Abstract
We have studied the functional and structural properties of nucleosomes reconstituted with H2BFWT, a recently identified putative histone variant of the H2B family with totally unknown function. We show that H2BFWT can replace the conventional histone H2B in the nucleosome. The presence of H2BFWT did not affect the overall structure of the nucleosome, and the H2BFWT nucleosomes exhibited the same stability as conventional nucleosomes. SWI/SNF was able to efficiently remodel and mobilize the H2BFWT nucleosomes. Importantly, H2BFWT, in contrast to conventional H2B, was unable to recruit chromosome condensation factors and to participate in the assembly of mitotic chromosomes. This was determined by the highly divergent (compared to conventional H2B) NH2 tail of H2BFWT. These data, in combination with the observations that H2BFWT was found by others in the sperm nuclei and appeared to be associated with the telomeric chromatin, suggest that H2BFWT could act as a specific epigenetic marker.
DNA is packaged in the somatic nucleus into chromatin. The nucleosome, the basal repeating unit of chromatin, is constituted of an octamer of the core histones (two of each H2A, H2B, H3, and H4), around which two superhelical turns of DNA are wrapped (48). The structures of both the histone octamer (5, 6) and the nucleosome (33) were solved by X-ray crystallography. Each histone within the octamer consists of a structured domain (the histone fold) and a nonstructured NH2 tail.
The NH2 tails, initially defined by their sensitivity to proteases, contain about 25% of the total mass of the core histones (5, 48). The tails are very basic, and UV laser protein-DNA cross-linking has shown that they bind to chromatin both in vitro and in vivo (36, 46). The NH2 tails are not required for either the assembly or the maintenance of the nucleosome particle, and their cleavage results in a very slight effect on the structural properties of the nucleosome (7, 24). However, the NH2 tails play an essential role in the folding and the maintenance of the 30-nm chromatin fiber (23). Both the compaction of the chromatin fiber and the orientation of the nucleosomes relative to the fiber axis are dependent on the integrity of the NH2 tails (1, 17). The tails of the individual histones appeared to be involved in distinct phenomena. For example, the integrity of the histone H4 tail is fully required for the folding of the 30-nm chromatin fiber (18), while the tail of the histone H2B is crucial for the assembly of mitotic chromosomes (13, 14). The NH2 tails of histones H3 and H4 are selectively required for the p300-dependent transcriptional activation of chromatin (2).
In addition to conventional histones, the cells express histone variants. The histone variants are nonallelic forms of the conventional histones, and they are present in the cell in a very small amount compared to the conventional ones (48). The histone variants participate in the regulation of different processes within the cell (26, 44). The best studied are the histone variants of the H2A and H3 families (22, 35, 42). The histone variant H2AX is involved in DNA repair and the maintenance of the stability of the genome (8, 9). Another histone variant, H2A.Z, appeared to be implicated in gene activation, gene silencing, and chromosome segregation (15, 41, 43). The inactive X chromosome is believed to be enriched in the histone variant macroH2A, which could play an essential role in its inactivation (12, 28). CENP-A, a universal histone variant of the H3 family, is specifically associated with the centromeric sequences and is crucial for the assembly and maintenance of the kinetochores (26, 27), while active genes are sites of replacement of H3.3, another universal histone H3 variant (26).
Few H2B variants have been identified to date, and their function is largely unknown (10, 21, 31). Recently, a novel putative H2B variant, H2BFWT, was cloned (10). This putative histone variant exhibits very low homology (45% identity) with the conventional H2B, with its NH2 tail showing the lowest homology. In this work we have studied the structural and functional properties of nucleosomes reconstituted with H2BFWT and compared them with the properties of conventional nucleosomes. Our data suggest that H2BFWT, in contrast to conventional H2B, is unable to recruit chromosome condensation factors and to assist mitotic chromosome assembly.
MATERIALS AND METHODS
Preparation of DNA probes.
The 152-bp fragment containing the Xenopus borealis 5S RNA gene was obtained from plasmid pXP10 as described previously (30). The 255-bp and 241-bp DNA fragments containing nucleosome-positioning sequence 601 (32), allowing reconstitution of end- or centrally positioned nucleosomes, were prepared by PCR amplification using 32P-labeled primers and plasmids pGEM3Z-601 and p199-1 (kind gifts from J. Widom and B. Bartholomew), respectively. Labeled DNA fragments were gel purified.
H2BFWT cloning and nucleosome reconstitution.
The H2BFWT coding sequence was amplified by PCR by using the EST IMAGE clone 5266336 cDNA and the primers 5′-CAGTGGCCATATGGCCACTGCCTCCGCC (forward) and 5′-TGAGGATCCTCACTTTCTCTGTTGCTGTATG (reverse). The PCR product was cloned in a pET3a vector (Novagen). H2BFWT was overexpressed in C41(DE3) cells (Avidis) and purified to homogeneity. The H2BFWT sequence (EST IMAGE clone 5266336 cDNA) contains two potential AUG start codons. We have used AUG start codon 67 (M23), which allows the translation of a shorter version of H2BFWT (153 amino acids), since we have observed this form of H2BFWT to be expressed in vivo (data not shown).
The green fluorescent protein (GFP)-H2BFWT and the hemagglutinin (HA)-H2BFWT (pcDNA/HA-H2BFWT) vectors, the chimeric histone NterH2B-FWT expression vector (which contains the sequence corresponding to the NH2 tail of the conventional human H2B [amino acids 1 to E36] in a fusion with the globular part of H2BFWT [from S80 to K175]), and the H3Nter-gH2A expression vector (which contains the sequence corresponding to the NH2 tail of H3 fused to the histone fold domain of H2A) were prepared using standard techniques.
Recombinant Xenopus laevis variant H2BFWT or mutant histones were expressed in bacteria and were purified to homogeneity (13). To reconstitute nucleosomes, an equimolar mixture of the histones was dialyzed overnight at 4°C against histone folding buffer (10 mM Tris [pH 7.5], 5 mM β-mercaptoethanol, 1 mM EDTA) containing 2.0 M NaCl. The histones were then mixed at a 0.8:1 molar ratio with a mixture of the 32P-labeled DNA fragment and nonlabeled nucleosomal size bulk DNA (at a ratio of 1:15) and stepwise dialyzed against decreasing concentration of NaCl down to 10 mM (36).
