Motil. Cytoskeleton 8:238-249. [DOI] [PubMed] [Google Scholar]
- 9.Cato, A. C., J. Weinmann, S. Mink, H. Ponta, D. Henderson, and A. Sonnenberg. 1989. The regulation of expression of mouse mammary tumor virus DNA by steroid hormones and growth factors. J. Steroid Biochem. 34:139-143. [DOI] [PubMed] [Google Scholar]
- 10.Chen, Z., V. B. Indjeian, M. McManus, L. Wang, and B. D. Dynlacht. 2002. CP110, a cell cycle-dependent CDK substrate, regulates centrosome duplication in human cells. Dev. Cell 3:339-350. [DOI] [PubMed] [Google Scholar]
- 11.Choi, Y. W., D. Henrard, I. Lee, and S. R. Ross. 1987. The mouse mammary tumor virus long terminal repeat directs expression in epithelial and lymphoid cells of different tissues in transgenic mice. J. Virol. 61:3013-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'Assoro, A. B., S. L. Barrett, C. Folk, V. C. Negron, K. Boeneman, R. Busby, C. Whitehead, F. Stivala, W. L. Lingle, and J. L. Salisbury. 2002. Amplified centrosomes in breast cancer: a potential indicator of tumor aggressiveness. Breast Cancer Res. Treat. 75:25-34. [DOI] [PubMed] [Google Scholar]
- 13.D'Assoro, A. B., R. Busby, K. Suino, E. Delva, G. J. Almodovar-Mercado, H. Johnson, C. Folk, D. J. Farrugia, V. Vasile, F. Stivala, and J. L. Salisbury. 2004. Genotoxic stress leads to centrosome amplification in breast cancer cell lines that have an inactive G1/S cell cycle checkpoint. Oncogene 23:4068-4075. [DOI] [PubMed] [Google Scholar]
- 14.D'Assoro, A. B., W. L. Lingle, and J. L. Salisbury. 2002. Centrosome amplification and the development of cancer. Oncogene 21:6146-6153. [DOI] [PubMed] [Google Scholar]
- 15.Deshpande, A., P. Sicinski, and P. W. Hinds. 2005. Cyclins and cdks in development and cancer: a perspective. Oncogene 24:2909-2915. [DOI] [PubMed] [Google Scholar]
- 16.Dougherty, M. K., J. Müller, D. A. Ritt, X. Z. Zhou, M. Zhou, T. D. Copeland, T. P. Conrads, T. Veenstra, K. P. Lu, and D. K. Morrison. 2005. Regulation of Raf-1 by direct feedback phosphorylation. Mol. Cell 17:215-224. [DOI] [PubMed] [Google Scholar]
- 17.Doxsey, S. 2002. Duplicating dangerously: linking centrosome duplication and aneuploidy. Mol. Cell 10:439-440. [DOI] [PubMed] [Google Scholar]
- 18.Doxsey, S., D. McCollum, and W. Theurkauf. 2005. Centrosomes in cellular regulation. Annu. Rev. Cell Dev. Biol. 21:411-434. [DOI] [PubMed] [Google Scholar]
- 19.Doxsey, S., W. Zimmerman, and K. Mikule. 2005. Centrosome control of the cell cycle. Trends Cell Biol. 15:303-311. [DOI] [PubMed] [Google Scholar]
- 20.Duensing, S., A. Duensing, C. P. Crum, and K. Munger. 2001. Human papillomavirus type 16 E7 oncoprotein-induced abnormal centrosome synthesis is an early event in the evolving malignant phenotype. Cancer Res. 61:2356-2360. [PubMed] [Google Scholar]
- 21.Duensing, S., L. Y. Lee, A. Duensing, J. Basile, S. Piboonniyom, S. Gonzalez, C. P. Crum, and K. Munger. 2000. The human papillomavirus type 16 E6 and E7 oncoproteins cooperate to induce mitotic defects and genomic instability by uncoupling centrosome duplication from the cell division cycle. Proc. Natl. Acad. Sci. USA 97:10002-10007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El-Deiry, W. S., T. Tokino, V. E. Velculescu, D. B. Levy, R. Parsons, J. M. Trent, D. Lin, W. E. Mercer, K. W. Kinzler, and B. Vogelstein. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817-825. [DOI] [PubMed] [Google Scholar]
