Abstract
The bZIP transcription factor Nrf2 controls a genetic program that protects cells from oxidative damage and maintains cellular redox homeostasis. Keap1, a BTB-Kelch protein, is the major upstream regulator of Nrf2. Keap1 functions as a substrate adaptor protein for a Cul3-dependent E3 ubiquitin ligase complex to repress steady-state levels of Nrf2 and Nrf2-dependent transcription. Cullin-dependent ubiquitin ligase complexes have been proposed to undergo dynamic cycles of assembly and disassembly that enable substrate adaptor exchange or recycling. In this report, we have characterized the importance of substrate adaptor recycling for regulation of Keap1-mediated repression of Nrf2. Association of Keap1 with Cul3 was decreased by ectopic expression of CAND1 and was increased by small interfering RNA (siRNA)-mediated knockdown of CAND1. However, both ectopic overexpression and siRNA-mediated knockdown of CAND1 decreased the ability of Keap1 to target Nrf2 for ubiquitin-dependent degradation, resulting in stabilization of Nrf2 and activation of Nrf2-dependent gene expression. Neddylation of Cul3 on Lys 712 is required for Keap1-dependent ubiquitination of Nrf2 in vivo. However, the K712R mutant Cul3 molecule, which is not neddylated, can still assemble with Keap1 into a functional ubiquitin ligase complex in vitro. These results provide support for a model in which substrate adaptor recycling is required for efficient substrate ubiquitination by cullin-dependent E3 ubiquitin ligase complexes.
Exposure of cells to reactive molecules, including reactive oxygen species and other chemically reactive compounds, can damage cellular macromolecules and compromise cellular functions (1, 2, 16, 20, 47). The coordinated induction of phase 2 genes provides an efficient mechanism for mammalian cells to neutralize reactive molecules, eliminate damaged macromolecules, and restore cellular redox homeostasis (8, 22, 26, 33). Transcription of phase 2 genes is controlled in large part by the Nrf2 transcription factor (22, 33). The ability of Nrf2 to accumulate in the nucleus and bind DNA appears to be the rate-limiting step that controls expression of phase 2 genes.
Nrf2 is a member of the CNC subfamily of bZIP transcription factors (32). Nrf2 contains an N-terminal regulatory domain (Neh2), a central transactivation domain, and a C-terminal bZIP domain that is responsible for both nuclear localization and DNA binding. The major upstream regulator of Nrf2 is a BTB-Kelch protein termed Keap1, which binds the Neh2 regulatory domain of Nrf2 via loops that extend out from the bottom face of the Kelch β-propeller domain (21, 27). Keap1 was initially regarded as a protein that sequestered Nrf2 in the cytoplasm (21). Subsequent work has revealed that Keap1 functions as a substrate adaptor protein for a Cul3-dependent E3 ubiquitin ligase that targets lysine residues within the Neh2 domain for ubiquitin conjugation (7, 14, 25, 26, 43). Ubiquitin conjugation onto specific N-terminal lysine residues marks Nrf2 for degradation by the 26S proteosome, such that Nrf2 is maintained at low steady-state levels under basal conditions (43). However, upon exposure to oxidative stress and reactive molecules, Nrf2 is no longer targeted for ubiquitin-dependent degradation (42, 43). Instead, Nrf2 accumulates in the nucleus and activates expression of phase 2 genes.
Cullin-dependent ubiquitin ligases are assembled around a common core complex consisting of one of six cullin proteins and a common RING finger protein, Rbx1 (also known as Roc1) (35). This core complex is used by a large number of substrate adaptor proteins that bring in specific substrates. Substrate adaptor proteins are modular in nature, having one domain that binds to the cullin protein and another domain that binds to a small number of specific substrate proteins. These two modules can be encoded by two different polypeptides, as in the case of the Skp1-F-box protein complexes that are substrate adaptors for Cul1. Alternatively, as exemplified by Keap1, a substrate adaptor for Cul3, these two domains can be encoded by a single polypeptide.
A model that describes how cullin-dependent ubiquitin ligases may be regulated by cycles of assembly and disassembly has been proposed (5). A central feature of this model is that the cullin-Rbx1 core complex cycles between active and inactive states. The active complex contains a substrate adaptor protein with its bound substrate docked at the N terminus of the cullin protein. In this active complex, the cullin protein is modified by Nedd8 conjugation on a conserved lysine residue (Lys 712 in Cul3). A ubiquitin-loaded E2 protein is recruited into the complex via protein-protein interactions with the Rbx1 subunit (9). The conjugated Nedd8 polypeptide may also facilitate E2 recruitment (24). The model proposes that the active complex is converted to an inactive complex in two steps. The first step is removal of the Nedd8 protein from the cullin protein by one or more deneddylases, such as the CSN5 subunit of the COP9 signalosome (CSN) (6, 15, 29, 40). The second step is the association of a protein known as CAND1 (also termed TIP120A) with the deneddylated cullin protein (17, 28, 45). CAND1 binds to both N-terminal and C-terminal sequences in Cul1 and blocks binding of the substrate adaptor protein (17, 19, 28, 30, 45). Subsequent conjugation of Nedd8 onto the cullin subunit by Ubc12, a Nedd8-specific E2 enzyme (5), is proposed to decrease the affinity of CAND1 for the cullin protein, enabling another substrate adaptor protein (presumably with its bound substrate) to displace CAND1 and initiate another cycle of substrate ubiquitination (5, 35). A recent crystal structure of the CAND1-Cul1 complex, which showed that Lys 720 in Cul1 is located at the CAND1:Cul1 interface, has provided support for the notion that neddylation of Cul1 and binding of CAND1 are antagonistic (17). However, it is not clear if CAND1 is a global regulator of all cullin-dependent ubiquitin ligase complexes (28, 30, 45).
Regulation of cullin-dependent ubiquitin ligases by cycles of assembly and disassembly provides an attractive paradigm for considering how substrate adaptor proteins target their specific substrates for ubiquitin-dependent degradation. In the case of Keap1, we have proposed that cycles of assembly and disassembly of the Keap1-Cul3-Rbx1 complex allow Keap1 to act in a catalytic manner to efficiently target Nrf2 for degradation (43). To test this hypothesis, we have examined the role of CAND1 in Keap1-dependent ubiquitination of Nrf2. Our results indicate that CAND1-dependent substrate adaptor recycling is required for efficient repression of Nrf2 by Keap1 and support the notion that CAND1 is a global regulator of cullin-dependent ubiquitin ligases.
MATERIALS AND METHODS
Construction of recombinant DNA molecules.
Plasmids expressing wild-type Keap1, chitin-binding domain (CBD)-tagged Keap1, Nrf2, wild-type Cul3, CBD-tagged gigaxonin (GAN), and sarcosin proteins have been previously described (43, 44). Mutant Cul3 cDNAs were generated by site-specific mutagenesis with standard overlap extension techniques. The myc-Rbx1 expression vector was a gift from Joan Conaway (23). The Flag-CAND1 expression vector was a gift from Cecile M. Pickart (41). All genes used in this study were sequenced in the context of the expression vectors used for the experiments.
Cell culture, transfections, and reporter gene assays.
COS1, MDA-MB-231, and HeLa cells were purchased from ATCC. CHO-K1 cells were obtained from Toshihiko Ezashi. ts41 cells were obtained from Chuck Sherr. Cells were maintained in Dulbecco's modified Eagle's medium, Eagle's minimal essential medium, or Ham's F-12 medium in the presence of 10% fetal bovine serum. Transfections of plasmid DNAs were performed with Lipofectamine Plus (Gibco BRL) according to the manufacturer's instructions. Transfections of small interfering RNA (siRNA) nucleotides were performed with Oligofectamine (Invitrogen) according to the manufacturer's instructions. All siRNA nucleotides used in this study were obtained from Dharmacon as purified and annealed duplexes. The antioxidant response element (ARE) TATA-Inr luciferase reporter plasmid pARE-Luc and a control plasmid encoding Renilla luciferase for transfection efficiency have been previously described (42). Reporter gene assays were performed using a Promega dual-light assay system as previously described (42).
Antibodies, immunoprecipitation, and immunoblot analysis.
The anti-Keap1 antibody has been described previously (42). Antibodies against Nrf2 (Santa Cruz), CAND1 (Santa Cruz), tubulin (Santa Cruz), chitin-binding domain (New England Biolabs), the Flag epitope (Sigma), the myc epitope (Santa Cruz), and the hemagglutinin (HA) epitope (Covance) were purchased from commercial sources.
