Abstract
We have taken a knockout approach to interrogate the function of protein kinase D (PKD) serine/threonine kinases in lymphocytes. DT40 B cells express two PKD family members, PKD1 and PKD3, which are both rapidly activated by the B-cell antigen receptor (BCR). DT40 cells with single or dual deletions of PKD1 and/or PKD3 were viable, allowing the role of individual PKD isoforms in BCR signal transduction to be assessed. One proposed downstream target for PKD1 in lymphocytes is the class II histone deacetylases (HDACs). Regulation of chromatin accessibility via class II histone deacetylases is an important mechanism controlling gene expression patterns, but the molecules that control this key process in B cells are not known. Herein, we show that phosphorylation and nuclear export of the class II histone deacetylases HDAC5 and HDAC7 are rapidly induced following ligation of the BCR or after treatment with phorbol esters (a diacylglycerol mimetic). Loss of either PKD1 or PKD3 had no impact on HDAC phosphorylation, but loss of both PKD1 and PKD3 abrogated antigen receptor-induced class II HDAC5/7 phosphorylation and nuclear export. These studies reveal an essential and redundant role for PKD enzymes in controlling class II HDACs in B lymphocytes and suggest that PKD serine kinases are a critical link between the BCR and epigenetic control of chromatin.
The protein kinase D (PKD) family comprises three different but closely related serine kinases, PKD1, PKD2, and PKD3, all of which have a highly conserved N-terminal regulatory domain containing two cysteine-rich diacylglycerol (DAG) binding domains and an autoinhibitory pleckstrin homology (PH) domain. PKD enzymes are highly expressed in hematopoietic cells, and they are selectively activated by the engagement of antigen receptors, including the B-cell antigen receptor (BCR), the T-cell receptor, and the FcɛR1 in B cells, T cells, and mast cells, respectively (22, 24). PKD family members are activated by a signaling pathway involving gamma phospholipase C activation, production of diacylglycerol, and activation of classical/novel PKCs. PKC-mediated phosphorylation of two conserved serine residues in the catalytic domains of PKDs is essential for their activation (13, 22, 45, 48, 49). In addition, binding of DAG to the regulatory domain of PKD contributes both to PKD1 activation (50) and to PKD subcellular localization (21, 23, 38-40).
PKD enzymes are predicted to play important functions in controlling lymphocyte biology, but most of the evidence that supports this hypothesis is indirect. For example, studies in transgenic mice have shown that constitutively active PKD1 can substitute for the pre-T-cell antigen receptor complex to regulate early thymocyte differentiation and proliferation (20). In other cell types, PKD enzymes have also been implicated in the regulation of Golgi organization and protein trafficking to the cell surface (1, 8, 12, 14, 18, 53), cell survival (44), NF-κΒ activation (30, 43, 44), glucose transport (5), and integrin activation/recycling (29, 51).
In addition, it has been proposed that PKD1 controls gene transcription via the regulation of class II histone deacetylases (HDACs) in T lymphocytes and in cardiac cells (7, 34, 46). Regulation of chromatin accessibility by acetylation/deacetylation of nucleosomal histones is a key mechanism used to modulate gene expression. Class II HDACs regulate chromatin structure by interacting with various transcription factors (including the myocyte enhancer factor 2 [MEF2] family) to repress their transcriptional activity (47). The repressive effects of class II HDACs on gene transcription are relieved by stimulatory signals that lead to the inducible phosphorylation of several highly conserved serine residues within the N termini of these HDACs. Phosphorylation at these regulatory sites (serine 259 and serine 498 in HDAC5) promotes the nuclear export of class II HDACs, thus disrupting HDAC-MEF2 interactions and allowing MEF2-dependent gene transcription to occur (27, 28).
Evidence that implicates PKD1 as a class II HDAC kinase comes from the observation that purified PKD1 can directly phosphorylate the regulatory serine residues of HDAC7 in vitro (7, 34). Furthermore, using overexpression of constitutively active and kinase-dead versions of PKD1, it has been shown that PKD1 can regulate the phosphorylation of the class II HDACs HDAC5 and HDAC7 in COS cells and in DO11.10 T-cell hybridoma cells (7, 34, 46). However, this approach has not shown whether PKD1 is essential for the regulation of class II HDACs in vivo. This is particularly important, as several other serine/threonine kinases with similar substrate specificities to PKDs can also phosphorylate class II HDACs at the putative PKD phosphorylation sites, including the calcium-calmodulin-regulated CaMKII and CaMKIV serine kinases (15, 19, 25-27) and the AMPK family kinase Mark2 (4).
In this context, current information about PKD1 effects on class II HDACs is based on overexpression experiments with active or kinase-dead mutants of PKD1 that either mimic or disrupt PKD-mediated signaling pathways, an approach that can have limitations. Recent studies have used RNA interference technology to knock down PKD1 expression in HeLa cells, L6 myotubes, and Jurkat cells to implicate PKD enzymes in the regulation of NF-κB activation, glucose transport, and Rap1 activation (5, 29, 44). However, RNA interference reduces but does not remove PKD1 expression. Furthermore, cells can coexpress more than one of the closely related PKD family members, and experiments so far have not addressed the potential for functional redundancy between PKD isoforms.
We have therefore taken a knockout approach to determine the functional requirement for a PKD enzyme(s) in lymphocytes. The avian DT40 B-cell line provides a robust, genetically manipulatable system that has been used extensively to characterize the function of individual BCR-regulated signaling molecules. DT40 B cells are highly proliferative and readily integrate exogenous DNA into specific gene loci by homologous recombination, thus allowing the rapid generation of knockout B-cell lines.
We have generated PKD1-, PKD3-, and double PKD1/3-deficient DT40 B cells, providing us with a unique set of resources with which to probe the role of PKD kinases in B lymphocytes. We have used these PKD-null cells to explore one proposed function for PKD, the regulation of class II HDACs. We show an essential role for PKD enzymes in the control of class II HDAC phosphorylation and function following BCR activation. In this respect, individual PKD isoforms are functionally redundant, with the loss of HDAC regulation only apparent following deletion of the entire cellular PKD protein pool. Significantly, PKD enzymes may regulate both housekeeping and stimulus-induced gene expression patterns in vivo since PKDs are essential for the regulation of both the basal and stimulus-induced phosphorylation of class II HDACs.
MATERIALS AND METHODS
Generation of PKD-deficient DT40 cell lines.