EMSA, DNase I footprinting, and exonuclease III mapping.
Electrophoretic mobility shift assay (EMSA) was carried out in a 5% polyacrylamide gel (acrylamide/bisacrylamide ratio, 29:1 [wt/wt]) in the presence of 1% glycerol (3). DNase I footprinting and exonuclease mapping of the nucleosome boundaries were done as described previously (3, 25). Nucleosome dilution experiments were performed by using histone H3 that was 32P labeled at serine 10 (11).
Histone transfer experiments.
For the histone transfer experiment, swapped-tail H3-H2A mutant histone (H3Nter-gH2A) was used. This allows the mutant H2A (H3Nter-gH2A) to be radioactively labeled at serine 10 by the Aurora A kinase (45) and its transfer to the H3-H4 tetrameric particle to be studied.
Briefly, conventional and H2BFWT nucleosomes were reconstituted by using 32P-labeled H3Nter-gH2A and unlabeled 255-bp 601 DNA fragment. Several reaction mixtures containing 14 ng of histone-labeled, centrally positioned nucleosomes in remodeling buffer together with a threefold molar excess of nonlabeled tetrameric H3-H4 particles (reconstituted on a 152-bp DNA fragment containing the 5S RNA gene of X. borealis) and 2 μl of SWI/SNF in a 10-μl final volume were prepared. The mixtures were incubated at 30°C for different times, the reactions were arrested, and the samples were stored on ice until being loaded on the gel. EMSA was carried out at 4°C (3).
Immunofluorescence and fluorescence recovery after photobleaching (FRAP).
HeLa or A431 cells were transfected with either GFP-H2A, GFP-H2BFWT, or HA-H2BFWT constructs by using either FuGENE 6 reagent (Roche) or Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's protocol. Nucleosomes were prepared by digestion of isolated nuclei from the transfected cells with micrococcal nuclease, followed by purification on 5 to 30% sucrose gradients containing 0.6 M NaCl (13). The immunoblotting was done according to the protocol described by Mutskov et al. (36). Stable transfected cells were selected with gentamicin (500 μg/ml). The positivity of the amplified cell clones was checked by fluorescence. Stable cell clones were established as described previously (20). Photobleaching and confocal microscopy were performed on a Zeiss LSM510 laser scanning confocal instrument as described previously (20).
Mitotic chromosome assembly.
Mitotic extracts from Xenopus laevis eggs were prepared as described previously (13). Assembly of mitotic chromosomes and nucleosome competition experiments were performed in a 50-μl final volume of reaction mixture exactly as described previously (13).
RESULTS
In vivo incorporation of HA-H2BFWT within the nucleosome.
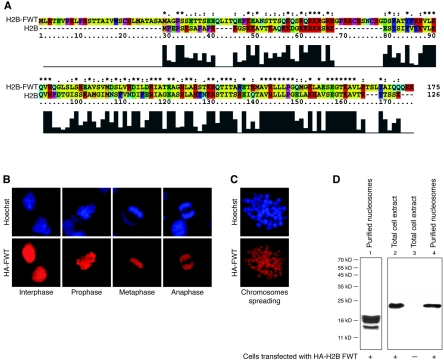
The putative histone variant H2BFWT exhibits a relatively low (45%) identity to the conventional H2B (Fig. 1A). This raises the question of whether H2BFWT is really a histone protein and whether it can be properly incorporated into the nucleosome. To address this question, we ectopically expressed HA-tagged H2BFWT in HeLa cells and visualized it by immunofluorescence microscopy (Fig. 1B and C) and Western blotting (Fig. 1D, lane 2) with anti-HA antibody. The Western blotting shows a specific reaction with the anti-HA antibody for extracts isolated from cells transfected with the HA-H2BFWT expression vector (Fig. 1D, compare lane 2 with lane 3). The anti-HA immunofluorescent staining shows that HA-H2BFWT is associated with chromatin throughout the cell cycle (Fig. 1B). This was further confirmed by staining of mitotic chromosome spreads with the anti-HA antibody (Fig. 1C). To analyze the incorporation of HA-H2BFWT into the nucleosome, nucleosomes were isolated from HeLa cells expressing HA-H2BFWT and purified through a sucrose gradient containing 0.6 M NaCl. The electrophoretic analysis shows that these highly purified nucleosomes contain only core histones and thus are free from nonhistone and nonspecifically associated proteins (Fig. 1D, lane 1). The Western blotting demonstrates that HA-H2BFWT was present within these nucleosomes (Fig. 1D, lane 4). We conclude that H2BFWT is a bona fide histone.
FIG. 1.
The histone variant H2BFWT is assembled into nucleosomes. (A) Sequence alignment of conventional H2B and the histone variant H2BFWT. The histograms under the sequences show the degree of homology between the two proteins. The amino acid residues are shown in different colors according to their properties. (B and C) In vivo association of H2BFWT with chromatin. HeLa cells were transiently transfected with a tagged HA-H2BFWT expression vector. The presence of the tagged HA-H2BFWT at different phases of the cell cycle (B) and in spread metaphase chromosomes (C) was detected with anti-HA antibody (red). Hoechst staining of DNA is shown in blue. (D) Incorporation of H2BFWT into the nucleosomes. Nuclei isolated from HA-H2BFWT-transfected cells were digested with microccocal nuclease, and nucleosomes were purified on a sucrose gradient containing 0.6 M NaCl. The presence of H2BFWT was detected by Western blotting by using anti-HA antibody. Lane 1, Coomassie blue staining of an 18% sodium dodecyl sulfate gel of the loaded nucleosomes. Lanes 2 to 4, Western blotting of extracts isolated from H2BFWT-transfected cells (lane 2), control nontransfected cells (lane 3), and isolated nucleosomes (lane 4). The positions of the molecular mass markers are shown on the left.