For detection of protein expression in total cell lysates, cells were lysed in sample buffer (50 mM Tris-HCl [pH 6.8], 2% sodium dodecyl sulfate [SDS], 10% glycerol, 100 mM dithiothreitol [DTT], 0.1% bromophenol blue) at 24 to 48 h posttransfection. For immunoprecipitation assays, cell extracts were prepared in lysis buffer (50 mM HEPES [pH 7.9], 250 mM NaCl, 5 mM EDTA, 0.1% Triton X-100) containing 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail (Sigma). In some cases, 10 mM N-ethylmaleimide (NEM) was included in the lysis solution to inactivate ubiquitin and Nedd8 hydrolase activities. Soluble cell lysates were incubated with 2 μg of affinity-purified antibodies for 2 h at 4°C, followed by incubation at 4°C with protein A-agarose beads (Sigma) for 2 h. Unbound proteins were removed by washing four times with lysis buffer. The immunoprecipitated proteins were eluted in sample buffer by boiling for 5 min, electrophoresed through SDS-polyacrylamide gels, transferred to nitrocellulose membranes, and subjected to immunoblot analysis.
In vivo ubiquitination assay.
For detection of ubiquitinated Nrf2 proteins in vivo, cells were transfected with expression vectors for HA-tagged ubiquitin, Gal4-Neh2, and the indicated proteins. Cells were lysed by boiling in a buffer containing 2% SDS, 10 mM NEM, 150 mM NaCl, and 10 mM Tris-HCl [pH 8.0]. This rapid lysis procedure inactivates cellular ubiquitin hydrolases and therefore preserves ubiquitin-substrate protein conjugates present in cells prior to lysis. Protein-protein interactions, including association of Nrf2 with Keap1, are also disrupted by this lysis procedure. For immunoprecipitation, these lysates were diluted with 4 volumes of a buffer containing 150 mM NaCl, 10 mM Tris-HCl [pH 8.0], 1 mM DTT, and 1% Triton X-100. The diluted lysates were precleared with protein A-agarose beads (Sigma) and incubated with 2 μg of anti-Gal4 antibodies. HA-ubiquitin conjugates in immunoprecipitated Gal4-Neh2 proteins were analyzed by immunoblot analysis with antibodies against the HA epitope.
In vitro ubiquitination assay.
For detection of ubiquitinated Nrf2 and Keap1 proteins in vitro, cells were transfected with expression vectors for HA-Nrf2, Keap1-CBD, myc-Rbx1, and the wild-type or mutant HA-Cul3 proteins. Cells were lysed in buffer B (15 mM Tris-HCl [pH 7.5], 500 mM NaCl, 0.25% NP-40) containing 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail (Sigma). The lysates were precleared with protein A-agarose beads prior to incubation with chitin beads (New England Biolabs) for 4 h at 4°C. Chitin beads were washed twice with buffer B, twice with buffer A (25 mM Tris-HCl [pH 7.5], 10% [vol/vol] glycerol, 1 mM EDTA, 0.01% NP-40, and 100 mM NaCl), and twice with reaction buffer (50 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 2 mM NaF, 0.6 mM DTT). The pellets were incubated with ubiquitin (300 pmol), E1 (2 pmol), E2-UbcH5a (10 pmol), and ATP (2 mM) in 1× reaction buffer in a total volume of 30 μl for 1 h at 37°C. Ubiquitin, E1, and E2-UbcH5a were purchased from Boston Biochem. The chitin beads were pelleted by centrifugation (3,000 × g); resuspended in 2% SDS, 150 mM NaCl, 10 mM Tris-HCl (pH 8.0), and 1 mM DTT; and boiled for 5 min to release bound proteins, inactivate any contaminating ubiquitin hydrolases, and disrupt protein-protein interactions. The supernatant was diluted fivefold with buffer lacking SDS prior to immunoprecipitation with anti-Nrf2 or anti-Keap1 antibodies. Immunoprecipitated proteins were subjected to immunoblot analysis with antiubiquitin antibodies (Sigma).
RESULTS
CAND1 competes with Keap1 for binding to Cul3.
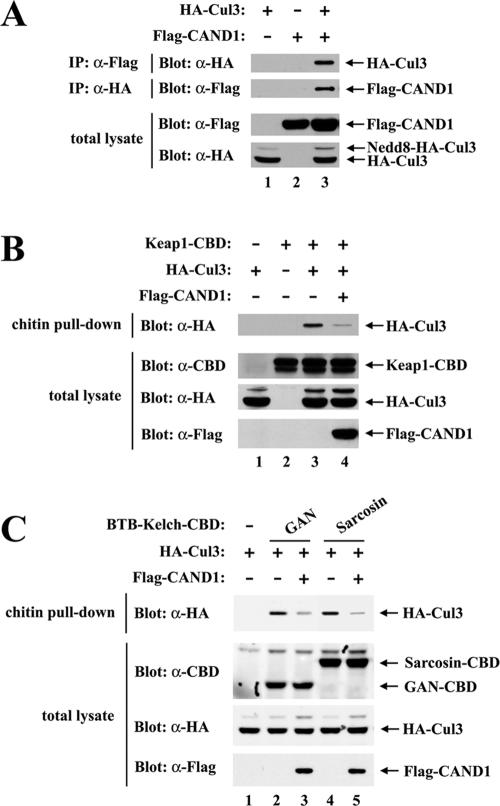
To determine if CAND1 functions as a regulator of Keap1-dependent ubiquitination of Nrf2, we first determined the ability of CAND1 to associate with Cul3. COS1 cells were transfected with expression vectors for Flag-tagged CAND1 and HA-tagged Cul3. Association between CAND1 and Cul3 was assessed by coimmunoprecipitation of cell lysates. The HA-tagged Cul3 protein was readily detected in anti-FLAG immunoprecipitates from cotransfected cells but not from singly transfected cells (Fig. 1A, top panel, lane 3). Similarly, the Flag-tagged CAND1 protein was observed only with anti-HA immunoprecipitates from cotransfected cells (Fig. 1A, second panel from top, lane 3). These results are consistent with the results reported by Min et al. indicating that CAND1 is able to associate with several different human cullin proteins, including Cul3 (30).
FIG. 1.
(A) Thirty-five-millimeter-diameter dishes of COS1 cells were transfected with 0.5 μg each of expression vectors for HA-Cul3 and Flag-CAND1 as indicated. Total cell lysates were analyzed by immunoblotting with anti-Flag (α-Flag) and α-HA antibodies (bottom two panels). α-Flag immunoprecipitates (IP) were subjected to immunoblot analysis using α-HA antibodies (top panel). α-HA IP were subjected to immunoblot analysis using α-Flag antibodies (second panel from the top). (B) Thirty-five-millimeter-diameter dishes of COS1 cells were transfected with 0.33 μg each of expression vectors for Keap1-CBD and HA-Cul3 as indicated. The Flag-CAND1 expression vector was either omitted (lanes 1 to 3) or included (lane 4). Total cell lysates were analyzed by immunoblotting with α-CBD, α-Flag, and α-HA antibodies (bottom three panels). The lysates were incubated with chitin beads, pelleted by centrifugation (3,000 × g), and washed three times in lysis buffer. Proteins that remained associated with the chitin beads were analyzed by immunoblotting with α-HA antibodies (top panel). (C) Thirty-five-millimeter-diameter dishes of COS1 cells were transfected with expression vectors for HA-Cul3 (lanes 1 to 5) and GAN-CBD (lanes 2 and 3) or sarcosin-CBD (lanes 4 and 5). The Flag-CAND1 expression vector was either omitted (lanes 1, 2, and 4) or included (lanes 3 and 5). Cell lysates were analyzed by immunoblotting with the indicated antibodies (bottom three panels) or incubated with chitin beads. Proteins that remained associated with the chitin beads after extensive washing were analyzed by immunoblotting with α-HA antibodies (top panel).
