Abstract
In addition to their essential roles in V(D)J recombination, the RAG proteins have been found to catalyze transposition in vitro, but it has been difficult to demonstrate transposition by the RAG proteins in vivo in vertebrate cells. As genomic instability and chromosomal translocations are common outcomes of transposition in other species, it is critical to understand if the RAG proteins behave as a transposase in vertebrate cells. To facilitate this, we have developed an episome-based assay to detect products of RAG-mediated transposition in the human embryonic kidney cell line 293T. Transposition events into the target episome, accompanied by characteristic target site duplications, were detected at a low frequency using RAG1 and either truncated “core” RAG2 or full-length RAG2. More frequently, insertion of the RAG-generated signal end fragment into the target was accompanied by deletions or more complex rearrangements, and our data indicate that these events occur by a mechanism that is distinct from transposition. An assay to detect transposition from an episome into the human genome failed to detect bona fide transposition events but instead yielded chromosome deletion and translocation events involving the signal end fragment mobilized by the RAG proteins. These assays provide a means of assessing RAG-mediated transposition in vivo, and our findings provide insight into the potential for the products of RAG-mediated DNA cleavage to cause genome instability.
Transposases are enzymes that catalyze the movement of a DNA segment, known as a transposable element or transposon, from one locus to another (4, 6, 8). Transposon-derived sequences form a significant portion of many genomes (15, 26, 54, 56), and the movement of transposons is thought to play an important role in genome evolution (4, 6). Some of the possible outcomes of transposase action are alterations in gene structure and expression, inversion, deletion, or translocation of chromosomal DNA, and genetic instability. Transposition can also result in the transduction of flanking DNA, further enhancing the potential for genetic rearrangements (32, 35). Thus, by virtue of the nature of the reactions catalyzed, transposases can both facilitate evolutionary change and have a detrimental effect on the host cell that harbors them. Interestingly, many present-day transposable elements, especially in higher eukaryotes, are inactive.
There exist situations in which host cells derive benefit from the activity of transposases present in their genome. Many bacterial transposable elements carry with them genes coding for antibiotic-resistant or virulence factors, and mobility of these elements can spread such genes in the bacterial gene pool. In vertebrates, the RAG proteins play a critical role in the generation of B- and T-cell antigen receptors and therefore in the development of B and T cells and the adaptive immune system. In addition, RAG1 and RAG2 have the ability to catalyze transposition in vitro (1, 19).
RAG1 and RAG2 together form the essential endonuclease that initiates VDJ recombination (37, 44), the process by which the variable region of antibody and T-cell receptor genes are assembled from different gene segments (49). The recombination event involves two cis-acting elements known as recombination signal sequences (RSSs) that lie adjacent to the participating gene segments. RSSs contain conserved heptamer and nonamer sequences separated by a spacer of either 12 or 23 bp (named the 12-RSS and 23-RSS, respectively). DNA cleavage by the RAG proteins is also thought to require the nonspecific DNA binding high-mobility group 1 or 2 protein (53). In the recombination process, RAG1 and RAG2 introduce nicks in the DNA immediately adjacent to the RSSs, generating a 3′ hydroxyl and a 5′ phosphate (12, 14, 30). The 3′ hydroxyls thus generated an attack of the antiparallel strand and led to the generation of two hairpin-coding ends and two blunt signal ends. All four DNA fragments remain bound to RAG proteins (18) and are processed by proteins belonging to the nonhomologous end-joining (NHEJ) pathway (12, 14). The hairpins are opened, nucleotides are added or deleted from the ends, and the two DNA-coding fragments are ligated to generate the coding joint. Signal ends are joined, typically without processing, to yield signal joints.
RAG protein-catalyzed transposition also begins with the generation of two double-stranded breaks by the same nick/hairpin process outlined above. The coding ends are joined quickly and are released. In contrast, signal joint formation is a comparatively slow process and signal ends remain bound to the RAG proteins in a signal end complex after the release of the coding ends (2, 18). The signal end complex can capture target DNA and transpose the signal end fragment. Target capture occurs with relatively little sequence specificity (1, 19) and requires prior release of coding ends (11). Certain DNA structures, particularly mismatches, are preferred targets for RAG-mediated transposition (50).
The full-length RAG1 and RAG2 polypeptides have been difficult to express and purify, and most of the biochemical characterization of the RAG proteins has been carried out using truncated versions with greater solubility. These “core” forms of RAG1 and RAG2 can catalyze V(D)J recombination of extrachromosomal substrates in transient transfection assays and hence are qualitatively similar to the full-length proteins (7, 42, 43, 45). However, there are significant differences between the core and full-length forms of the proteins. The core RAG proteins perform V(D)J recombination less efficiently than the full-length proteins (3, 9, 24, 28) and generate aberrant products at an elevated rate (48). The noncore regions have been associated with roles in many aspects of the reaction, including RSS recognition/cleavage, end processing/joining, and the regulation of the turnover of the proteins (41). Activities that have been identified in the noncore regions are an E3 ubiquitin ligase activity in the N-terminal region of RAG1 (21, 57), a phosphorylation site in the C terminus of RAG2 that controls cell cycle-dependent degradation of the protein (27), and phospholipid (10) and histone binding in the RAG2 C-terminal domain (55). Finally, and of particular relevance here, the noncore C-terminal region of RAG2 has been demonstrated to contribute to the suppression of transposition in vitro, although the mechanism by which it does so is controversial (11, 47, 52).
The fact that DNA cleavage by the RAG proteins is indispensable to lymphocyte physiology raises the question of how the potential detrimental consequences of the transposition activity of the proteins are prevented in vivo. One possibility is that conditions or factors in the vertebrate cell prevent RAG-mediated transposition without perturbing V(D)J recombination. Another possibility is that the C-terminal domain of RAG2 plays an important role in suppressing transposition. To date, no in vivo assay in vertebrate cells has been available with which to investigate these possibilities or to determine what the suppressive conditions or factors might be. Intramolecular transposition mediated by the RAG proteins has been detected in yeast (5), and a single RAG-mediated transposition event has been reported in a human T-cell line (33).
Here, we describe an episome-based assay to detect intermolecular transposition catalyzed by the RAG proteins in the embryonic kidney cell line 293T. Our results show that the RAG proteins do indeed catalyze transposition in vivo, albeit at a low frequency. More frequently, the signal end fragment generated by the RAG proteins becomes inserted into the target episome with accompanying deletions or rearrangements in a process that is mechanistically distinct from transposition. In an attempt to detect RAG-mediated transposition into the genome, we isolated two aberrant events in which the integration of a signal end fragment into the human genome was accompanied by large-scale chromosomal deletion or translocation. RAG-generated signal end fragments can therefore be involved in processes, distinct from transposition, that lead to major chromosomal aberrations.
MATERIALS AND METHODS
Plasmids.
The pEBB expression vector (29) was used to express the RAG proteins (39). The BglII site was removed from pECFP-1 to generate pECFP-1*. The donor plasmid, pTetRSS*, was constructed from pTetRSS (1) by insertion of the rpsL and sacB genes, generously provided by N. Grindley. The sequence of 12- and 23-RSSs are as follows: 12-RSS, 5′CACAGTGATACAGCCCTTAACAAAAACC3′, and 23-RSS, 5′CACAGTGATGCAGGCCAAGTGTGAAGCCATACAAAAACC3′. pBSK-12puro23-2DT, the donor plasmid for the genomic transposition assay, was created by inserting a puromycin resistance (puror) gene flanked by RSSs into pBluescript SK(+). The sequences of the 12- and 23-RSSs were identical to those specified above. The diphtheria toxin (DT) expression cassette, kindly provided by U. Grawunder and M. Lieber (16), was cloned on either side of the RSSs to generate the pBSK-12puro23-2DT plasmid.
Plasmid transposition assay.