Remodeling and mobilization of H2BFWT nucleosomes by SWI/SNF.
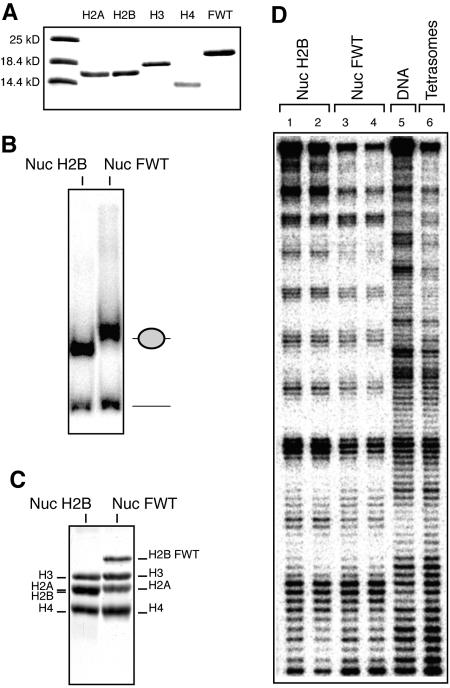
We have shown recently that incorporation of the histone variants macroH2A and H2ABbd results in alterations in the nucleosome structure, which, in turn, interferes with nucleosome variant remodeling (3, 20). Since H2BFWT exhibits a quite high divergence in its primary structure compared to the conventional H2B, one may think that the variant H2BFWT nucleosome would behave similarly to the macroH2A and H2ABbd nucleosomes. To test this hypothesis, we purified to homogeneity conventional core histones and the histone variant H2BFWT (Fig. 2A) and used them, together with a 32P-labeled 152-bp DNA fragment containing the X. borealis 5S rRNA gene, to reconstitute positioned nucleosomes (Fig. 2B and C). In agreement with the in vivo data (Fig. 1), the EMSA (Fig. 2B) and the histone composition analysis (Fig. 2C) showed an efficient incorporation of H2BFWT into the nucleosome. We next analyzed the structure of the variant H2BFWT nucleosome in solution by DNase I footprinting (Fig. 2D). The DNase I cutting patterns of the conventional (Fig. 2D, lanes 1 and 2) and the H2BFWT (Fig. 2D, lanes 3 and 4) nucleosomes, in contrast to the digestion pattern of the (H3-H4)2 tetramer (Fig. 2D, lane 6), are essentially the same. This somewhat surprising result strongly suggests that the structure of the H2BFWT nucleosome in solution is very close to that of the conventional nucleosome.
FIG. 2.
The presence of H2BFWT has no effect on the structure of the nucleosome. An equimolar mixture of purified-to-homogeneity recombinant core histones H2A, H3, and H4 and either conventional H2A or histone variant H2BFWT and a 152-bp EcoRI-RsaI 32P-end-labeled DNA fragment containing the X. borealis 5S RNA gene were used to reconstitute nucleosomes. (A) An 18% sodium dodecyl sulfate electrophoresis of conventional core histones and the histone variant H2BFWT. The first lane shows the molecular mass markers. (B) EMSA of the reconstituted conventional (Nuc) and histone variant H2BFWT (Nuc FWT) nucleosomes. The positions of the nucleosomes and the free DNA are shown on the right. (C) Histone composition of the reconstituted nucleosomes. Preparative EMSA was carried out, the conventional and H2BFWT nucleosome bands were cut from the gel, and the nucleoproteins were eluted. Histones were isolated and separated by 18% sodium dodecyl sulfate electrophoresis, and the gel was stained with Coomassie blue. The positions of the histones are indicated. (D) DNase I footprinting of reconstituted conventional H2AB nucleosomes (in duplicate, lanes 1 and 2), variant H2AB nucleosomes (in duplicate, lanes 3 and 4), and tetrasomal particles containing only the (H3-H4)2 tetramer (lane 6). Lane 5, footprinting of naked DNA.
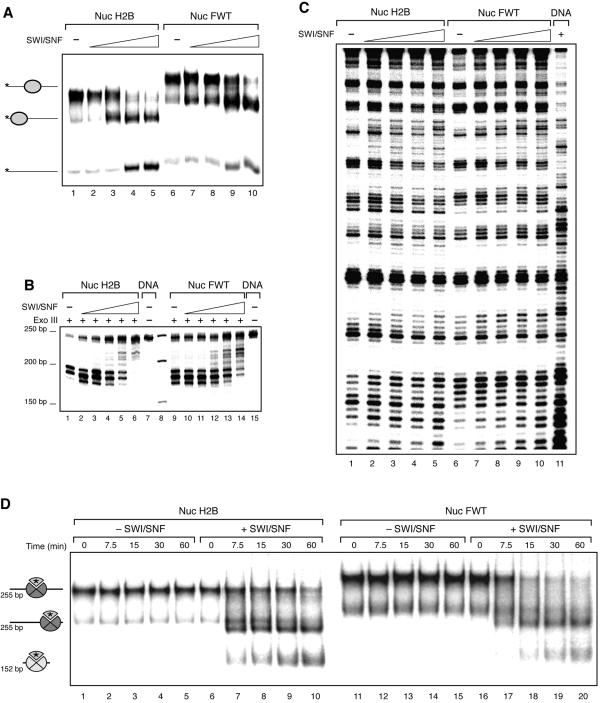
To study the mobilization of the H2BFWT nucleosomes, we used centrally positioned nucleosome particles reconstituted on a 255-bp 601 DNA fragment (Fig. 3A). Incubation of both conventional and H2BFWT nucleosomal particles with increasing amounts of SWI/SNF in the presence of ATP resulted in efficient mobilization of both histone octamers and the formation of end-positioned nucleosomes (Fig. 3A, compare lanes 2 to 5 with lanes 7 to 10). This was further confirmed by the exonuclease III mapping data (Fig. 3B). Indeed, these data clearly show that upon increasing the amount of added SWI/SNF, both the conventional and the H2BFWT variant octamers slide from a central to an end position (Fig. 3B, compare lanes 2 to 6 with lanes 10 to 14).