CAND1 has been proposed to sequester the Cul1 protein in an inactive form that is unable to bind substrate adaptor proteins (5, 17, 45). Therefore, we determined if overexpression of CAND1 would perturb the ability of Keap1 to associate with Cul3. For these experiments, a Keap1 protein containing a C-terminal chitin-binding domain was expressed in COS1 cells along with Cul3 and association of Cul3 with Keap1 was measured following affinity purification of Keap1 on chitin beads. The level of Keap1-associated Cul3 was markedly reduced in cells cotransfected with an expression vector for Flag-CAND1 (Fig. 1B, top panel, compare lanes 3 and 4). These results are consistent with the notion that CAND1 sequesters Cul3 away from Keap1.
Keap1 is one of 49 BTB-Kelch proteins encoded by the human genome (37). Several other BTB-Kelch proteins, including GAN and sarcosin, are able to associate with Cul3 and Rbx1 to form functional ubiquitin ligase complexes (13, 44). The ability of CAND1 to compete with either GAN or sarcosin for binding to Cul3 was determined. For these experiments, CBD-tagged GAN or sarcosin proteins were expressed in COS1 cells along with Cul3. Association of Cul3 with either GAN or sarcosin was measured following affinity purification with chitin beads. The amount of Cul3 that copurified with either CBD-GAN or CBD-sarcosin was markedly reduced in cells cotransfected with an expression vector for Flag-CAND1 (Fig. 1C, top panel, lanes 3 and 5). These results are consistent with the notion that CAND1 is a general regulator of Cul3-dependent E3 ubiquitin ligase complexes.
Increased CAND1 expression reduces Keap1-dependent ubiquitination of Nrf2 and increases steady-state levels of Nrf2.
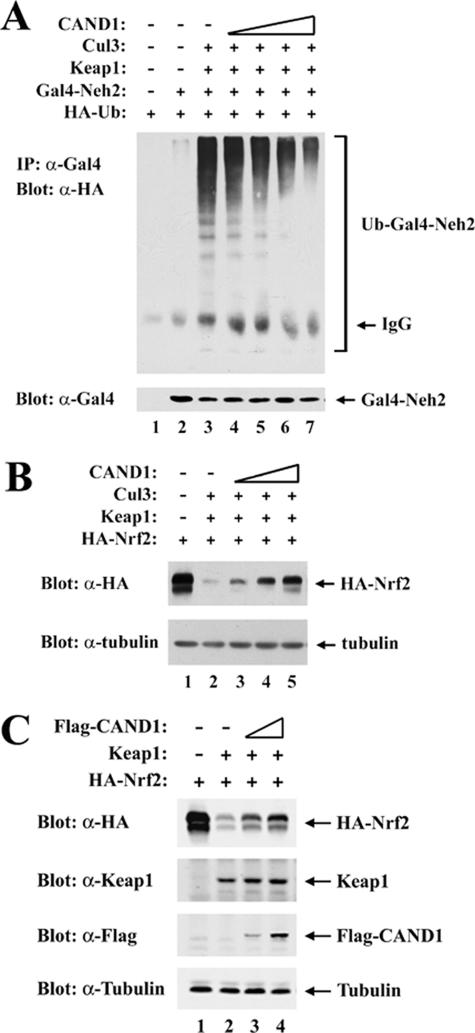
Keap1, as a substrate adaptor for Cul3, targets specific lysine residues within the N-terminal Neh2 domain of its substrate, the Nrf2 transcription factor, for ubiquitin conjugation (43). We therefore determined if increased levels of CAND1, which reduce association between Keap1 and Cul3, would also reduce Keap1-dependent ubiquitination of Nrf2. In the presence of Keap1 and Cul3, strong ubiquitin conjugation onto a Gal4-Neh2 fusion protein was observed with transfected MDA-MB-231 cells (Fig. 2A, top panel, lane 3). However, levels of ubiquitin conjugation onto Gal4-Neh2 were markedly reduced in the presence of increasing amounts of a cotransfected expression vector for CAND1 (Fig. 2A, top panel, lanes 4 to 7). In parallel experiments, the effect of CAND1 overexpression on steady-state levels of Nrf2 was determined. In both MDA-MB-231 cells (Fig. 2B, top panel) and HeLa cells (Fig. 2C, top panel), increased expression of CAND1 markedly increased steady-state levels of Nrf2 in the presence of coexpressed Keap1. Ectopic expression of CAND1 did not reduce binding of Nrf2 to Keap1 (data not shown). Rather, these results indicate that increased CAND1 expression, which reduces the ability of Keap1 to associate with Cul3, interferes with the ability of Keap1 to target Nrf2 for ubiquitin-dependent degradation.
FIG. 2.
(A) Thirty-five-millimeter-diameter dishes of MDA-MB-231 cells were transfected with expression vectors for HA-Ub (0.4 μg), Gal4-Neh2 (0.4 μg, lanes 2 to 7), Keap1 (0.1 μg, lanes 3 to 7), Cul3 (0.1 μg, lanes 3 to 7), and CAND1 (from 0.0125 to 0.1 μg, lanes 4 to 7). The transfected cells were treated with MG132 for 5 h prior to cell lysis. Total cell lysates were analyzed by immunoblotting with anti-Gal4 (α-Gal4) antibodies (bottom panel). α-Gal4 immunoprecipitates (IP) were analyzed by immunoblotting with α-HA antibodies (top panel). IgG, immunoglobulin G. (B) Twenty-four-well plates of MDA-MB-231 cells were transfected with expression vectors for HA-Nrf2 (0.18 μg), Keap1 (0.018 μg, lanes 2 to 5), Cul3 (0.07 μg, lanes 2 to 5), and CAND1 (from 0.035 to 0.14 μg, lanes 3 to 5). Total cell lysates were subjected to immunoblot analysis with α-HA (top panel) and α-tubulin (bottom panel) antibodies. (C) Twenty-four-well plates of HeLa cells were transfected with expression vectors for HA-Nrf2 (0.18 μg), Keap1 (0.018 μg, lanes 2 to 4), and Flag-CAND1 (0.035 to 0.14 μg, lanes 3 and 4). Total cell lysates were subjected to immunoblot analysis with α-HA, α-Keap1, α-Flag, and α-tubulin antibodies as indicated.
CAND1 is required for efficient repression of Nrf2 by Keap1.
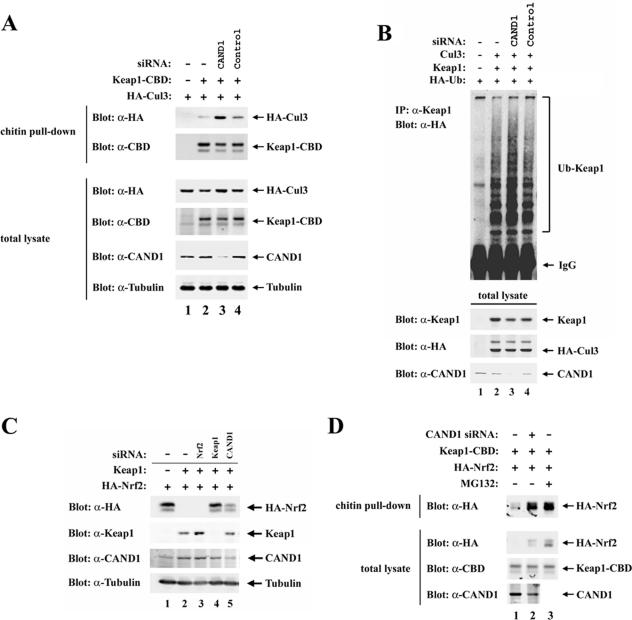
To further evaluate the functional significance of CAND1 for Keap1-mediated regulation of Nrf2, a siRNA molecule against CAND1 was used to reduce levels of the endogenous CAND1 protein. Immunoblot analysis using an anti-CAND1 antibody confirmed that the anti-CAND1 siRNA molecule was effective in reducing expression of the endogenous CAND1 protein (Fig. 3A, second panel from bottom, lane 3). Increased association of Cul3 with Keap1 was observed with HeLa cells transfected with the anti-CAND1 siRNA compared to cells transfected with a control siRNA molecule (Fig. 3A, top panel, compare lanes 2 to 4). The steady-state level of Keap1 was slightly reduced in the presence of the anti-CAND1 siRNA molecule (Fig. 3A, third panel from bottom, lane 3) and ubiquitin conjugation onto Keap1 was slightly increased (Fig. 3B, top panel, lane 3), presumably a consequence of increased association of Keap1 with Cul3. These results demonstrate that while ectopic overexpression of CAND1 results in reduced levels of association between Keap1 and Cul3 (Fig. 1B), siRNA-mediated knockdown of endogenous CAND1 results in increased levels of association between Keap1 and Cul3 (Fig. 3A).