Plasmid transposition assays (see Fig. 1) were carried out by transfecting 293T cells with 4 μg each of the pEBB-RAG1 and -RAG2-expressing plasmids, 6 μg of donor plasmid, and 10 μg of target plasmid by calcium phosphate precipitation (23). Twenty-four hours posttransfection, cells were washed with phosphate-buffered saline (PBS) and fresh medium was added. Forty-eight hours after transfection, cells were washed extensively with PBS and DNA was isolated by rapid alkaline lysis (17). Expression of RAG1 and RAG2 was confirmed by Western blot analysis. The presence of signal joints in the DNA preparations, as detected by PCR, using primers 5′GATGCGTCCGGCGTAGAGGATCC3′ and 5′GATGCGTCCGGCGTAGAGGATCC3′, was used to confirm RAG protein activity. The purified DNA preparation was treated with BglII (to digest donor pTetRSS*) and RNase A. The digested DNA was transformed into electrocompetent MC1061 bacterial cells, and 100 μl of 1:10,000 diluted transformation mix was plated on a Luria-Bertani (LB) agar plate containing 30 μg/ml kanamycin (KAN). The remaining undiluted culture was plated on an LB agar plate containing 30 μg/ml kanamycin, 12 μg/ml tetracycline (TET), and 30 μg/ml streptomycin (a triple selection hereafter referred to as KTS). The number of colonies on both plates was counted, and plasmid DNA was isolated from colonies on KTS plates and digested with BglII, ApaLI, or FspI. Plasmid DNA resistant to BglII, linearized by ApaLI and cleaved twice by FspI, was sequenced to assess whether the plasmid had a structure consistent with transposition and, if so, the length of target size duplication. The GC content of transposition target sites was calculated based on the sequence of the target site duplication and the adjacent 5 bp on either side, as depicted in Table S1 in the supplemental material.
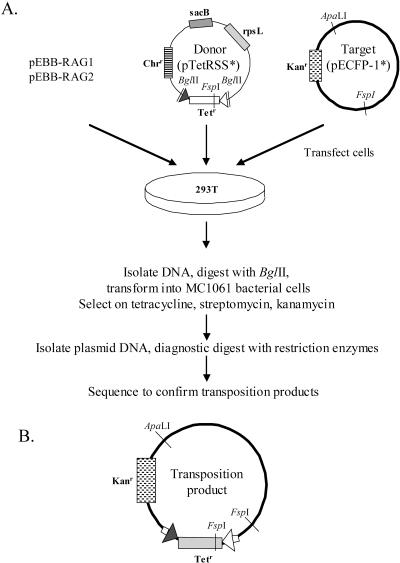
FIG. 1.
(A) Episomal assay for RAG-mediated transposition. 293T cells were transfected with four plasmids: RAG1 and RAG2 expression vectors, a donor plasmid harboring two RSSs flanking a tetracycline resistance gene (pTetRSS*), and a target plasmid (pECFP-1*). RSSs are indicated as triangles and genes as rectangles. DNA isolated from transfected cells was digested with BglII (which cuts only in the donor backbone), transformed into bacteria, and grown on agar plates containing KTS. Under these conditions, only bacteria harboring a target plasmid into which the Tetr gene of the donor was inserted should survive. Bacteria harboring both the donor and target plasmids should be eliminated by streptomycin by virtue of the rpsL gene of the donor. Plasmid DNA extracted from KTSr bacterial colonies was analyzed by restriction digestion and sequenced to determine the nature and site of signal end integration into the target plasmid. (B) Schematic representation of the transposition product.
Mixing experiments.
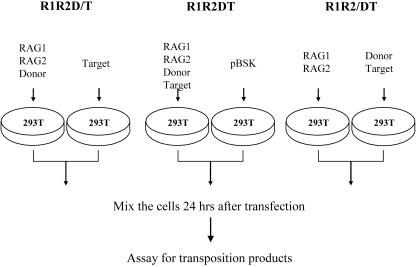
293T cells were transfected with various combinations of RAG protein expression plasmids and donor and target DNA as shown in Fig. 3. Each mixing experiment was comprised of three reactions, each of which consisted of two plates of 293T cells transfected with different sets of plasmids. Twenty-four hours posttransfection, cells were washed twice with medium and trypsinized and the cells from the two plates mixed, replated on two dishes, and allowed to grow for another 24 h before harvesting and processing as described above. The different reactions are as follows: in R1R2D/T, plate 1 transfected with RAG1 and RAG2 expression plasmids along with the donor plasmid and plate 2 transfected with target DNA; in R1R2DT, plate 1 transfected with RAG1 and RAG2 expression plasmids, donor, and target and plate 2 transfected with pBluescript SK(+); and in R1R2/DT, plate 1 transfected with RAG1 and RAG2 expression plasmids and plate 2 transfected with donor and template. All of the transfection mixes had appropriate amounts of pBluescript SK(+) DNA added so that the total amount of DNA in each transfection reaction was the same (24 μg).
FIG. 3.
Schematic representation of the mixing experiment. Three different types of mixing experiments were performed and labeled R1R2D/T, R1R2DT, and R1R2/DT. Each oval depicts a plate of 293T cells transfected with the indicated plasmids. Twenty-four hours after transfection, cells from the two plates were harvested, mixed, and replated and after another 24 h, harvested and processed as indicated in Fig. 1. See text, especially Materials and Methods, for details. R1, RAG1; R2, RAG2; D, donor; T, target; pBSK, pBluescript SK(+).
Transformation of the signal end fragment and target DNA into bacteria.
The signal end fragment was generated by PCR using pTetRSS* as template and primers TERSFSH, 5′CACAGTGATGGAAGCTCAATCTGAACTCTGACAAAAACC3′, and TERSRSH, 5′CACAGTGATACAGACCTTAACAAAAACCAATTCTTGGA3′, using Pfu polymerase. The fragment was purified, and DNA concentration was measured. Two reactions were carried out wherein 1 μg of target plasmid was transformed into electrocompetent MC1061 along with 0.5 μg of signal end fragment. Next, 100 μl of transformation mix diluted 1:10,000 was plated on an LB agar plate containing 30 μg/ml kanamycin. The remaining undiluted culture was plated on an LB agar plate containing 30 μg/ml kanamycin, 12 μg/ml tetracycline, and 30 μg/ml streptomycin. The number of colonies on both plates was counted. Plasmid DNA was isolated from colonies on KTS plates and analyzed by restriction digestion using enzyme BglII, ApaLI, or FspI as described above.
Genomic transposition assay.
The genomic transposition assay was carried out by transfecting 12 μg pBSK-12puro23-2DT into 293T cells along with 5 μg each of RAG1 and RAG2 expression plasmids by the calcium phosphate precipitation method (23). After 2 days, cells were trypsinized and put in medium containing 0.5 μg/ml of puromycin. After 3 weeks of selection, 150 puromycin-resistant colonies were isolated and expanded. Genomic DNA was isolated from these individual colonies. A PCR strategy was designed to distinguish putative transposition products from products of random integration of the plasmid. PCRs were carried out with three pairs of primers: the first pair (primers C2 and E3) (Fig. 4) was specific to the puromycin-resistant gene (puror), while the other two primer pairs (T7-2 and IE, T3-2 and IC) each contained one primer located within the signal end fragment (IE and IC) and the other located in the vector sequences outside the signal end fragment (T7-2 and T3-2). Transposition should result in separation of the signal end fragment from flanking vector sequences while maintaining the integrity of puror. PCRs using C2 and E3 confirmed the presence and integrity of the puror gene in each colony. Genomic DNA from puromycin-resistant colonies that yielded amplification products with primer pairs T7-2 and IE or T3-2 and IC were interpreted to have acquired the puror gene by random integration of the pBSK-12puro23-2DT plasmid. Colonies that were negative in both of these PCR assays were expected to have the signal end fragment separated from the rest of the donor plasmid and hence were candidates for harboring a transposition product. Digestion-circularization PCR (DC-PCR) was performed using genomic DNA isolated from clones harboring putative transposition events to recover the genomic sequences flanking the transposed fragments. For DC-PCR, DNA was digested with a restriction enzyme and then ligated under a condition favoring intramolecular ligation. The ligated DNA was used to perform PCRs with primers IE and IC (Fig. 4) or other primer pairs. The PCR products were cloned and sequenced to determine the sequence of the site of integration of the puror gene.
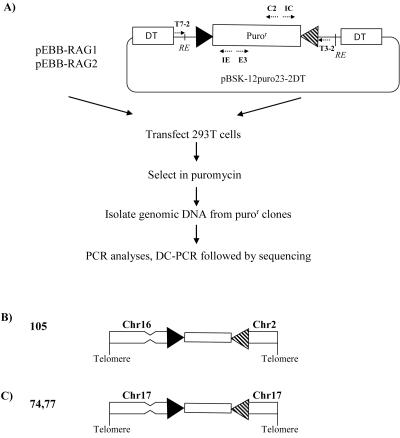
FIG. 4.