A 780-bp chicken expressed sequence tag (EST) corresponding to the C-terminal 217 residues and 3′ untranslated region (UTR) of chicken PKD3 was identified from the Bursal EST Database (http://genetics.hpi.uni-hamburg.de/dt40est.html). Additional chicken PKD3 cDNA (corresponding to residues 460 to 700 of human PKD3) was obtained by 3′ rapid amplification of cDNA ends using a DT40 cDNA library directionally cloned in a λZAPII vector (gift of T. Kurosaki, Kansai University, Kansai, Japan) and nested primers (5′-CACAGTGCACAATATTCTTGAAGTGTAAATTCCTCAA-3′ and 5′-CACACAGCTTCACTTGAGGGAATGGCTCTGCTGATGC-3′). Primers based on the cPKD3 cDNA sequence were used to PCR amplify a 4.3-kb fragment from wild-type DT40 genomic DNA that contained the C-terminal region and 3′ UTR of the chicken PKD3 gene (see Fig. 1A). Targeting vectors pSL1180-cPKD3-hisD and pSL1180-cPKD3-hygro were generated by subcloning the PKD3 genomic fragment and the pSL1180 cloning vector and replacing the exons corresponding to human PKD3 residues 667 to 750 with histidinol or hygromycin resistance cassettes. The targeting vector pSL1180-cPKD3-hisD was linearized and introduced into wild-type DT40 cells (stably expressing the TeT repressor protein) by electroporation (950 μF, 250 V). Transfectants were selected in the presence of 1 mg/ml histidinol, and resistant clones were screened by genomic PCR. A second round of transfection followed where the linearized pSL1180-PKD3-hygro targeting vector was electroporated into a correctly targeted histidinol-resistant clone, and resulting clones were selected with both 1 mg/ml histidinol and 2 mg/ml hygromycin.
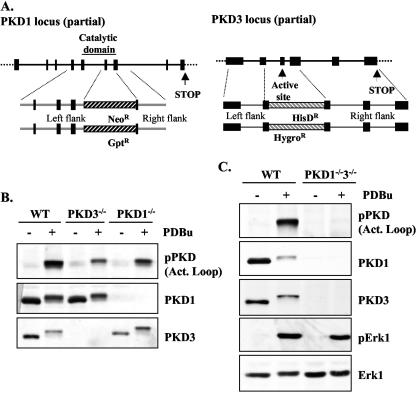
FIG. 1.
Generation of the single and double PKD1 and PKD3 knockout DT40 B cells. (A) Chicken PKD1 and PKD3 genomic loci (partial) were characterized as described in Materials and Methods. (B) Expression and phosphorylation of PKD enzymes in wild-type, PKD1−/−, and PKD3−/− DT40 B cells (either untreated or treated with 25 ng/ml PDBu for 10 min) by Western blotting of whole-cell extracts with the indicated antibodies. (C) Expression and activation of PKD1, PKD3, and Erk1 protein levels in wild-type (WT) and double deficient PKD1/3 DT40 B cells (either untreated or treated with 25 ng/ml PDBu for 10 min) was analyzed by Western blotting of whole-cell extracts with the indicated antibodies. Note: in response to activation, phosphorylated PKD isoforms display reduced immunoreactivity for PKD1 and PKD3 antibodies.
A 575-bp chicken cDNA fragment containing the C-terminal 95 residues and 3′ UTR of chicken PKD1 was identified from the Bursal EST Database. This information, together with sequence information obtained from the chicken genome project, was used to predict the intron-exon structure of the C terminus and 3′ UTR of the cPKD1 gene. PCR primers were used to amplify a 4.2-kb fragment from wild-type DT40 genomic DNA (see Fig. 1A). The targeting vectors pSL1180-cPKD1-Neo and pSL1180-cPKD1-GPT were generated by subcloning the cPKD1 genomic fragment into pSL1180 and replacing the genomic region corresponding to human PKD1 residues 690 to 787 with a neomycin resistance cassette or with a cassette containing the bacterial gpt gene, which confers resistance to mycophenolic acid. Transfections and clonal selection were performed, as described for PKD3, using 2 mg/ml G418 and 10 to 25 μg/ml mycophenolic acid as required.
To generate PKD1/3 double knockout DT40 cells, a PKD3−/− line was transfected with a doxycycline-inducible Flag-tagged PKD3 expression construct to generate subclones that expressed FLAG-PKD3 under tight control of the doxycycline-inducible promoter to generate a PKD3−/− Flag-PKD3+ cell line. We then sequentially targeted the PKD1 locus of this cell line with the above PKD1-Neo- and PKD1-GPT-targeting constructs to obtain PKD1/3 double knockout cells. During the final transfection process, PKD3−/− PKD1−/− Flag-PKD3+ clones were selected for in the presence of doxycycline to maintain Flag-PKD3 expression. No PKD1 protein was detectable in PKD1−/− PKD3−/− cells (hereafter referred to as PKD1−/−3−/− cells) grown under any conditions, but in the presence of doxycycline, the Flag-PKD3 transgene was expressed at levels comparable to the endogenous PKD3 present in wild-type DT40 cells (data not shown). Expression of Flag-PKD3 protein was significantly reduced within 48 h after the removal of doxycycline and was completely lost after 72 h, resulting in a total PKD-null phenotype (see Fig. 1B and data not shown). PKD1−/−3−/− cells washed out of doxycycline for at least 5 days were used for all experiments described here unless otherwise indicated.
Cell culture.
Murine A20 B cells (BALB/c B-cell lymphoma) were cultivated in RPMI 1640 medium supplemented with 10% fetal calf serum, 10 U/ml penicillin-streptomycin, 2 mM glutamine, and 50 μM 2-β-mercaptoethanol at 37°C in 5% CO2. Chicken DT40 B cells stably expressing the TeT repressor protein (Invitrogen) were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum, 1% chicken serum, 10 U/ml penicillin-streptomycin, 2 mM glutamine, and 20 μg/ml blasticidin.
Vectors and cDNA constructs.
The pSL1180 cloning vector was from Promega, and the green fluorescent protein (GFP)-HDAC5 plasmid was previously described (46). The doxycycline-inducible Flag-PKD3 construct and the various GFP-PKD1/GFP-PKD3 constructs have previously been described (22) or were generated by subcloning the various cDNA fragments into the pEGFPC1 vector (Clontech). The GFP-HDAC7 plasmid was generated by PCR amplification of the entire murine HDAC7 coding sequence from Image Clone 6826982 (MRC GeneService) using Pfu DNA polymerase (Roche Diagnostics). Forward (5′-GACTAGATCTCACAGCCCCGGCGCGGGCTG) and reverse (5′-GATCGTCGACCTAGAGGTTCATGGGTTCTTCC) primers contained BglII and SalI restriction sites, respectively, and the resulting PCR product was cloned in-frame into the pEGFPC1 vector (Clontech). The integrity of the GFP-HDAC7 coding sequence was verified by automated ABI DNA sequence analysis (The Sequencing Service, School of Life Sciences, University of Dundee).
Maxipreps and transient transfections.
Maxipreps of plasmid DNA were generated by cesium chloride density centrifugation. For transient transfections, 1 × 107 cells were resuspended in serum-free RPMI, mixed with 10 μg plasmid DNA, and transferred to 4-mm electroporation cuvettes (Bio-Rad GenePulser Xcell). Electroporation was carried out at 310 V/950 μF, and in the case of DT40 cells, cuvettes were placed on ice for 5 min before and after electroporation.
Cell stimulation and lysis.