- 23.Fisk, H. A., and M. Winey. 2001. The mouse Mps1p-like kinase regulates centrosome duplication. Cell 106:95-104. [DOI] [PubMed] [Google Scholar]
- 24.Fujimori, F., K. Takahashi, C. Uchida, and T. Uchida. 1999. Mice lacking Pin1 develop normally, but are defective in entering cell cycle from G0 arrest. Biochem. Biophys. Res. Commun. 265:658-663. [DOI] [PubMed] [Google Scholar]
- 25.Fukasawa, K., T. Choi, R. Kuriyama, S. Rulong, and G. F. Vande Woude. 1996. Abnormal centrosome amplification in the absence of p53. Science 271:1744-1747. [DOI] [PubMed] [Google Scholar]
- 26.Ghadimi, B. M., D. L. Sackett, M. J. Difilippantonio, E. Schrock, T. Neumann, A. Jauho, G. Auer, and T. Ried. 2000. Centrosome amplification and instability occurs exclusively in aneuploid, but not in diploid colorectal cancer cell lines, and correlates with numerical chromosomal aberrations. Genes Chromosomes Cancer 27:183-190. [PMC free article] [PubMed] [Google Scholar]
- 27.Goepfert, T. M., Y. E. Adigun, L. Zhong, J. Gay, D. Medina, and W. R. Brinkley. 2002. Centrosome amplification and overexpression of aurora A are early events in rat mammary carcinogenesis. Cancer Res. 62:4115-4122. [PubMed] [Google Scholar]
- 28.Harper, J. W., G. R. Adami, N. Wei, K. Keyomarsi, and S. J. Elledge. 1993. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75:805-816. [DOI] [PubMed] [Google Scholar]
- 29.Reference deleted.
- 30.Hennighausen, L. 2000. Mouse models for breast cancer. Breast Cancer Res. 2:2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hennighausen, L., R. J. Wall, U. Tillmann, M. Li, and P. A. Furth. 1995. Conditional gene expression in secretory tissues and skin of transgenic mice using the MMTV-LTR and the tetracycline responsive system. J. Cell Biochem. 59:463-472. [DOI] [PubMed] [Google Scholar]
- 32.Hinchcliffe, E. H., C. Li, E. A. Thompson, J. L. Maller, and G. Sluder. 1999. Requirement of Cdk2-cyclin E activity for repeated centrosome reproduction in Xenopus egg extracts. Science 283:851-854. [DOI] [PubMed] [Google Scholar]
- 33.Hinchcliffe, E. H., and G. Sluder. 2001. “It takes two to tango”: understanding how centrosome duplication is regulated throughout the cell cycle. Genes Dev. 15:1167-1181. [DOI] [PubMed] [Google Scholar]
- 34.Katayama, H., K. Sasai, H. Kawai, Z. M. Yuan, J. Bondaruk, F. Suzuki, S. Fujii, R. B. Arlinghaus, B. A. Czerniak, and S. Sen. 2004. Phosphorylation by aurora kinase A induces Mdm2-mediated destabilization and inhibition of p53. Nat. Genet. 36:55-62. [DOI] [PubMed] [Google Scholar]
- 35.Kuriyama, R., S. Dasgupta, and G. G. Borisy. 1986. Independence of centriole formation and initiation of DNA synthesis in Chinese hamster ovary cells. Cell Motil. Cytoskeleton 6:355-362. [DOI] [PubMed] [Google Scholar]