Assay for RAG-mediated transposition into the genome. (A) The donor plasmid, pBSK-12puro23-2DT, contained a puromycin resistance gene (Puror) flanked by two RSSs (triangles) and two DT expression cassettes. 293T cells were transfected with pBSK-12puro23-2DT and RAG expression vectors and subsequently grown in puromycin to select for acquisition of the puror gene. Random integration of the donor plasmid was selected against by the DT cassettes. In puror colonies, PCR with primers E3 and C2 was used to confirm the presence of puror while PCR with T7-2 and IE or with T3-2 and IC was used to detect the presence of the vector flanks (which should be absent in cells that have taken up the signal end fragment due to transposition or to insertion). For DC-PCR, restriction enzyme-digested genomic DNA was recircularized under conditions favoring intramolecular ligation and PCR amplified as described in Materials and Methods. (B and C) Schematic representations of the signal end fragment insertion events in clones 105 and 74/77.
RESULTS
Episome-to-episome transposition assay.
To provide a sensitive means of detecting transposition mediated by the RAG proteins in vivo, we created donor and recipient plasmids that could be introduced, together with RAG expression vectors, into the highly transfectable human embryonic kidney cell line 293T (Fig. 1). The donor plasmid, pTetRSS*, contains a tetracycline resistance gene flanked by consensus 12- and 23-RSSs and two negative selection markers (sacB and rpsL, although in the experiments described here, we took advantage only of rpsL). The target plasmid, pECFP-1*, contains a kanamycin resistance marker and a significant proportion (>60%) of nonessential DNA into which transposition could occur without interfering with plasmid replication or the expression of Kanr in bacteria. Transient transfection of cells with the four plasmids shown in Fig. 1A resulted in the RAG-mediated excision of a signal end DNA fragment containing the Tetr gene from the donor plasmid. In a typical V(D)J recombination reaction, the NHEJ machinery reseals the donor plasmid (coding joint formation) and circularizes the excised signal end fragment (signal joint formation).
The excised signal end fragment may also undergo transposition into another DNA molecule, which could be any of the transfected plasmids or the host genome. Transposition into the intended target episome (pECFP-1*) creates a plasmid containing both the Kanr and Tetr genes (Fig. 1B). Two days after transfection, episomal DNA was harvested from the transfected 293T cells, digested with BglII (which cleaves the donor plasmid backbone but not the recipient plasmid), and transformed into bacteria. The bacteria were plated on medium containing kanamycin alone (to measure the total recovery of recipient plasmids, as a means of estimating transfection efficiency) or on KTS. TET and KAN allow selection for the transposition product, and streptomycin selects against donor plasmids which contain the streptomycin sensitivity gene rpsL. Pilot experiments demonstrated that both digestion with BglII and selection on streptomycin were necessary to prevent the isolation of large numbers of bacterial colonies cotransformed with unaltered donor and target plasmids (data not shown).
Several different forms of the murine RAG proteins were used in various combinations in the transposition assays: untagged full-length RAG1 [RAG1FL(UN); amino acids (aa) 1 to 1040], glutathione S-transferase-tagged core RAG1 [RAG1C(GST); aa 384 to 1008], untagged full-length RAG2 [RAG2FL(UN); aa 1 to 537], and polyhistidine- and myc-tagged core RAG2 [RAG2C(HM); aa 1 to 387]. RAG expression was driven by the strong elongation factor 1α promoter of the pEBB expression vector, as described previously (13, 39, 51). Thirty-two independent assays (each representing an independent transfection) were performed using the combination RAG1FL and RAG2C(HM), six with RAG1C(GST) and RAG2C(HM), and nine with RAG1FL(UN) and RAG2FL(UN) (Tables 1 and 2).
TABLE 1.
Summary of results of transposition assays
| Parameter | RAG protein combination
|
|||
|---|---|---|---|---|
| RAG1FL(UN), RAG2C(HM) | RAG1C(GST), RAG2C(HM) | RAG1FL(UN), RAG2FL(UN) | RAG1, RAG2 | |
| No. of assays | 32 | 6 | 9 | 10j |
| Mean no. of Kanr colonies (106)a | 7.5 | 3.3 | 12.6 | 2.0 |
| Mean no. of KTSr coloniesb | 6.0 | 2.6 | 2.9 | 0 |
| No. of Tnp-positive assaysc | 11 | 2 | 2 | 0 |
| No. of plasmids sequencedd | 178 | 15 | 29 | 0 |
| No. of Tnp eventse | 17 | 2 | 2 | 0 |
| No. of simple insertionsf | 94 | 9 | 12 | 0 |
| No. of complex insertionsg | 67 (15, 52) | 4 (2, 2) | 15 (5, 10) | 0 |
| Incidence of Tnph | 0.5 | 0.3 | 0.2 | 0 |
| Mean frequency of Tnpi | 1.7 × 10−7 | 1.2 × 10−7 | 0.06 × 10−7 | 0 |
Mean number of Kanr colonies obtained in assays with the indicated combination of RAG proteins.
Mean number of triple resistant (KTSr) colonies obtained.
Number of assays that yielded one or more transposition (Tnp) events.
Total number of plasmids sequenced from KTSr colonies.
Number of transposition events identified by sequencing.
Number of simple insertion events identified by sequencing.
Number of complex insertion events identified by sequencing (number of complex insertions with inversion and duplication of target DNA, number of complex insertions with loss of signal end fragment integrity).
Incidence of transposition, defined as the number of events/number of assays.
Mean frequency of transposition, defined as the number of events/number of Kanr colonies.
Includes six R1R2/DT reactions, as defined in Fig. 3.
TABLE 2.
Transposition products observed in transposition-positive reactions
| RAG1 | RAG2 | No. of:
|
||
|---|---|---|---|---|
| Kanr colonies (106)a | KTSr coloniesb | Tnp eventsc | ||
| RAG1FL(UN) | RAG2C(HM) | 4.2 | 11 | 1 |
| RAG1FL(UN) | RAG2C(HM) | 0.48 | 1 | 1 |
| RAG1FL(UN) | RAG2C(HM) | 40 | 12 | 3 |
| RAG1FL(UN) | RAG2C(HM) | 1.2 | 3 | 1 |
| RAG1FL(UN) | RAG2C(HM) | 3.9 | 29 | 3 |
| RAG1FL(UN) | RAG2C(HM) | 3.5 | 14 | 1 |
| RAG1FL(UN) | RAG2C(HM) | 5.2 | 9 | 3 |
| RAG1FL(UN) | RAG2C(HM) | 5.6 | 20 | 1 |
| RAG1FL(UN) | RAG2C(HM) | 21.6 | 63 | 1 |
| RAG1FL(UN) | RAG2C(HM) | 12.4 | 18 | 1 |
| RAG1FL(UN) | RAG2C(HM) | 7.1 | 7 | 1 |
| RAG1C(GST) | RAG2C(HM) | 1.4 | 1 | 1 |
| RAG1C(GST) | RAG2C(HM) | 7.6 | 6 | 1 |
| RAG1FL(UN) | RAG2FL(UN) | 2.8 | 6 | 1 |
| RAG1FL(UN) | RAG2FL(UN) | 7.1 | 2 | 1 |
Number of Kanr colonies obtained in this assay with the indicated combination of RAG proteins.
Number of triple resistant (KTSr) colonies obtained.
Number of transposition (Tnp) events identified by sequencing.
Typically, a plasmid transposition reaction yielded a small number of KTSr colonies (averages ranged from 2.6 to 6) and 3.3 × 106 to 12.6 × 106 Kanr colonies (Table 1). Importantly, no KTSr colonies were obtained when RAG1 or RAG2 or both were omitted from the reaction (Table 1), demonstrating that the appearance of triple-resistant colonies in this assay is RAG dependent.
DNA isolated from KTSr colonies was digested with BglII, ApaLI, and FspI. Transposition products were expected to be resistant to BglII, linearized by ApaLI, and cleaved twice by FspI. Since there is one FspI recognition site in the target DNA and one in the signal end fragment, the sizes of the fragments generated by FspI digestion were dependent on the site of integration of the signal end fragment into the target DNA. This restriction enzyme analysis revealed that more than 95% of the plasmids isolated from KTSr colonies fulfilled the above criteria for being candidate transposition products. The remaining (<5%) KTSr colonies contained plasmids with restriction digestion profiles that could not be resolved or contained both the donor and the target DNA. These were not analyzed further. All plasmids that satisfied the restriction digestion criteria (222 in total) were sequenced to determine whether they exhibited the hallmark features of RAG-mediated transposition: an inserted signal end fragment with intact RSSs flanked by target site duplications of 3 to 5 bp.