A20 B cells and DT40 B cells were resuspended in fresh RPMI 1640 medium supplemented with 10% fetal calf serum, 10 U/ml penicillin-streptomycin, and 2 mM glutamine at a concentration of 1 × 107 cells/ml before stimulation. After stimulation, the cells were harvested on ice, washed rapidly in ice-cold phosphate buffered saline (PBS), and lysed on ice in NP-40 lysis buffer (100 mM HEPES [pH 7.4], 150 mM NaCl, 1% Nonidet P-40, 20 mM sodium fluoride, 20 mM iodoacetamide, 2 mM EDTA, 40 mM β-glycerophosphate, protease inhibitors, 1 mM phenylmethylsulfonyl fluoride). After clarification by centrifugation at 16,000 × g for 15 min at 4°C, soluble proteins were precipitated with 1.5 volumes of ice-cold acetone. In some experiments, GFP-HDAC7 proteins were immunoprecipitated from cell lysates by using a GFP polyclonal antibody (BD Biosciences) and protein G Sepharose beads. Protein samples were separated by sodium dodecyl sulfate-8% polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membranes, and detected by Western blot analysis with the indicated antibodies by using standard techniques.
Chloramphenicol acetyltransferase assays.
A class II HDAC-repressed reporter construct containing a 3.8-kb Tth1 fragment of the Nur77 promoter cloned into pCAT-3L was provided by A. Winoto (University of California, Berkeley, Calif.). For reporter assays, cells were lysed in 150 μl of a buffer containing 0.65% (vol/vol) NP-40, 10 mM Tris (pH 8.0), 1 mM EDTA, and 150 mM NaCl for 20 min on ice. Lysates were then transferred to a 68°C water bath for 10 min. Cell debris was pelleted, and aliquots of lysate were removed to a fresh tube in an assay volume of 100 μl, to which 40 μl of a start solution containing 0.5 mM acetyl coenzyme A, 5 mM chloramphenicol, 0.5 M Tris (pH 8.0), and 1 μl per point of 50 μCi/ml [14C]acetyl coenzyme A was added. The assay mixture was incubated for 16 h at 37°C before chloramphenicol was extracted using 150 μl ethyl acetate per point. The amount of radioactivity in the acetylated product (100 μl top phase) and nonacetylated substrate (50 μl bottom phase) for each reaction was determined by liquid scintillation counting of organic and aqueous phases, respectively. Results were calculated as the percent conversion of chloramphenicol to the acetylated form.
Reagents and antibodies.
Complete mini EDTA-free protease inhibitor tablets were from Roche Diagnostics, the M4 monoclonal chicken anti-immunoglobulin M hybridoma cell line was obtained from Riken (Japan), and M4 was purified using standard techniques. A monoclonal GFP antibody was from CRUK (London, England), the phospho-S259 HDAC5 antibody was previously described (46), and a PKD3 polyclonal antibody was previously described (22). All other antibodies were obtained from Cell Signaling Technology Inc. Other reagents were of the highest quality available.
Microscopy.
DT40 B cells expressing either GFP-HDAC5 or GFP-HDAC7 were resuspended in phenol red-free RPMI-0.5% fetal bovine serum and allowed to attach to HCl-rinsed glass coverslips at 37°C for 20 min. Subsequently, the culture medium (containing nonadherent cells) was removed and replaced with medium containing different stimuli, as indicated in the figures. After 2 h of incubation at 37°C, the medium was removed and the cells were fixed in freshly prepared 4% paraformaldehyde-2% sucrose in PBS for 10 min at room temperature. Fixation was stopped by washing one time with PBS-0.1 M ammonium chloride and three times with PBS. Nuclei were stained with 1 μg/ml DAPI in 0.1% Triton X-100-PBS for 5 min at room temperature, and the coverslips were then washed three times in PBS and one time in distilled H2O before mounting them onto microscopy slides using ProLong Antifade (Molecular Probes).
Images were acquired on a Leica SP2 confocal scanning microscope using a 63× Plan Apochromat 1.4 objective lens. Excitation/emission settings were 405 nm/410 to 500 nm for DAPI and 488/505 to 550 nm for GFP. Image conversion and overlay were performed with Adobe Photoshop CS software. Cells were scored as positive or negative for nuclear localized GFP HDAC5/7 (with the nuclear compartment defined as positive for DAPI staining) in a blind manner.
All experiments shown are representative of at least three independent experiments unless otherwise indicated.
RESULTS
We set out to generate a powerful genetic knockout system for studying PKD function in lymphocytes. As previously described, both PKD1 and PKD3 family members are expressed in DT40 B cells, where they are strongly and rapidly activated in response to BCR engagement (22, 42). We could not detect expression of the PKD2 protein in DT40 cell extracts, however (data not shown). Therefore, we made DT40 B cells lacking either PKD1 or PKD3 by targeting critical exons encoding key motifs essential for enzymatic activity (Fig. 1A and B) (see Materials and Methods). As both PKD1 and PKD3 can phosphorylate similar peptide substrates in vitro, raising the possibility of functional redundancy in vivo, we also generated PKD1/PKD3 double deficient DT40 B cells (Fig. 1C) (see Materials and Methods).
Integration of the targeting constructs into correct gene loci was confirmed by the absence of detectable PKD1 and/or PKD3 proteins in the resulting knockout cell lines, as shown by Western blot analysis of whole-cell extracts with isoform-specific antibodies (Fig. 1B and C). The PKD1 polyclonal antibody used was directed against a C-terminal epitope of PKD1/2 that is not present in PKD3. As the amino acid sequence of this epitope is fully conserved between PKD1 and PKD2, this confirmed the absence of PKD2 expression in the DT40 B cell line. As shown in Fig. 1B, loss of either PKD1 or PKD3 partially reduces the amount of detectable, phosphorylated PKD in phorbol ester-treated cell extracts, indicating that PKD1 and PKD3 both make significant contributions to the total PKD pool present in DT40 B cells, whereas no activated PKD was detectable in the PKD1/PKD3 double deficient DT40 B cells (Fig. 1C). These data reveal that the expression and activation of PKD1 and PKD3 are independently regulated in vivo, as the expression and phorbol ester-induced phosphorylation of PKD1 was intact in the PKD3−/− cells and vice versa.
Initial characterization revealed that the PKD1−/− and PKD3−/− DT40 B cells were generally normal: cell surface BCR levels were similar to those of wild-type DT40 B cells, and population-doubling times were similar to those of wild-type cells (within the range of 11 to 13 h) (data not shown). In addition, long-term culture of PKD1−/−3−/− cells showed that the absence of any PKD expression had no adverse effect on cellular growth rates (data not shown), indicating that loss of the total cellular PKD pool does not effect basal DT40 cell proliferation/survival. Further characterization of the PKD1−/−3−/− cells will be described elsewhere.
To date, one proposed downstream target for PKDs in lymphocytes is the class II HDACs (7, 34). The BCR regulates key transcriptional changes in B lymphocytes that control effector function, and a large body of work has focused on the direct regulation of transcription factor activity by the BCR. However, little is known about BCR-mediated signaling events that regulate chromatin acetylation/deacetylation. Inhibitors of HDACs have indicated a role for HDACs in controlling B-cell differentiation and survival (2, 11, 17, 35, 41). HDACs are also implicated in the repression of specific lytic viral genes and upregulation of latent membrane protein 1 during latent Epstein-Barr virus infection of B cells (3, 9, 10, 32, 33, 37). However, little is known about how HDACs are regulated in B lymphocytes; in particular, it is unclear whether the BCR is functionally coupled to class II HDACs in vivo.