FIG. 3.
(A) Thirty-five-millimeter-diameter dishes of HeLa cells were transfected with control (lane 4) or anti-CAND1 (α-CAND1) (lane 3) siRNA nucleotides (300 nM), allowed to recover for 24 h, and transfected with 0.5 μg each of expression vectors for HA-Cul3 and Keap1-CBD (lanes 2 to 4). Cell lysates were collected after an additional 24 h and immunoblotted with the indicated antibodies (bottom four panels) or incubated with chitin beads. Proteins that remained bound to the chitin beads after extensive washing were analyzed with the indicated antibodies (top two panels). (B) HeLa cells were transfected with control (lane 4) or α-CAND1 (lane 3) siRNA nucleotides (100 nM), allowed to recover for 24 h, and transfected with expression vectors for HA-Ub, Keap1, and Cul3 as indicated. Cell lysates were collected after an additional 24 h and immunoblotted with the indicated antibodies (bottom three panels). α-Keap1 immunoprecipitates (IP) were analyzed by immunoblotting with α-HA antibodies (top panel). IgG, immunoglobulin G. (C) Twenty-four-well plates of HeLa cells were transfected with α-Nrf2 (lane 3), α-Keap1 (lane 4), or α-CAND1 (lane 5) siRNA nucleotides (50 nM), allowed to recover for 24 h, and then transfected with 0.2 μg each of expression vectors for HA-Nrf2 and Keap1 (lanes 2 to 5). Total cell lysates were collected and subjected to immunoblot analysis with the indicated antibodies. (D) Thirty-five-millimeter-diameter dishes of HeLa cells were first mock transfected (lanes 1 and 3) or transfected with α-CAND1 siRNA nucleotides (300 nM), allowed to recover for 24 h, and then transfected with 0.5 μg each of expression vectors for HA-Nrf2 and Keap1-CBD. The transfected cells were either untreated (lanes 1 and 2) or treated with MG132 for 5 h prior to cell lysis. The lysates were incubated with chitin beads, and levels of HA-Nrf2 proteins that remained bound to the chitin beads were determined by immunoblotting with α-HA antibodies (top panel). Total lysates were analyzed by immunoblotting with the indicated antibodies (bottom three panels).
Increased association between Keap1 and Cul3 might increase the ability of Keap1 to target Nrf2 for ubiquitin-dependent degradation. Therefore, steady-state levels of Nrf2 were determined in the absence or presence of siRNA molecules directed against CAND1. HeLa cells were transfected with expression vectors for Keap1 and Nrf2 along with siRNA molecules against Nrf2, Keap1, or CAND1. Coexpression of Keap1 and Nrf2 markedly reduced steady-state levels of Nrf2 (Fig. 3C, top panel, lanes 1 and 2). As expected, steady-state levels of Nrf2 were fully recovered in the presence of a siRNA molecule against Keap1 (Fig. 3C, top panel, lane 4). However, a significant increase in steady-state levels of Nrf2 was also observed following siRNA-mediated knockdown of endogenous CAND1 expression (Fig. 3C, top panel, lane 5). Thus, although reduced levels of CAND1 result in increased association of Keap1 with Cul3, the ability of Keap1 to efficiently target Nrf2 for ubiquitin-dependent degradation is decreased.
To determine if increased steady-state levels of Nrf2 in the presence of the anti-CAND1 siRNA results from decreased binding of Nrf2 to Keap1, the level of Keap1-associated Nrf2 was determined in the absence and presence of the anti-CAND1 siRNA molecule. Increased steady-state levels of Nrf2 were again observed in the presence of the anti-CAND1 siRNA molecule (Fig. 3D, second panel from top, compare lanes 1 and 2). A marked increase in Keap1-associated Nrf2 was also observed (Fig. 3D, top panel, compare lanes 1 and 2). Thus, decreased levels of CAND1 do not disrupt the ability of Keap1 to bind Nrf2. Rather, these results indicate that decreased levels of CAND1 disrupt the ability of Keap1 to efficiently target Nrf2 for ubiquitin-dependent degradation and thereby increase steady-state levels of Keap1-associated Nrf2.
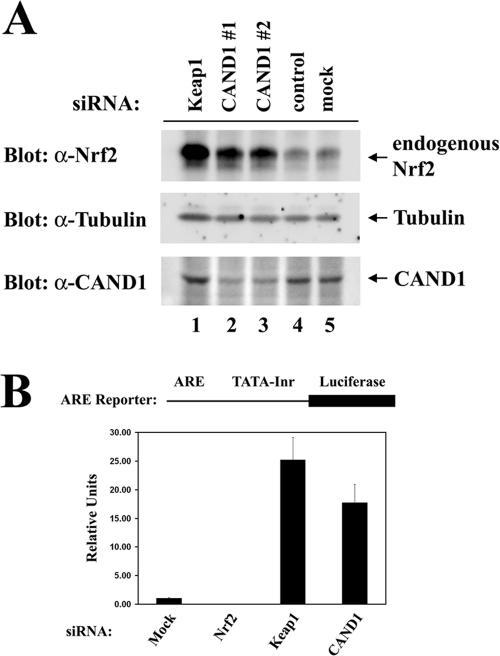
To confirm the importance of CAND1 for regulation of steady-state levels of Nrf2, steady-state levels of the endogenous Nrf2 protein in HeLa cells transfected with siRNA molecules against Keap1 or CAND1 were measured. As expected, the siRNA molecule against Keap1 markedly increased steady-state levels of the endogenous Nrf2 protein (Fig. 4A, top panel, compare lanes 1 and 5). A marked increase in the steady-state levels of the endogenous Nrf2 protein was also observed in the presence of two different siRNA molecules against CAND1 (Fig. 4A, top panel, lanes 2 and 3). A control siRNA had no effect on steady-state levels of Nrf2 (Fig. 4A, top panel, lane 4).
FIG. 4.
(A) Twenty-four-well plates of HeLa cells were transfected with 100 nM of control siRNA (lane 4) or siRNA nucleotides targeting Keap1 (lane 1) or CAND1 at different regions in CAND1 mRNA (lanes 2 and 3). Levels of endogenous Nrf2 were determined by immunoblot analysis with anti-Nrf2 (α-Nrf2) antibodies (top panel). Levels of CAND1 and tubulin were analyzed by immunoblotting with the indicated antibodies (bottom two panels). (B) Twenty-four-well plates of HeLa cells were transfected with α-Nrf2, α-Keap1, or α-CAND1 siRNA nucleotides (300 nM) as indicated and with an ARE-dependent firefly luciferase reporter gene construct (100 ng). A plasmid encoding Renilla luciferase (10 ng) was included as a control for transfection efficiency. The data shown represent the means and standard deviations of results from three independent experiments.
Reporter gene assays were performed to determine if increased steady-state levels of Nrf2 resulted in increased Nrf2-dependent gene expression. A luciferase reporter gene controlled by a minimal promoter containing four copies of an ARE was transfected into HeLa cells in the absence or presence of cotransfected siRNA molecules against Nrf2, Keap1, or CAND1. A low level of Nrf2-dependent reporter gene expression was observed with the cells transfected only with the reporter plasmid (Fig. 4B). This basal level of reporter gene expression was reduced by cotransfection of siRNA against Nrf2, confirming the validity of this assay as a measurement of transcription driven by the endogenous Nrf2 protein. Levels of reporter gene expression were increased 25-fold in the presence of a siRNA molecule against Keap1, while a siRNA molecule against CAND1 resulted in a 15-fold increase in Nrf2-dependent reporter gene expression (Fig. 4B).
Our results indicate a critical role for CAND1 in regulation of Keap1-mediated control over steady-state levels of Nrf2 and Nrf2-dependent transcription. Ectopic expression of CAND1 decreased the ability of Keap1 to assemble with Cul3 into a functional E3 ubiquitin ligase complex, resulting in increased steady-state levels of Nrf2. On the other hand, decreased CAND1 expression, which increased complex formation between Keap1 and Cul3, also increased steady-state levels of Nrf2 and Nrf2-dependent gene expression. Taken together, these results indicate that an optimal level of CAND1 is required for efficient down-regulation of Nrf2 by Keap1. These results are consistent with a model in which CAND1-mediated cycles of assembly and disassembly of the Keap1-Cul3-Rbx1 E3 ubiquitin ligase complex are required for efficient control by Keap1 over steady-state levels of Nrf2 and Nrf2-dependent transcription.