The sequence analysis revealed that only 10% (21 of 222) of plasmids from KTSr colonies contained the expected 3- to 5-bp target size duplication. Of the 47 assays performed, only 15 yielded bona fide transposition products, with one to three transposition events detected in the positive reactions (Tables 1 and 2). In the 21 distinct transposition products obtained (Table 2), the sites of integration of the signal end fragment were concentrated in the permissible regions of the target plasmid (Fig. 2). Five-base-pair target site duplications were most commonly observed, followed by 4- and 3-bp duplications (see Table S1 in the supplemental material). Moreover, target sites were found to be enriched for G and C residues (64% GC compared to 53% for the available target regions of the target plasmid, P < 0.05). These observations are strikingly similar to results obtained from in vitro transposition experiments performed with purified RAG proteins (1, 19, 50), arguing strongly that the plasmids isolated with our assay contain bona fide transposition events. The failure to detect transposition in the other 32 assays was not a result of poor transfection efficiency: transposition-positive and -negative reactions yielded equivalent numbers of Kanr and KTSr colonies and similar levels of RAG proteins (as assessed by Western blot analysis) (data not shown) and RAG activity (as judged by levels of signal joints) (data not shown).
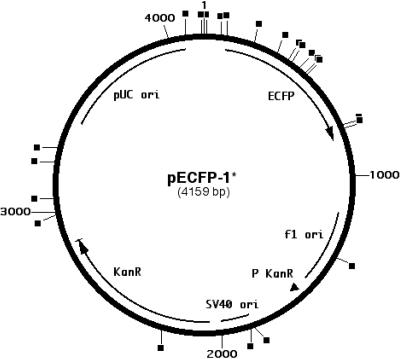
FIG. 2.
Map of sites of transposition in the episomal assay. Transposition sites are marked with black squares. ECFP, promoterless enhanced cyan fluorescent protein gene; SV40 ori, SV40 origin of replication; f1 ori, f1 single-stranded DNA origin; P KanR, bacterial promoter for expression of the Kanr gene; KanR, kanamycin resistance gene; pUC ori, pUC plasmid replication origin. The nucleotides of the target plasmid are numbered as indicated outside the circle. The basis of kanamycin resistance for the transposition event that occurred within KanR is unclear.
About half of the plasmids analyzed from KTSr colonies (51%; 115 of 222) contained an insertion of the entire signal end fragment (with intact RSSs) into the target DNA but without a flanking target site duplication. We hereafter refer to these events as simple insertions to distinguish them from transposition events. Most of the simple insertion plasmids exhibited the deletion of target plasmid sequences at the site of insertion of the signal end fragment, with the length of deletion ranging from a few nucleotides up to 2 kb. Six simple insertion plasmids had precise insertion of the signal end fragment without any nucleotide loss or addition.
The remaining 38% of the plasmids (86 of 222) had more complex rearrangements associated with the acquisition of Tetr sequences (complex insertions). Many (64 of 222) lacked some or all of one or both RSSs, as well as variable amounts of the internal signal end fragment sequence. The rest (22 of 222) retained both RSSs but contained inversions and duplications of target DNA (remote from the site of insertion) as well as deletions and duplications of target sequences immediately flanking the site of insertion. Overall, these results suggest that the signal end fragment generated by the RAG proteins can be inserted into a target DNA molecule by at least two pathways: RAG-mediated transposition, which maintains the structural integrity of the target, and insertion, which almost invariably results in deletions or other rearrangements of target DNA. The distinct nature of these two pathways and their dependence on the RAG proteins is explored further below.
Importantly, transposition products were generated by the full-length RAG proteins as well as by the core proteins (Tables 1 and 2), supporting the conclusion that the full-length RAG proteins can catalyze transposition in mammalian cells (33) and demonstrating that the C-terminal domain of RAG2 is not able to suppress transposition completely. While the frequency of transposition appeared to be higher with core RAG2 than with full-length RAG2 (mean frequencies of transposition being 1.7 × 10−7 and 0.06 × 10−7, respectively) (Table 1), the difference was not statistically significant (P = 0.44). Moreover, the low total number of transposition events detected and the known variations in protein expression and activity of these two forms of RAG2 make it difficult to assess the significance of the differences in the mean transposition frequencies. Core RAG2 accumulates to higher levels than the full-length protein because it lacks a degradation motif present near the C terminus (27), and signal ends accumulate to higher levels with core RAG2 than with full-length RAG2 (46) for reasons that are not completely understood but which may relate to the efficiency with which the signal end complex is remodeled to allow signal joint formation. We conclude that both core and full-length RAG2 can support transposition in vivo. However, it remains to be determined whether the C-terminal domain of RAG2 suppresses in vivo transposition activity appreciably.
Overall, the data in Tables 1 and 2 indicate that the episome-based assay provides a sensitive method for detecting RAG-mediated transposition in mammalian cells and that transposition by the RAG proteins, while very inefficient, occurs at detectable levels.
Are transposition and insertion products generated in vivo? Mixing experiments.
To confirm that transposition and insertion products obtained in this assay are formed inside transfected cells rather than through cell death and release of active signal end complexes and target DNA into the culture media, several mixing experiments were performed (Fig. 3). In these, subsets of the four plasmids were transfected into two separate plates, and after 1 day, the transfected cells from two plates were trypsinized, combined, replated, and harvested 1 day later (see Materials and Methods). In one group (R1R2D/T), RAG1 and RAG2 were expressed together with the donor plasmid in one plate of cells while target DNA was transfected into the other. In a second group (R1R2/DT), the two RAG proteins were expressed in one plate while the donor and target plasmids were introduced into the other. And in a third, positive-control group (R1R2DT), all four plasmids were transfected into the same cells and the companion plate was transfected with an irrelevant plasmid (Fig. 3). In the first two groups, the RAG proteins and the target DNA were in separate cells and transposition products were not expected to be produced unless transposition products could be generated through mixing of the contents of different cells.
Six independent mixing experiments were performed, each containing one each of R1R2D/T, R1R2/DT, and R1R2DT (Table 3). Two out of six experiments yielded one transposition product, and both were derived from instances in which all four DNA molecules were transfected into the same cells (R1R2DT). Neither insertion nor transposition products (nor any KTSr colonies) were observed in any of the reactions in which the RAG proteins and the target plasmid were in different cells (R1R2D/T and R1R2/DT), arguing against the possibility that the bona fide transposition products or the insertions were generated as a result of the mixing of cellular contents. As expected, PCR analyses carried out to detect signal joints showed the presence of such joints in cells containing all four plasmids or containing RAG1 and RAG2 together with donor DNA, but not in cells in which the donor and RAG proteins were in different cells (R1R2/DT) (data not shown).
TABLE 3.
Transposition products observed in mixing experiments
| Reaction no. | RAG protein combination | Reaction type | No. of:
|
||
|---|---|---|---|---|---|
| Kanr colonies (106)a | KTSr coloniesb | Tnp eventsc | |||
| 1 | RAG1C(GST), RAG2C(HM) | R1R2D/T | 0.08 | 0 | 0 |
| R1R2DT | 0.8 | 1 | 0 | ||
| R1R2/DT | 0.2 | 0 | 0 | ||
| 2 | RAG1C(GST), RAG2C(HM) | R1R2D/T | 0.48 | 0 | 0 |
| R1R2DT | 3 | 1 | 0 | ||
| R1R2/DT | 1.4 | 0 | 0 | ||
| 3 | RAG1FL(UN), RAG2C(HM) | R1R2D/T | 0.8 | 0 | 0 |
| R1R2DT | 1.2 | 5 | 1d | ||
| R1R2/DT | 0.3 | 0 | 0 | ||
| 4 | RAG1FL(UN), RAG2C(HM) | R1R2D/T | 1.7 | 0 | 0 |
| R1R2DT | 0.8 | 0 | 0 | ||
| R1R2/DT | 1.3 | 0 | 0 | ||
| 5 | RAG1FL(UN), RAG2C(HM) | R1R2D/T | 5.7 | 0 | 0 |
| R1R2DT | 5.6 | 4 | 1d | ||
| R1R2/DT | 9.8 | 0 | 0 | ||
| 6 | RAG1FL(UN), RAG2C(HM) | R1R2D/T | 1 | 0 | 0 |
| R1R2DT | 0.8 | 0 | 0 | ||
| R1R2/DT | 0.4 | 0 | 0 | ||
Number of Kanr colonies obtained in this assay with the indicated combination of RAG proteins.
Number of triple resistant (KTSr) colonies obtained.
Number of transposition (Tnp) events identified by sequencing.
Overall frequency of transposition in R1R2DT reactions, 1.7 × 10−7 Tnp per Kanr colony.
Are transposition or insertion products generated in bacteria?