- 36.Lacey, K. R., P. K. Jackson, and T. Stearns. 1999. Cyclin-dependent kinase control of centrosome duplication. Proc. Natl. Acad. Sci. USA 96:2817-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lamb, J., S. Ramaswamy, H. L. Ford, B. Contreras, R. V. Martinez, F. S. Kittrell, C. A. Zahnow, N. Patterson, T. R. Golub, and M. E. Ewen. 2003. A mechanism of cyclin D1 action encoded in the patterns of gene expression in human cancer. Cell 114:323-334. [DOI] [PubMed] [Google Scholar]
- 38.Lingle, W. L., S. L. Barrett, V. C. Negron, A. B. D'Assoro, K. Boeneman, W. Liu, C. M. Whitehead, C. Reynolds, and J. L. Salisbury. 2002. Centrosome amplification drives chromosomal instability in breast tumor development. Proc. Natl. Acad. Sci. USA 99:1978-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lingle, W. L., W. H. Lutz, J. N. Ingle, N. J. Maihle, and J. L. Salisbury. 1998. Centrosome hypertrophy in human breast tumors: implications for genomic stability and cell polarity. Proc. Natl. Acad. Sci. USA 95:2950-2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lingle, W. L., and J. L. Salisbury. 1999. Altered centrosome structure is associated with abnormal mitoses in human breast tumors. Am. J. Pathol. 155:1941-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liou, Y. C., R. Ryo, H. K. Huang, P. J. Lu, R. Bronson, F. Fujimori, U. Uchidafl, T. Hunter, and K. P. Lu. 2002. Loss of Pin1 function in the mouse causes phenotypes resembling cyclin D1-null phenotypes. Proc. Natl. Acad. Sci. USA 99:1335-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu, K. P. 2004. Pinning down cell signaling, cancer and Alzheimer's disease. Trends Biochem. Sci. 29:200-209. [DOI] [PubMed] [Google Scholar]
- 43.Lu, K. P. 2003. Prolyl isomerase Pin1 as a molecular target for cancer diagnostics and therapeutics. Cancer Cell 4:175-180. [DOI] [PubMed] [Google Scholar]
- 44.Lu, K. P., S. D. Hanes, and T. Hunter. 1996. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature 380:544-547. [DOI] [PubMed] [Google Scholar]
- 45.Lu, K. P., Y. C. Liou, and X. Z. Zhou. 2002. Pinning down the proline-directed phosphorylation signaling. Trends Cell Biol. 12:164-172. [DOI] [PubMed] [Google Scholar]
- 46.Lu, P. J., X. Z. Zhou, Y. C. Liou, J. P. Noel, and K. P. Lu. 2002. Critical role of WW domain phosphorylation in regulating its phosphoserine-binding activity and the Pin1 function. J. Biol. Chem. 277:2381-2384. [DOI] [PubMed] [Google Scholar]
- 47.Lu, P. J., X. Z. Zhou, M. Shen, and K. P. Lu. 1999. A function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science 283:1325-1328. [DOI] [PubMed] [Google Scholar]
- 48.Mantovani, F., S. Piazza, M. Gostissa, S. Strano, P. Zacchi, R. Mantovani, G. Blandino, and G. Del Sal. 2004. Pin1 links the activities of c-Abl and p300 in regulating p73 function. Mol. Cell 14:625-636. [DOI] [PubMed] [Google Scholar]
- 49.Matsumoto, Y., K. Hayashi, and E. Nishida. 1999. Cyclin-dependent kinase 2 (Cdk2) is required for centrosome duplication in mammalian cells. Curr. Biol. 9:429-432. [DOI] [PubMed] [Google Scholar]