Neddylation of Cul3 on Lys 712 is required for efficient Keap1-dependent ubiquitination of Nrf2 in vivo.
Cyclical assembly and disassembly of cullin-containing E3 ubiquitin ligase complexes has been suggested to involve both CAND1 and site-specific modification of the cullin protein by the small ubiquitin-like protein, Nedd8 (5). The neddylated form of Cul3, which exhibits a slower mobility on SDS-polyacrylamide gels, can be readily observed following immunoblot detection of Cul3 in whole-cell lysates (Fig. 1A, bottom panel). Neddylation of Cul3 occurs at a conserved C-terminal lysine residue (Lys 712 in human Cul3), and the biological importance of Nedd8 modification has been demonstrated for the Cul3 homologs in Drosophila melanogaster and Caenorhabditis elegans (36, 46).
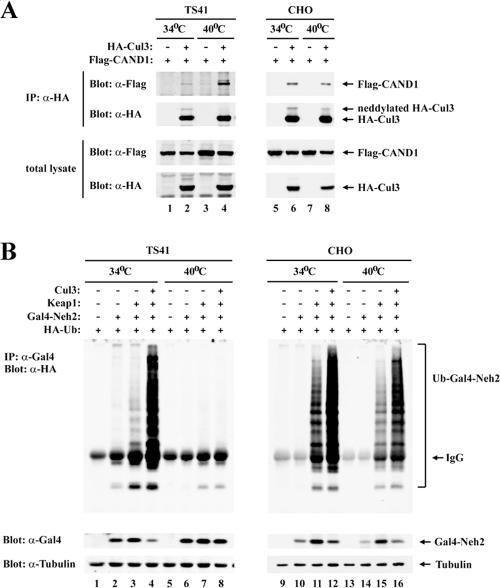
To determine if neddylation of Cul3 is required for Keap1-dependent ubiquitination of Nrf2, the ability of Keap1 to target Nrf2 for ubiquitination was characterized for the ts41 hamster ovary cell line. The ts41 cell line contains a temperature-sensitive defect in the Nedd8 E1 activation enzyme (3, 34). As expected, the slower-migrating form of Cul3 was not detected in ts41 cells following a shift to the restrictive temperature of 40°C (Fig. 5A, second panel from top, lane 4). Increased association of CAND1 with Cul3 was also observed with ts41 cells at the restrictive temperature (Fig. 5A, top panel, lanes 2 and 4), consistent with the notion that CAND1 preferentially binds to the nonneddylated form of Cul3 (17, 19, 28, 45). In contrast, the neddylated form of Cul3 was readily detectable in wild-type Chinese hamster ovary cells grown at either 34°C or 40°C, and no changes in CAND1 association with Cul3 were observed (Fig. 5A, top panel, lanes 6 and 8). Ectopic expression of Keap1 and Cul3 markedly increased ubiquitination onto the Gal4-Neh2 fusion protein in both ts41 and wild-type cells grown at 34°C and in the wild-type cells grown at 40°C (Fig. 5B, lanes 1 to 4 and 9 to 16). However, Keap1-dependent ubiquitination of the Gal4-Neh2 protein was abolished in ts41 cells shifted to 40°C (Fig. 5B, lanes 5 to 8).
FIG. 5.
(A) Sixty-millimeter-diameter dishes of ts41 cells or wild-type CHO cells were cotransfected with 1.0 μg each of expression vectors for HA-Cul3 and Flag-CAND1 as indicated. The transfected cells were either kept at 34°C (lanes 1, 2, 5, and 6) or shifted to 40°C (lanes 3, 4, 7, and 8) for 24 h. Total cell lysates were analyzed by immunoblotting with anti-Flag (α-Flag) and α-HA antibodies (bottom two panels). α-HA immunoprecipitates (IP) were subjected to immunoblot analysis using α-Flag and α-HA antibodies (top two panels). (B) Sixty-millimeter-diameter dishes of ts41 cells or wild-type CHO cells were cotransfected with expression vectors for HA-Ub (0.7 μg), Gal4-Neh2 (0.8 μg), Keap1 (0.2 μg), and Cul3 (0.3 μg) as indicated. The transfected cells were either kept at 34°C (lanes 1 to 4 and 9 to 12) or shifted to 40°C (lanes 5 to 8 and 13 to 16) for 24 h, and all were treated with MG132 for 2.5 h prior to cell lysis. Total cell lysates were analyzed by immunoblotting with α-Gal4 and α-tubulin antibodies (bottom two panels). α-Gal4 IP were analyzed by immunoblotting with α-HA antibodies (top panel). IgG, immunoglobulin G.
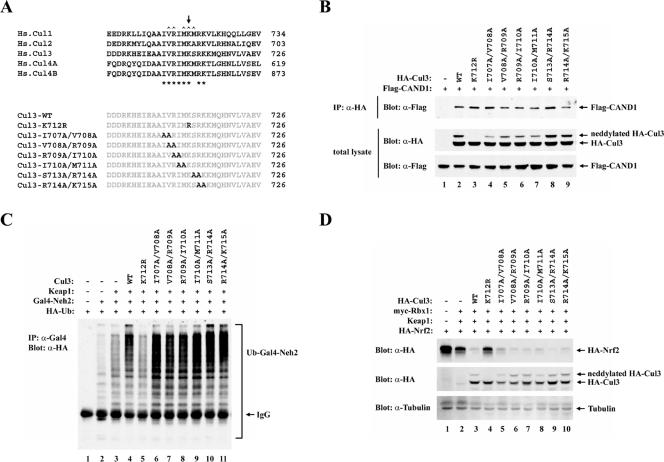
Neddylation of the human Cul3 protein is predicted to occur at Lys 712, located within a short stretch of amino acids that are highly conserved in other cullin proteins (Fig. 6A). Several amino acids in this region, including Lys 712, participate in direct contacts between Cul1 and CAND1 (17), suggesting that there may be an antagonistic relationship between modification of cullin proteins by Nedd8 and binding of cullin proteins to CAND1 (4, 17, 28, 31, 45). To further explore the functional significance of Cul3 neddylation and CAND1 association for efficient Keap1-dependent ubiquitination of Nrf2, a series of mutant Cul3 proteins were constructed. These mutations introduced either a single arginine substitution in place of Lys 712 or two alanine substitutions at adjacent residues flanking Lys 712 (Fig. 6A). Modification of Cul3 by neddylation was abolished only by the K712R substitution (Fig. 6B, middle panel, lane 3). However, all of the mutant Cul3 proteins, including the K712R protein, were able to associate with CAND1 (Fig. 6B, upper panel), indicating that single or double amino acid substitutions within this conserved region are not sufficient to disrupt binding of Cul3 to CAND1.
FIG. 6.
(A) Sequence alignment of neddylation sites among five members of the cullin superfamily is shown. The conserved Lys residue for Nedd8 conjugation in the cullin proteins is indicated above the alignment by an arrow (39). Conserved residues in this region are indicated below the alignment by asterisks. Residues in this region of Cul1 that make direct contacts with CAND1 are indicated above the alignment by carets (17). The conserved residues that were substituted with alanine or arginine in the Cul3 mutant proteins utilized in this report are illustrated. Hs., Homo sapiens. (B) Thirty-five-millimeter-diameter dishes of COS1 cells were cotransfected with 0.5 μg each of expression vectors for Flag-CAND1 and the mutant or wild-type (WT) HA-Cul3 proteins as indicated. Cell lysates were collected in the presence of 10 mM NEM and analyzed by immunoblotting with anti-Flag (α-Flag) and α-HA antibodies (bottom two panels). α-HA immunoprecipitates (IP) were subjected to immunoblot analysis using α-Flag antibodies (top panel). (C) Thirty-five-millimeter-diameter dishes of MDA-MB-231 cells were transfected with expression vectors for HA-Ub (0.4 μg), Gal4-Neh2 (0.4 μg, lanes 2 to 11), Keap1 (0.1 μg, lanes 3 to 11), and the WT or mutant Cul3 proteins (0.1 μg, lanes 4 to 11). The transfected cells were treated with MG132 for 2.5 h prior to cell lysis. α-Gal4 IP from cell lysates were analyzed by immunoblotting with α-HA antibodies. IgG, immunoglobulin G. (D) Twenty-four-well plates of MDA-MB-231 cells were transfected with expression vectors for HA-Nrf2 (0.18 μg), myc-Rbx1 (0.11 μg, lanes 2 to 10), Keap1 (0.018 μg, lanes 2 to 10), and the WT or mutant Cul3 proteins (0.11 μg, lanes 3 to 10). Total cell lysates were subjected to immunoblot analysis with α-HA (top and middle panels) and α-tubulin (bottom panel) antibodies.