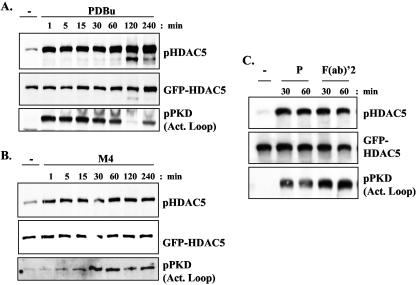
We therefore initially investigated whether activation of the BCR complex or DAG signals (which would activate a PKC-PKD signaling pathway) could regulate class II HDACs in B cells. To assess class II HDAC regulation, we transiently expressed a GFP-tagged HDAC5 protein in avian DT40 B cells or in murine A20 B cells and used a phosphospecific HDAC5 antibody that specifically recognizes HDAC5 molecules phosphorylated at the regulatory serine 259 site (46). Low-level basal phosphorylation of HDAC5 at this key site was consistently observed in resting DT40 B cells, which was rapidly and strongly enhanced after BCR cross-linking or following stimulation with the DAG-mimetic PDBu, and remained elevated over a prolonged time period (Fig. 2A and B). Both the basal and BCR-/PDBu-induced phosphorylations of HDAC5 were lost when a HDAC5 mutant (in which the regulatory serine residues were replaced by nonphosphorylatable alanine residues) was expressed in DT40 cells (data not shown). Antigen receptor activation, or DAG signals alone, also induced robust and sustained phosphorylation of HDAC5 in a murine A20 B-cell line (Fig. 2C).
FIG. 2.
Regulation of HDAC5 phosphorylation by the B-cell antigen receptor complex. (A and B) Avian DT40 B cells or (C) murine A20 B cells were transfected with a GFP-HDAC5 expression construct as described in Materials and Methods. Twenty-four hours after transfection, the cells were left unstimulated (−) or were stimulated with either 10 μg/ml M4 monoclonal antibody (to cross-link the avian BCR), 10 μg/ml of a rabbit anti-mouse F(ab)′2 fragment of immunoglobulin G (to cross-link the murine BCR), or 20 ng/ml PDBu (P) as indicated for the specified times. In all cases, cell extracts were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting with an antibody that specifically recognizes HDAC5 molecules phosphorylated at the regulatory S259 site. Reprobing with an anti-GFP monoclonal antibody determined HDAC5 expression levels. A phosphospecific antibody that recognizes phosphorylated PKD was used to demonstrate the activation of PKD family members.
Class II HDAC-repressed gene targets in B cells are unknown. Therefore, to investigate the role of PKD enzymes in the regulation of class II HDACs, we used a reporter construct linked to the Nur77 promoter (52), which is regulated by class II HDACs following antigen receptor triggering in T cells (7, 34). As demonstrated in Fig. 3A, the activity of the Nur77 reporter was low in untreated wild-type DT40 B cells and was strongly induced in response to either phorbol ester treatment or BCR cross-linking (Fig. 3A). Strikingly, activation of the Nur77 reporter in response to either phorbol esters or BCR triggering was clearly defective in PKD1−/−3−/− B cells (Fig. 3A). These data suggest that PKD enzymes are required for the correct regulation of class II HDACs in DT40 B cells. Currently, class II HDAC-repressed gene targets in B cells that are regulated by PKDs are unknown, but this is under investigation.
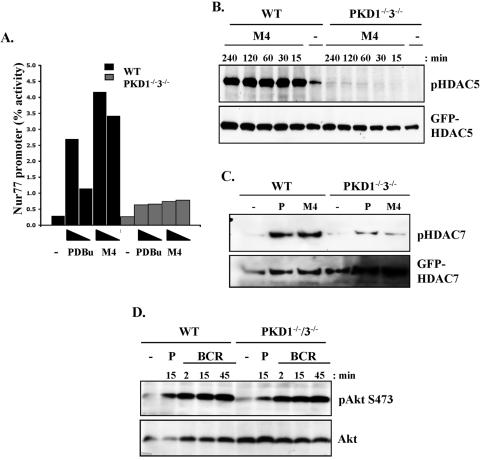
FIG. 3.
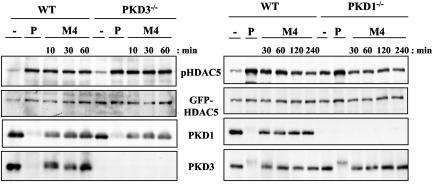
Analysis of class II HDAC function and phosphorylation in PKD1−/−3−/− double knockout DT40 B-cell lines. (A) PKD1−/−PKD3−/− double knockout cells, transfected with the Nur77-CAT reporter construct, were left unstimulated or were stimulated with either PDBu (25 or 10 ng/ml) or the M4 monoclonal antibody (10 or 5 μg/ml) for 6 h. Transactivation of the reporter gene was measured as described in Materials and Methods. WT, wild type. (B) Wild-type and knockout DT40 cells were transfected with a GFP-HDAC5 expression construct, were left unstimulated (−), or were stimulated with 10 μg/ml M4 for the indicated times. HDAC5 phosphorylation and expression were analyzed as described in the legend to Fig. 2. (C) Wild-type and knockout DT40 cells, transfected with a GFP-HDAC7 expression construct, were left unstimulated (−) or were stimulated with 10 μg/ml M4 or with 25 ng/ml PDBu (P) for 30 min. HDAC7 phosphorylation in GFP immunoprecipitates was analyzed using the pHDAC5 S259 antibody since the phosphospecific epitope is highly conserved in HDAC7 (46). (D) Activation of the Akt/PKB serine kinase in untreated (−), PDBu-stimulated (P), or antigen receptor-activated (M4) wild-type and knockout DT40 cells was analyzed by Western blotting whole-cell extracts with the indicated antibodies.
Many signaling molecules other than class II HDACs have been shown to regulate the Nur77 promoter, including Creb and NF-κB (6, 36). Therefore, to determine if PKD enzymes are indeed required to directly regulate class II HDACs, we investigated whether PKD enzymes were required for the regulation of HDAC5 phosphorylation in DT40 B cells. As discussed previously, inducible phosphorylation of the regulatory serine residues in class II HDACs is required to relieve their repressive effects on gene transcription. As observed above, low basal phosphorylation of HDAC5 was detectable in untreated, wild-type DT40 B cells, which was rapidly enhanced in a sustained manner following BCR stimulation (Fig. 3B). Strikingly, both the basal and BCR-induced HDAC5 phosphorylations were almost completely abolished in PKD1−/−3−/− B cells, even after prolonged stimulation (Fig. 3C). Phorbol ester-induced HDAC5 phosphorylation was also almost completely abolished in the PKD1−/−3−/− B cells (see Fig. 5B and data not shown). We went on to investigate the requirement for PKD enzymes in the regulation of another class II HDAC, HDAC7. Similar to our observations for HDAC5, basal phosphorylation of HDAC7 was consistently observed in wild-type DT40 B cells, which was enhanced in response to phorbol ester treatment or BCR triggering (Fig. 3C). Again, both basal and stimulus-induced phosphorylations of HDAC7 were significantly reduced in PKD1−/−3−/− B cells (Fig. 3C). However, the BCR could regulate other downstream signals in PKD1−/− PKD3−/− B cells, as assessed by the activation of the Akt/PKB serine kinase (Fig. 3D).