FIG. 3.
SWI/SNF remodels and mobilizes conventional and variant H2BFWT nucleosomes. (A) A 255-bp DNA fragment containing the positioning sequence 601 was used to reconstitute conventional and variant H2BFWT centrally positioned nucleosomes. Both types of nucleosomes were incubated with increasing amounts of SWI/SNF in the presence of ATP for 30 min at 30°C. The mobility of the histone octamers was assessed on a 5.5% native polyacrylamide gel. On the left are indicated the positions of the centrally and end-positioned nucleosomes and free DNA. (B) Exonuclease III mapping of the positions of conventional and H2BFWT nucleosomes. Both types of nucleosomes were incubated with SWI/SNF as described for panel A and, after the reaction was arrested, digested with exonuclease III. (C) DNase I footprinting of conventional (lanes 1 to 5) and variant H2BFWT (lanes 6 to 10) nucleosomes, reconstituted on the 152-bp EcoRI-RsaI 32P-end-labeled DNA fragment containing the X. borealis 5S RNA. The nucleosomes were incubated with SWI/SNF as for panel A and then subjected to DNase I digestion. Lane 11, DNase I digestion pattern of naked DNA. (D) The H2A-H2BFWT dimer is transferred efficiently to an (H3-H4)2 tetrameric particle. Conventional and H2BFWT variant nucleosomes were reconstituted on a 255-bp DNA fragment which contains the 601 positioning sequence by using radioactively labeled H2A. Histone H3-H4 particles were reconstituted on a 152-bp fragment containing the X. borealis 5S gene. The nucleosomes were incubated in the presence of a threefold molar excess of tetrameric particles at 30°C for the indicated times in the presence or absence of SWI/SNF. The positions of the centrally and end-positioned 255-bp nucleosomes and of the 152-bp nucleosomes are indicated. The asterisk indicates the radioactively labeled H2A. Note that the conventional H2A-H2B and the variant H2A-H2BFWT dimers exhibit very similar efficiencies of transfer to the tetrameric particles.
The ability of SWI/SNF to remodel H2BFWT variant nucleosomes was studied by using DNase I footprinting (Fig. 3C). In the presence of ATP, increasing amount of SWI/SNF in the reaction mixture containing either conventional or H2BFWT nucleosomes resulted in dramatic changes in the DNase I cutting patterns of both particles (Fig. 3C, compare lanes 1 to 5 with lanes 6 to 10), and with the larger amount of SWI/SNF used, the patterns were very similar to that of naked DNA (lane 11). These changes reflect the alterations in the histone-DNA interactions generated by the action of SWI/SNF. Therefore, SWI/SNF was able, as in the case of conventional nucleosomes, to remodel the variant H2BFWT particle. In agreement with this, we found that SWI/SNF has the capacity to induce an efficient transfer of the variant H2A-H2BFWT dimer to a (H3-H4)2 tetrameric particle (Fig. 3D). In this experiment, we used centrally positioned conventional and variant H2BFWT nucleosomes reconstituted on a 255-bp fragment containing the 601 positioning sequence by using 32P-radiolabeled H2A. The tetrameric particle was reconstituted on a 152-bp fragment which contains the Xenopus borealis 5S rRNA gene. When these nucleosomes were mixed with the nonlabeled tetrameric particles in the presence of ATP and SWI/SNF, both labeled octamers were mobilized, and then the labeled conventional H2A-H2B or H2A-H2BFWT variant dimers were transferred efficiently to the tetrameric particles (Fig. 3D, lanes 6 to 10 and lanes 16 to 20). This transfer is dependent on SWI/SNF, since in the absence of the remodeler, no transfer is observed (Fig. 3D, lanes 1 to 5 and lanes 11 to 15). These data demonstrate that H2BFWT does not affect the SWI/SNF-dependent transfer of the heterotypic dimer to the tetrameric particle.
Stability of the H2BFWT variant nucleosome.
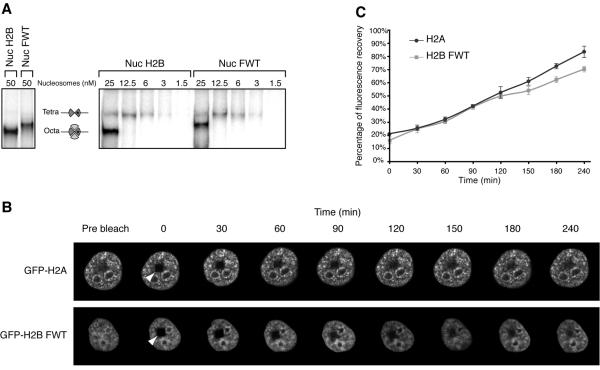
Does the presence of H2BFWT affect the stability of the variant nucleosome? This question was addressed both in vitro and in vivo (Fig. 4). We have recently demonstrated that at a very low nucleosome concentration a selective release of the heterotypic H2A-H2B dimer from the nucleosome particle occurs (11). This release results in the formation of a tetrameric (H3-H4)2 particle and reflects the strength of the interaction of the H2A-H2B dimer with both the (H3-H4)2 tetramer and the nucleosomal DNA, i.e., the internal stability of the nucleosome (11). To study the dissociation of H2A-H2BFWT from the nucleosome at a very low nucleosome concentration, we reconstituted both conventional and H2BFWT variant nucleosomes by using 32P-labeled histone H3 and a 241-bp DNA fragment containing the 601 positioned sequence (11) (the use of 32P-labeled H3 was crucial in this experiment, since this has allowed us to visualize the particles at a very low concentration). EMSA was then carried out to follow the release of the heterotypic conventional and H2A-H2BFWT dimers from the nucleosome upon successive twofold dilutions in the range of 50 to 1.5 nM nucleosome particles (Fig. 4A). Upon dilution, in addition to the nucleosome band, a second band corresponding to the tetrameric (H3-H4)2 particle was observed (Fig. 4A) (it should be noted that identical volumes of individual samples were loaded on the gel, and each successively loaded sample contains half as much radioactivity as the previous one). Interestingly, at the same nucleosome dilution, the extents of dissociation of the conventional H2A-H2B- and variant H2A-H2BFWT dimers were very similar, thus suggesting very similar in vitro stabilities of conventional and H2BFWT variant nucleosomes (Fig. 4A).