Since the transposition assay requires bacterial transformation, it was necessary to test if products resembling transposition or insertion events could be formed in bacteria into which the signal end fragment and target plasmid were introduced (note that in the standard assay, DNA samples are carefully deproteinized prior to transformation into bacteria). To this end, bacterial cells were cotransformed with an artificially generated signal end fragment and target DNA and selected on KTS (see Materials and Methods). While KTSr colonies were obtained, restriction enzyme analysis of DNA isolated from these colonies showed that they were neither transposition products nor insertions (data not shown). Most of the KTSr cells harbored unaltered target DNA, and it is likely that integration of the signal end fragment into the bacterial genome enabled the growth of these cells. It is therefore unlikely that the transposition or insertion products we isolated were generated in bacteria.
Are the insertions generated by a mechanism involving transposition?
Our previous in vitro experiments with short oligonucleotides and purified RAG proteins suggested that, with certain types of targets, transposition products lacking a target site duplication can be generated (50). It was therefore possible that the insertion events we observed were generated by a mechanism involving transposition (usually with the accompanying deletion of target sequences). To test this possibility, in vivo transposition reactions were performed with RAG1-S723C, a mutant RAG1 protein that is severely defective for transposition but which retains the ability to perform DNA cleavage at RSSs (51). If insertions involve a transposition-like step, this mutant should be severely defective in their generation. This was not the case. In two independent assays, full-length, untagged RAG1-S723C together with RAG2C(HM) yielded a total of 11 KTSr colonies, 7 of which had a restriction enzyme digestion pattern consistent with transposition or insertion. Sequence analysis of these seven revealed two simple and five complex insertions and, as expected, no transposition events. As with wild-type RAG1, the simple insertion plasmids generated by S723C had deletions of target sequences at the site of integration of the signal end fragment. Therefore, a transposition-defective mutant of RAG1 is competent to generate insertions.
Because the detection of insertions required expression of the RAG proteins, we then considered the possibility that the function of the RAG proteins was to cleave the donor to generate the signal end fragment, after which general cellular activities could generate the observed insertion events. As a first test of this idea, an artificially generated signal end fragment was cotransfected into 293T cells with the target plasmid in the presence or absence of the RAG expression vectors (two assays of each type were performed). In all four assays, KTSr colonies were obtained, and a subset was screened for transposition/insertion events as described above (see Table S2 in the supplemental material). No transposition events were observed. However, both simple and complex insertions of the signal end fragment were observed in all four reactions. Therefore, the integration of signal end DNA into the target to generate insertion events can occur in the absence of the RAG proteins.
Overall, these results indicate that the insertions observed in the episomal transposition assay are unlikely to be generated by RAG-mediated transposition; rather, they may be generated by the illegitimate recombination of RAG-generated signal end fragments with the target DNA. Furthermore, the role of the RAG proteins in generating insertions may be confined to the DNA cleavage event that liberates the signal end fragment from the donor vector.
Genomic transposition assay.
A natural extension of the plasmid transposition assay was to use the genome as the target for transposition/insertion by the RAG proteins. For this assay, a new donor plasmid (pBSK-12puro23-2DT) was constructed, harboring a puror gene flanked by 12- and 23-RSSs (Fig. 4A). In this case, transposition of the signal end fragment into the genome by the RAG proteins allows cells to survive in the presence of puromycin. Two copies of a DT expression cassette were engineered into the donor plasmid to select against its random integration into the genome (a strategy used previously for gene targeting by homologous recombination [16]). We reasoned that random integration events would be unlikely to disrupt expression of both DT cassettes and that integration of even one intact DT cassette would be sufficient to kill the host cell.
RAG expression plasmids and pBSK-12puro23-2DT were cotransfected into 293T cells, and 150 puror single-cell colonies were isolated. Genomic DNA prepared from the puror colonies was screened with three different PCRs (see Materials and Methods for details). One reaction (primers C2 and E3) (Fig. 4A) confirmed the presence of the puror gene, whereas the other two reactions (primers T7-2 and IE or T3-2 and IC) tested for the presence of intact donor DNA flanking the signal end fragment. Transposition/insertion should separate the signal end fragment from vector sequences at both flanks, leading to a negative result in both of the latter two reactions, whereas a positive signal in one or both of these reactions indicated that the genome had acquired the puror gene by random integration of the plasmid and not by transposition. These PCR analyses revealed that 121 of the puror colonies had vector sequences flanking the signal end fragment, and such colonies were not analyzed further.
The remaining 29 colonies were negative in both of the vector flank PCR assays and hence were candidates for harboring a transposition (or insertion) product. DC-PCR was then performed with DNA from these colonies to characterize the structure of vector sequences containing puror and to isolate the flanking genomic DNA. Genomic DNA was digested with one of several different restriction enzymes, ligated under conditions favoring intramolecular ligation, and amplified with various pairs of PCR primers depending on the restriction enzyme used. PCR products, where obtained, were cloned and sequenced. For each colony, we first attempted to obtain a DC-PCR product containing both flanks by cleaving with one of several restriction enzymes that did not cut within the signal end fragment and using primers IE and IC. If that was unsuccessful, we then attempted to identify the two flanks independently in two different DC-PCRs (for example, using a restriction enzyme that cleaved near one end of the signal end fragment and using primers IC and C2 or IE and E3).
This yielded information about the sequences at one or both flanks of the signal end fragment for 24 of the 29 colonies. Of these 24 colonies, 15 contained vector sequences attached to the RSSs on one or both sides; it is not clear why these colonies were negative in the donor flank PCR assays described above. Of the remaining nine colonies, six were found to have human genomic sequences immediately flanking an intact RSS on one flank, but information about the other flank could not be obtained. In the final three colonies (numbers 74, 77, and 105), sequence information for both flanks was obtained, revealing two unique events (numbers 74 and 77 were identical and presumably clonally related). For these colonies, the two flanks were obtained from a single DC-PCR by using primers IE and IC and restriction enzyme AseI (number 105) or XbaI (number 74/77).
These two informative puror colonies, numbers 105 and 74/77, were of special interest due to the nature of the insertion of the signal end fragment. In neither case was a target site duplication found. In clone 105, the two ends of the signal end fragment were joined to two different chromosomes (chromosomes 2 and 16, at position 4948093, under accession no. NT_005058.14, and at position 19424825, under accession no. NT_010498.15, respectively), indicating that the signal end fragment is located at a chromosomal translocation breakpoint (Fig. 4B). In clone 74/77, the signal end fragment was inserted into chromosome 17. One end of the signal end fragment was joined to a site within the colony-stimulating factor 3 gene (position 1898098, accession no. NT_010755) on this chromosome. The other end mapped within an AluSp repeat sequence (position 7953577, accession no. NT_010641) located approximately 36 Mbp away, suggesting that insertion of the signal end fragment was accompanied by a large deletion of chromosomal DNA (Fig. 4C). Based on these findings, we conclude that acquisition of the signal end fragment in these colonies was the result of insertion and not transposition. The association of signal end insertion with a chromosomal translocation and a large chromosomal deletion indicates that signal end fragments can participate in processes that contribute to genomic instability.
DISCUSSION
We have established an assay that, for first time, provides a systematic means of generating and characterizing RAG-mediated transposition events in mammalian cells. This assay involves the jumping of a fragment of DNA, flanked by RSSs, from one episome (the donor) to another (the target) in transfected 293T cells and takes advantage of both positive (resistance to KAN and TET) and negative (sensitivity to streptomycin) selections in bacteria to eliminate the recovery of nonrecombinant substrate plasmids. Other factors that contribute to the success of the assay are the high transfectability of the host 293T cells and the high-level expression of the RAG proteins that can be achieved in them. The triple KTS selection, combined with restriction digestion that cuts the donor plasmid, yields bacterial colonies which in almost all cases (>95%) contain a recombinant plasmid consisting of the target plasmid plus the RAG-generated signal end fragment. These recombinants are of three types: transposition events (10%), in which no sequences are lost from the target or signal end fragment and a characteristic target site duplication is observed immediately flanking the RSSs; simple insertions (51%), in which integration of the signal end fragment is almost invariably accompanied by the deletion of target sequences; and complex insertions (39%), with deletion of one or both RSSs or the inversion/duplication/deletion of plasmid sequences.