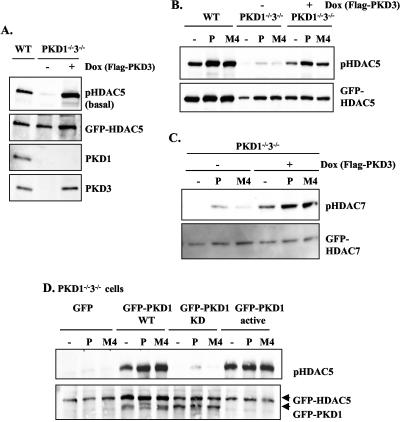
FIG. 5.
Expression of either PKD1 or PKD3 is sufficient to rescue HDAC5 and HDAC7 phosphorylation in PKD1−/−3−/− double knockout DT40 B cells. (A) Basal phosphorylation of HDAC5 is restored in PKD1−/−3−/− cells cultured in doxycycline-containing medium to induce expression of the FLAG-PKD3 transgene (+Dox) but is lost when PKD1−/−3−/− cells were washed out of doxycycline for 5 days (−Dox). The various DT40 cell lines were transfected with a GFP-HDAC5 expression construct, and basal HDAC5 phosphorylation and expression were analyzed as described in the legend to Fig. 2. WT, wild-type. (B) BCR and phorbol ester-induced phosphorylation of HDAC5 is rescued in PKD1−/−3−/− cells cultured in doxycycline-containing medium that express the FLAG-PKD3 transgene (+Dox) but are lost when PKD1−/−3−/− cells were washed out of doxycycline for 5 days (−Dox, no Flag-PKD3 expression). The various DT40 cell lines were transfected with a GFP-HDAC5 expression construct, were left unstimulated (−), or were stimulated with either 10 μg/ml M4 or 20 ng/ml PDBu (P) as indicated for 30 min. HDAC5 phosphorylation and expression was analyzed as described in the text. (C) PKD1−/−3−/− cells cultured with or without doxycycline-containing medium were transfected with a GFP-HDAC7 expression construct and subsequently left unstimulated (−) or were stimulated with 10 μg/ml M4 or with 25 ng/ml PDBu (P) for 30 min. HDAC7 phosphorylation was analyzed as described in the text. (D) Rescue of basal and induced HDAC5 phosphorylation by transient transfection of PKD1 expression constructs in PKD1−/−3−/− cells. PKD1−/−3−/− cells were cotransfected with GFP-HDAC5, together with either a control GFP vector or the indicated GFP-PKD1 expression constructs (KD, kinase-dead; active, isolated catalytic domain). Twenty-four hours after transfection, the cells were left unstimulated (−) or were treated with either 20 ng/ml PDBu or 10 μg/ml M4 for 30 min, as indicated in the figure. HDAC5 phosphorylation and expression were analyzed as described previously.
Given the dysregulation of HDAC5 and HDAC7 in PKD-null DT40 B cells, we went on to address the issue of functional redundancy between PKD1 and PKD3 in this context. To date, the majority of work indicating functional roles for PKD enzymes in vivo have focused on the PKD1 isoform. Although all three PKD isoforms exhibit similar sequence specificity requirements in vitro, they may have distinct functions in vivo, particularly as they can be found localized in distinct subcellular compartments within the same cell (reference 40 and data not shown [M. Spitaler and D. A. Cantrell]). We initially analyzed HDAC5 phosphorylation in single-knockout DT40 B cells lacking either PKD1 or PKD3. We observed that BCR- and phorbol ester-induced phosphorylation of HDAC5 in the PKD3−/− B cells was comparable to that observed in wild-type DT40 cells (Fig. 4). HDAC5 phosphorylation in the PKD1−/− cells appeared elevated basally and was not further enhanced in response to BCR triggering, although the stronger stimulus PDBu did increase HDAC5 phosphorylation in these cells (Fig. 4). These data indicate that loss of a single PKD isoform does not affect HDAC5 phosphorylation: the essential role for PKDs as HDAC kinases in DT40 B cells is revealed only by the loss of both PKD1 and PKD3.
FIG. 4.
HDAC5 phosphorylation remains intact in single PKD1−/− or PKD3−/− knockout DT40 B cells. Wild-type (WT) and PKD1−/− or PKD3−/− knockout DT40 cells were transfected with a GFP-HDAC5 expression construct, were left unstimulated (−), or were stimulated with either 25 ng/ml PDBu (P) for 60 min or 10 μg/ml M4 for the indicated times. HDAC5 phosphorylation and expression were analyzed as described in the legend to Fig. 1. Expression of residual PKD1 or -3 in the PKD3 and PKD1 single-knockout cell lines was analyzed by Western blotting with the indicated antibodies.
To further investigate the functional redundancy of PKD1 and PKD3 in the control of HDAC5 phosphorylation, we expressed a PKD3 transgene (under the control of a doxycycline-inducible promoter) in our PKD1−/−3−/− B cells. We observed that basal phosphorylation of HDAC5 was restored in PKD1−/−3−/− cells that inducibly expressed the Flag-PKD3 transgene but not in PKD1−/−3−/− cells that were cultured in the absence of doxycycline and thus lacked Flag-PKD3 expression (Fig. 5A, −Dox, +Dox). Importantly, phorbol ester- and BCR-induced HDAC5 phosphorylation was also restored to wild-type levels in PKD1−/−3−/− DT40 cells that inducibly expressed the Flag-PKD3 transgene (Fig. 5B, +Dox) but again, not in PKD1−/−3−/− cells cultured in the absence of doxycycline (Fig. 5B, −Dox). We also observed that doxycycline-induced expression of the Flag-PKD3 transgene was sufficient to increase basal and BCR/phorbol ester-induced phosphorylation of HDAC7 in the PKD1−/−3−/− cells (Fig. 5C, −Dox, +Dox).
Transient reconstitution of the PKD1−/−3−/− double knockout cells with wild-type GFP-tagged PKD1 also rescued both basal and phorbol ester/BCR-induced HDAC5 phosphorylation compared to control, GFP-transfected cells (Fig. 5D). Strikingly, expression of kinase-dead PKD1 in the double knockout cells did not rescue HDAC5 phosphorylation (either in untreated or in stimulated cells), whereas expression of constitutively activated PKD1 resulted in a high basal phosphorylation of HDAC5 that could not be further enhanced by external stimuli (Fig. 5D). Identical results were observed when the PKD1/3 double knockout cells were transfected with activated and kinase-dead versions of PKD3 (data not shown).