- 50.McDonald, K. 1984. Osmium ferricyanide fixation improves microfilament preservation and membrane visualization in a variety of animal cell types. J. Ultrastruct Res. 86:107-118. [DOI] [PubMed] [Google Scholar]
- 51.Meraldi, P., R. Honda, and E. A. Nigg. 2002. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53−/− cells. EMBO J. 21:483-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meraldi, P., J. Lukas, A. M. Fry, J. Bartek, and E. A. Nigg. 1999. Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A. Nat. Cell Biol. 1:88-93. [DOI] [PubMed] [Google Scholar]
- 53.Meraldi, P., and E. A. Nigg. 2002. The centrosome cycle. FEBS Lett. 521:9-13. [DOI] [PubMed] [Google Scholar]
- 54.Muller, W. J., E. Sinn, P. K. Pattengale, R. Wallace, and P. Leder. 1988. Single-step induction of mammary adenocarcinoma in transgenic mice bearing the activated c-neu oncogene. Cell 54:105-115. [DOI] [PubMed] [Google Scholar]
- 55.Nelsen, C. J., R. Kuriyama, B. Hirsch, V. C. Negron, W. L. Lingle, M. M. Goggin, M. W. Stanley, and J. H. Albrecht. 2005. Short term cyclin D1 overexpression induces centrosome amplification, mitotic spindle abnormalities, and aneuploidy. J. Biol. Chem. 280:768-776. [DOI] [PubMed] [Google Scholar]
- 56.Nigg, E. A. 2002. Centrosome aberrations: cause or consequence of cancer progression? Nat. Rev. Cancer 2:815-825. [DOI] [PubMed] [Google Scholar]
- 57.Okuda, M., H. F. Horn, P. Tarapore, Y. Tokuyama, A. G. Smulian, P. K. Chan, E. S. Knudsen, I. A. Hofmann, J. D. Snyder, K. E. Bove, and K. Fukasawa. 2000. Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell 103:127-140. [DOI] [PubMed] [Google Scholar]
- 58.Pihan, G., and S. J. Doxsey. 2003. Mutations and aneuploidy: co-conspirators in cancer? Cancer Cell 4:89-94. [DOI] [PubMed] [Google Scholar]
- 59.Pihan, G. A., and S. J. Doxsey. 1999. The mitotic machinery as a source of genetic instability in cancer. Semin. Cancer Biol. 9:289-302. [DOI] [PubMed] [Google Scholar]
- 60.Pihan, G. A., A. Purohit, J. Wallace, H. Knecht, B. Woda, P. Quesenberry, and S. J. Doxsey. 1998. Centrosome defects and genetic instability in malignant tumors. Cancer Res. 58:3974-3985. [PubMed] [Google Scholar]
- 61.Pihan, G. A., A. Purohit, J. Wallace, R. Malhotra, L. Liotta, and S. J. Doxsey. 2001. Centrosome defects can account for cellular and genetic changes that characterize prostate cancer progression. Cancer Res. 61:2212-2219. [PubMed] [Google Scholar]
- 62.Pihan, G. A., J. Wallace, Y. Zhou, and S. J. Doxsey. 2003. Centrosome abnormalities and chromosome instability occur together in pre-invasive carcinomas. Cancer Res. 63:1398-1404. [PubMed] [Google Scholar]
- 63.Ranganathan, R., K. P. Lu, T. Hunter, and J. P. Noel. 1997. Structural and functional analysis of the mitotic peptidyl-prolyl isomerase Pin1 suggests that substrate recognition is phosphorylation dependent. Cell 89:875-886. [DOI] [PubMed] [Google Scholar]
- 64.Rippmann, J. F., S. Hobbie, C. Daiber, B. Guilliard, M. Bauer, J. Birk, H. Nar, P. Garin-Chesa, W. J. Rettig, and A. Schnapp. 2000. Phosphorylation-dependent proline isomerization catalyzed by Pin1 is essential for tumor cell survival and entry into mitosis. Cell Growth Differ. 11:409-416. [PubMed] [Google Scholar]