FIG. 4.
In vitro and in vivo stabilities of the variant H2BFWT nucleosome. (A) The H2BFWT variant nucleosome exhibits in vitro the same stability as the conventional H2A nucleosome. Conventional and variant H2BFWT nucleosomes and tetrameric (H3-H4)2 particles were reconstituted by using 32P-labeled histone H3. The samples were diluted to the indicated concentrations with 1× phosphate-buffered saline and run on a native 5.5% polyacrylamide gel. On the left are shown the conventional and H2BFWT nucleosomes at 50 nM. The positions of the intact nucleosomes and tetrameric particles are indicated. Note that upon dilution, the conventional H2A-H2B and the variant H2A-H2BFWT dimers are released with essentially the same efficiency from the reconstituted nucleosomes. (B) FRAP analysis of the mobility of the GFP-H2BFWT variant fusion. Cells stably expressing either GFP-H2A or GFP-H2BFWT were photobleached, and images were recorded at the indicated times postbleaching. The rectangular bleached area is designated by an arrowhead. Images of the cell before bleaching (prebleach) are also shown. (C) Quantification of the data shown in panel B. The means and standard deviations for 45 nuclei from five independent experiments are shown.
The in vivo stability of the H2BFWT nucleosome was analyzed in a series of FRAP experiments by using stable cell lines expressing either GFP-H2A or GFP-H2BFWT (Fig. 4B). In these cell lines the GFP fusions were assembled into nucleosomes, and no free fusions were present (reference 20 and results not shown). A small rectangular area of the nucleus of a single cell expressing either GFP-H2A or GFP-H2BFWT was photobleached (Fig. 4B). Pictures were taken before the photobleaching and then at the indicated times during the recovery of fluorescence (Fig. 4B). The results show that the recovery kinetics are very similar for both fusions (Fig. 4B and C), showing that GFP-H2BFWT exchanges as quickly as the conventional fusion, GFP-H2A. Since the histone exchange rate directly reflects the stability of the nucleosome particle (29), the above-described FRAP data allow the conclusion that the conventional and the H2BFWT nucleosomes exhibit very similar stability.
H2BFWT and assembly of mitotic chromosomes.
The NH2 tail of the conventional histone H2B is crucial for condensation and assembly of mitotic chromosomes (13). The reported data suggest that conserved factors, essential for chromosome condensation, specifically recognize the tail of histone H2B and assist the assembly of mitotic chromosomes (13, 14). The NH2 tail of H2BFWT is, however, the most divergent H2BFWT domain compared to the respective domains of conventional H2B (Fig. 1A). This suggests that H2BFWT may not be able to play a role similar to that of H2B in chromosome condensation. We tested this hypothesis by the approach we have applied to demonstrate the role of H2B in chromosome condensation in a series of nucleosome competition experiments (13).
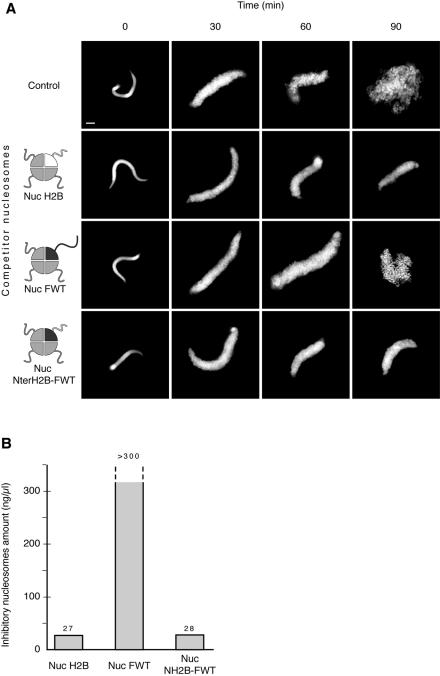
We used Xenopus egg extract and demembranated Xenopus sperm nuclei to assemble mitotic chromosomes (14). Incubation of the sperm in the extract was initially accompanied by a very rapid massive decondensation (Fig. 5A), which reflects the release of the protamine-like proteins from the sperm and their replacement with histones (16, 40). Further incubation resulted in condensation of chromatin and finally in the formation of well-defined chromosomes (Fig. 5A, control) (14). In agreement with reports in the literature (13), when exogenous reconstituted conventional nucleosomes at a concentration of 30 ng/μl were added to the reaction mixture, the assembly was inhibited and the process was arrested at the decondensation state (Fig. 5A, Nuc H2B). This phenomenon is associated with the titration of chromosome assembly factors by the NH2 tails of the exogenous nucleosomes and in particular by the NH2 tail of histone H2B (13, 14). As a result of the titration, these chromosome assembly factors are no longer available for the decondensed sperm nuclei, and the nuclei fail to further assemble into chromosomes (13, 14). The addition of the same concentration of H2BFWT variant nucleosomes, however, had no effect on the assembly of chromosomes (Fig. 5A, Nuc FWT). Indeed, after 90 min of incubation of the sperm nuclei in the extract in the presence of H2BFWT nucleosomes, piles of well-defined chromosomes (Fig. 5A, Nuc FWT), indistinguishable from the assembled ones in the control reaction (Fig. 5A, control), were observed. Importantly, even an increase in the added H2BFWT nucleosomes in the chromosome assembly reaction of one order of magnitude did not affect the assembly process (Fig. 5B). Therefore, the variant H2BFWT nucleosomes, in contrast to the conventional nucleosomes, were unable to interfere with the mitotic chromosome assembly. Chromosome condensation was also completely inhibited when a mixture of conventional and H2BFWT nucleosomes was used in the competition experiments (results not shown), showing that the H2BFWT nucleosomes do not contain contaminants which could interact with inhibitors of chromosome condensation and affect their ability to function properly.