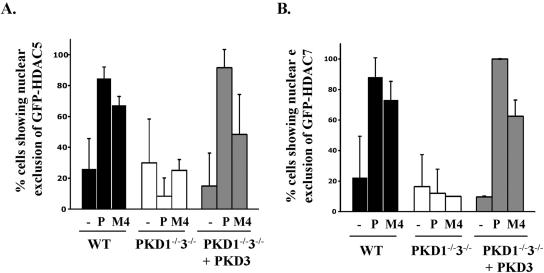
Phosphorylation of class II HDACs controls their nuclear export and thus dictates whether HDACs can exert their repressive effects on target gene expression. Having shown that PKDs were required for class II HDAC phosphorylation, we therefore looked to see if there was a role for PKDs in controlling HDAC cellular localization. We quantified the subcellular distribution of GFP-tagged HDAC5 or HDAC7 in unstimulated and stimulated wild-type and PKD1−/−3−/− DT40 B cells by confocal microscopy. Predominantly, unstimulated wild-type DT40 B cells showed nuclear localization of GFP-HDAC5 and GFP-HDAC7 (Fig. 6). Following BCR engagement or phorbol ester treatment, both HDAC5 and HDAC7 were observed to consistently redistribute from the nucleus to the cytosol. However, there was no significant nuclear exclusion of HDAC5 or HDAC7 in BCR- or phorbol ester-stimulated PKD1−/−3−/− DT40 B cells (Fig. 6). The data in Fig. 6 thus show that in PKD1/3-null B cells, HDAC5 and HDAC7 do not redistribute from the nucleus to the cytosol in response to BCR or phorbol ester signaling.
FIG. 6.
Class II HDAC nuclear export in PKD1−/−3−/− double knockout DT40 B-cell lines. Wild-type (WT) or PKD1−/−3−/− double knockout cells (cultured with or without doxycycline to induce Flag-PKD3 expression or not) were transfected with GFP-HDAC5 (A) or GFP-HDAC7 (B) expression constructs before the cells were left unstimulated or were stimulated with either PDBu (P, 25 ng/ml) or the M4 monoclonal antibody (10 μg/ml) for 2 h. The cells were then fixed and processed for microscopy as described in Materials and Methods. The data are presented as the percentage of cells showing nuclear exclusion of GFP-HDAC5 or GFP-HDAC7 from two to four separate experiments.
We went on to determine whether reintroduction of PKD3 in the PKD1/3 knockout cells could restore normal HDAC5/7 localization patterns. As shown in Fig. 6, triggering of the BCR or phorbol ester stimulation in PKD1−/−3−/− B cells that expressed a PKD3 transgene could now induce nuclear exclusion of HDAC5 and HDAC7. Thus, the rescue of HDAC5 and HDAC7 phosphorylation after reexpression of PKD3 in the double knockout cells was accompanied by normal nuclear exclusion of these class II HDACs in response to phorbol ester or BCR stimulation (Fig. 5 versus Fig. 6).
DISCUSSION
The serine kinase PKD1 has been implicated in the regulation of gene transcription by controlling the phosphorylation status of class II HDACs in T lymphocytes and cardiac cells (7, 34, 46). Class II HDACs function to suppress specific gene transcription, and regulation of their function via phosphorylation is a key mechanism to control gene expression patterns in response to extracellular signals. We have shown that the B-cell antigen receptor regulates both the phosphorylation status and nuclear localization of the class II HDACs HDAC5 and HDAC7, as well as the activity of a class II HDAC-repressed reporter gene in DT40 B lymphocytes. To date, three classes of HDAC kinases have been identified, namely, the CaMKs, Mark2, and PKD1, all of which are capable of phosphorylating the signal-responsive phosphorylation motifs in class II HDACs in vitro in a variety of cell types. Herein, we show that PKD1 can regulate HDAC phosphorylation in B cells but is not essential for BCR or phorbol ester-induced HDAC phosphorylation. Hence, in the absence of PKD1, the other PKD family isoform expressed in B cells, PKD3, can compensate and mediate BCR-induced HDAC5 phosphorylation. PKD-1/3 double deficient B cells appear to proliferate and survive normally in culture. However, phorbol ester-induced and BCR-induced phosphorylation/nuclear export of HDAC5 and HDAC7, as well as the activation of a class II HDAC-repressed reporter gene, is abolished in PKD1/3-deficient B cells. PKD family kinases are thus the key kinases that regulate the phosphorylation and subcellular localization of class II HDACs in DT40 B cells and here, PKD1 and PKD3 have functionally redundant roles.
Most studies of PKD family serine kinases describe these as playing a role in the PLC/DAG-induced signaling pathways triggered by cell surface receptors. However, we show that low-level basal phosphorylation of HDAC5 occurs in B cells in the absence of antigen receptor stimulation, which is also absolutely dependent on PKD expression. Importantly, PKD activity is required for this basal HDAC5 phosphorylation, as expression of catalytically inactive PKD1 or PKD3 molecules in PKD1−/−3−/− B cells is not sufficient to rescue basal HDAC5 phosphorylation. Thus, PKD enzymes are essential to regulate HDAC5 both during conditions of basal cell growth and also during acute cellular activation. Recently, the significance of ligand-independent tonic signaling via the BCR has gained prominence and appears to be important for controlling B-cell survival (16, 31). Our data would indicate that low-level tonic PKD activity is also present in B cells, and this may function to modulate housekeeping gene expression patterns via the control of class II HDACs. Whether this basal PKD activity is dependent on tonic BCR signaling or whether it is due to other factors is unknown at present.
To date, the question of functional redundancy among PKD family members has not been addressed. This is important, as different PKD isoforms can be found differentially localized in distinct subcellular compartments within the same cell type (39, 40). As PKD kinases are activated by a common DAG/PKC signaling cascade to phosphorylate similar peptide sequence motifs in vitro, this observation has led to the hypothesis that different PKD isoforms may have distinct roles or phosphorylate different substrates in vivo. We have now shown that PKD1 and PKD3 are functionally redundant in regards to the regulation of class II HDAC5 and HDAC7: loss of single PKD isoforms has no impact on HDAC5 phosphorylation, and either PKD1 or PKD3 can restore HDAC5/7 phosphorylation in PKD1−/−3−/− B cells. Of note, exogenous expression of the third PKD isoform, PKD2, which is not normally expressed in DT40 B cells, also rescues normal HDAC5 phosphorylation in PKD-null cells (data not shown). Whether this functional redundancy extends to all biological functions of PKD1, PKD2, and PKD3 enzymes in vivo remains to be determined. If PKD enzymes are fully redundant in vivo, it is striking that the genomes of higher organisms encode multiple PKD isoforms and that many cell types express more than one family member.
In summary, our data demonstrate a specific and essential role for PKD family kinases for the regulation of gene transcription via control of class II HDAC phosphorylation and nuclear export in B lymphocytes and suggest a critical link between the BCR, PKD activity, and epigenetic control of chromatin structure and gene expression. Calcium/calmodulin-regulated CaMKs and the LKB1-regulated kinase Mark2 have also been proposed to be capable of phosphorylating the signal-responsive phosphorylation motifs in class II HDACs. Although PKD isoforms are essential for the regulation of class II HDAC kinases in B lymphocytes in response to antigen receptor signals, functional roles for other proposed HDAC kinases in different cellular/receptor-signaling contexts in different cell lineages cannot be ruled out.
Acknowledgments
We thank members of the Cantrell lab for useful discussions and R. Chamberlyne for providing the GFP-PKD3 kinase-dead and constitutively active constructs.
This work was supported by a Royal Society Dorothy Hodgkin Research Fellowship (S. A. Matthews), The Wellcome Trust (D. A. Cantrell), and NIH grant AI45901 (A. M. Scharenberg).