- 65.Ryo, A., Y. C. Liou, K. P. Lu, and G. Wulf. 2003. Prolyl isomerase Pin1: a catalyst for oncogenesis and a potential therapeutic target in cancer. J. Cell Sci. 116:773-783. [DOI] [PubMed] [Google Scholar]
- 66.Ryo, A., Y. C. Liou, G. Wulf, N. Nakamura, S. W. Lee, and K. P. Lu. 2002. PIN1 is an E2F target gene essential for the Neu/Ras-induced transformation of mammary epithelial cells. Mol. Cell. Biol. 22:5281-5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ryo, A., N. Nakamura, G. Wulf, Y. C. Liou, and K. P. Lu. 2001. Pin1 regulates turnover and subcellular localization of beta-catenin by inhibiting its interaction with APC. Nature Cell Biol. 3:793-801. [DOI] [PubMed] [Google Scholar]
- 68.Ryo, A., F. Suizu, Y. Yoshida, K. Perrem, Y. C. Liou, G. Wulf, R. Rottapel, S. Yamaoka, and K. P. Lu. 2003. Regulation of NF-κB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol. Cell 12:1413-1426. [DOI] [PubMed] [Google Scholar]
- 69.Saavedra, H. I., B. Maiti, C. Timmers, R. Altura, Y. Tokuyama, K. Fukasawa, and G. Leone. 2003. Inactivation of E2F3 results in centrosome amplification. Cancer Cell 3:333-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Salisbury, J. L., A. B. D'Assoro, and W. L. Lingle. 2004. Centrosome amplification and the origin of chromosomal instability in breast cancer. J. Mammary Gland Biol. Neoplasia 9:275-283. [DOI] [PubMed] [Google Scholar]
- 71.Sato, N., K. Mizumoto, M. Nakamura, N. Maehara, Y. A. Minamishima, S. Nishio, E. Nagai, and M. Tanaka. 2001. Correlation between centrosome abnormalities and chromosomal instability in human pancreatic cancer cells. Cancer Genet. Cytogenet. 126:13-19. [DOI] [PubMed] [Google Scholar]
- 72.Sicinski, P., J. L. Donaher, S. B. Parker, T. Li, A. Fazeli, H. Gardner, S. Z. Haslam, R. T. Bronson, S. J. Elledge, and R. A. Weinberg. 1995. Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell 82:621-630. [DOI] [PubMed] [Google Scholar]
- 73.Sinn, E., W. Muller, P. Pattengale, I. Tepler, R. Wallace, and P. Leder. 1987. Coexpression of MMTV/v-Ha-ras and MMTV/c-myc genes in transgenic mice: synergistic action of oncogenes in vivo. Cell 49:465-475. [DOI] [PubMed] [Google Scholar]
- 74.Stukenberg, P. T., and M. W. Kirschner. 2001. Pin1 acts catalytically to promote a conformational change in Cdc25. Mol. Cell 7:1071-1083. [DOI] [PubMed] [Google Scholar]
- 75.Tarapore, P., H. F. Horn, Y. Tokuyama, and K. Fukasawa. 2001. Direct regulation of the centrosome duplication cycle by the p53-p21Waf1/Cip1 pathway. Oncogene 20:3173-3184. [DOI] [PubMed] [Google Scholar]
- 76.Tutt, A., A. Gabriel, D. Bertwistle, F. Connor, H. Paterson, J. Peacock, G. Ross, and A. Ashworth. 1999. Absence of Brca2 causes genome instability by chromosome breakage and loss associated with centrosome amplification. Curr. Biol. 9:1107-1110. [DOI] [PubMed] [Google Scholar]
- 77.Wang, T. C., R. D. Cardiff, L. Zukerberg, E. Lees, A. Arnold, and E. V. Schmidt. 1994. Mammary hyperplasia and carcinoma in MMTV-cyclin D1 transgenic mice. Nature 369:669-671. [DOI] [PubMed] [Google Scholar]