FIG. 5.
The assembly of mitotic chromosomes in Xenopus egg extract is not inhibited by the variant H2BFWT nucleosomes, in contrast to conventional nucleosomes. (A) Demembranated sperm nuclei were incubated in the extract for the times indicated in the absence (control) or in the presence of either conventional H2B (Nuc H2B), variant H2BFWT (Nuc FWT), or fusion NterH2B-histone fold H2BFWT (Nuc NterH2B-FWT) nucleosomes at a concentration of 30 ng per μl of extract. After fixation and staining with Hoechst 33258, the assembled structures were observed by fluorescence microscopy. The reconstituted particles are schematically presented on the left. Conventional H2B is in white, while H2BFWT is in black. Note that both the conventional and the fusion NterH2B-histone fold H2BFWT nucleosomes inhibit chromosome assembly, while the variant H2BFWT nucleosomes do not. Bar, 5 μm. (B) Quantification of the minimal nucleosome amount that is able to inhibit mitotic chromosome condensation. Increasing amount of conventional H2B (Nuc H2B), variant H2BFWT (Nuc FWT), or fusion NterH2B-histone fold H2BFWT (Nuc NterH2B-FWT) nucleosomes were added to the chromosome assembly reaction mixture, and the minimal concentration of nucleosomes sufficient to inhibit chromosome condensation was measured. Note that even the use of 300 ng/μl of competitor H2BFWT nucleosomes (the highest nucleosomal concentration possible to be used in the experiments) was unable to inhibit the assembly process.
Bearing in mind that (i) the NH2 tail of conventional H2B is crucial for the assembly of mitotic chromosomes and (ii) the NH2 tail of H2B is quite divergent from that of H2BFWT, these results indicate that the H2BFWT tail could be responsible for the inability of H2BFWT nucleosomes to inhibit chromosome assembly. If this was correct, a chimeric nucleosome containing, instead of H2BFWT, a fusion of the tail of conventional H2B with the histone fold motif of H2BFWT (NterH2B-FWT) would inhibit the condensation process. We found that this was actually the case, since the addition of only 30 ng/μl of NterH2B-FWT nucleosomes completely rescued the inhibition and the assembly process was arrested at the decondensation state (Fig. 5A and B).
DISCUSSION
The incorporation of a histone variant into the histone octamer may create a nucleosome with a different structure and distinct properties. For example, the incorporation of either macroH2A or H2ABbd into the nucleosome resulted in alterations in its structure, which appeared to interfere with both nucleosome remodeling and mobilization (3, 4). H2ABbd nucleosomal arrays are more easily transcribed than their conventional counterparts (4), while the presence of macroH2A may interfere with transcription (39). In addition, H2ABbd nucleosomes showed lower stability both in vitro and in vivo (20). The structure of the variant H2A.Z nucleosome exhibited an extended acidic surface across the face of the H2A.Z octamer (47), which could be important for chromatin fiber folding (19). Thus, the incorporation of variants of the histone H2A family within the histone octamer results in alterations in the nucleosome structure, which may have functional consequences.
Here we have carried out a detailed structural and functional analysis of a nucleosome reconstituted with H2BFWT, a recently cloned putative histone variant showing only 45% identity with conventional H2B (10). We have demonstrated that H2BFWT is a bona fide histone. The reconstituted H2BFWT nucleosomes, in contrast to the H2A variant nucleosomes, were both in vitro and in vivo structurally and dynamically indistinguishable from those reconstituted with conventional histones. In addition, the H2BFWT nucleosomes, unlike the H2ABbd and macroH2A nucleosomes, were efficiently mobilized and remodeled by SWI/SNF. These structural and functional properties of the H2BFWT nucleosomes are determined mainly by the structure of the histone fold domain of the whole variant octamer and its interaction with DNA, which should be very similar to those of the conventional octamer. Interestingly, the structural properties of the hTSH2B nucleosome, the only other histone H2B variant nucleosome studied, were also reported to be the same as those of the conventional nucleosome (31). This indicates that in general the incorporation of an H2B variant within the histone octamer may not alter the properties of the nucleosome, which depends on the structure of the histone fold domain of the octamer.
The NH2 tail of H2BFWT is very divergent from the conventional NH2 tail of H2B (Fig. 1A), suggesting that the H2BFWT nucleosome may exhibit distinct NH2 tail-dependent properties. The NH2 tail of H2B is crucial for mitotic chromosome assembly, since conserved (between species) chromosome assembly factors interact specifically with it and this allows chromosome condensation to proceed (Fig. 5A) (13, 14). Our competition experiments with Xenopus egg extract showed that H2BFWT nucleosomes do not appear to be able to interact with and to titrate these factors from the extract, since the addition of one order of magnitude more of H2BFWT nucleosomes compared to the amount of conventional nucleosomes did not inhibit the assembly of mitotic chromosomes (Fig. 5B). This distinct property of H2BFWT resides in its NH2 tail, as nucleosomes reconstituted with the fusion NterH2B-FWT were found to be indistinguishable from the conventional ones in their ability to inhibit chromosome assembly (Fig. 5A and B).
Recent experiments with somatic cells, possessing very large telomeric sequences, indicate that the telomere interstitial blocks might be enriched in H2BFWT (10). The telomere chromatin exhibits a specific chromatin structure (34), and closed chromatin loops are present at the end of the chromosomes (37); thus, H2BFWT may assist in the preservation of this structure during mitosis and consequently its conservation and transmission during the cell cycle. This could be essential for the proper function of the telomeres. Importantly, H2BFWT was found in sperm nuclei, and the reported data suggest that H2BFWT colocalized again with the telomeric sequences (10). We speculate that H2BFWT, like CENP-A (which remains associated with the centromeres in mature spermatozoa [38]), is an epigenetic marker required for telomeric identity and necessary for the transmission of specific chromatin structure (the telomeric chromatin) through generations.