The assay has a high degree of sensitivity, able to detect one transposition event from among approximately 107 recovered target molecules (one recovered transposition product per ∼107 Kanr colonies). Despite this, only about one-third of assays yields one or more transposition products, demonstrating that even when cells harbor high levels of substrate, target, and active RAG proteins, transposition is an infrequent and inefficient process. Previous studies also found a low level of RAG-mediated transposition in human (33) and yeast (5) cells. The first study reported a transposition event in an adult human peripheral T-cell clone wherein a T-cell receptor alpha signal end fragment was found integrated into the hypoxanthine phosphoribosyltransferase (HPRT) locus (33). A second event was also described in which insertion of a T-cell receptor alpha signal end fragment was accompanied by duplication and inversion of flanking HPRT sequences. In the second study, expression of the RAG proteins in yeast cells led to the cleavage of pairs of RSSs to generate signal ends and to intramolecular transposition, but examples of intermolecular transposition were not observed. The estimated frequencies of transposition in the human and yeast studies were 1.7 × 10−7 and 1.4 ×10−8 per cell, respectively. We note that it remains unclear how active the RAG proteins are when expressed in yeast and that the human study yielded only a single, unambiguous transposition event. Our findings extend the prior studies by demonstrating that RAG-mediated transposition can be detected in a reproducible fashion in mammalian cells and that the reaction must be very tightly regulated.
A number of factors have been suggested to suppress RAG-mediated transposition in vivo, although in the absence of an assay for mammalian cells, they have been difficult to test. Perhaps the most obvious candidate is the formation of signal joints, a process thought to generate dead end products in which the two RSSs are joined together, but a recent study demonstrates that signal joints can be cleaved and the resulting signal end fragment can be transposed by the RAG proteins (36). Under certain in vitro conditions, the RAG proteins can catalyze “disintegration,” which reverses the transposition process (31). Transposition is inhibited in vitro by the ubiquitous guanine nucleotide GTP (52) and by coding end DNA remaining bound to the RAG proteins (11). Finally, several studies have reported that the C-terminal region of RAG2, absent from the core RAG2 protein used in most biochemical studies, strongly suppresses transposition in vitro (11, 47, 52). The relevance of these in vitro findings for in vivo transposition was recently called into question, however, by the finding from the Desiderio laboratory that in vivo-generated signal end complexes containing either core or full-length RAG2 transpose equally efficiently in vitro (20).
Here, we have been able to test the ability of core and full-length RAG2 to mediate transposition in vivo in mammalian cells. We find, consistent with the results of Jiang and colleagues (20), that both core and full-length RAG2 catalyze transposition in vivo. While the frequency of transposition appears to be lower with full-length RAG2 than with the core protein, this finding should be interpreted cautiously for at least two reasons. First, the number of transposition events that we were able to score was quite small. And second, as noted above, both RAG2 protein levels and steady-state signal end fragment levels are lower when the full-length protein is used (28). It remains to be determined whether there exists a quantitative difference in the transposition potential of the core and full-length RAG2 proteins. Our results, and the finding of transposition in a human T-cell clone (33), demonstrate that the full-length RAG2 protein retains some transposition activity in vivo. Our data also demonstrate, as was the case in vitro (47), that core and full-length forms of RAG1 have approximately equivalent transposition activities (P = 0.78), although, again, only a small number of events were obtained for full-length RAG1 (Table 1).
One can envision several explanations for our inability to detect transposition into the genome of transfected 293T cells. First, it is possible that some feature of chromatin renders it a poor target for RAG-mediated transposition. Second, mammalian cells may have mechanisms to sense the immediate product of transposition (a stable protein-DNA complex referred to as the strand transfer product [2]) and prevent the final steps of transposition or perhaps trigger cell death. And third, the sensitivity of the genomic transposition assay may not be adequate. This assay yielded a high frequency (∼90%) of puror clones that contained random integrations of the donor plasmid, despite the fact that the plasmid contained two negative selection cassettes. This high background of undesired events suggests that the third possibility, inadequate sensitivity, was a significant factor in our failure to detect RAG-mediated transposition into the genome. The finding of a transposition event in a human T-cell clone demonstrates that the process can occur with chromosomal DNA as target (33).
In both the episomal and genomic transposition assays, signal end insertion occurs more readily than does transposition. Our data demonstrate that such insertions are often accompanied by deletions, inversions, or other more complex rearrangements of target sequences. In the case of the two genomic insertions, we were able to characterize fully either a large deletion of many megabases or a chromosomal translocation directly associated with signal end insertion. These findings indicate that the signal ends generated by the RAG proteins can participate in processes that threaten genomic stability.
We also provide evidence that signal end insertion occurs via a pathway distinct from that of transposition; insertions are generated efficiently both by a mutant form of RAG1 that is defective for transposition and in the complete absence of the RAG proteins if an artificially generated signal end fragment is provided. Our data are consistent with the idea that the role of the RAG proteins in signal end insertion is primarily or exclusively to serve as the endonuclease that initially generates the signal end fragment. Because the RAG proteins bind tightly to signal ends and shield them from nucleases and ligases (2, 22), it is likely that insertions are generated only after release of the signal end fragment from the RAG proteins. Therefore, it appears that both full-length and core RAG proteins can release the signal end fragment, presumably before they have been properly recognized by the NHEJ repair machinery. The high levels of DNA substrate and RAG proteins present in the transiently transfected cells in our assay may contribute to this.
Based on our results, we propose two distinct mechanisms by which signal end fragments contribute to genetic instability. The first, transposition, requires the participation of the RAG proteins, while the second, insertion, occurs in a RAG-independent manner (both pathways require the RAG proteins to generate the signal end fragment in the first place). In the case of insertions, a critical initiating event is likely to be the generation of a DNA double-stranded break in the target molecule, which could occur by a number of different (RAG-independent) mechanisms. Repair of the break can then result in the capture of filler DNA (such as the signal end fragment) at the site of rejoining, a well-known phenomenon associated with NHEJ and chromosomal translocation breakpoints (40). In this case, the signal end fragment is a passive bystander in the generation of insertions. Another possible mechanism for the generation of insertions involves the RAG-mediated transposition of one signal end into the target followed by the illegitimate recombination to join the other signal end to the target. Transposition of one signal end is a well-characterized feature of RAG-mediated transposition in vitro (19). While it is difficult to test this idea, one prediction of such a mechanism is that the target sequences immediately adjacent to the RSSs in the insertion products might be enriched in GC nucleotides. To investigate this, we analyzed the GC content of the 5 bp adjacent to 12- and 23-RSSs in all 115 simple insertion events (see Table S3 in the supplemental material). In contrast to transposition events, no GC bias was observed in the simple insertions (53% to 54% GC in the simple insertion flanks compared to 53% GC in the target plasmid). Therefore, while we cannot rule out a contribution of one-ended transposition to the generation of insertions, we have no evidence in favor of such a process.
While the RAG proteins have been linked to a number of aberrant chromosomal rearrangements and translocations (25, 34, 38), neither transposition nor the signal end fragment has been directly implicated in any of them, with the exception of the two events involving the HPRT locus noted above. Interestingly, some of the complex rearrangements that we found associated with signal end insertion closely resembled the HPRT locus signal end insertion event that was accompanied by sequence inversion/duplication (33). Our data demonstrate that RAG-mediated transposition is a reproducible phenomenon in mammalian cells and highlight the association of chromosomal deletions and translocations with signal end fragment insertion.
Supplementary Material
Acknowledgments
We thank N. Grindley for the plasmid containing the rpsL and sacB genes, U. Grawunder and M. Lieber for the diphtheria toxin expression cassette, S. Fugmann for early contributions to the design of the assays, O. Yarborough and M. Miller for experimental contributions, members of the Schatz laboratory for helpful discussions, and anonymous reviewers for insightful comments.
This work was supported by grant AI32524 from the National Institutes of Health. M.C. is a recipient of the Anna Fuller Fund Postdoctoral Fellowship, and D.G.S. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Agrawal, A., Q. M. Eastman, and D. G. Schatz. 1998. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature 394:744-751. [DOI] [PubMed] [Google Scholar]
- 2.Agrawal, A., and D. G. Schatz. 1997. RAG1 and RAG2 form a stable postcleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell 89:43-53. [DOI] [PubMed] [Google Scholar]
- 3.Akamatsu, Y., R. Monroe, D. D. Dudley, S. K. Elkin, F. Gartner, S. R. Talukder, Y. Takahama, F. W. Alt, C. H. Bassing, and M. A. Oettinger. 2003. Deletion of the RAG2 C terminus leads to impaired lymphoid development in mice. Proc. Natl. Acad. Sci. USA 100:1209-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berg, D., and M. Howe (ed.). 1989. Mobile DNA. American Society for Microbiology, Washington, D.C.
- 5.Clatworthy, A. E., M. A. Valencia, J. E. Haber, and M. A. Oettinger. 2003. V(D)J recombination and RAG-mediated transposition in yeast. Mol. Cell 12:489-499. [DOI] [PubMed] [Google Scholar]
- 6.Craig, N. L., R. Craigie, M. Gellert, and A. M. Lambowitz (ed.). 2002. Mobile DNA II. American Society for Microbiology, Washington, D.C.