All of the mutant Cul3 proteins were able to associate with Keap1 and Rbx1 (Fig. 7A and data not shown). Coexpression of the wild-type Cul3 protein with Keap1 markedly enhanced ubiquitination of the Gal4-Neh2 substrate protein (Fig. 6C, lanes 2 to 4) and markedly reduced steady-state levels of Nrf2 (Fig. 6D, lanes 1 to 3). However, only low levels of ubiquitin transfer onto the Gal4-Neh2 substrate protein were observed in the presence of the mutant K712R Cul3 protein (Fig. 6C, lane 5). Elevated steady-state levels of Nrf2 were observed in the presence of the mutant K712R Cul3 protein (Fig. 6D, lane 4). Two other mutant Cul3 proteins, containing Ala or Met, respectively, at residue 712, were also impaired in their ability to repress steady-state levels of Nrf2 (data not shown). All of the other mutant Cul3 proteins were as effective as the wild-type Cul3 protein for supporting Keap1-dependent ubiquitination of Nrf2 and Keap1-mediated repression of steady-state levels of Nrf2 (Fig. 6C, lanes 6 to 11, and Fig. 6D, lanes 5 to 10).
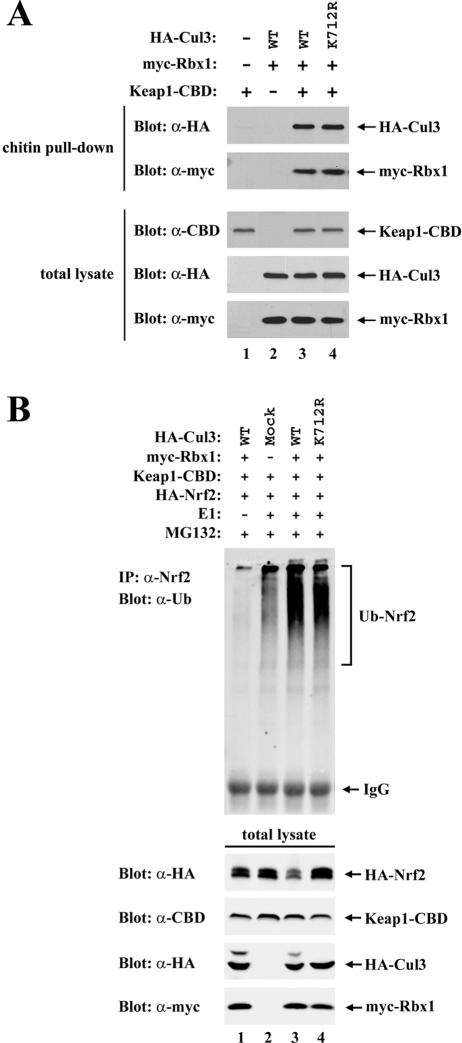
FIG. 7.
(A) Thirty-five-millimeter-diameter dishes of COS1 cells were cotransfected with expression vectors for Keap1-CBD (0.3 μg, lanes 1, 3, and 4), myc-Rbx1 (0.1 μg, lanes 2 to 4), and the wild-type (WT) or mutant HA-Cul3 proteins (0.3 μg, lanes 2 to 4) as indicated. Cell lysates were analyzed by immunoblotting with the indicated antibodies (bottom three panels) or incubated with chitin beads. Proteins that remained bound to the chitin beads after extensive washing were analyzed by immunoblotting with the indicated antibodies (top two panels). (B) Sixty-millimeter-diameter dishes of COS1 cells were transfected with 0.5 μg each of the expression vectors for HA-Nrf2, Keap1-CBD, myc-Rbx1, and the WT or mutant HA-Cul3 proteins, as indicated in lanes 1 to 4. The transfected cells were treated with MG132 for 5 h prior to cell lysis. Lysates from three 60-mm-diameter dishes were pooled for each sample. One percent of the cell lysates were analyzed by immunoblotting with the indicated antibodies (bottom four panels), and the rest of the lysates were incubated with chitin beads. After washing, the chitin beads were incubated with E1, E2-UbcH5a, ubiquitin, and ATP. The E1 enzyme was omitted from one sample (lane 1). Subsequently, the chitin beads were pelleted and washed, and proteins that were eluted from the beads after boiling under denaturing conditions were immunoprecipitated with anti-Nrf2 (α-Nrf2) antibodies and then analyzed by immunoblotting with antiubiquitin antibodies (top panel). IP, immunoprecipitate; IgG, immunoglobulin G.
To confirm that the K712R mutant Cul3 protein is able to assemble into a functional E3 ubiquitin ligase complex with Keap1, in vitro ubiquitination assays were carried out following affinity purification of the complex from transfected cells. Both the wild-type and K712R mutant Cul3 proteins were able to form a complex with Keap1 and Rbx1 that could be affinity purified from transfected cells (Fig. 7A, top two panels, lanes 3 and 4). Furthermore, following affinity purification of the quaternary Nrf2-Keap1-Cul3-Rbx1 complex from transfected cells, both the K712R mutant and the wild-type Cul3 proteins were able to support ubiquitin transfer onto Nrf2 in vitro (Fig. 7B, lanes 3 and 4). Ubiquitin transfer onto Keap1 was also readily observed in vitro in the presence of either the K712R mutant or wild-type Cul3 (data not shown). Ubiquitin conjugation onto Nrf2 was not observed with reactions carried out in the absence of the E1 ubiquitin conjugation enzyme (Fig. 7B, lane 1). Importantly, only low levels of ubiquitin conjugation onto Nrf2 were observed in the absence of the coexpressed Cul3-Rbx1 subcomplex (Fig. 7B, lane 2), indicating that the robust ubiquitin conjugation activity observed in the presence of the K712R mutant protein is not due simply to copurification of the endogenous Cul3-Rbx1 subcomplex with ectopically expressed Keap1-CBD. Thus, under the conditions of the in vitro assay, neddylation of Lys 712 is not required for the assembly of Cul3 into a functional ubiquitin ligase complex (Fig. 7B). However, neddylation of Cul3 is required to support efficient Keap1-dependent ubiquitination of Nrf2 in vivo (Fig. 6C).
DISCUSSION
The ability of Keap1 to function as a substrate adaptor protein for Cul3 and target Nrf2 for ubiquitin-dependent degradation has emerged as a major regulatory mechanism that regulates expression of cytoprotective phase 2 genes (7, 14, 25, 43). Cullin-dependent E3 ubiquitin ligases have been proposed to undergo cycles of assembly and disassembly that allow substrate adaptor recycling. Cyclical assembly and disassembly of cullin-dependent E3 ubiquitin ligase complexes is mediated, in part, by the antagonistic actions of Nedd8 modification of the cullin protein and association of cullin proteins with CAND1 (17, 31, 35). In the present work, we have used several independent experimental approaches to examine the role of substrate adaptor recycling for Keap1-mediated repression of Nrf2. Ectopic expression of CAND1 reduced the level of complex formation between Keap1 and Cul3, while siRNA-mediated knockdown of endogenous CAND1 expression increased complex formation between Keap1 and Cul3. However, both ectopic CAND1 expression and siRNA-mediated knockdown of endogenous CAND1 decreased the ability of Keap1 to target Nrf2 for ubiquitin-dependent degradation. Notably, a marked increase in Nrf2-dependent gene expression was observed following siRNA-mediated knockdown of CAND1 expression. Modification of Cul3 by Nedd8 at a conserved Lys acceptor residue is also required for Keap1-dependent ubiquitination in vivo. Taken together, these results are consistent with a model in which the ability of Keap1 to participate in multiple cycles of substrate adaptor exchange is a critical regulatory aspect of Keap1-mediated repression of Nrf2-dependent gene expression.