Acknowledgments
This work was supported by CNRS, INSERM, Région Rhône-Alpes, and grants from the Ministère de la Recherche (ACI Biologie Cellulaire Moléculaire et Structurale [BCM0070]; ACI Interface Physique-Chimie-Biologie: Dynamique et réactivité des Assemblages Biologiques [DRAB], 2004, no. 04 2 136; and ANR project no. NT05-1_41978).
REFERENCES
- 1.Allan, J., N. Harborne, D. C. Rau, and H. Gould. 1982. Participation of core histone “tails” in the stabilization of the chromatin solenoid. J. Cell Biol. 93:285-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.An, W., V. B. Palhan, M. A. Karymov, S. H. Leuba, and R. G. Roeder. 2002. Selective requirements for histone H3 and H4 N termini in p300-dependent transcriptional activation from chromatin. Mol. Cell 9:811-821. [DOI] [PubMed] [Google Scholar]
- 3.Angelov, D., A. Molla, P. Y. Perche, F. Hans, J. Cote, S. Khochbin, P. Bouvet, and S. Dimitrov. 2003. The Histone variant macroH2A interferes with transcription factor binding and SWI/SNF nucleosome remodeling. Mol. Cell 11:1033-1041. [DOI] [PubMed] [Google Scholar]
- 4.Angelov, D., A. Verdel, W. An, V. Bondarenko, F. Hans, C. M. Doyen, V. M. Studitsky, A. Hamiche, R. G. Roeder, P. Bouvet, and S. Dimitrov. 2004. SWI/SNF remodeling and p300-dependent transcription of histone variant H2ABbd nucleosomal arrays. EMBO J. 23:3815-3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arents, G., R. W. Burlingame, B.-C. Wang, W. E. Love, and E. N. Moudrianakis. 1991. The nucleosomal core histone octamer at 3.1 A resolution: a tripartite protein assembly and a left-handed superhelix. Proc. Natl. Acad. Sci. USA 88:10148-10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arents, G., and E. N. Moudrianakis. 1995. The histone fold: a ubiquitous architectural motif utilized in DNA compaction and protein dimerization. Proc. Natl. Acad. Sci. USA 92:11170-11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ausio, J., F. Dong, and K. E. van Holde. 1989. Use of selectively trypsinized nucleosome core particles to analyze the role of the histone “tails” in the stabilization of the nucleosome. J. Mol. Biol. 206:451-463. [DOI] [PubMed] [Google Scholar]
- 8.Bassing, C. H., H. Suh, D. O. Ferguson, K. F. Chua, J. Manis, M. Eckersdorff, M. Gleason, R. Bronson, C. Lee, and F. W. Alt. 2003. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell 114:359-370. [DOI] [PubMed] [Google Scholar]
- 9.Celeste, A., S. Difilippantonio, M. J. Difilippantonio, O. Fernandez-Capetillo, D. R. Pilch, O. A. Sedelnikova, M. Eckhaus, T. Ried, W. M. Bonner, and A. Nussenzweig. 2003. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell 114:371-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Churikov, D., J. Siino, M. Svetlova, K. Zhang, A. Gineitis, E. Morton Bradbury, and A. Zalensky. 2004. Novel human testis-specific histone H2B encoded by the interrupted gene on the X chromosome. Genomics 84:745-756. [DOI] [PubMed] [Google Scholar]
- 11.Claudet, C., D. Angelov, P. Bouvet, S. Dimitrov, and J. Bednar. 2005. Histone octamer instability under single molecule experiment conditions. J. Biol. Chem. 280:19958-19965. [DOI] [PubMed] [Google Scholar]
- 12.Costanzi, C., and J. R. Pehrson. 1998. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature 393:599-601. [DOI] [PubMed] [Google Scholar]
- 13.de la Barre, A. E., D. Angelov, A. Molla, and S. Dimitrov. 2001. The N-terminus of histone H2B, but not that of histone H3 or its phosphorylation, is essential for chromosome condensation. EMBO J. 20:6383-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de la Barre, A. E., V. Gerson, S. Gout, M. Creaven, C. D. Allis, and S. Dimitrov. 2000. Core histone N-termini play an essential role in mitotic chromosome condensation. EMBO J. 19:379-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhillon, N., and R. T. Kamakaka. 2000. A histone variant, Htz1p, and a Sir1p-like protein, Esc2p, mediate silencing at HMR. Mol. Cell 6:769-780. [DOI] [PubMed] [Google Scholar]
- 16.Dimitrov, S., M. C. Dasso, and A. P. Wolffe. 1994. Remodeling sperm chromatin in Xenopus laevis egg extracts: the role of core histone phosphorylation and linker histone B4 in chromatin assembly. J. Cell Biol. 126:591-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dimitrov, S. I., T. M. Apostolova, V. L. Makarov, and I. G. Pashev. 1986. Chromatin superstructure. A study with an immobilized trypsin. FEBS Lett. 200:322-326. [DOI] [PubMed] [Google Scholar]
- 18.Dorigo, B., T. Schalch, K. Bystricky, and T. J. Richmond. 2003. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J. Mol. Biol. 327:85-96. [DOI] [PubMed] [Google Scholar]
- 19.Fan, J. Y., D. Rangasamy, K. Luger, and D. J. Tremethick. 2004. H2A.Z alters the nucleosome surface to promote HP1alpha-mediated chromatin fiber folding. Mol. Cell 16:655-661. [DOI] [PubMed] [Google Scholar]
- 20.Gautier, T., D. W. Abbott, A. Molla, A. Verdel, J. Ausio, and S. Dimitrov. 2004. Histone variant H2ABbd confers lower stability to the nucleosome. EMBO Rep. 5:715-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gineitis, A. A., I. A. Zalenskaya, P. M. Yau, E. M. Bradbury, and A. O. Zalensky. 2000. Human sperm telomere-binding complex involves histone H2B and secures telomere membrane attachment. J. Cell Biol. 151:1591-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Govin, J., C. Caron, S. Rousseaux, and S. Khochbin. 2005. Testis-specific histone H3 expression in somatic cells. Trends Biochem. Sci. 30:357-359. [DOI] [PubMed] [Google Scholar]