- 7.Cuomo, C. A., and M. A. Oettinger. 1994. Analysis of regions of RAG-2 important for V(D)J recombination. Nucleic Acids Res. 22:1810-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curcio, M. J., and K. M. Derbyshire. 2003. The outs and ins of transposition: from mu to kangaroo. Nat. Rev. Mol. Cell Biol. 4:865-877. [DOI] [PubMed] [Google Scholar]
- 9.Dudley, D. D., J. Sekiguchi, C. Zhu, M. J. Sadofsky, S. Whitlow, J. DeVido, R. J. Monroe, C. H. Bassing, and F. W. Alt. 2003. Impaired V(D)J recombination and lymphocyte development in core RAG1-expressing mice. J. Exp. Med. 198:1439-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elkin, S. K., D. Ivanov, M. Ewalt, C. G. Ferguson, S. G. Hyberts, Z. Y. Sun, G. D. Prestwich, J. Yuan, G. Wagner, M. A. Oettinger, and O. P. Gozani. 2005. A PHD finger motif in the C terminus of RAG2 modulates recombination activity. J. Biol. Chem. 280:28701-28710. [DOI] [PubMed] [Google Scholar]
- 11.Elkin, S. K., A. G. Matthews, and M. A. Oettinger. 2003. The C-terminal portion of RAG2 protects against transposition in vitro. EMBO J. 22:1931-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fugmann, S. D., A. I. Lee, P. E. Shockett, I. J. Villey, and D. G. Schatz. 2000. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu. Rev. Immunol. 18:495-527. [DOI] [PubMed] [Google Scholar]
- 13.Fugmann, S. D., I. J. Villey, L. M. Ptaszek, and D. G. Schatz. 2000. Identification of two catalytic residues in RAG1 that define a single active site within the RAG1/RAG2 protein complex. Mol. Cell 5:97-107. [DOI] [PubMed] [Google Scholar]
- 14.Gellert, M. 2002. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu. Rev. Biochem. 71:101-132. [DOI] [PubMed] [Google Scholar]
- 15.Goff, S. A., D. Ricke, T. H. Lan, G. Presting, R. Wang, M. Dunn, J. Glazebrook, A. Sessions, P. Oeller, H. Varma, D. Hadley, D. Hutchison, C. Martin, F. Katagiri, B. M. Lange, T. Moughamer, Y. Xia, P. Budworth, J. Zhong, T. Miguel, U. Paszkowski, S. Zhang, M. Colbert, W. L. Sun, L. Chen, B. Cooper, S. Park, T. C. Wood, L. Mao, P. Quail, R. Wing, R. Dean, Y. Yu, A. Zharkikh, R. Shen, S. Sahasrabudhe, A. Thomas, R. Cannings, A. Gutin, D. Pruss, J. Reid, S. Tavtigian, J. Mitchell, G. Eldredge, T. Scholl, R. M. Miller, S. Bhatnagar, N. Adey, T. Rubano, N. Tusneem, R. Robinson, J. Feldhaus, T. Macalma, A. Oliphant, and S. Briggs. 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296:92-100. [DOI] [PubMed] [Google Scholar]
- 16.Grawunder, U., D. Zimmer, S. Fugmann, K. Schwarz, and M. R. Lieber. 1998. DNA ligase IV is essential for V(D)J recombination and DNA double-strand break repair in human precursor lymphocytes. Mol. Cell 2:477-484. [DOI] [PubMed] [Google Scholar]
- 17.Hesse, J. E., M. R. Lieber, M. Gellert, and K. Mizuuchi. 1987. Extrachromosomal DNA substrates in pre-B cells undergo inversion or deletion at immunoglobulin V-(D)-J joining signals. Cell 49:775-783. [DOI] [PubMed] [Google Scholar]
- 18.Hiom, K., and M. Gellert. 1998. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol. Cell 1:1011-1019. [DOI] [PubMed] [Google Scholar]
- 19.Hiom, K., M. Melek, and M. Gellert. 1998. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell 94:463-470. [DOI] [PubMed] [Google Scholar]
- 20.Jiang, H., A. E. Ross, and S. Desiderio. 2004. Cell cycle-dependent accumulation in vivo of transposition-competent complexes between recombination signal ends and full-length RAG proteins. J. Biol. Chem. 279:8478-8486. [DOI] [PubMed] [Google Scholar]
- 21.Jones, J. M., and M. Gellert. 2003. Autoubiquitylation of the V(D)J recombinase protein RAG1. Proc. Natl. Acad. Sci. USA 100:15446-15451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones, J. M., and M. Gellert. 2001. Intermediates in V(D)J recombination: a stable RAG1/2 complex sequesters cleaved RSS ends. Proc. Natl. Acad. Sci. USA 98:12926-12931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kingston, R. E. 1997. Calcium phosphate transfection, p. 9.0.1-9.0.5. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 2. John Wiley & Sons, Inc., New York, N.Y. [Google Scholar]
- 24.Kirch, S. A., G. A. Rathbun, and M. A. Oettinger. 1998. Dual role of RAG2 in V(D)J recombination: catalysis and regulation of ordered Ig gene assembly. EMBO J. 17:4881-4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuppers, R., and R. Dalla-Favera. 2001. Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene 20:5580-5594. [DOI] [PubMed] [Google Scholar]
- 26.Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody, J. Baldwin, K. Devon, K. Dewar, M. Doyle, W. FitzHugh, R. Funke, D. Gage, K. Harris, A. Heaford, J. Howland, L. Kann, J. Lehoczky, R. LeVine, P. McEwan, K. McKernan, J. Meldrim, J. P. Mesirov, C. Miranda, W. Morris, J. Naylor, C. Raymond, M. Rosetti, R. Santos, A. Sheridan, C. Sougnez, N. Stange-Thomann, N. Stojanovic, A. Subramanian, D. Wyman, J. Rogers, J. Sulston, R. Ainscough, S. Beck, D. Bentley, J. Burton, C. Clee, N. Carter, A. Coulson, R. Deadman, P. Deloukas, A. Dunham, I. Dunham, R. Durbin, L. French, D. Grafham, S. Gregory, T. Hubbard, S. Humphray, A. Hunt, M. Jones, C. Lloyd, A. McMurray, L. Matthews, S. Mercer, S. Milne, J. C. Mullikin, A. Mungall, R. Plumb, M. Ross, R. Shownkeen, S. Sims, R. H. Waterston, R. K. Wilson, L. W. Hillier, J. D. McPherson, M. A. Marra, E. R. Mardis, L. A. Fulton, A. T. Chinwalla, K. H. Pepin, W. R. Gish, S. L. Chissoe, M. C. Wendl, K. D. Delehaunty, T. L. Miner, A. Delehaunty, J. B. Kramer, L. L. Cook, R. S. Fulton, D. L. Johnson, P. J. Minx, S. W. Clifton, T. Hawkins, E. Branscomb, P. Predki, P. Richardson, S. Wenning, T. Slezak, N. Doggett, J. F. Cheng, A. Olsen, S. Lucas, C. Elkin, E. Uberbacher, M. Frazier, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 27.Li, Z., D. I. Dordai, J. Lee, and S. Desiderio. 1996. A conserved degradation signal regulates RAG-2 accumulation during cell division and links V(D)J recombination to the cell cycle. Immunity 5:575-589. [DOI] [PubMed] [Google Scholar]
- 28.Liang, H. E., L. Y. Hsu, D. Cado, L. G. Cowell, G. Kelsoe, and M. S. Schlissel. 2002. The “dispensable” portion of RAG2 is necessary for efficient V-to-DJ rearrangement during B and T cell development. Immunity 17:639-651. [DOI] [PubMed] [Google Scholar]
- 29.Mayer, B. J., H. Hirai, and R. Sakai. 1995. Evidence that SH2 domains promote processive phosphorylation by protein-tyrosine kinases. Curr. Biol. 5:296-305. [DOI] [PubMed] [Google Scholar]
- 30.McBlane, J. F., D. C. van Gent, D. A. Ramsden, C. Romeo, C. A. Cuomo, M. Gellert, and M. A. Oettinger. 1995. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell 83:387-395. [DOI] [PubMed] [Google Scholar]
- 31.Melek, M., and M. Gellert. 2000. RAG1/2-mediated resolution of transposition intermediates: two pathways and possible consequences. Cell 101:625-633. [DOI] [PubMed] [Google Scholar]