REFERENCES
- 1.Baron, C. L., and V. Malhotra. 2002. Role of diacylglycerol in PKD recruitment to the TGN and protein transport to the plasma membrane. Science 295:325-328. [DOI] [PubMed] [Google Scholar]
- 2.Bereshchenko, O. R., W. Gu, and R. Dalla-Favera. 2002. Acetylation inactivates the transcriptional repressor BCL6. Nat. Genet. 32:606-613. [DOI] [PubMed] [Google Scholar]
- 3.Bryant, H., and P. J. Farrell. 2002. Signal transduction and transcription factor modification during reactivation of Epstein-Barr virus from latency. J. Virol. 76:10290-10298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, S., S. Bezprozvannaya, S. Li, and E. N. Olson. 2005. An expression screen reveals modulators of class II histone deacetylase phosphorylation. Proc. Natl. Acad. Sci. USA 102:8120-8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, J., G. Lu, and Q. J. Wang. 2005. Protein kinase C-independent effects of protein kinase D3 in glucose transport in L6 myotubes. Mol. Pharmacol. 67:152-162. [DOI] [PubMed] [Google Scholar]
- 6.Darragh, J., A. Soloaga, V. A. Beardmore, A. Wingate, G. R. Wiggin, M. Peggie, and J. S. Arthur. 2005. MSKs are required for the transcription of the nuclear orphan receptors Nur77, Nurr1 and Nor1 downstream of MAPK signalling. Biochem. J. 390:749-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dequiedt, F., J. Van Lint, E. Lecomte, V. Van Duppen, T. Seufferlein, J. R. Vandenheede, R. Wattiez, and R. Kettmann. 2005. Phosphorylation of histone deacetylase 7 by protein kinase D mediates T cell receptor-induced Nur77 expression and apoptosis. J. Exp. Med. 201:793-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diaz Anel, A. M., and V. Malhotra. 2005. PKCeta is required for beta1gamma2/beta3gamma2- and PKD-mediated transport to the cell surface and the organization of the Golgi apparatus. J. Cell Biol. 169:83-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gradoville, L., D. Kwa, A. El-Guindy, and G. Miller. 2002. Protein kinase C-independent activation of the Epstein-Barr virus lytic cycle. J. Virol. 76:5612-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gruffat, H., E. Manet, and A. Sergeant. 2002. MEF2-mediated recruitment of class II HDAC at the EBV immediate early gene BZLF1 links latency and chromatin remodeling. EMBO Rep. 3:141-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo, F., C. Sigua, J. Tao, P. Bali, P. George, Y. Li, S. Wittmann, L. Moscinski, P. Atadja, and K. Bhalla. 2004. Cotreatment with histone deacetylase inhibitor LAQ824 enhances Apo-2L/tumor necrosis factor-related apoptosis inducing ligand-induced death inducing signaling complex activity and apoptosis of human acute leukemia cells. Cancer Res. 64:2580-2589. [DOI] [PubMed] [Google Scholar]
- 12.Hausser, A., P. Storz, S. Martens, G. Link, A. Toker, and K. Pfizenmaier. 2005. Protein kinase D regulates vesicular transport by phosphorylating and activating phosphatidylinositol-4 kinase IIIbeta at the Golgi complex. Nat. Cell Biol. 7:880-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iglesias, T., R. T. Waldron, and E. Rozengurt. 1998. Identification of in vivo phosphorylation sites required for protein kinase D activation. J. Biol. Chem. 273:27662-27667. [DOI] [PubMed] [Google Scholar]
- 14.Jamora, C., N. Yamanouye, J. Van Lint, J. Laudenslager, J. R. Vandenheede, D. J. Faulkner, and V. Malhotra. 1999. Gbetagamma-mediated regulation of Golgi organization is through the direct activation of protein kinase D. Cell 98:59-68. [DOI] [PubMed] [Google Scholar]
- 15.Kao, H. Y., A. Verdel, C. C. Tsai, C. Simon, H. Juguilon, and S. Khochbin. 2001. Mechanism for nucleocytoplasmic shuttling of histone deacetylase 7. J. Biol. Chem. 276:47496-47507. [DOI] [PubMed] [Google Scholar]
- 16.Kraus, M., M. B. Alimzhanov, N. Rajewsky, and K. Rajewsky. 2004. Survival of resting mature B lymphocytes depends on BCR signaling via the Igalpha/beta heterodimer. Cell 117:787-800. [DOI] [PubMed] [Google Scholar]
- 17.Lee, S. C., A. Bottaro, and R. A. Insel. 2003. Activation of terminal B cell differentiation by inhibition of histone deacetylation. Mol. Immunol. 39:923-932. [DOI] [PubMed] [Google Scholar]
- 18.Liljedahl, M., Y. Maeda, A. Colanzi, I. Ayala, J. Van Lint, and V. Malhotra. 2001. Protein kinase D regulates the fission of cell surface destined transport carriers from the trans-Golgi network. Cell 104:409-420. [DOI] [PubMed] [Google Scholar]
- 19.Lu, J., T. A. McKinsey, R. L. Nicol, and E. N. Olson. 2000. Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc. Natl. Acad. Sci. USA 97:4070-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marklund, U., K. Lightfoot, and D. Cantrell. 2003. Intracellular location and cell context-dependent function of protein kinase D. Immunity 19:491-501. [DOI] [PubMed] [Google Scholar]
- 21.Matthews, S., T. Iglesias, D. Cantrell, and E. Rozengurt. 1999. Dynamic re-distribution of protein kinase D (PKD) as revealed by a GFP-PKD fusion protein: dissociation from PKD activation. FEBS Lett. 457:515-521. [DOI] [PubMed] [Google Scholar]
- 22.Matthews, S. A., R. Dayalu, L. J. Thompson, and A. M. Scharenberg. 2003. Regulation of protein kinase Cnu by the B-cell antigen receptor. J. Biol. Chem. 278:9086-9091. [DOI] [PubMed] [Google Scholar]
- 23.Matthews, S. A., T. Iglesias, E. Rozengurt, and D. Cantrell. 2000. Spatial and temporal regulation of protein kinase D (PKD). EMBO J. 19:2935-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matthews, S. A., E. Rozengurt, and D. Cantrell. 2000. Protein kinase D. A selective target for antigen receptors and a downstream target for protein kinase C in lymphocytes. J. Exp. Med. 191:2075-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKinsey, T. A., C. L. Zhang, J. Lu, and E. N. Olson. 2000. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 408:106-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKinsey, T. A., C. L. Zhang, and E. N. Olson. 2000. Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proc. Natl. Acad. Sci. USA 97:14400-14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKinsey, T. A., C. L. Zhang, and E. N. Olson. 2001. Identification of a signal-responsive nuclear export sequence in class II histone deacetylases. Mol. Cell. Biol. 21:6312-6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKinsey, T. A., C. L. Zhang, and E. N. Olson. 2002. Signaling chromatin to make muscle. Curr. Opin. Cell Biol. 14:763-772. [DOI] [PubMed] [Google Scholar]