- 23.Hansen, J. C. 2002. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu. Rev. Biophys. Biomol. Struct. 31:361-392. [DOI] [PubMed] [Google Scholar]
- 24.Hayes, J. J., D. J. Clark, and A. P. Wolffe. 1991. Histone contributions to the structure of DNA in the nucleosome. Proc. Natl. Acad. Sci. USA 88:6829-6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayes, J. J., and K. M. Lee. 1997. In vitro reconstitution and analysis of mononucleosomes containing defined DNAs and proteins. Methods 12:2-9. [DOI] [PubMed] [Google Scholar]
- 26.Henikoff, S., and K. Ahmad. 2005. Assembly of Variant Histones into Chromatin. Annu. Rev. Cell Dev. Biol. 21:133-153. [DOI] [PubMed] [Google Scholar]
- 27.Henikoff, S., and Y. Dalal. 2005. Centromeric chromatin: what makes it unique? Curr. Opin. Genet. Dev. 15:177-184. [DOI] [PubMed] [Google Scholar]
- 28.Hernandez-Munoz, I., A. H. Lund, P. van der Stoop, E. Boutsma, I. Muijrers, E. Verhoeven, D. A. Nusinow, B. Panning, Y. Marahrens, and M. van Lohuizen. 2005. Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc. Natl. Acad. Sci. USA 102:7635-7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimura, H., and P. R. Cook. 2001. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 153:1341-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, K.-M., and J. J. Hayes. 1998. Linker DNA and H1-dependent reorganization of histone-DNA interactions within the nucleosome. Biochemistry 37:8622-8628. [DOI] [PubMed] [Google Scholar]
- 31.Li, A., A. H. Maffey, W. D. Abbott, N. Conde e Silva, A. Prunell, J. Siino, D. Churikov, A. O. Zalensky, and J. Ausio. 2005. Characterization of nucleosomes consisting of the human testis/sperm-specific histone H2B variant (hTSH2B). Biochemistry 44:2529-2535. [DOI] [PubMed] [Google Scholar]
- 32.Lowary, P. T., and J. Widom. 1998. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276:19-42. [DOI] [PubMed] [Google Scholar]
- 33.Luger, K., A. W. Mäder, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 34.Makarov, V. L., S. Lejnine, J. Bedoyan, and J. P. Langmore. 1993. Nucleosomal organization of telomere-specific chromatin in rat. Cell 73:775-787. [DOI] [PubMed] [Google Scholar]
- 35.Malik, H. S., and S. Henikoff. 2003. Phylogenomics of the nucleosome. Nat. Struct. Biol. 10:882-891. [DOI] [PubMed] [Google Scholar]
- 36.Mutskov, V., D. Gerber, D. Angelov, J. Ausio, J. Workman, and S. Dimitrov. 1998. Persistent interactions of core histone tails with nucleosomal DNA following acetylation and transcription factor binding. Mol. Cell. Biol. 18:6293-6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nikitina, T., and C. L. Woodcock. 2004. Closed chromatin loops at the ends of chromosomes. J. Cell Biol. 166:161-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palmer, D. K., K. O'Day, and R. L. Margolis. 1990. The centromere specific histone CENP-A is selectively retained in discrete foci in mammalian sperm nuclei. Chromosoma 100:32-36. [DOI] [PubMed] [Google Scholar]
- 39.Perche, P., C. Vourch, C. Souchier, M. Robert-Nicoud, S. Dimitrov, and C. Khochbin. 2000. Higher concentrations of histone macroH2A in the Barr body are correlated with higher nucleosome density. Curr. Biol. 10:1531-1534. [DOI] [PubMed] [Google Scholar]
- 40.Philpott, A., G. H. Leno, and R. A. Laskey. 1991. Sperm decondensation in Xenopus egg cytoplasm is mediated by nucleoplasmin. Cell 65:569-578. [DOI] [PubMed] [Google Scholar]
- 41.Rangasamy, D., I. Greaves, and D. J. Tremethick. 2004. RNA interference demonstrates a novel role for H2A.Z in chromosome segregation. Nat. Struct. Mol. Biol. 11:650-655. [DOI] [PubMed] [Google Scholar]
- 42.Redon, C., D. Pilch, E. Rogakou, O. Sedelnikova, K. Newrock, and W. Bonner. 2002. Histone H2A variants H2AX and H2AZ. Curr. Opin. Genet. Dev. 12:162-169. [DOI] [PubMed] [Google Scholar]
- 43.Santisteban, M. S., T. Kalashnikova, and M. M. Smith. 2000. Histone H2A.Z regulates transcription and is partially redundant with nucleosome remodeling complexes. Cell 103:411-422. [DOI] [PubMed] [Google Scholar]
- 44.Sarma, K., and D. Reinberg. 2005. Histone variants meet their match. Nat. Rev. Mol. Cell Biol. 6:139-149. [DOI] [PubMed] [Google Scholar]
- 45.Scrittori, L., F. Hans, D. Angelov, M. Charra, C. Prigent, and S. Dimitrov. 2001. pEg2 aurora-A kinase, histone H3 phosphorylation, and chromosome assembly in Xenopus egg extract. J. Biol. Chem. 276:30002-30010. [DOI] [PubMed] [Google Scholar]
- 46.Stefanovsky, V., S. I. Dimitrov, V. R. Russanova, D. Angelov, and I. G. Pashev. 1989. Laser-induced crosslinking of histones to DNA in chromatin and core particles: implications in studying histone-DNA interactions. Nucleic Acids Res. 17:10069-10081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suto, R. K., M. J. Clarkson, D. J. Tremethick, and K. Luger. 2000. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat. Struct. Biol. 7:1121-1124. [DOI] [PubMed] [Google Scholar]
- 48.van Holde, K. 1988. Chromatin. Springer-Verlag KG, Berlin, Germany.