A key feature of this model is that physical release of Nrf2 from Keap1 is not required for activation of Nrf2-dependent transcription. Indeed, knockdown of CAND1 markedly increases the level of Keap1-associated Nrf2 yet also increases Nrf2-dependent transcription. Thus, decreasing the ability of Keap1 to efficiently target Nrf2 for ubiquitin-dependent degradation is sufficient to activate Nrf2-dependent gene expression. We propose that a decreased ability of Keap1 to target Nrf2 for ubiquitin-dependent degradation results in the accumulation of an excess of Nrf2 relative to Keap1 such that free Nrf2 proteins are able to localize to the nucleus and activate Nrf2-dependent gene expression.
The ability of Keap1 to act in a catalytic manner and target multiple Nrf2 proteins for ubiquitin-dependent degradation has important implications for how Nrf2 is able to escape Keap1-mediated repression following exposure of cells to oxidative stress and chemopreventive compounds. As Keap1 is able to sequester Nrf2 in the cytoplasm, it has been suggested that reactive molecules bind to Keap1 and cause the physical release of Nrf2 from Keap1. Consistent with this suggestion, direct binding of chemically reactive molecules to Keap1 has been demonstrated both in vitro and in vivo (10, 18). However, the importance of physical release of Nrf2 from Keap1 has not been clearly established. For example, although one report suggested that chemical inducers of Nrf2-dependent gene expression dissociate the Keap1-Nrf2 complex in vitro (10), a subsequent report suggested that reactive molecules simply alter the mobility of the complex in native polyacrylamide gels without causing the physical release of Nrf2 (12). In our previous work, we have demonstrated that association between Nrf2 and Keap1 in vivo is not decreased but is actually increased following exposure of cells to inducers of Nrf2-dependent transcription (43). In a similar vein, we find that knockdown of CAND1, which markedly increases Nrf2-dependent gene expression, also increases the level of Keap1-associated Nrf2. Taken together, the available experimental evidence is most consistent with a model in which reactive molecules do not cause the physical release of Nrf2 from Keap1 but interfere with the ability of Keap1 to act in a catalytic manner to efficiently target Nrf2 for ubiquitin-dependent degradation.
Keap1 has been proposed to be a sensor of reactive molecules and oxidative stress, such that a redox-dependent modification of one or more cysteine residues in Keap1 is coupled to increased expression of Nrf2-dependent genes. Indeed, multiple cysteine residues in Keap1, including Cys 151, Cys 273, and Cys 288, can be modified by reactive molecules either in vivo or in vitro (10, 12, 18). Mutational analysis has revealed that Cys 273 and Cys 288, which are located in the central linker domain and participate in binding Zn2+ (11), are required for repression of Nrf2-dependent gene expression. In contrast, Cys 151, located in the N-terminal BTB domain, is specifically required for increased Nrf2-dependent gene expression following exposure of cells to oxidative stress (38, 42, 43). Mutant Keap1 proteins containing serine or alanine substitutions at Cys 151, Cys 273, or Cys 288 are still able to bind Nrf2 (42). However, serine substitutions at Cys 273 and Cys 288 compromise the ability of Keap1 to target Nrf2 for ubiquitination because they reduce the ability of Keap1 to bind Cul3 (unpublished data). On the other hand, serine substitution at Cys 151 renders Keap1-dependent ubiquitination of Nrf2 resistant to inhibition by oxidative stress or chemopreventive compounds, presumably because modification of Cys 151 by chemopreventive compounds, such as sulforaphane, decreases the ability of Keap1 to associate with Cul3 (43). Taken together, the current evidence is most consistent with a model in which modification of these (and perhaps other) cysteine residues in Keap1 would perturb the ability of Keap1 to assemble with Cul3 and target Nrf2 for ubiquitin-dependent degradation rather than inducing the release of Nrf2 from Keap1. An attractive feature of a model in which Keap1 acts in a catalytic manner to efficiently target Nrf2 for ubiquitin-dependent degradation is that modulating the catalytic efficiency of Keap1 by modification of one or more cysteine residues in Keap1 would allow a graded response of Nrf2-dependent gene expression following exposure of cells to oxidative stress and chemically reactive molecules.
Acknowledgments
We thank Joan Conaway, Stowers Institute for Medical Research, for help and advice with the in vitro ubiquitination assays and for her gift of the myc-Rbx1 expression plasmid. We thank Chuck Sherr, St. Jude Children’s Research Hospital, for his gift of ts41 cells and Toshihiko Ezashi for his gift of the CHO-K1 cell line. We thank Shrikesh Sachdev and Donna Zhang for thoughtful discussions.
This work was supported by the University of Missouri Molecular Biology Program, the University of Missouri Food for the 21st Century program, a research grant from NIH to M.H. (CA106593), and a development project in CA103130.
REFERENCES
- 1.Ames, B. N., and M. K. Shigenaga. 1993. DNA and free radicals, p. 1-15. In B. Halliwell and O. I. Aruoma (ed.), Oxidants are a major contributor to cancer and aging. Ellis Horwood, New York, N.Y.
- 2.Ceconi, C., A. Boraso, A. Cargnoni, and R. Ferrari. 2003. Oxidative stress in cardiovascular disease: myth or fact? Arch. Biochem. Biophys. 420:217-221. [DOI] [PubMed] [Google Scholar]
- 3.Chen, Y., D. L. McPhie, J. Hirschberg, and R. L. Neve. 2000. The amyloid precursor protein-binding protein APP-BP1 drives the cell cycle through the S-M checkpoint and causes apoptosis in neurons. J. Biol. Chem. 275:8929-8935. [DOI] [PubMed] [Google Scholar]
- 4.Chuang, H. W., W. Zhang, and W. M. Gray. 2004. Arabidopsis ETA2, an apparent ortholog of the human cullin-interacting protein CAND1, is required for auxin responses mediated by the SCF(TIR1) ubiquitin ligase. Plant Cell 16:1883-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cope, G. A., and R. J. Deshaies. 2003. COP9 signalosome: a multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell 114:663-671. [DOI] [PubMed] [Google Scholar]
- 6.Cope, G. A., G. S. Suh, L. Aravind, S. E. Schwarz, S. L. Zipursky, E. V. Koonin, and R. J. Deshaies. 2002. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science 298:608-611. [DOI] [PubMed] [Google Scholar]
- 7.Cullinan, S. B., J. D. Gordan, J. Jin, J. W. Harper, and J. A. Diehl. 2004. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol. Cell. Biol. 24:8477-8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies, K. J. 2000. Oxidative stress, antioxidant defenses, and damage removal, repair, and replacement systems. IUBMB Life 50:279-289. [DOI] [PubMed] [Google Scholar]
- 9.Deffenbaugh, A. E., K. M. Scaglione, L. Zhang, J. M. Moore, T. Buranda, L. A. Sklar, and D. Skowyra. 2003. Release of ubiquitin-charged Cdc34-S-Ub from the RING domain is essential for ubiquitination of the SCF(Cdc4)-bound substrate Sic1. Cell 114:611-622. [DOI] [PubMed] [Google Scholar]
- 10.Dinkova-Kostova, A. T., W. D. Holtzclaw, R. N. Cole, K. Itoh, N. Wakabayashi, Y. Katoh, M. Yamamoto, and P. Talalay. 2002. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc. Natl. Acad. Sci. USA 99:11908-11913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dinkova-Kostova, A. T., W. D. Holtzclaw, and N. Wakabayashi. 2005. Keap1, the sensor for electrophiles and oxidants that regulates the phase 2 response, is a zinc metalloprotein. Biochemistry 44:6889-6899. [DOI] [PubMed] [Google Scholar]