- 78.Winkler, K. E., K. I. Swenson, S. Kornbluth, and A. R. Means. 2000. Requirement of the prolyl isomerase Pin1 for the replication checkpoint. Science 287:1644-1647. [DOI] [PubMed] [Google Scholar]
- 79.Wulf, G., G. Finn, F. Suizu, and K. P. Lu. 2005. Phosphorylation-specific prolyl isomerization: is there an underlying theme? Nature Cell Biol. 7:435-441. [DOI] [PubMed] [Google Scholar]
- 80.Wulf, G., P. Garg, Y. C. Liou, D. Iglehart, and K. P. Lu. 2004. Modeling breast cancer in vivo and ex vivo reveals an essential role of Pin1 in tumorigenesis. EMBO J. 23:3397-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wulf, G. M., Y. C. Liou, A. Ryo, S. W. Lee, and K. P. Lu. 2002. Role of Pin1 in the regulation of p53 stability and p21 transactivation, and cell cycle checkpoints in response to DNA damage. J. Biol. Chem. 277:47976-47979. [DOI] [PubMed] [Google Scholar]
- 82.Wulf, G. M., A. Ryo, G. G. Wulf, S. W. Lee, T. Niu, and K. P. Lu. 2001. Pin1 is overexpressed in breast cancer and potentiates the transcriptional activity of phosphorylated c-Jun towards the cyclin D1 gene. EMBO J. 20:3459-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xu, X., Z. Weaver, S. P. Linke, C. Li, J. Gotay, X. W. Wang, C. C. Harris, T. Ried, and C. X. Deng. 1999. Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol. Cell 3:389-395. [DOI] [PubMed] [Google Scholar]
- 84.Yaffe, M. B., M. Schutkowski, M. Shen, X. Z. Zhou, P. T. Stukenberg, J. Rahfeld, J. Xu, J. Kuang, M. W. Kirschner, G. Fischer, L. C. Cantley, and K. P. Lu. 1997. Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science 278:1957-1960. [DOI] [PubMed] [Google Scholar]
- 85.Yeh, E., M. Cunningham, H. Arnold, D. Chasse, T. Monteith, G. Ivaldi, W. C. Hahn, P. T. Stukenberg, S. Shenolikar, T. Uchida, C. M. Counter, J. R. Nevins, A. R. Means, and R. Sears. 2004. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat. Cell Biol. 6:308-318. [DOI] [PubMed] [Google Scholar]
- 86.Yu, Q., Y. Geng, and P. Sicinski. 2001. Specific protection against breast cancers by cyclin D1 ablation. Nature 411:1017-1021. [DOI] [PubMed] [Google Scholar]
- 87.Zacchi, P., M. Gostissa, T. Uchida, C. Salvagno, A. Avolio, S. Voliniak, Z. Ronai, G. Blandino, C. Schneider, and G. Del Sal. 2002. The prolyl isomerase Pin1 reveals a mechanism to control p53 functions after genotoxic insults. Nature 419:853-857. [DOI] [PubMed] [Google Scholar]
- 88.Zheng, H., H. You, X. Z. Zhou, S. A. Murray, T. Uchida, G. Wulf, L. Gu, X. Tang, K. P. Lu, and Z. X. J. Xiao. 2002. The prolyl isomerase Pin1 is a regulator of p53 in genotoxic response. Nature 419:849-853. [DOI] [PubMed] [Google Scholar]
- 89.Zhou, H., J. Kuang, L. Zhong, W. L. Kuo, J. W. Gray, A. Sahin, B. R. Brinkley, and S. Sen. 1998. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat. Genet. 20:189-193. [DOI] [PubMed] [Google Scholar]
- 90.Zhou, X. Z., O. Kops, A. Werner, P. J. Lu, M. Shen, G. Stoller, G. Küllertz, M. Stark, G. Fischer, and K. P. Lu. 2000. Pin1-dependent prolyl isomerization regulates dephosphorylation of Cdc25C and tau proteins. Mol. Cell 6:873-883. [DOI] [PubMed] [Google Scholar]