- 32.Mendiola, M. V., I. Bernales, and F. de la Cruz. 1994. Differential roles of the transposon termini in IS91 transposition. Proc. Natl. Acad. Sci. USA 91:1922-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Messier, T. L., J. P. O'Neill, S. M. Hou, J. A. Nicklas, and B. A. Finette. 2003. In vivo transposition mediated by V(D)J recombinase in human T lymphocytes. EMBO J. 22:1381-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mills, K. D., D. O. Ferguson, and F. W. Alt. 2003. The role of DNA breaks in genomic instability and tumorigenesis. Immunol. Rev. 194:77-95. [DOI] [PubMed] [Google Scholar]
- 35.Moran, J. V., R. J. DeBerardinis, and H. H. Kazazian, Jr. 1999. Exon shuffling by L1 retrotransposition. Science 283:1530-1534. [DOI] [PubMed] [Google Scholar]
- 36.Neiditch, M. B., G. S. Lee, L. E. Huye, V. L. Brandt, and D. B. Roth. 2002. The V(D)J recombinase efficiently cleaves and transposes signal joints. Mol. Cell 9:871-878. [DOI] [PubMed] [Google Scholar]
- 37.Oettinger, M. A., D. G. Schatz, C. Gorka, and D. Baltimore. 1990. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science 248:1517-1523. [DOI] [PubMed] [Google Scholar]
- 38.Raghavan, S. C., P. C. Swanson, X. Wu, C. L. Hsieh, and M. R. Lieber. 2004. A non-B-DNA structure at the Bcl-2 major breakpoint region is cleaved by the RAG complex. Nature 428:88-93. [DOI] [PubMed] [Google Scholar]
- 39.Roman, C. A., S. R. Cherry, and D. Baltimore. 1997. Complementation of V(D)J recombination deficiency in RAG-1(−/−) B cells reveals a requirement for novel elements in the N-terminus of RAG-1. Immunity 7:13-24. [DOI] [PubMed] [Google Scholar]
- 40.Roth, D. B., X. B. Chang, and J. H. Wilson. 1989. Comparison of filler DNA at immune, nonimmune, and oncogenic rearrangements suggests multiple mechanisms of formation. Mol. Cell. Biol. 9:3049-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sadofsky, M. J. 2004. Recombination-activating gene proteins: more regulation, please. Immunol. Rev. 200:83-89. [DOI] [PubMed] [Google Scholar]
- 42.Sadofsky, M. J., J. E. Hesse, and M. Gellert. 1994. Definition of a core region of RAG-2 that is functional in V(D)J recombination. Nucleic Acids Res. 22:1805-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sadofsky, M. J., J. E. Hesse, J. F. McBlane, and M. Gellert. 1993. Expression and V(D)J recombination activity of mutated RAG-1 proteins. Nucleic Acids Res. 21:5644-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schatz, D. G., M. A. Oettinger, and D. Baltimore. 1989. The V(D)J recombination activating gene, RAG-1. Cell 59:1035-1048. [DOI] [PubMed] [Google Scholar]
- 45.Silver, D. P., E. Spanopoulou, R. C. Mulligan, and D. Baltimore. 1993. Dispensable sequence motifs in the RAG-1 and RAG-2 genes for plasmid V(D)J recombination. Proc. Natl. Acad. Sci. USA 90:6100-6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steen, S. B., J. O. Han, C. Mundy, M. A. Oettinger, and D. B. Roth. 1999. Roles of the “dispensable” portions of RAG-1 and RAG-2 in V(D)J recombination. Mol. Cell. Biol. 19:3010-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swanson, P. C., D. Volkmer, and L. Wang. 2004. Full-length RAG-2, and not full-length RAG-1, specifically suppresses RAG-mediated transposition, but not hybrid joint formation or disintegration. J. Biol. Chem. 279:4034-4044. [DOI] [PubMed] [Google Scholar]
- 48.Talukder, S. R., D. D. Dudley, F. W. Alt, Y. Takahama, and Y. Akamatsu. 2004. Increased frequency of aberrant V(D)J recombination products in core RAG-expressing mice. Nucleic Acids Res. 32:4539-4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tonegawa, S. 1983. Somatic generation of antibody diversity. Nature 302:575-581. [DOI] [PubMed] [Google Scholar]
- 50.Tsai, C. L., M. Chatterji, and D. G. Schatz. 2003. DNA mismatches and GC-rich motifs target transposition by the RAG1/RAG2 transposase. Nucleic Acids Res. 31:6180-6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsai, C. L., A. H. Drejer, and D. G. Schatz. 2002. Evidence of a critical architectural function for the RAG proteins in end processing, protection, and joining in V(D)J recombination. Genes Dev. 16:1934-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsai, C. L., and D. G. Schatz. 2003. Regulation of RAG1/RAG2-mediated transposition by GTP and the C-terminal region of RAG2. EMBO J. 22:1922-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Gent, D. C., K. Hiom, T. T. Paul, and M. Gellert. 1997. Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J. 16:2665-2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Waterston, R. H., K. Lindblad-Toh, E. Birney, J. Rogers, J. F. Abril, P. Agarwal, R. Agarwala, R. Ainscough, M. Alexandersson, P. An, S. E. Antonarakis, J. Attwood, R. Baertsch, J. Bailey, K. Barlow, S. Beck, E. Berry, B. Birren, T. Bloom, P. Bork, M. Botcherby, N. Bray, M. R. Brent, D. G. Brown, S. D. Brown, C. Bult, J. Burton, J. Butler, R. D. Campbell, P. Carninci, S. Cawley, F. Chiaromonte, A. T. Chinwalla, D. M. Church, M. Clamp, C. Clee, F. S. Collins, L. L. Cook, R. R. Copley, A. Coulson, O. Couronne, J. Cuff, V. Curwen, T. Cutts, M. Daly, R. David, J. Davies, K. D. Delehaunty, J. Deri, E. T. Dermitzakis, C. Dewey, N. J. Dickens, M. Diekhans, S. Dodge, I. Dubchak, D. M. Dunn, S. R. Eddy, L. Elnitski, R. D. Emes, P. Eswara, E. Eyras, A. Felsenfeld, G. A. Fewell, P. Flicek, K. Foley, W. N. Frankel, L. A. Fulton, R. S. Fulton, T. S. Furey, D. Gage, R. A. Gibbs, G. Glusman, S. Gnerre, N. Goldman, L. Goodstadt, D. Grafham, T. A. Graves, E. D. Green, S. Gregory, R. Guigo, M. Guyer, R. C. Hardison, D. Haussler, Y. Hayashizaki, L. W. Hillier, A. Hinrichs, W. Hlavina, T. Holzer, F. Hsu, A. Hua, T. Hubbard, A. Hunt, I. Jackson, D. B. Jaffe, L. S. Johnson, M. Jones, T. A. Jones, A. Joy, M. Kamal, E. K. Karlsson, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520-562. [DOI] [PubMed] [Google Scholar]
- 55.West, K. L., N. C. Singha, P. De Ioannes, L. Lacomis, H. Erdjument-Bromage, P. Tempst, and P. Cortes. 2005. A direct interaction between the RAG2 C terminus and the core histones is required for efficient V(D)J recombination. Immunity 23:203-212. [DOI] [PubMed] [Google Scholar]
- 56.Yu, J., S. Hu, J. Wang, G. K. Wong, S. Li, B. Liu, Y. Deng, L. Dai, Y. Zhou, X. Zhang, M. Cao, J. Liu, J. Sun, J. Tang, Y. Chen, X. Huang, W. Lin, C. Ye, W. Tong, L. Cong, J. Geng, Y. Han, L. Li, W. Li, G. Hu, J. Li, Z. Liu, Q. Qi, T. Li, X. Wang, H. Lu, T. Wu, M. Zhu, P. Ni, H. Han, W. Dong, X. Ren, X. Feng, P. Cui, X. Li, H. Wang, X. Xu, W. Zhai, Z. Xu, J. Zhang, S. He, J. Xu, K. Zhang, X. Zheng, J. Dong, W. Zeng, L. Tao, J. Ye, J. Tan, X. Chen, J. He, D. Liu, W. Tian, C. Tian, H. Xia, Q. Bao, G. Li, H. Gao, T. Cao, W. Zhao, P. Li, W. Chen, Y. Zhang, J. Hu, S. Liu, J. Yang, G. Zhang, Y. Xiong, Z. Li, L. Mao, C. Zhou, Z. Zhu, R. Chen, B. Hao, W. Zheng, S. Chen, W. Guo, M. Tao, L. Zhu, L. Yuan, and H. Yang. 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296:79-92. [DOI] [PubMed] [Google Scholar]
- 57.Yurchenko, V., Z. Xue, and M. Sadofsky. 2003. The RAG1 N-terminal domain is an E3 ubiquitin ligase. Genes Dev. 17:581-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.