- 12.Eggler, A. L., G. Liu, J. M. Pezzuto, R. B. van Breemen, and A. D. Mesecar. 2005. Modifying specific cysteines of the electrophile-sensing human Keap1 protein is insufficient to disrupt binding to the Nrf2 domain Neh2. Proc. Natl. Acad. Sci. USA 102:10070-10075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furukawa, M., Y. J. He, C. Borchers, and Y. Xiong. 2003. Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat. Cell Biol. 5:1001-1007. [DOI] [PubMed] [Google Scholar]
- 14.Furukawa, M., and Y. Xiong. 2005. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the cullin 3-Roc1 ligase. Mol. Cell. Biol. 25:162-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gan-Erdene, T., K. Nagamalleswari, L. Yin, K. Wu, Z. Q. Pan, and K. D. Wilkinson. 2003. Identification and characterization of DEN1, a deneddylase of the ULP family. J. Biol. Chem. 278:28892-28900. [DOI] [PubMed] [Google Scholar]
- 16.Golden, T. R., D. A. Hinerfeld, and S. Melov. 2002. Oxidative stress and aging: beyond correlation. Aging Cell 1:117-123. [DOI] [PubMed] [Google Scholar]
- 17.Goldenberg, S. J., T. C. Cascio, S. D. Shumway, K. C. Garbutt, J. Liu, Y. Xiong, and N. Zheng. 2004. Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell 119:517-528. [DOI] [PubMed] [Google Scholar]
- 18.Hong, F., K. R. Sekhar, M. L. Freeman, and D. C. Liebler. 2005. Specific patterns of electrophile adduction trigger Keap1 ubiquitination and Nrf2 activation. J. Biol. Chem. 280:31768-31775. [DOI] [PubMed] [Google Scholar]
- 19.Hwang, J. W., K. W. Min, T. A. Tamura, and J. B. Yoon. 2003. TIP120A associates with unneddylated cullin 1 and regulates its neddylation. FEBS Lett. 541:102-108. [DOI] [PubMed] [Google Scholar]
- 20.Imlay, J. A. 2003. Pathways of oxidative damage. Annu. Rev. Microbiol. 57:395-418. [DOI] [PubMed] [Google Scholar]
- 21.Itoh, K., N. Wakabayashi, Y. Katoh, T. Ishii, K. Igarashi, J. D. Engel, and M. Yamamoto. 1999. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 13:76-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaiswal, A. K. 2004. Nrf2 signaling in coordinated activation of antioxidant gene expression. Free Radic. Biol. Med. 36:1199-1207. [DOI] [PubMed] [Google Scholar]
- 23.Kamura, T., D. M. Koepp, M. N. Conrad, D. Skowyra, R. J. Moreland, O. Iliopoulos, W. S. Lane, W. G. Kaelin, Jr., S. J. Elledge, R. C. Conaway, J. W. Harper, and J. W. Conaway. 1999. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science 284:657-661. [DOI] [PubMed] [Google Scholar]
- 24.Kawakami, T., T. Chiba, T. Suzuki, K. Iwai, K. Yamanaka, N. Minato, H. Suzuki, N. Shimbara, Y. Hidaka, F. Osaka, M. Omata, and K. Tanaka. 2001. NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 20:4003-4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobayashi, A., M. I. Kang, H. Okawa, M. Ohtsuji, Y. Zenke, T. Chiba, K. Igarashi, and M. Yamamoto. 2004. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 24:7130-7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kobayashi, M., and M. Yamamoto. 2005. Molecular mechanisms activating the Nrf2-Keap1 pathway of antioxidant gene regulation. Antioxid. Redox Signal. 7:385-394. [DOI] [PubMed] [Google Scholar]
- 27.Li, X., D. Zhang, M. Hannink, and L. J. Beamer. 2004. Crystal structure of the Kelch domain of human Keap1. J. Biol. Chem. 279:54750-54758. [DOI] [PubMed] [Google Scholar]
- 28.Liu, J., M. Furukawa, T. Matsumoto, and Y. Xiong. 2002. NEDD8 modification of CUL1 dissociates p120(CAND1), an inhibitor of CUL1-SKP1 binding and SCF ligases. Mol. Cell 10:1511-1518. [DOI] [PubMed] [Google Scholar]
- 29.Lyapina, S., G. Cope, A. Shevchenko, G. Serino, T. Tsuge, C. Zhou, D. A. Wolf, N. Wei, and R. J. Deshaies. 2001. Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science 292:1382-1385. [DOI] [PubMed] [Google Scholar]
- 30.Min, K. W., J. W. Hwang, J. S. Lee, Y. Park, T. A. Tamura, and J. B. Yoon. 2003. TIP120A associates with cullins and modulates ubiquitin ligase activity. J. Biol. Chem. 278:15905-15910. [DOI] [PubMed] [Google Scholar]
- 31.Min, K. W., M. J. Kwon, H. S. Park, Y. Park, S. K. Yoon, and J. B. Yoon. 2005. CAND1 enhances deneddylation of CUL1 by COP9 signalosome. Biochem. Biophys. Res. Commun. 334:867-874. [DOI] [PubMed] [Google Scholar]
- 32.Motohashi, H., T. O'Connor, F. Katsuoka, J. D. Engel, and M. Yamamoto. 2002. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene 294:1-12. [DOI] [PubMed] [Google Scholar]
- 33.Nguyen, T., P. J. Sherratt, and C. B. Pickett. 2003. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu. Rev. Pharmacol. Toxicol. 43:233-260. [DOI] [PubMed] [Google Scholar]
- 34.Osaka, F., H. Kawasaki, N. Aida, M. Saeki, T. Chiba, S. Kawashima, K. Tanaka, and S. Kato. 1998. A new NEDD8-ligating system for cullin-4A. Genes Dev. 12:2263-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petroski, M. D., and R. J. Deshaies. 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6:9-20. [DOI] [PubMed] [Google Scholar]
- 36.Pintard, L., T. Kurz, S. Glaser, J. H. Willis, M. Peter, and B. Bowerman. 2003. Neddylation and deneddylation of CUL-3 is required to target MEI-1/Katanin for degradation at the meiosis-to-mitosis transition in C. elegans. Curr. Biol. 13:911-921. [DOI] [PubMed] [Google Scholar]
- 37.Stogios, P. J., G. S. Downs, J. J. S. Jauhal, S. K. Nandra, and G. G. Privé. 2005. Sequence and structural analysis of BTB domain proteins. Genome Biol. 6:R82.1-R82.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wakabayashi, N., A. T. Dinkova-Kostova, W. D. Holtzclaw, M. I. Kang, A. Kobayashi, M. Yamamoto, T. W. Kensler, and P. Talalay. 2004. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc. Natl. Acad. Sci. USA 101:2040-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu, K., A. Chen, and Z. Q. Pan. 2000. Conjugation of Nedd8 to CUL1 enhances the ability of the ROC1-CUL1 complex to promote ubiquitin polymerization. J. Biol. Chem. 275:32317-32324. [DOI] [PubMed] [Google Scholar]
- 40.Yamoah, K., K. Wu, and Z. Q. Pan. 2005. In vitro cleavage of Nedd8 from cullin 1 by COP9 signalosome and deneddylase 1. Methods Enzymol. 398:509-522. [DOI] [PubMed] [Google Scholar]
- 41.You, J., M. Wang, T. Aoki, T. A. Tamura, and C. M. Pickart. 2003. Proteolytic targeting of transcriptional regulator TIP120B by a HECT domain E3 ligase. J. Biol. Chem. 278:23369-23375. [DOI] [PubMed] [Google Scholar]
- 42.Zhang, D. D., and M. Hannink. 2003. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol. Cell. Biol. 23:8137-8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang, D. D., S. C. Lo, J. V. Cross, D. J. Templeton, and M. Hannink. 2004. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell. Biol. 24:10941-10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, D. D., S. C. Lo, Z. Sun, G. M. Habib, M. W. Lieberman, and M. Hannink. 2005. Ubiquitination of Keap1, a BTB-Kelch substrate adaptor protein for Cul3, targets Keap1 for degradation by a proteasome-independent pathway. J. Biol. Chem. 280:30091-30099. [DOI] [PubMed] [Google Scholar]
- 45.Zheng, J., X. Yang, J. M. Harrell, S. Ryzhikov, E. H. Shim, K. Lykke-Andersen, N. Wei, H. Sun, R. Kobayashi, and H. Zhang. 2002. CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Mol. Cell 10:1519-1526. [DOI] [PubMed] [Google Scholar]
- 46.Zhu, S., R. Perez, M. Pan, and T. Lee. 2005. Requirement of Cul3 for axonal arborization and dendritic elaboration in Drosophila mushroom body neurons. J. Neurosci. 25:4189-4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu, X., A. K. Raina, H. G. Lee, G. Casadesus, M. A. Smith, and G. Perry. 2004. Oxidative stress signalling in Alzheimer's disease. Brain Res. 1000:32-39. [DOI] [PubMed] [Google Scholar]