- 29.Medeiros, R. B., D. M. Dickey, H. Chung, A. C. Quale, L. R. Nagarajan, D. D. Billadeau, and Y. Shimizu. 2005. Protein kinase D1 and the beta1 integrin cytoplasmic domain control beta1 integrin function via regulation of rap1 activation. Immunity 23:213-226. [DOI] [PubMed] [Google Scholar]
- 30.Mihailovic, T., M. Marx, A. Auer, J. Van Lint, M. Schmid, C. Weber, and T. Seufferlein. 2004. Protein kinase D2 mediates activation of nuclear factor kappaB by Bcr-Abl in Bcr-Abl+ human myeloid leukemia cells. Cancer Res. 64:8939-8944. [DOI] [PubMed] [Google Scholar]
- 31.Monroe, J. G. 2004. Ligand-independent tonic signaling in B-cell receptor function. Curr. Opin. Immunol. 16:288-295. [DOI] [PubMed] [Google Scholar]
- 32.Nishikawa, J., L. L. Kis, A. Liu, X. Zhang, M. Takahara, K. Bandobashi, C. Kiss, N. Nagy, K. Okita, G. Klein, and E. Klein. 2004. Upregulation of LMP1 expression by histone deacetylase inhibitors in an EBV carrying NPC cell line. Virus Genes 28:121-128. [DOI] [PubMed] [Google Scholar]
- 33.Park, J. H., and D. V. Faller. 2002. Epstein-Barr virus latent membrane protein-1 induction by histone deacetylase inhibitors mediates induction of intercellular adhesion molecule-1 expression and homotypic aggregation. Virology 303:345-363. [DOI] [PubMed] [Google Scholar]
- 34.Parra, M., H. Kasler, T. A. McKinsey, E. N. Olson, and E. Verdin. 2005. Protein kinase D1 phosphorylates HDAC7 and induces its nuclear export after T-cell receptor activation. J. Biol. Chem. 280:13762-13770. [DOI] [PubMed] [Google Scholar]
- 35.Pasqualucci, L., O. Bereschenko, H. Niu, U. Klein, K. Basso, R. Guglielmino, G. Cattoretti, and R. Dalla-Favera. 2003. Molecular pathogenesis of non-Hodgkin's lymphoma: the role of Bcl-6. Leuk. Lymphoma 44(Suppl. 3):S5-S12. [DOI] [PubMed] [Google Scholar]
- 36.Pei, L., A. Castrillo, M. Chen, A. Hoffmann, and P. Tontonoz. 2005. Induction of NR4A orphan nuclear receptor expression in macrophages in response to inflammatory stimuli. J. Biol. Chem. 280:29256-29262. [DOI] [PubMed] [Google Scholar]
- 37.Radkov, S. A., R. Touitou, A. Brehm, M. Rowe, M. West, T. Kouzarides, and M. J. Allday. 1999. Epstein-Barr virus nuclear antigen 3C interacts with histone deacetylase to repress transcription. J. Virol. 73:5688-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rey, O., S. H. Young, D. Cantrell, and E. Rozengurt. 2001. Rapid protein kinase D translocation in response to G protein-coupled receptor activation. Dependence on protein kinase C. J. Biol. Chem. 276:32616-32626. [DOI] [PubMed] [Google Scholar]
- 39.Rey, O., J. Yuan, and E. Rozengurt. 2003. Intracellular redistribution of protein kinase D2 in response to G-protein-coupled receptor agonists. Biochem. Biophys. Res. Commun. 302:817-824. [DOI] [PubMed] [Google Scholar]
- 40.Rey, O., J. Yuan, S. H. Young, and E. Rozengurt. 2003. Protein kinase C nu/protein kinase D3 nuclear localization, catalytic activation, and intracellular redistribution in response to G protein-coupled receptor agonists. J. Biol. Chem. 278:23773-23785. [DOI] [PubMed] [Google Scholar]
- 41.Sakajiri, S., T. Kumagai, N. Kawamata, T. Saitoh, J. W. Said, and H. P. Koeffler. 2005. Histone deacetylase inhibitors profoundly decrease proliferation of human lymphoid cancer cell lines. Exp. Hematol. 33:53-61. [DOI] [PubMed] [Google Scholar]
- 42.Sidorenko, S. P., C. L. Law, S. J. Klaus, K. A. Chandran, M. Takata, T. Kurosaki, and E. A. Clark. 1996. Protein kinase C mu (PKC mu) associates with the B cell antigen receptor complex and regulates lymphocyte signaling. Immunity 5:353-363. [DOI] [PubMed] [Google Scholar]
- 43.Storz, P., H. Döppler, and A. Toker. 2004. Protein kinase Cδ selectively regulates protein kinase D-dependent activation of NF-κB in oxidative stress signaling. Mol. Cell. Biol. 24:2614-2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Storz, P., and A. Toker. 2003. Protein kinase D mediates a stress-induced NF-kappaB activation and survival pathway. EMBO J. 22:109-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sturany, S., J. Van Lint, F. Muller, M. Wilda, H. Hameister, M. Hocker, A. Brey, U. Gern, J. Vandenheede, T. Gress, G. Adler, and T. Seufferlein. 2001. Molecular cloning and characterization of the human protein kinase D2. A novel member of the protein kinase D family of serine threonine kinases. J. Biol. Chem. 276:3310-3318. [DOI] [PubMed] [Google Scholar]
- 46.Vega, R. B., B. C. Harrison, E. Meadows, C. R. Roberts, P. J. Papst, E. N. Olson, and T. A. McKinsey. 2004. Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Mol. Cell. Biol. 24:8374-8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Verdin, E., F. Dequiedt, and H. G. Kasler. 2003. Class II histone deacetylases: versatile regulators. Trends Genet. 19:286-293. [DOI] [PubMed] [Google Scholar]
- 48.Waldron, R. T., T. Iglesias, and E. Rozengurt. 1999. Phosphorylation-dependent protein kinase D activation. Electrophoresis 20:382-390. [DOI] [PubMed] [Google Scholar]
- 49.Waldron, R. T., O. Rey, T. Iglesias, T. Tugal, D. Cantrell, and E. Rozengurt. 2001. Activation loop Ser744 and Ser748 in protein kinase D are transphosphorylated in vivo. J. Biol. Chem. 276:32606-32615. [DOI] [PubMed] [Google Scholar]
- 50.Wood, C. D., U. Marklund, and D. A. Cantrell. 2005. Dual phospholipase C/diacylglycerol requirement for protein kinase D1 activation in lymphocytes. J. Biol. Chem. 280:6245-6251. [DOI] [PubMed] [Google Scholar]
- 51.Woods, A. J., D. P. White, P. T. Caswell, and J. C. Norman. 2004. PKD1/PKCmu promotes alphavbeta3 integrin recycling and delivery to nascent focal adhesions. EMBO J. 23:2531-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woronicz, J. D., A. Lina, B. J. Calnan, S. Szychowski, L. Cheng, and A. Winoto. 1995. Regulation of the Nur77 orphan steroid receptor in activation-induced apoptosis. Mol. Cell. Biol. 15:6364-6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yeaman, C., M. I. Ayala, J. R. Wright, F. Bard, C. Bossard, A. Ang, Y. Maeda, T. Seufferlein, I. Mellman, W. J. Nelson, and V. Malhotra. 2004. Protein kinase D regulates basolateral membrane protein exit from trans-Golgi network. Nat. Cell Biol. 6:106-112. [DOI] [PMC free article] [PubMed] [Google Scholar]